Abstract
CD4+ helper T cells are crucial for autoimmune and infectious diseases; however, the recognition of the many, diverse fates available continues unabated. Precisely what controls specification of helper T cells and preserves phenotypic commitment is currently intensively investigated. In this review, we will discuss the major factors that impact helper T cell fate choice, ranging from cytokines and the microbiome to metabolic control and epigenetic regulation. We will also discuss the technological advances along with the attendant challenges presented by “big data”, which allow the understanding of these processes on comprehensive scales.
1. Introduction
The generation of diverse types of effector CD4 T cells and the balance with production of various types of regulatory T cells is crucial for the protection against infections, but is also critical for a variety of autoimmunity diseases (Reiner et al. 2007). In fact, understanding how CD4 T cells differentiate into these diverse fates has already provided insights not only into immunopathogenesis but also has facilitated the development of new therapies. CD4 T cell fate choice has been recognized since the late 1980's, but the remarkable complexity of options available to these cells continue to be elucidated. Aside from T helper 1 (Th1) cells and Th2 cells, subsets termed Th17, Th22, Th9 and follicular T helper (Tfh) cells (Zhou et al. 2009a) have been recognized. Equally relevant for the pathogenesis of autoimmune disease are the mechanisms that lead to different types of regulatory T cells, including those that express Foxp3 and those that do not (Rudensky 2011) (Ohkura et al. 2013) (Awasthi et al. 2007) (Gregori et al. 2012). But even among these defined subsets, we also appreciate considerable heterogeneity and plasticity (Cannons et al. 2013) (O'Shea and Paul 2010) (Coomes et al. 2013) (Yamane and Paul 2012) (Dong 2011) (Zhu and Paul 2010). Consequently, the previous 1:1:1 model of differentiation (one lineage/function, one signature cytokine and one master regulator transcription) has given way to a more nuanced view of specification (Crotty 2012), and the plasticity versus stability of these subsets, both effector and regulatory continues to be intensively investigated. Thus, more sophisticated understanding of helper T cell differentiation will surely continue to be useful for immunologists both in terms of understanding and treating disease.
In this review, we will discuss the current views of helper T cell diversity and evolving insights into the mechanisms that underlie their differentiation. The appreciation of the enormous range of T cells fates has occurred at a time when our basic understanding of the regulation of gene expression is changing and new techniques are being devised. The impact of the epigenome on cell fate determination is being re-examined as new technologies to measure these changes also emerge. Indeed, the more flexible view of cell fate has been a general lesson of cell biology, well beyond immune cells. It is premature at this time to propose a unifying framework of how networks of transcription factors and epigenomic changes converge to drive helper T cell fate choice while maintaining opportunities for plasticity. Nor can we hope to be comprehensive in covering all of these topics in a single review. Rather, we will try to provide a few illustrative examples of molecular mechanisms that can promote flexibility in the context of cellular differentiation. We will try to explain how new technologies have modified our views of the CD4 T cells biology and their capacity for plasticity in response to a constantly changing environment.
2. Old and new players in lineage specification of helper T cells
Based on their function and cytokine expression, activated CD4+ T helper (Th) cells were initially classified into two subsets (Mosmann and Coffman 1989): Th1 cells that produce Interferon-γ (IFN-γ) and Th2 cells that produce interleukin (IL)-4, IL-5 and IL-13 as their respective signature effector cytokines. In this way, CD4 T cells orchestrate the type of immune response that ensues upon encounter of diverse microbial pathogens. Regulated cytokine production is required for the proper elimination of microbial pathogens: Th1 cells for intracellular microbes and Th2 cell for helminthes (Abbas et al. 1996). Extrinsic factors, especially cytokines, are also critical in that they activate transcription factors especially members of the signal transducer and activator of transcription (STAT) family, which in turn control helper cell differentiation. IFN-γ and IL-12 activate STAT1 and STAT4 whereas IL-4 activates STAT6. Th2 cells lose their sensitivity to the Th1 cell-inducing cytokine IL-12 via downregulation of IL-12 receptor ß2 (IL-12R ß2) and STAT4 expression (Szabo et al. 1997) (Usui et al. 2003). In addition to STAT4 and STAT6, STAT5 plays a critical role for both Th1 and Th2 cell differentiation, transmitting IL-2-dependent signals (Liao et al. 2011). Differentiation leads to high expression of transcription factors termed master regulators, which when over-expressed are sufficient to induce signature cytokines. For instance, T cell receptor (TCR) and STAT signals induce the T-box-containing protein, T-bet (encoded by Tbx21) (Szabo et al. 2000). Conversely, GATA-binding protein-3 (GATA-3) is critical for the development of the Th2 cell lineage (Zhang et al. 1997) and overexpression is sufficient to drive Th2 differentiation. Unlike T-bet though, GATA-3 also has other functions in thymic development.
While the appreciation of diverse CD4 helper cell fates was a breakthrough in elucidation of immunoregulatory mechanisms, it also became clear that a dichotomous view of Th1 and Th2 cells failed to explain many aspects of immune responses and especially autoimmunity. The discovery of another subset of cells, Th17 cells, subsequently provided a number of insights (Yang et al. 2008b) (Stockinger et al. 2007) (Korn et al. 2009) (Weaver et al. 2007) (Steinman 2007). Th17 cells are characterized by the production of IL-17A and IL-17F, but can also produce IL-21 and IL-22. Th17 cells are an important component of the adaptive immune response to certain microbes, particularly extracellular bacteria and fungi (McGeachy and Cua 2008). A number of transcription factors including retinoic acid-related orphan nuclear hormone receptor RORγτ, STAT3, Batf and IRF4 are involved in the differentiation of Th17 cells (Acosta-Rodriguez et al. 2007b) (Huber et al. 2008) (Yang et al. 2011) (Lohoff et al. 2002) (Brustle et al. 2007) (Biswas et al. 2010). A variety of cytokines also contribute to the differentiation of Th17 cells including transforming growth factor (TGF)-β, IL-1, IL-6, IL-23 (Mangan et al. 2006) and IL-21 (Korn et al. 2007) (Zhou et al. 2007) (Mangan et al. 2006). However, it is becoming increasingly clear that “Th17” cells represent a heterogeneous collection of cells, which may or may not be pathogenic (Ghoreschi et al.) (Zielinski et al. 2012) (Wu et al. 2013) (Yosef et al. 2013) (Lee et al. 2012). IL-17 and IL-22 production is also influenced by environmental factors such as aryl hydrocarbon receptor (AHR) ligands (Veldhoen et al. 2009) (Veldhoen et al. 2008a), which include environmental pollutants. Dietary components can also activate AHR and IL-17 is even influenced by dietary salt (Yosef et al. 2013) (Wu et al. 2013) (Lee et al. 2012) (Kleinewietfeld et al. 2013). In this context, it needs to be emphasized however, that CD4+ Th17 cells are by no means the only source of IL-17 and IL-22 cytokines.
In addition to Th1, Th2 and Th17 cells, there are subsets of CD4 T cells that preferentially produce IL-9 and IL-22 (Veldhoen et al. 2008b) (Dardalhon et al. 2008) (Duhen et al. 2009) (Trifari et al. 2009). These are referred to as Th9 and Th22 cells; however, there are circumstances in which these cytokines are not produced exclusively, but are produced with other effector cytokines. IL-4 together with TGF-β induce a population of IL-9+IL-10+ T cells, which don't express Foxp3 but induced colitis and peripheral neuritis after transfer into RAG-1-deficient mice (Dardalhon et al. 2008). TGF-β can reprogram committed Th2 cells to switch to IL-9 secretion and therefore drives the differentiation of Th9 cells (b).
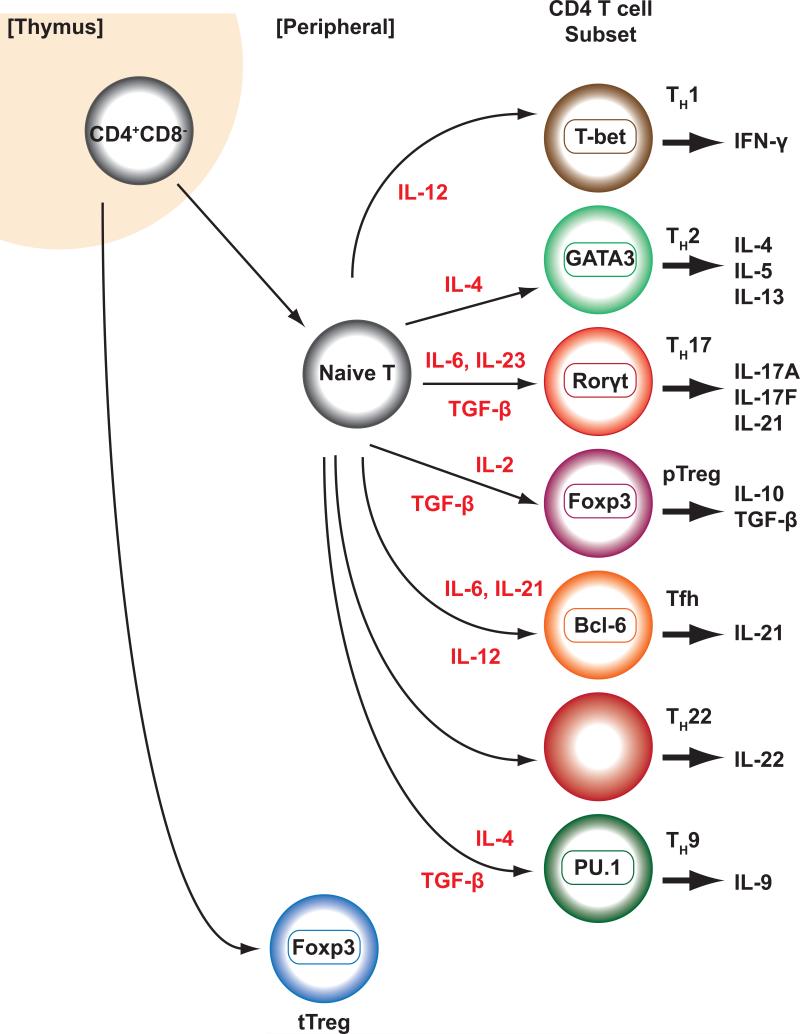
Adding to the complexity of CD4+ T cells are follicular helper T cells, which provide help to B cells and are found within B-cell follicles or germinal centers. They express CXCR5, which contributes to their localization, PD-1 and IL-21; however, PD-1 and IL-21 are not exclusively expressed by Tfh. Moreover, in addition to IL-21 Tfh cells can produce IL-4, IL-17 and IL-10 (Crotty 2011) (Reinhardt et al. 2009). The “master regulator” for Tfh is the transcription factor Bcl-6 (Crotty 2011) (Johnston et al. 2009) (Yu et al. 2009) (Nurieva et al. 2009). The factors responsible for Tfh differentiation remain perplexing and the search for unique inducers has been frustrating; rather, it appears that multiple factors can contribute to Tfh differentiation. That is, STAT1, STAT3, and STAT4 all contribute to Tfh cell development whereas STAT5 supresses formation of this subset (Choi et al. 2014) (Johnston et al. 2012) (Yang et al. 2008a) (Nurieva et al. 2012) (Ma et al. 2012) (Nakayamada et al. 2011). Cytokines that activate these STATs, including IL-6, IL-21, IL-27 and IL-12, have all been implicated in driving Tfh cell differentiation, whereas IL-2 has a negative effect (Eto et al. 2011) (Schmitt et al. 2013) (Poholek et al. 2010) (Harker et al. 2011) (Batten et al. 2006) (Ballesteros-Tato et al. 2012). A summary of diverse T cell subsets can be found in Figure 1.
Figure 1. Classical view of helper T cell differentiation.
Helper T cells can differentiate from naïve T cells into diverse helper T cells, including Th1, Th2, Th17, T follicular helper (Tfh) cells, Th9 and Th22 cells and peripheral derived regulatory T cells (pTreg) and in contrast to thymically derived regulatory T cells (tpTreg) cells that develop in the thymus.
Traditionally, helper T cell differentiation has been viewed as a largely irreversible process akin to terminal differentiation of other specialized cells. In this way, naive CD4 T cells can be regarded as a pluripotent cell, specification of which is initiated by environmental triggers to generate distinctive “lineages” in which different master regulator transcription factors are induced that in turn supervise unique transcriptional programs. However, as the recognition of the number of different “lineages” and fate choices available for CD4 T cells continue to expand, the extent to which this view is apt has been re-examined. Moreover, considerable evidence continues to mount that argues for a view in which differentiated CD4+ T cells retain the capacity to redirect their functional programs in response to changing milieus. The complexity of helper cell fate determination is made further complicated by the appreciation of overlap between “effector” and “regulatory” functions.
2.1 The Diversity of Regulatory T cells
In addition to orchestrating immune responses by production of key effector cytokines, diverse types of regulatory CD4+ T (Treg) cells are essential for maintenance of immune self-tolerance and homeostasis by suppressing excessive immune responses (Sakaguchi et al. 2008). A subset of Treg cells is defined by their expression of the transcription factor forkhead boxp3 (FoxP3), which is necessary for the development and maintenance of these key cells (Zheng and Rudensky 2007). Mutations of FOXP3 underlie the disorder termed immunodysregulation polyendocrinopathy eneropathy X-linked (IPEX) syndrome (Bennett et al. 2001). Likewise, mice that carry a mutation or deletion of FoxP3 develop a fatal systemic autoimmune disease (Brunkow et al. 2001) (Fontenot et al. 2003). Overexpression of FoxP3 confers suppressive activity to conventional T cells, leading to the conclusion that FoxP3 serves as the master regulator and a lineage-specifying transcription factor for Treg cells (Hori et al. 2003). Equally though, it is also recognized that an array of other transcription factors contribute to the phenotype of Treg cells; indeed, FoxP3-dependent and FoxP3-independent factors are important for the phenotype of regulatory cells (Fu et al. 2012).
Treg cells that are derived from the neonatal thymus in response to self antigens have been termed thymic derived regulatory (tTreg) T cells (Shevach 2000). Their program of specification is critically dependent upon TCR signals that lead to induction of FoxP3 (Ohkura et al. 2012). Peripheral derived (p)Treg cells, by contrast, develop in the periphery in response to environmental self-antigens (Chen et al. 2003) (Horwitz et al. 2008) (Abbas et al. 2013). Induced Treg (iTreg) cells can be generated from naïve precursor cells in vitro through activation and cytokine receptor engagement (Figure 1). More specific markers, like the Ikaros family transcription factor Helios, or neuropilin-1, which are thought to distinguish tTreg and pTreg cells, have been described recently (Thornton et al. 2010) (Yadav et al. 2012) (Hansen et al. 2012) (Delgoffe et al. 2013). IL-2 signals through STAT5, which in turn binds directly to the FoxP3 promoter. Continued stable expression of Foxp3 is influenced by IL-2 receptor expression, which presumably invokes STAT5 in maintenance of FoxP3 expression (Miyao et al. 2012). Conversely STAT3-dependent signals can destabilize Foxp3 expression (Laurence et al. 2012). Stable Foxp3 expression depends on DNA methylation of a selected 5`intronic region, called Treg-specific determining regions (TSDR) of the Foxp3 gene (Huehn et al. 2009). All trans Retinoid acid (RA) has been shown to augment TGF-beta and TCR induced effects (Coombes et al. 2007) (Mucida et al. 2007). RA facilitates the binding of pSMAD3 to the enhancer of Foxp3 (Xu et al. 2010).
FoxP3-expressing Treg cells, however, are not the only regulatory T cells. On the contrary, multiple types of regulatory T cells exist and go by a variety of names such as Th3, Tr1 or Tr35 cells. These cells produce critical anti-inflammatory cytokines like TGFβ, IL-10 and IL-35 (Weiner et al. 2011) (Battaglia et al. 2006) (Collison et al. 2007) (Gagliani et al. 2013) (Awasthi et al. 2007). Absence of TGF-β or its receptors results in lethal autoimmunity. Many cells produce TGF-β but T cell expression of TGF-β and its receptors is critical (Li and Flavell 2008) (Oh and Li 2013) (Li et al. 2007a) (Li et al. 2006). Similarly, IL-10 is another key immunoregulatory cytokine. Mutations and polymorphisms of IL-10 or its receptor are associated with inflammatory bowel disease (Glocker et al. 2009) (Engelhardt et al. 2013). IL-22 is yet another critical cytokine; in certain circumstances, it too has essential anti-inflammatory properties (Nakagome et al. 2011). Finally, IL-35 is a regulatory cytokine that is able to generate a population of cells with suppressive activity called Tr35 cells (Collison et al. 2007). Added to this is recognition that cytotoxic effector cells, including NK cells that express perforin, can have regulatory functions by limiting T and myeloid cell function (Magnani et al. 2011) (Crome et al. 2013).
Thus, it should be clear that what comprises the constellation of regulatory T cells and how they exert their immunosuppressive effects is anything but simple.
3. Blurring the lines of “lineages”
Despite views of different T cell subsets as stable terminally differentiated lineages, many lines of evidences argue for a more fluid view of regulation (Zhou et al. 2009a) (O'Shea and Paul). There are many examples at this point, so just a few illustrative examples will be provided: For instance, extended culture of Th17 cells leads to IFN-γ production (Mukasa et al.) and the IFN-γ+ cells that arise also under Th17-inducing conditions can have properties distinct from conventional Th1 cells (Boniface et al.). IL17/IFN-γ double positive T cells are found under normal and inflammatory conditions in mouse and man (Annunziato et al. 2007) (McGeachy et al. 2007) (Acosta-Rodriguez et al. 2007a) (Chen et al. 2007) (Lee et al. 2009) (Bending et al. 2009). Recent data show that co-expression of T-bet and Runx1 or Runx3 is required for the generation of pathogenic IFN-γ producing Th17 cells. At this point, the notion that Th17 cells are heterogeneous collection of cells is no longer a subject of debate.
Many cells also produce IL-10, not just Tr1 cells. In fact, IL-10 was first noted in Th2 cells and Th17 cells also make IL-10. In S. aureus-primed cultures, Th17 cells produce IL17 and IL-10 (Zielinski et al. 2012). Again, it needs to be remembered that cells other than T cells clearly produce IL-10; this cytokine is widely produced by myeloid, B and NK cells in addition to T cells.
Th2 cells have a certain degree of plasticity too. Infection with lymphocytic choriomeningitis virus can reprogram Th2 cells into GATA-3+T-bet+ cells, which gain the capability to produce IL-4 and IFN-γ (Hegazy et al.). This reprogramming is mediated by IFN-α and STAT1. Hybrid Th1/Th2 cells can develop in response to IFN-γ, IL-12 and IL-4 (Peine et al. 2013).
Tfh cells are one of the hardest cells to pigeonhole as a “lineage”, as they do not have a unique pattern of signature cytokine secretion and have the ability to produce cytokines of other lineages. IL-21-producing Tfh-like cells generated in vitro can be re-differentiated upon re-culture with the appropriate cytokines to make IFN-γ, IL-4, or IL-17 (Lu et al. 2011). In addition, Tfh features are present early in Th1 differentiation but as differentiation proceeds, Tfh characteristics are repressed and Th1 features dominate (Nakayamada et al. 2011). This flexibility is not limited to in vitro differentiation. In vivo, Tfh cells express the cytokines that are signatures of a given infection. During helminth infection, for example, all the IL-4-producing cells in the lymph nodes are located in germinal centers, suggesting they are in fact Tfh cells (King and Mohrs 2009) (Glatman Zaretsky et al. 2009) (Reinhardt et al. 2009). Similarly, during a Th1-type bacterial infection, Tfh cells express IFN-γ (Reinhardt et al. 2009). Given this flexibility, it is hard to view Tfh cells as a classic lineage analogous to Th1 or Th2 cells. An alternative model that would incorporate inherent flexibility would be to differentiate between cells that are retained in the lymph nodes versus those that exit: only the former would be bonafide Tfh cells and those that migrate to the tissues would be viewed as Th1 or Th2 cells. In this way, the “lineage” would be defined by localization and not production of signature cytokines. T cells retained in the lymph node would likely receive additional signals from B cells, which would further enforce or stabilize the Tfh features of the T cell, while permitting acquisition of some features of Th1 or Th2 cell (Crotty 2011) (Deenick et al. 2010) (Goenka et al. 2011) (Cannons et al. 2013). Until the specific signals to drive Tfh cells are worked out, this will likely remain an area of active investigation.
While it is accepted that iTreg cells are prone to Foxp3 instability and can produce effector cytokines (Yang et al. 2008a), the debate to what extent tTreg cells are plastic or not is still ongoing (Bailey-Bucktrout and Bluestone 2011) (Hori 2011). Using Foxp3 lineage reporter mice, two groups reported that tTreg cells were stable (Rubtsov et al. 2010) (Miyao et al. 2012). Using a different model, it has been reported that Treg cells can lose Foxp3 expression, gain effector attributes and become drivers of inflammation (Zhou et al. 2009b) (Bailey-Bucktrout et al. 2013) (Komatsu et al. 2014).
3.1 Responding to more than one Master
The recognition of substantial plasticity of differentiated helper T cells begs the question as to what mechanisms drive stability and flexibility of phenotype. In the next sections, we will review these mechanisms. In broad, non-mutually exclusive terms can be influenced by flexible expression of “master regulators” and other transcription factors, which themselves are regulated by post-translational regulation of transcription factors and exert their effect in conjunction with metabolic, dietary and microbial factors. Along with well-known factors like cytokines, all these signals are integrated to modify diverse genomic switches in developing CD4 T cells and techniques are rapidly being devised to measure the impact of transcription factors and epigenomic changes on global, comprehensive scales. We will start the discussion with insights into flexible and complex expression of master regulator transcription factors.
One important insight that has emerged in recent years is recognition of the limitations of viewing helper T cells from the perspective of a single “master regulator” effecting lineage commitment. Perhaps such a model is reasonably apt for some sessile tissues, like muscle, in which MyoD can appropriately be visualized as a true master regulator. Though the classical view of helper T cells as a homogenous population that expressed a signature cytokine in response to a single master regulator transcription factor was a useful construct, the limitations of this “fallacy” has also become increasingly apparent (Crotty 2012). A major limitation to a monolithic view of CD4 lineages is the proliferation of observations that helper cells can express multiple “master regulators”. T-bet, for example is now appreciated to be expressed in FoxP3+-Treg, Bcl6+-Tfh, Rorγt+-Th17 and even GATA3+-Th2 cells (Koch et al. 2009) (Oldenhove et al. 2009) (Ghoreschi et al.) (Cosmi et al. 2011) (Nistala et al. 2010) (Nistala et al. 2008).
In some cases, key transcription factors work in opposition, titering functionality. On the one hand, T-bet can recruit the transcriptional repressor Bcl-6 (Oestreich et al. 2011); at low cellular concentrations of Bcl-6 the formation of a T-bet-BCL6 complex can block Bcl6 from repressing its target genes (Oestreich et al. 2012). Analogously, FoxP3 represses RORγτ and both transcription factors can be co-expressed (Zhou et al. 2008) (Yang et al. 2008a) (Lochner et al. 2008) (Voo et al. 2009). In some circumstances, Treg cells can produce effector cytokines (Stock et al. 2004) (Sawitzki et al. 2005) (Wei et al. 2009) and in humans some IL17-producing cells arise from cells that expressed FoxP3 (Beriou et al. 2009). This is important to keep in mind insofar as FOXP3 is generally more fluid in its expression in human cells and is broadly induced in activated T cells and is not necessarily associated with suppressor cell functionality (Wang et al. 2007) (Roncador et al. 2005).
Another view is that expression of more than one “master regulator” can help provide specialized functions. Like other helper cell subsets, Treg cells are also heterogeneous. Co-expression of FoxP3 with T-bet, GATA3, Bcl6 or STAT3 has been argued to produce specialized subsets of Treg cells that control Th1, Th2, Tfh or Th17 responses (Oldenhove et al. 2009) (Koch et al. 2009) (Koch et al. 2012) (Wang et al. 2011d) (Chung et al. 2011) (Wohlfert et al. 2011) (Chaudhry et al. 2009). Similarly, Treg cell flexibility can also be controlled by transcription factors such as Eos (IKZF4), a known corepressor for FoxP3 (Sharma et al. 2013). The marked heterogeneity of Treg cells is further illustrated by the identification of populations of Treg cells in nonlymphoid tissues that can be characterized by differential chemokine receptor expression (Burzyn et al. 2013a) (Burzyn et al. 2013b). In adipose Treg cells, PPAR-γ collaborates with Foxp3 to impose on naïve T cells the distinctive transcriptional profile (Cipolletta et al. 2012).
Adding to this complexity is the increasing recognition that master regulators work in concert with an array of other transcription factors. Even though overexpression of a factor might be associated with a phenotype, the array of other factors present in the cell is certainly not irrelevant. Indeed, in Treg cells much of the transcriptional signature is not ascribable to FoxP3 but rather to other factors and other signalling pathways (Fu et al. 2012). Another good example of a critical transcription factor that preserves immune tolerance is Bach2, which pervasively limits effector cell differentiation. Bach2 is important for homeostasis of both Treg and conventional T cells. In reality, a cadre of factors are important for helper T cells including Runx1, Runx3, Bcl6, c-Maf, Blimp1 (encoded by Prdm1), IRF4, Batf, NFκb, IKZF2, IKZF3 and many others. Exactly how these factors all work together is very poorly appreciated. What is clear is that along with GATA3 and Rorγt, which have functions at multiple points in development, networks of transcription factors may not neatly track with a single lineage. IRF4 is an obvious example – it is important for Th2, Th17, Th9, iTreg and TFH cells (Staudt et al. 2010) (Ahyi et al. 2009) (Brustle et al. 2007) (Kwon et al. 2009).
A technology that is revolutionizing how we think about transcription factor action is chromatin immunoprecipitation and massive parallel sequencing. This technique, developed in 2007, allows the enumeration of genome-wide binding sites for DNA binding proteins (Rivera and Ren 2013). This information is rapidly accumulating thanks to efforts like Encode and Roadmap (http://www.encode-roadmap.org); however, our understanding of the functional consequence of the binding of all these different transcription factors in distinct states of T cell activation is in its infancy. Even more challenging though is trying to make sense of this onslaught of data. As will be discussed below, we have some sense of what different transcription factors are doing with respect to organizing the epigenome and the results are surprising. Despite the daunting prospect of this task, it is now feasible to rigorously map binding of the major factors and try to impute their functions.
3.2. Transcription factors are regulated post-translationally
Another mechanism that needs to be borne in mind regarding transcription factors, including so-called master regulators is that they can be regulated post-transcriptionally. In other words, mere presence of transcription factor may not be sufficient for action of a transcription factor; covalent modifications of diverse types can positively and negatively regulate function. For instance, master regulators like T-bet are regulated by phosphorylation (Hwang et al. 2005a) (Hwang et al. 2005b) (Jang et al. 2013). Linking all the relevant pathways that might impinge upon this modification is important, but this area is surprisingly understudied and the number of potential modifications beyond phosphorylation is immense.
Also, like other proteins, transcription factors can be actively degraded and this offers another molecular mechanism for regulating phenotypic stability. This appears to be the case for FoxP3; the levels of this key protein can be controlled in a number of ways. HIF-1 attenuates Treg cell development by binding FoxP3 and targeting it for proteasomal degradation. Importantly, this regulation occurs under both normoxic and hypoxic conditions and HIF-1α-deficient T cells have reduced pathogenic potential (Dang et al. 2011). FoxP3 can be also regulated by polyubiquination of multiple lysine residues (van Loosdregt et al. 2013). Stress signals, like proinflammatory cytokines or lipopolysaccherids can also lead to the degradation of FoxP3 through the ligase Stub1 (Chen et al. 2013). Degradation of key factors like FoxP3 and other “master regulators” obviously can have profound effects on helper cell function and stability of phenotype.
3.3. Control of helper cell fate by metabolic cues
We have discussed the importance of exposure to cytokines in the determination of how T helper cells differentiate after exposure to cognate peptide. In addition to cues from co-receptors and cytokines, the availability of nutrients, oxygen and even the balance of salts in their local environment influence T cell differentiation.
A resting naïve T cell required little energy and what little metabolism occurs does so aerobically. By contrast, an activated T lymphoblast switches on metabolic pathways, similar to cancer cells, catabolizing glucose anaerobically and using the metabolites to generate building blocks of protein, lipid and DNA synthesis (Fox et al. 2005). The serine kinase mTOR (mammalian Target of Rapamycin) acts as a nutrient sensor: it is inhibited by a deficiency in the availability of amino acids particularly leucine and glutamine (Christie et al. 2002) (Nicklin et al. 2009) or by a decrease in the ATP:AMP ratio (Gwinn et al. 2008). By contrast, mTOR is activated by T cell receptor, CD28 co-receptor stimulation and cytokines known to support T cell proliferation via their ability to activate Phospho-inositol phosphatidyl 3’ kinase (PI3K) (Brennan et al. 1997) (Lafont et al. 2000) (Kane et al. 2001). mTOR in the centerpiece of two signaling complexes: mTOR complex 1 (mTORC1) and complex 2 (mTORC2). mTORC1 is critical for the translation of many proteins including both metabolic enzymes and the transporters required for cells to take up local nutrients (Gordan et al. 2007) (Wang et al. 2011c). mTORC2 has a specialized role in activating members of the AGC serine kinase family including protein kinases B and C (PKB/Akt and PKC) and the serum and glucocorticoid inducible kinase-1 (SGK-1) (Guertin et al. 2006) (Garcia-Martinez and Alessi 2008). Complete inhibition of mTOR by the immunosuppressive rapamycin, blocks T effector cell differentiation and facilitates the development of FoxP3+ Treg cells (Delgoffe et al. 2009) in part through its regulation of expression of HIF1a (Quinlan and Hall 2010).
Using genetic models it is possible to see the contributions of the individual mTOR complexes; loss of mTORC1 prevents Th1 and peripheral Th17 differentiation (Delgoffe et al. 2011) whereas loss of mTORC2 prevents Th2 differentiation and Th17 differentiation within the Thymus (Delgoffe et al. 2011) (Kim et al. 2013). Recent work has linked SGK-1 with IL-23 dependent Th17 differentiation, a process that is influenced by the sodium concentration of the environment (Kleinewietfeld et al. 2013) (Wu et al. 2013).
3.4. Control of helper cell differentiation by the microbiome
In thinking about factors that drive helper cell differentiation, it cannot be ignored that all barrier sites are inhabited by a community of commensal organisms, which greatly out-numbers the host in number of cells and genes (Grice et al. 2009) (Ley et al. 2006) (Beck et al. 2012). Our knowledge of composition of the microbiota has grown exponentially over the past decade as sequencing technology has allowed us to capture a better picture of the diversity amongst various commensal communities. What has also emerged it is not just pathogens that drive helper T cells specification – commensals are also critical factors. The commensal microbiota of the GI tract in particular contributes to host health via the provision of key metabolites and nutrients derived from food that is otherwise indigestible by the host. However, since commensals are microbes, they have the potential to induce inflammatory immune responses. Therefore, the mammalian immune system has evolved complex mechanisms to prevent untoward activation at barrier sites (Hooper and Macpherson 2010). For instance, the immune system has evolved to receive homeostatic cues from the microbiota, as it has long been noted that the immune system does not properly develop in germ-free animal models (Smith et al. 2007) (Bauer et al. 1963). Multiple studies have revealed that some, but not all commensal strains can compliment certain immune defects associated with germ-free development, implying that the immune system interacts with specific commensals and their products (Mazmanian et al. 2005) (Umesaki et al. 1999) (Hall et al. 2008). Perhaps the best example of this phenomenon is the effect of segmented filamentous bacteria (SFB) on the development of Th17 T cells in the GI tract. The direct role for SFB in T cell differentiation came from studies of genetically identical animals from two different vendors that either lacked or possessed Th17 T cells in their GI tissue and the presence of SFB was shown to be sufficient for the differentiation of Th17 cells (Ivanov et al. 2008) (Ivanov et al. 2009) (Gaboriau-Routhiau et al. 2009). In contrast, bacteria of the Clostridia class have been shown to be associated with the differentiation of Treg cells in the colon (Atarashi et al. 2011) (Atarashi et al. 2013). Interestingly, these bacteria are particularly adept at the production of Short-Chain Fatty Acids (SCFA) and three recent studies have shown that the presence of SCFA powerfully supports the production and maintenance of Tregs in the GI tract (Smith et al. 2013) (Furusawa et al. 2013) (Arpaia et al. 2013) (Pryde et al. 2002). Since the production of SCFA from dietary fiber requires multiple bacterial strains, perhaps SCFA is a key metabolite via which the immune system can ascertain the health of the microbiota. It should be noted that while SFB and Clostridia are sufficient for the production of Th17 cells and Treg cells respectively, they are not strictly necessary as Treg cells are present in reduced numbers in the total absence of the microbiota (Atarashi et al. 2011). As well, some strains of Bacteroides fragilis are potent inducers of Treg cells in the GI tract (Round and Mazmanian 2010). Finally, Th17 T cells can be induced in some gnotobiotic mice that lack SFB entirely And Th17 cells at other barrier sites, such as the skin, rely on local bacterial populations and are independent of the GI microbiota (Geuking et al. 2011) (Naik et al. 2012).
One of the crucial unanswered questions in mucosal immunology is how any given T cell is directed to differentiate to a specific state, given the multitude of coincident signals associated with a diverse commensal microbiota. During infection, T cells specific to commensal antigens have been shown to differentiate according to the dominant inflammatory milieu, but at homeostasis where multiple signals may compete for prominence, how this works is less clear (Hand et al. 2012). Perhaps as a result of multiple signals, the barrier surfaces are home to many T cells that co-express transcriptional modules associated with different states. For example, Treg cells that co-express the transcription factor Rorγt, T-bet and GATA-3 are commonly found at barrier sites (Wohlfert et al. 2011) (Zhou et al. 2008) (Koch et al. 2009) (Wang et al. 2011d). As well, under inflammatory conditions ‘pathogenic’ Th17 cells that co-express IL-17A and IFN-γ are commonly found in the GI tract (Ahern et al. 2010). While it is clear that commensal microbiota along with pathogenic microbes have a major impact on helper T cell differentiation and plasticity, where are really only just beginning to understand the rules. Suffice it to say that in considering all the environmental factors from cytokines to nutrients and pollutants that provide signals to helper T cells, the enormous diversity of microbial ecosystem is undoubtedly a major factor.
4. Gene Expression, Genomes and Chromatin Organization
It should be very evident at this point that the selective pattern of gene expression that occurs in differentiating helper T cells is the result of numerous extrinsic and intrinsic cues. Precisely how the cell interprets these cues to induce and maintain characteristic gene expression is obviously the critical question. However, the issues of selective gene expression that accompanies lineage commitment and terminal differentiation along with the capacity of differentiated cells to retain plasticity obviously go far beyond T cells. This is a fundamental issue in developmental biology. Accordingly, lessons learned from many systems have implications for understanding these processes in T cells. Reconsidering T cell plasticity is timely insofar as completion of the human and other genomes and advances in sequencing technologies are dramatically changing how fundamental biological problems are viewed (Rivera and Ren 2013).
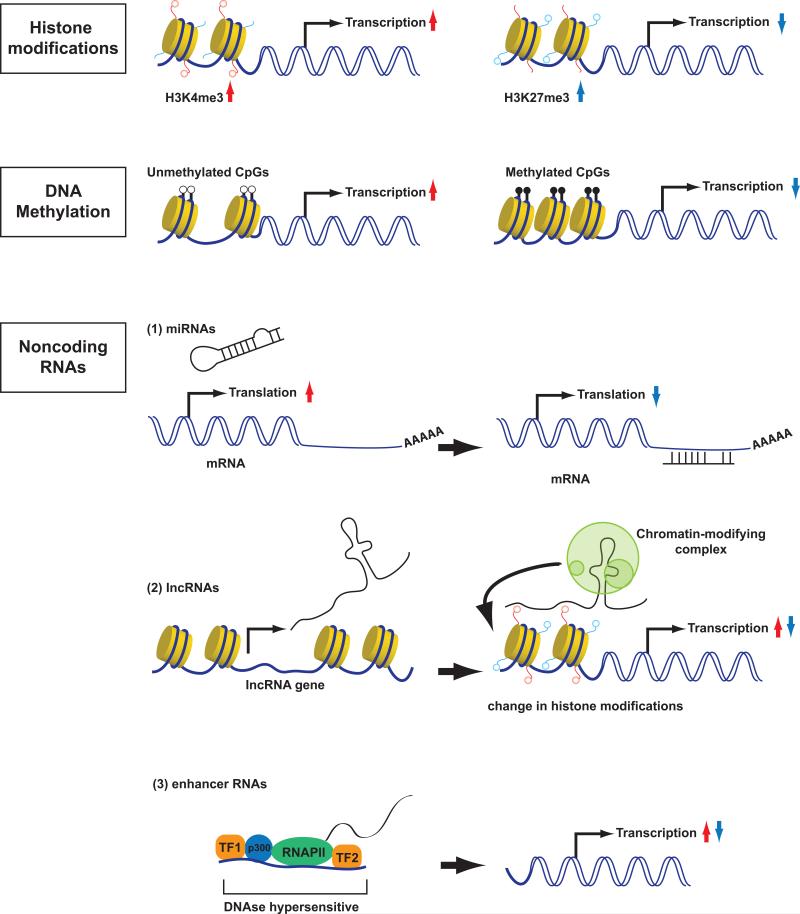
A surprising finding resulting from the elucidation of human and other genomes was the paucity of “genes” within the genome. Of the roughly three billion of nucleotides in the genome, there are only on the order of 2 × 105 genes in mammals. Despite the paucity of classical genes, as defined by the central dogma of molecular biology (DNA transcribed to RNA translated to protein), it is now recognized that consideration of selective gene expression needs is controlled by a vast number of distal “switches” and that the majority of the genome is transcribed. How accessibility of genes is controlled both locally and by largescale changes in genomic architecture is currently the focus of intense investigation. In this context, it is worth reviewing some of the newer insights and how these might relate to helper T cell diversity. Like other specialized cells, for the most part (TCR locus excluded), changes in DNA sequence (genome) is not responsible for differential functions; rather it is the “epigenome” that changes dramatically. The epigenome refers to a variety of alterations in chromatin including histone tail modifications, DNA methylation, nucleosome compaction and long-range chromatin interactions. Production of long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) are additional mechanisms that to contribute to gene regulation, as do distal elements termed enhancers transcribed to produce eRNAs. Better understanding of the dynamic epigenomes of helper T cells have provided useful insights into our notions of plasticity, but this is certainly an area that will receive increasingly greater attention (Wilson et al. 2009) (Ansel et al. 2003) (Kanno et al. 2012) (Zhu et al. 2010). We will start by briefly summarizing the most important epigenetic modifications (Figure 2).
Fig 2. Epigenetic mechanisms that modify gene transcription.
Specific alterations in chromatin including histone tail modifications, DNA methylation, production of long noncoding RNAs (lncRNAs), microRNAs (miRNAs) and enhancer RNAs (eRNAs) are mechanisms that to contribute to gene regulation.
4.1 DNA methylation
The classic modification associated with silencing of gene expression is DNA methylation (Figure 2). DNA modification occurs at cytosine-guanine (CpG) dinucleotides, which are often clustered within promoters and are associated with chromatin-remodelling factors such as methyl-DNA-binding proteins that promote the condensation and inaccessibility of chromatin. In contrast, demethylation of CpG motifs leads to a relaxation of chromatin and an increased accessibility of target sequences, thereby allowing the binding of specific transcription factors. Therefore CpG methylation at promoters is generally associated with transcriptional repression. CpG methylation is maintained by DNA methyltransferases (DNMTs), which use S-adenosylmethionine as the methyl group donor (Goll and Bestor 2005). Maintenance of a pre-existing methylation pattern is catalysed by Dnmt1; Dnmt1 recognizes a hemi-methylated DNA and introduces methyl-groups on the unmethylated strand (Bestor et al. 1988). In contrast, de novo DNA methylation is mediated by Dnmt3 methyltrasferases, (Okano et al. 1998) (Okano et al. 1999) (Bestor et al. 1988). Both processes are important for stabilizing helper cell phenotypes (Okano et al. 1999) (Young et al. 1994) (Winders et al. 2004) (Thomas et al. 2012) (Lee et al. 2001) (Makar et al. 2003).
As mentioned above FoxP3 expression and stability of Treg cells is also regulated by the DNA methylation status at several key genomic loci called the Treg-specific demethylation region (TSDR) (Floess et al. 2007) (Baron et al. 2007) (Lal et al. 2009) (Janson et al. 2008) (Polansky et al. 2008) (Zheng et al. 2010) (Huehn et al. 2009).
Furthermore recent data show that methylation of tTreg cells is necessary for FoxP3 to acquire a genome-wide Treg cell-type gene expression pattern, lineage stability and suppressive activity. The Treg-cell specific hypomethylation pattern is dependent on T cell receptor stimulation and Foxp3 independent (Ohkura et al. 2012).
While DNA methylation is important for the maintenance of T cell lineage stability, it is also true that massive demethylation occurs during T cell activation and is selective with specification of subsets. For instance, the Ifng locus is demethylated during a Th1 differentiation, whereas differentiated Th2 or Th17 cells sustain their methylation status. Similarly, the Il4, IL13 and IL17a loci are methylated in naïve T cells and are demethylated during Th2 and Th17 differentiation respectively (Santangelo et al. 2009) (Schoenborn et al. 2007) (Aune et al. 2013). The ability of Th17 to express IL-17 and IFN-γ concomitantly is associated with hypomethylation of the IFN-γ, IL17a and Rorc promoters (Cohen et al. 2011). DNA demethylation of Rorc under inflammatory conditions can lead to IL-17 production in Treg cells (Schmidl et al. 2011). Although this is a widely appreciated event, we have almost no understanding of the molecular basis of these changes. On one hand, the massive proliferation of T cells that occurs with activation is no doubt an important factor; what is less clear is how this process can be so selective.
Global dysregulation of DNA methylation has long been recognized as a potential contributor to autoimmune disease including RA and SLE (Richardson et al. 1990) (Corvetta et al. 1991) (Absher et al. 2013) (Altorok et al. 2013). To some degree it is hard to separate cause and effect since these diseases are associated with T cell activation; however, the fact that drugs known to affect DNA methylation are associated with lupus suggest at least some degree of causality. DNA methylation inhibitors like 5-azacytidine can induce T-cell autoreactivity and lupus symptoms in mice (Quddus et al. 1993) and drug induced lupus is associated with reduced DNA methylation (Cornacchia et al. 1988).
Over the past decade the global DNA methylation technologies have rapidly evolved; instead of measuring small, localized changes in DNA methylation, it is now possible to measure genomewide changes. At present though, the quality as compared to the average cost has to be taken into account and the high costs have limited acquisition of data for complete analysis of methylomes. Techniques like endonuclease digestion-based DNA methylation assays (MRE-seq) or affinity-based enrichment assays (MeDIP-seq) for example have moderate costs; although the resolution is comparatively low compared to MethylC-seq or reduced representation bisulfite sequencing (RRBS). Newer techniques like oxidative bisulfite sequencing (oxBS-seq) and TET-assisted bisulfite sequencing (TAB-sequ) yield single base resolution methods, but these remain costly (Rivera and Ren 2013). Array-based methylation assays (such as the Illumina HM450 array), however, can detect DNA methylation at single nucleotide resolution with low cost by only targeting known CpG islands using locus specific probes. As the costs of sequencing continue to decline, the expectation is that DNA methylation data sets will become far more commonplace. This is welcome as this epigenetic modification is thought to be one of the more stable marks.
4.2 Enumerating Global Histone Modifications
Another important factor that dictates accessibility of genes and therefore transcription is that DNA is wrapped into nucleosomes that consist of histone octamers. Histones can be modified by many different post-translational modifications including acetylation, methylation, phosphorylation, ubiquination, sumoylation, etc, which all contribute to define the overall structure of chromatin (Rivera and Ren 2013) (Kanno et al. 2012) (Figure 2); in fact, more than 130 different histone modifications have been described. In the interest of brevity, we will only discuss a few major modifications. Both active (euchromatin) and silenced (heterochromatin) configurations are associated with certain combination of histone modifications. For example, histone H3 lysine 4 tri-methylation (H3K4me3) is associated with active, accessible promoters whereas H3K36 tri-methylation is associated with actively transcribed coding regions. Enhancers are marked by histone H3 lysine 4 mono- and di-methylation (H3K4me1 and H3K4me2). H3 lysine 27 acetylation (H3K27Ac) in the context of H3K4me1 is a mark of an active enhancer; without this mark, H3K4me1 alone indicates a “poised” enhancer. In contrast, histone H3 lysine 27 tri-methylation (H3K27me3) and histone 3 lysine 9 tri-methylation (H3K9me3) are present in broad domains that encompass inactive genes. H3K4me3 and H3K27me3 modifications can both be present at some genomic regions, and these bivalent modifications have been suggested to poise genes ready for either activation or repression during differentiation.
H3K27 trimethylation is mediated by polycomb proteins, which comprise two major repressive complexes: polycomb repressive complex 1 (PRC1) and PRC2. The polycomb proteins Enhancer of Zeste Homolog 1 (Ezh1) and Ezh2 form two closely related PRC2 complexes that can trimethylate H3K27 and are required for maintenance of cellular identity at several stages of development. Ezh2 constrains differentiation and plasticity of Th1 and Th2 cells, by controlling the expression of Tbx21 and Gata3 in developing Th1 and Th2 cells and inhibiting spontaneous generation of IFNγ-producing cells via repression of Eomes (Tumes et al. 2013). Interestingly, in Th9 differentiation, TGF-β acting via Smad2 and Smad4 serve to displace Ezh2 from the Il9 locus (Wang et al. 2013).
The global enumeration of histone marks for helper T cells has been accomplished and the findings help to explain some of their apparent plasticity (Wei et al. 2009). One striking finding is genes that encode key regulatory transcription factors including Tbx21, Gata3, Rorc, Prdm1, and others. The presence of bivalent poised domains suggests a potential for phenotypic plasticity even in terminally differentiated cells. This helps explain the previously unanticipated breadth of expression of “lineage-defining master regulators”. Assessment of histone modifications reveal that Bcl6 is accessible in all subsets, thus explaining the flexibility of Tfh cells (Nakayamada et al. 2011). Thus, the epigenetic landscape of genes encoding master regulators may allow flexibility in expression and thereby permit the blurring lineages, allow fine tuning or provide subspecialization.
It is important to note that histone modifications can be targeted therapeutically. HDAC inhibitors are the best studied epigenetic therapeutic agents and have been proven to reduce the levels of proinflammatory cytokines (Leoni et al. 2005) (Leoni et al. 2002). Beneficial effects of HDAC inhibitors have been initially described in animal models of arthritis (Nishida et al. 2004) (Lin et al. 2007) (Nasu et al. 2008) (Saouaf et al. 2009) (Joosten et al. 2011). In humans 18 HDACs have been described and further characterized (Wang et al. 2009b). In line with this, it has been reported that blocking histone deacetylase is efficient in treatment of juvenile idiopathic arthritis (Vojinovic et al. 2011). However, the data on T cells are very limited. First of all, some HDAC inhibitors are not very specific and can target other transcription factors like NF-ΚB or STAT3, which are also important in inflammation. Second, most data come from other cell types like fibroblasts or macrophages. Studies from in vivo models, like CIA, point to a role of Treg cells, whose number and function was reported to be increased after treatment with valproic acid (Saouaf et al. 2009) (Wang et al. 2009a).
An additional level of gene regulation derives from proteins that “read” histone and DNA modifications, such as bromodomain-containing proteins that bind acetylated histones including BRD2, BRD3, and BRD4. The BET family member, BRD4 has a unique C-terminal domain that binds to the positive transcription elongation factor b (P-TEFb; composed of the cyclin-dependent kinase CDK9 and its partner, cyclin T1) complex. BRD4 recruits P-TEFb to acetylated histones, promoting phosphorylation of paused RNA polymerase II (Pol II) and the repressive complexes DSIF and NELF by CDK9, thereby allowing productive mRNA elongation. Of note, BET inhibitors appear to have efficacy in preclinical models of autoimmune disease (Bandukwala et al. 2012). In addition BRD2 and BRD4 associate with the Il17 locus and control the Th17 differentiation (Mele et al. 2013).
4.3 Defining Enhancers
Many aspects of human biology depend on the tissue-specific control of gene expression (Noonan and McCallion 2010). Regulatory information creating such complex gene expression patterns is accommodated across the non-coding part of genome in the form of cis-regulatory elements. The key point is that only a subset of these cis-regulatory elements is brought into play in every cell type in a temporal and spatial-dependent manner.
Among many types of regulatory elements, enhancers play essential roles in cell-type-specific gene expression. Enhancers are non-coding DNA sequences that “enhance” transcription of cognate target genes, regardless of their location or orientation. The Ifng and Il4 loci are good examples of the complicated architecture of finely regulated lineage-specific genes (Wilson et al. 2009) (Ansel et al. 2006). For instance, a conserved noncoding sequence in the distal site of the Il4 locus is critical for IL-4 expression in Tfh cells but not in Th2 cells (Harada et al. 2012). Likewise, a FoxP3 enhancer (CNS1) is essential for peripheral Treg cells but dispensable for natural Treg cell generation; notably, this element is only active in placental mammals (Zheng et al. 2010) (Samstein et al. 2012).
A major breakthrough for enhancer biology was the discovery of the chromatin signatures of enhancers, facilitated by next-generation sequencing technology. It is now believed that presence of a unique combination of histone modifications map the enhancer elements of each cell type. Pioneering work identified H3K4me1-high, H3K4me3-low as a chromatin signature of enhancers in human cells (Heintzman et al. 2009). Not all H3K4me1-high, H3K4me3-low regions are functionally active and these regions are representatives of “poised” enhancers. Additional marks such as the binding of acetyltransferase p300 or H3K27Ac have been related to “active enhancers” in many studies.
It has been argued that the prominent action of master regulator factors supervise the creation of the enhancer landscape (Natoli 2010). Among key transcription factors in the immune system, early studies characterized the role of PU.1, encoded by Sfpi1, as an organizer of enhancer elements (Ghisletti et al. 2010) (Heinz et al. 2010). These studies suggested that master regulators of other cell types might have similar pervasive impacts on the enhancer cohorts. In the context of CD4+ T cell subset differentiation, master regulators are defined as required and sufficient transcription factors for programming a T cell subset. Another aspect of this definition is that the master regulator has a restricted expression pattern, which means that it is only found in cells of a specific fate. As mentioned above FoxP3 has been argued to be necessary and sufficient for Treg cell development. However, FoxP3 has limited impacts on the enhancer landscape of Treg cells (Samstein et al. 2012). The major changes in the epigenetic landscape of tTregs are FoxP3-independent and are created by TCR-dependent signals. Likewise, the master regulators of Th1, Th2 and Th17 cells, have also limited impacts on key enhancer cohorts. Taken together, these studies provide compelling evidence that the master regulators involved in helper cell specification are not the factors that shape the enhancer landscape of CD4+ T cell subsets. It remains a question then how the many T-cell expressed transcription factors work in concert to create the genomic enhancer landscape of T cells.
A key aspect in the acquisition of distinct T helper cell phenotypes is the cytokine milieu. Upon encounter of diverse microbial pathogens, dendritic cells and other cells of the innate and adaptive immune system produce cytokines, which serve to instruct distinct T cell fates. The major specifying cytokines exert their effect through STAT family transcription factors. Strikingly, the majority of differentially active enhancers in Th1 and Th2 cells and Th17 cells are STAT-dependent and many are direct STAT targets of STATs (Hirahara et al.). Importantly, reconstitution of STAT4- and STAT6-deficient cells with the master regulators T-bet and GATA3 failed to recover the active enhancer landscapes, again arguing for a primary role of environmental sensors in dictating global landscapes.
A new subset of enhancers termed “super” or “stretch” enhancers has recently been appreciated. Super-enhancers regions have been suggested to have crucial functions in defining cell identity. In embryonic stem cells for instance, super enhancer domains are associated with genes that play essential roles in ESC biology. Transcription at super-enhancer genes is also disproportionately sensitive to any perturbations. The superenhancer landscapes of helper T cells has not been catalogued, but this is obviously an area of considerable interest.
Transcription abounds
A recent surprise that even though only 2% of the genome represents standard, protein-coding genes, most of the genome is active and roughly 80% of the genome transcribed despite the fact that no protein is produced. RNA-seq technology and other technologies have permit this abundance of transcription, including the identification of the repertoire of non-coding RNAs that include microRNAs (miR), enhancer RNA (eRNA) and long noncoding RNA (lncRNA) (Birney et al. 2007) (Kapranov et al. 2007) (Kim et al. 2010). Work on eRNAs and lncRNAs is just beginning, although we already know that eRNAs are functionally important for promoting accessibility of canonical protein-coding genes (Mousavi et al. 2013) (Hu et al. 2013a).
4.4 microRNA and helper T cells
MiRNAs are transcribed by RNA polymerase II as long precursor transcripts termed pri-miRNAs, which are then processed into premiRNAs by the enzyme complex including the RNase III type endonuclease Drosha and its partner DiGeorge syndrome critical region gene 8 (DGCR8). In the cytoplasm, premiRNAs are further cleaved by the RNase III endonuclease Dicer to generate a miRNA-induced silencing complex (miRISC), which destabilizes target mRNA and reduces their translation into proteins (Bartel 2009) (Fabian et al. 2010). Loss of Drosha and Dicer can lead to instability of Th cells and cause autoimmunity (O'Connell et al. 2010b) (Bartel 2004) (Chong et al. 2008) (Liston et al. 2008) (Baumjohann and Ansel 2013). By targeting crucial T cell lineage commitment genes, miRNAs are involved in regulation of gene expression network that determines T cell identity, plasticity and function (Figure 2).
Individually, a variety of miRNAs have been shown to influence effector cell differentiation and stability. Once again, this is a fast moving field and our goal is not to be comprehensive, but rather to give a few relevant examples of factors that influence gene expression. For instance, MiRNA-155 is a multi-functional miRNA that regulates a number of aspects of B and T cell functions. Its effects include the development of Th1, Th17 cells and miR-155 deficient mice are also protected from EAE and CIA, (Murugaiyan et al. 2011) (Bluml et al. 2011) (O'Connell et al. 2010a). MiR-155 is also expressed in Tregs and miR155 deficient mice have lower numbers of Treg cells (Lu et al. 2009) and develop enteric and lung inflammation (Rodriguez et al. 2007). Mir-155 is regulated by several transcription factors, such as STAT3 (Escobar et al. 2013) and FoxP3 (Zheng et al. 2007) (Marson et al. 2007), which is compatible with miR-155 expression in Th1, Th17, and Treg cells.
One important target of MiR-155 is suppressor of cytokine signaling 1 (Socs1) (Lu et al. 2009), which negatively regulates IL-2 signalling, and is important for the maintenance of Treg cells. MiR-155 also targets SMAD2 (Louafi et al. 2010), SMAD5 (Rai et al. 2010), and alters TGF-β signaling. In effector T cells, Socs1 and SH2 domain-containing inositol polyphosphate 5′ phosphatase 1 (Ship1), which is negative regulator of cytokine signaling in addition to Socs1, are important targets of miR-155 as well as Ifngr1 (Huffaker et al. 2012) (Banerjee et al. 2010). Recently, miR-155 directly targets Ets1, a negative regulator of Th17 differentiation, and subsequence down regulation of Th17-specific genes, such as IL-17F, IL-22, and IL-23R (Hu et al. 2013b).
Mir146a is another multi-functional MiRNA. Mir-146a-deficient T cells show increased IFN-γ production (Yang et al. 2012) (Huffaker et al. 2012) (Lu et al. 2010) because miR-146a regulates Th1 differentiation by targeting Stat1 (Lu et al. 2010). MiR-146a also targets IL-1 receptor-associated kinase 1 (IRAK1) and TNF receptor-associated factor 6 (TRAF6), and derepression of IRAK1 and TRAF6 increase activation of NFκB. This leads to TCR hyper-responsiveness and subsequent upregulation of IFN-γ expression from effector T cells (Yang et al. 2012). In addition, miR-146a seems to have an effect on Treg cell function (Lu et al. 2010).
Mir-181 also augments TCR signalling. Mir-181a targets several phosphatases involved in proximal TCR signalling, such as tyrosine phosphatases H2 domain-containing protein tyrosine phosphatase 2 (Shp2) and protein tyrosine phosphatase non-receptor type 22 (PTPN22) and inactivator of MAP kinases dual-specificity protein phosphatase 5 (Dusp5) and Dusp6 (Li et al. 2007b). Skewed TCR signalling by inhibition of miR-181a impairs sensitivity of double positive cells in thymus and obstruct positive and negative selection (Li et al. 2007b). Mir-181a is also important in the peripheral CD4+ T cell response. Expression of miR-181a is high in human neonatal naïve CD4+ T cells, and age-associated decrease of miR-181a increased DUSP6 expression, and diminished ERK phosphorylation via TCR stimulation (Palin et al. 2013). Reconstitution of miR-181a lowered DUSP6 expression, and restored CD4+ T cell responses (Li et al. 2012).
The miR-17~92 cluster regulates several aspects of T cell activation but importantly, regulates Tfh differentiation, relevant targets being Rora (Baumjohann et al. 2013) and CXCR5 (Yu et al. 2009).
Mir-10a attenuates the conversion of iTregs into Tfh cells by targeting Bcl6; miR-10a can also limit Th17 differentiation (Takahashi et al. 2012).
MiRNAs regulate broad range of gene modification in various immune cells including Th cells. The role of miRNAs in different diseases has been studied extensively. Especially, the role of miR-155 and miR-146 has been best described as pro- and anti-inflammatory genes in both in infectious and autoimmune diseases in human and mouse. Silencing of miR-155 reduced the susceptibility of LPS treatment in murine model (Androulidaki et al. 2009). Contrary, lack of miR-146a exhibited exaggerated inflammatory response to LPS (Boldin et al. 2011). MiR-155 and miR-326 are overexpressed in active multiple sclerosis (MS) brain lesions, and mice lacking or silencing of these miRNAs leads to ameliorate the symptom of EAE, the mouse model of MS (Junker 2011). In another autoimmune disease, systemic lupus erythematosus (SLE), underexpression of miR-146a, a negative regulator of interferon signalling, has a negative correlation with the disease activity (Tang et al. 2009). The involvement of MiR-155 and miR-146 is also reported in rheumatoid arthritis (Kurowska-Stolarska et al. 2011) (Nakasa et al. 2008) (Pauley et al. 2008), atopic dermatitis (Sonkoly et al. 2010), Sjoegren's syndrome, and IgA nephropathy (Wang et al. 2011b). Modulation of miRNAs might be an effective therapeutic approach. Recent reports have shown experimental evidences of the efficacy about blocking miRNA. Antagonizing miR-126 reduce the Th2 response in asthma model (Collison et al. 2011).
4.5 long non-coding RNAs
More recently long non-coding RNAs (lncRNAs) have taken the center stage within the non-coding world as critical regulators of cellular identity. Dramatically, germline knockouts of several lncRNAs have proven to be embryonic or postnatal lethal, emphasizing that lncRNA are not simply transcriptional noise (Sauvageau et al. 2013) (Marahrens et al. 1997). Work with ES and iPS cells have shown that lncRNAs are highly dynamic and play an integral role in both maintaining pluripotency and promoting differentiation, so it's attractive to envision that they would be essential players in the face of a dynamic CD4+ immune response (Guttman et al. 2011) (Sheik Mohamed et al. 2010) (Loewer et al. 2010) (Figure 2).
lncRNAs are a remarkably diverse class of transcripts that together are defined as being larger than 200 nucleotides and lacking a functional open reading frame. Like mRNA, lncRNAs may be transcribed by RNA polymerase II, 5’ capped, spliced, 3’ polyadenylated, and bound by ribosomes (Ingolia et al. 2011). Several large scale studies have been conducted to identify lncRNAs resulting in an overwhelming number of transcripts: 9277 human lncRNA genes were identified through the ENCODE project (Derrien et al. 2012); using cDNA sequencing FANTOM found 34,030 lncRNA transcripts (Maeda et al. 2006) (Carninci et al. 2005); 1586 mouse and 1833 human novel lncRNAs were identified using adjacent H3K4me3 and H3K36me3 chromatin signature signifying activated genes (Guttman et al. 2009) (Khalil et al. 2009); and 4662 lncRNAs were established using RNA-seq across 24 human tissues and cell types (Cabili et al. 2011). Two studies have examined the lncRNA population with the adaptive immune compartment. The first used custom microarrays to identify ~1200 lncRNAs within human and mouse CD8+ T cell with differential lncRNA expression between the naïve/memory and effector populations (Pang et al. 2009). Recently Keji Zhao's group through an impressive tour de force cataloged the both the poly-A and total lncRNA transcriptome from murine thymocytes and Th1, Th2, Th17, and iTreg differentiated CD4+ T cells across a two-week time course (Hu et al. 2013a). 1524 lncRNA-expressing genomic regions were identified, the majority of which were polyadenylated, with 464 regions in the double-negative thymocytes, 515 in the double- and single- positive thymocytes, and 646 in the naïve / differentiated CD4+ helper T cell subsets. The lncRNAs were dynamically regulated both temporally and across the various cell types; with the T helper cells subsets only 15.5% of the lncRNAs were shared, in contrast to 78% of the 13,934 mRNA transcripts.
lncRNAs have been shown to act through an assortment of differing mechanisms. A strong trend that has emerged is that lncRNAs play a prominent role forming ribonucleoprotein complexes that mediate epigenetic regulation and transcriptional expression of highly specified target genes (Mattick et al. 2009) (Bernstein and Allis 2005) (Rodriguez-Campos and Azorin 2007). Not only is RNA is an integral component of chromatin but many histone methyltransferases lack DNA binding properties, instead possessing RNA binding motifs, indicating that lncRNAs may play a much more pervasive role that currently recognized (Rodriguez-Campos and Azorin 2007) (Khalil et al. 2009). One such example is TMEVPG1 (NeST; LincR-Ifng-3’AS), a lncRNA expressed in Th1 and CD8+, but not NK cells (Vigneau et al. 2003) (Collier et al. 2012) (Gomez et al. 2013). This lncRNA protects against persistent Theiler's virus infection by regulating the epigenetics of Inf-γ through association with WDR5, a scaffolding protein for the SET1 family of methyltransferases.
In addition to acting as scaffolds for histone modifying proteins, lncRNAs have been reported to regulate multi-protein complexes responsible for transcription factor activity. NRON (ncRNA repressor of the nuclear factor of activated T cells), a lncRNA expressed in both mice and humans, acts to bring together NFAT, IQGAP1, and the NFAT kinases CK1, GSK3, and DYRK, thereby maintaining NFAT in a phosphorylated, inactive state (Willingham et al. 2005) (Sharma et al. 2011). Depletion of NRON resulted in enhanced NFAT dephosphorylation, nuclear transport, and increased IL-2 production. None of the proteins within the NRON-NFAT complex contained an identified RNA-binding domain, suggesting that lncRNAs may play a wider role in transcription factor regulation than previous appreciated.
lncRNAs can also serve as direct regulators of transcription, as is with the case of the lncRNA T early alpha (TEA). The act of transcribing TEA regulates transcription across the Jα segments of the TCRα by activating the Jα58, Jα57, and Jα59 promoters while also opening the chromatin associated with the promoter-less Jα61, Jα53, and Jα52 regions. Knocking out TEA, or inhibiting its transcription through the insertion of a transcription terminator cassette abrogates germline transcription across these segments (Abarrategui and Krangel 2006) (Abarrategui and Krangel 2007) (Corcoran 2010).
lncRNAs can also regulate T cell function by acting as decoy targets for protein and miRNAs. GAS5 represses the glucocorticoid pathway by acting to inhibit glucocorticoid receptor (GR) transcriptional regulation of glucocorticoid-responsive genes (Kino et al. 2010). Repression is mediated through a hairpin structure on GAS5 that resembles the glucocorticoid response element (GRE) found within DNA. The GAS5 GRE-decoy binds to the GR (Kd ~30 nM, compared to ~60 nM for DNA-GRE binding to GR), thereby preventing GR binding to the DNA. The lncRNA contains a 5’ terminal oligopyrimidine tract (5’ TOP) that results in active degradation by nonsense-mediated decay in proliferating cells but confers transcript stability under conditions that result in growth arrest such as serum starvation, loss of growth factors, or pharmacological inhibition of the mTOR pathway (Schneider et al. 1988) (Coccia et al. 1992). Depletion of GAS5 by siRNA in T cells prompted continued proliferation in otherwise exhausted cultures, while over-expression of GAS5 attenuated proliferation (Mourtada-Maarabouni et al. 2008). lncRNAs can also function as miRNA decoys (Poliseno et al. 2010) (Salmena et al. 2011). Termed competing endogenouse RNAs (ceRNAs), they can act as miRNA sponges to sequester miRNAs and protect protein-coding mRNAs from degradation. Recently a new class of circularized miRNA sinks (cirRNAs) has been identified with increased stability and a higher capacity to sequester miRNAs than ceRNAs (Hansen et al. 2013) (Memczak et al. 2013).
An exciting new area of RNA biology is the discovery that active enhancers produce non-polyadenylated RNA transcripts, called eRNA (enhancer RNA) (Djebali et al. 2012). eRNAs are bi-directionally transcribed from the enhancer and function to promote chromatin looping and formation of the RNAPII transcription machinery (Mousavi et al. 2013) (Melo et al. 2013) (Li et al. 2013). Notably, the presence of eRNAs appear to have a strong association with active enhancers, and may prove to be a better indicator of enhancer activity than currently identified epigenetic markers (Wang et al. 2011a) (Figure 2).
Non-coding RNA is highly dynamic both in its expression and functionality. Given the central role that it plays in regulating epigenetic states and transcriptional networks, we predict that it will be increasingly found to play a pervasive role in the regulation of helper T cell plasticity.
4.6 Higher-order chromatin organization for cell identity
As mentioned previously, enhancers are the key to tune the cell-type specific gene expression profile. It is believed that enhancers act through physical contacts with target gene promoters. Therefore, higher-order chromatin conformation that forms loops to facilitate enhancer-promoter interactions and to exclude intervening genes has become a critical aspect for studying gene regulation. Indeed, the genome-wide mapping of promoter-enhancer interactomes reveals that global gene expression is fine-tuned by tissue-specific enhancers, even for those genes that are not cell-type specific (Kieffer-Kwon et al. 2013). For instance, within near five thousand promoter interactions shared by B cells and ES cells, up to 90% use at least one cell type-specific enhancer, indicating genome-wide dynamics of enhancer landscape during development and differentiation (Kieffer-Kwon et al. 2013).
In nucleus, the genome is folded complexly in a hierarchical manner (Gibcus and Dekker 2013). Instead of randomly tangling with each other, chromosomes tend to occupy their own territories that contain euchromatin and heterochromatin compartments for aggregation of active and inactive genes, respectively (Lieberman-Aiden et al. 2009). Using 3C-based assays, megabase-scaled topologically associated domains (TADs) are found in both euchromatin and heterochromatin compartments. The boundaries of TADs are invariant between different cell types and are conserved between distinct species (Dixon et al. 2012) (Nora et al. 2012). However, submegabase-sized interactions that reside within the TADs reveal cell-type specificity (Phillips-Cremins et al. 2013).
The DNA looping within sub-TADs can be partially explained by the interplay between three well-recognized chromatin organizers, CCCTC-binding factor (CTCF), Mediator and cohesin. It is appreciated that CTCF and cohesin colocalize to form long-range structural loops that may support short-range enhancer-promoter interactions bridged by Mediator and cohesin (Phillips-Cremins et al. 2013). Master transcription factors and Polycomb proteins also play essential roles for the formation of cell-type specific chromatin architecture. For instance, in mouse pluripotent stem cells, lineage-specific master transcription factors, Nanog, Sox2, and Oct4, binding sites preferentially interact with each other and frequently colocalize with Polycomb proteins. Depletion of one master regulator or Polycomb subunit disrupts their associated DNA contacts, but not the overall chromosome topology (de Wit et al. 2013) (Denholtz et al. 2013).
Studies have identified cell type-specific and stimulus-inducible chromatin architecture on immune-related genes; most of them focus on uni-gene regulation. For instance, Ifng locus forms a chromatin hub specifically in Th1 cells but not in Th2 or naïve CD4+ T cells. This Th1-specific chromatin hub is CTCF/cohesin/T-bet-dependent and is conserved between human and mouse (Sekimata et al. 2009) (Hadjur et al. 2009). Similarly, the contacts across Th2 cytokine (IL-4, Il-5 and Il-13) locus appear preferentially in T and NK cells but not in B cells or fibroblast (Spilianakis and Flavell 2004). Upon activation of Th2 cells, the Th2 cytokine locus transforms from a “poised” status to a more complicated “cage-like” configuration with multiple loops along with gene upregulation (Cai et al. 2006). Intriguingly, in primary human fibroblast cells, the enhancers of TNF-α responding genes already interact with their target promoters before stimulation (Jin et al. 2013), suggesting the cell-type specific chromosome topology can also set the stage for inducible genes to be rapidly activated.
Inter-chromosomal interactions also play a role in hierarchical gene regulation and co-regulation. During CD4+T cells differentiation, Ifn-γ locus on chromosome 10 and Th2 cytokine on chromosome 11 interact with each other specifically in naïve CD4+ T cells while they are inactive and then dissociates during differentiation, suggesting a co-regulation or “poised” nuclear organization for lineage-specific genes (Spilianakis et al. 2005). To sum up, the cell-type specific networking of chromatin architecture may determine cell identity by framing cis-regulatory element activity for gene expression.
5 Conclusions
What should be obvious at this point is that there is no shortage of mechanisms that can promote flexibility of responses in CD4 T cells. Indeed, recent advances have shaken fundamental views of terminal differentiation and lineage commitment – indeed, thanks to the pioneering work of Yamanaka and colleagues, it is commonplace to reprogram fully differentiated cells with four transcription factors and/or a cocktail of drugs (Yamanaka and Blau 2010) (Malik and Rao 2013) (Obokata et al. 2014) and most recently by incubating cells in acidic medium. If lymphocytes turn into stem cells by brief exposures to relatively innocuous stressors such as low pH or pipetting, it is increasingly difficult to argue that helper T cell subsets are immune from changing phenotypes. A few years ago, we would have said with certainty that CD4 and CD8 cells are unquestionably stable lineages; however, recent work points to the fact that even such cells can alter their transcriptional program (Mucida et al. 2013). In view of these developments along with much experimental data in CD4 T cells, it seems appropriate to recognize that terms like Th1, Th2 and Th17 cells, should really be thought of as a shorthand that approximates functionality. But we should not deceive ourselves: this convenient, but artificial shorthand should by no means be interpreted to suggest that populations of T cells are terminally differentiated and unable to acquire new functionalities.
In this review, we have tried to emphasize that helper T cells express a network of transcription factors that can work in concert, in opposition or to allow for specialized function. T cell master regulators, if they exist, work with a vast array of other factors. Transcription factors can be covalently modified to activate or inactivate their function and can be degraded. The epigenetic modifications of many of these key transcription factors permit flexible expression of master regulator transcription factors, even when modifications of effector cytokine genes appear “fixed”. Formation of the epigenetic landscape that modulates the accessibility of transcription factors and other key genes is profoundly affected by signals emanating from the T cell receptor and cytokine receptors. Moreover, we now recognize that cell signalling is intimately linked to cellular metabolism and microbiome. Epigenomic landscapes integrate all these environmental signals and thereby can be considered as an extension of signal transduction.
The good news is that we can now measure it all: genomes, epigenomes, metabolomes, microbiomes and transcriptomes. The bad news is that we can measure it all. How do we understand the complexity of multilayer networks built on top of each other? What tools do we need to develop to assist in providing models that explain the biology of CD4 T cells in health and disease? Moving ahead, all T cell biologists will need to adapt “systems biology” approach in some way. Although hypothesis-driven studies with a focus on a few select genes remain useful, one needs to bear in mind all the things that could, but are not being measured would influence the experimental outcome. As our annotation of the human genome becomes more complete and we develop more sophisticated understanding of the impact of human genetic variation, we are approaching a new era that can redefine T cell function in health and disease by employing “omic” views to comprehensively quantitate states of dynamic cellular activities.
References
- Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nature immunology. 2006;7(10):1109–1115. doi: 10.1038/ni1379. doi:10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. Embo J. 2007;26(20):4380–4390. doi: 10.1038/sj.emboj.7601866. doi:10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S, Jiang S, Kuchroo VK, Mathis D, Roncarolo MG, Rudensky A, Sakaguchi S, Shevach EM, Vignali DA, Ziegler SF. Regulatory T cells: recommendations to simplify the nomenclature. Nature immunology. 2013;14(4):307–308. doi: 10.1038/ni.2554. doi:10.1038/ni.2554. [DOI] [PubMed] [Google Scholar]
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383(6603):787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS genetics. 2013;9(8):e1003678. doi: 10.1371/journal.pgen.1003678. doi:10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007a;8(9):942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007b;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33(2):279–288. doi: 10.1016/j.immuni.2010.08.010. doi:10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. Journal of immunology. 2009;183(3):1598–1606. doi: 10.4049/jimmunol.0803302. doi:10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altorok N, Coit P, Hughes T, Koelsch KA, Stone DU, Rasmussen A, Radfar L, Scofield RH, Sivils KL, Farris AD, Sawalha AH. Genome-wide DNA methylation patterns in naive CD4 T cells from patients with primary Sjogren's syndrome. Arthritis and rheumatism. 2013 doi: 10.1002/art.38264. doi:10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31(2):220–231. doi: 10.1016/j.immuni.2009.06.024. doi:10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E, Romagnani S. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204(8):1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual review of immunology. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. doi:10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4(7):616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. doi:10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. doi:10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. doi:10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune TM, Collins PL, Collier SP, Henderson MA, Chang S. Epigenetic Activation and Silencing of the Gene that Encodes IFN-gamma. Front Immunol. 2013;4:112. doi: 10.3389/fimmu.2013.00112. doi:10.3389/fimmu.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, Weiner HL. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nature immunology. 2007;8(12):1380–1389. doi: 10.1038/ni1541. doi:10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- Bailey-Bucktrout SL, Bluestone JA. Regulatory T cells: stability revisited. Trends in immunology. 2011;32(7):301–306. doi: 10.1016/j.it.2011.04.002. doi:10.1016/j.it.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Bucktrout SL, Martinez-Llordella M, Zhou X, Anthony B, Rosenthal W, Luche H, Fehling HJ, Bluestone JA. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 2013;39(5):949–962. doi: 10.1016/j.immuni.2013.10.016. doi:10.1016/j.immuni.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36(5):847–856. doi: 10.1016/j.immuni.2012.02.012. doi:10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AM, Smithers N, Lee K, Witherington J, Tough DF, Prinjha RK, Peters B, Rao A. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(36):14532–14537. doi: 10.1073/pnas.1212264109. doi:10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. European journal of immunology. 2010;40(1):225–231. doi: 10.1002/eji.200939381. doi:10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J, Thiel A, Boeld TJ, Hoffmann P, Edinger M, Turbachova I, Hamann A, Olek S, Huehn J. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. doi:10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Gregori S, Bacchetta R, Roncarolo MG. Tr1 cells: from discovery to their clinical application. Seminars in immunology. 2006;18(2):120–127. doi: 10.1016/j.smim.2006.01.007. doi:10.1016/j.smim.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, Lee J, de Sauvage FJ, Ghilardi N. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat Immunol. 2006;7(9):929–936. doi: 10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. The American journal of pathology. 1963;42:471–483. [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nature reviews Immunology. 2013;13(9):666–678. doi: 10.1038/nri3494. doi:10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nature immunology. 2013;14(8):840–848. doi: 10.1038/ni.2642. doi:10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160(4):258–266. doi: 10.1016/j.trsl.2012.02.005. doi:10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bending D, De La Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009 doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nature genetics. 2001;27(1):20–21. doi: 10.1038/83713. doi:10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Beriou G, Costantino CM, Ashley CW, Yang L, Kuchroo VK, Baecher-Allan C, Hafler DA. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113(18):4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes & development. 2005;19(14):1635–1655. doi: 10.1101/gad.1324305. doi:10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas PS, Gupta S, Chang E, Song L, Stirzaker RA, Liao JK, Bhagat G, Pernis AB. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. The Journal of clinical investigation. 2010;120(9):3280–3295. doi: 10.1172/JCI42856. doi:10.1172/JCI42856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluml S, Bonelli M, Niederreiter B, Puchner A, Mayr G, Hayer S, Koenders MI, van den Berg WB, Smolen J, Redlich K. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis and rheumatism. 2011;63(5):1281–1288. doi: 10.1002/art.30281. doi:10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. The Journal of experimental medicine. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. doi:10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniface K, Blumenschein WM, Brovont-Porth K, McGeachy MJ, Basham B, Desai B, Pierce R, McClanahan TK, Sadekova S. Malefyt RD Human Th17 Cells Comprise Heterogeneous Subsets including IFN-{gamma}-Producing Cells with Distinct Properties from the Th1 Lineage. J Immunol. doi: 10.4049/jimmunol.1000366. [DOI] [PubMed] [Google Scholar]
- Brennan P, Babbage JW, Burgering BM, Groner B, Reif K, Cantrell DA. Phosphatidylinositol 3-kinase couples the interleukin-2 receptor to the cell cycle regulator E2F. Immunity. 1997;7(5):679–689. doi: 10.1016/s1074-7613(00)80388-x. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nature genetics. 2001;27(1):68–73. doi: 10.1038/83784. doi:10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nature immunology. 2007;8(9):958–966. doi: 10.1038/ni1500. doi:10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nature immunology. 2013a;14(10):1007–1013. doi: 10.1038/ni.2683. doi:10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013b;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. doi:10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. doi:10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature genetics. 2006;38(11):1278–1288. doi: 10.1038/ng1913. doi:10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cannons JL, Lu KT, Schwartzberg PL. T follicular helper cell diversity and plasticity. Trends in immunology. 2013;34(5):200–207. doi: 10.1016/j.it.2013.01.001. doi:10.1016/j.it.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. doi:10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. doi:10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. doi:10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Barbi J, Bu S, Yang HY, Li Z, Gao Y, Jinasena D, Fu J, Lin F, Chen C, Zhang J, Yu N, Li X, Shan Z, Nie J, Gao Z, Tian H, Li Y, Yao Z, Zheng Y, Park BV, Pan Z, Dang E, Wang H, Luo W, Li L, Semenza GL, Zheng SG, Loser K, Tsun A, Greene MI, Pardoll DM, Pan F, Li B. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity. 2013;39(2):272–285. doi: 10.1016/j.immuni.2013.08.006. doi:10.1016/j.immuni.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Tato CM, Muul L, Laurence A, O'Shea JJ. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. 2007;56(9):2936–2946. doi: 10.1002/art.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jang H, Kim H, Lee JH, Kim ST, Cho EJ, Youn HD. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic acids research. 2014;42(1):224–234. doi: 10.1093/nar/gkt873. doi:10.1093/nar/gkt873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. The Journal of experimental medicine. 2008;205(9):2005–2017. doi: 10.1084/jem.20081219. doi:10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM. Intracellular sensing of amino acids in Xenopus laevis oocytes stimulates p70 S6 kinase in a target of rapamycin-dependent manner. The Journal of biological chemistry. 2002;277(12):9952–9957. doi: 10.1074/jbc.M107694200. doi:10.1074/jbc.M107694200. [DOI] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17(8):983–988. doi: 10.1038/nm.2426. doi:10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. doi:10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L, Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Molecular and cellular biology. 1992;12(8):3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Crome SQ, MacDonald KG, Dai EL, Mager DL, Levings MK. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. Journal of immunology. 2011;187(11):5615–5626. doi: 10.4049/jimmunol.1101058. doi:10.4049/jimmunol.1101058. [DOI] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. Journal of immunology. 2012;189(5):2084–2088. doi: 10.4049/jimmunol.1200774. doi:10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. doi:10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450(7169):566–569. doi: 10.1038/nature06306. doi:10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. The Journal of experimental medicine. 2007;204(8):1757–1764. doi: 10.1084/jem.20070590. doi:10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes SM, Pelly VS, Wilson MS. Plasticity within the alphabeta(+)CD4(+) T-cell lineage: when, how and what for? Open Biol. 2013;3(1):120157. doi: 10.1098/rsob.120157. doi:10.1098/rsob.120157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Seminars in immunology. 2010;22(6):353–361. doi: 10.1016/j.smim.2010.08.001. doi:10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. Journal of immunology. 1988;140(7):2197–2200. [PubMed] [Google Scholar]
- Corvetta A, Della Bitta R, Luchetti MM, Pomponio G. 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J Chromatogr. 1991;566(2):481–491. doi: 10.1016/0378-4347(91)80265-e. [DOI] [PubMed] [Google Scholar]
- Cosmi L, Cimaz R, Maggi L, Santarlasci V, Capone M, Borriello F, Frosali F, Querci V, Simonini G, Barra G, Piccinni MP, Liotta F, De Palma R, Maggi E, Romagnani S, Annunziato F. Evidence of the transient nature of the Th17 phenotype of CD4+CD161+ T cells in the synovial fluid of patients with juvenile idiopathic arthritis. Arthritis and rheumatism. 2011;63(8):2504–2515. doi: 10.1002/art.30332. doi:10.1002/art.30332. [DOI] [PubMed] [Google Scholar]
- Crome SQ, Lang PA, Lang KS, Ohashi PS. Natural killer cells regulate diverse T cell responses. Trends in immunology. 2013;34(7):342–349. doi: 10.1016/j.it.2013.03.002. doi:10.1016/j.it.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH). Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. doi:10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Crotty S. The 1-1-1 fallacy. Immunological reviews. 2012;247(1):133–142. doi: 10.1111/j.1600-065X.2012.01117.x. doi:10.1111/j.1600-065X.2012.01117.x. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. doi:10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, Khoury S, Oukka M, Kuchroo VK. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nature immunology. 2008;9(12):1347–1355. doi: 10.1038/ni.1677. doi:10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, Heard E, Geijsen N, Poot RA, Chambers I, de Laat W. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501(7466):227–231. doi: 10.1038/nature12420. doi:10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- Deenick EK, Chan A, Ma CS, Gatto D, Schwartzberg PL, Brink R, Tangye SG. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity. 2010;33(2):241–253. doi: 10.1016/j.immuni.2010.07.015. doi:10.1016/j.immuni.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832–844. doi: 10.1016/j.immuni.2009.04.014. doi:10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature immunology. 2011;12(4):295–303. doi: 10.1038/ni.2005. doi:10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D, Bonnevier J, Workman CJ, Vignali DA. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501(7466):252–256. doi: 10.1038/nature12428. doi:10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13(5):602–616. doi: 10.1016/j.stem.2013.08.013. doi:10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. doi:10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. doi:10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. doi:10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. Genetic controls of Th17 cell differentiation and plasticity. Exp Mol Med. 2011;43(1):1–6. doi: 10.3858/emm.2011.43.1.007. doi:10.3858/emm.2011.43.1.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nature immunology. 2009;10(8):857–863. doi: 10.1038/ni.1767. doi:10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Engelhardt KR, Shah N, Faizura-Yeop I, Kocacik Uygun DF, Frede N, Muise AM, Shteyer E, Filiz S, Chee R, Elawad M, Hartmann B, Arkwright PD, Dvorak C, Klein C, Puck JM, Grimbacher B, Glocker EO. Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2013;131(3):825–830. doi: 10.1016/j.jaci.2012.09.025. doi:10.1016/j.jaci.2012.09.025. [DOI] [PubMed] [Google Scholar]
- Escobar T, Yu CR, Muljo SA, Egwuagu CE. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2013;54(6):4017–4025. doi: 10.1167/iovs.13-11937. doi:10.1167/iovs.13-11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One. 2011;6(3):e17739. doi: 10.1371/journal.pone.0017739. doi:10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. doi:10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4(4):330–336. doi: 10.1038/ni904. doi:10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nature reviews Immunology. 2005;5(11):844–852. doi: 10.1038/nri1710. doi:10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nature immunology. 2012;13(10):972–980. doi: 10.1038/ni.2420. doi:10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. doi:10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. doi:10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, Guo B, Herbert DR, Bulfone A, Trentini F, Di Serio C, Bacchetta R, Andreani M, Brockmann L, Gregori S, Flavell RA, Roncarolo MG. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nature medicine. 2013;19(6):739–746. doi: 10.1038/nm.3179. doi:10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J. 2008;416(3):375–385. doi: 10.1042/BJ20081668. doi:10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. doi:10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–328. doi: 10.1016/j.immuni.2010.02.008. doi:10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W. O'Shea JJ Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. doi: 10.1016/j.molcel.2013.02.011. doi:10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatman Zaretsky A, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. The Journal of experimental medicine. 2009;206(5):991–999. doi: 10.1084/jem.20090303. doi:10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361(21):2033–2045. doi: 10.1056/NEJMoa0907206. doi:10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka R, Barnett LG, Silver JS, O'Neill PJ, Hunter CA, Cancro MP, Laufer TM. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. Journal of immunology. 2011;187(3):1091–1095. doi: 10.4049/jimmunol.1100853. doi:10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. doi:10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. doi:10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. doi:10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori S, Goudy KS, Roncarolo MG. The cellular and molecular mechanisms of immuno-suppression by human type 1 regulatory T cells. Front Immunol. 2012;3:30. doi: 10.3389/fimmu.2012.00030. doi:10.3389/fimmu.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. doi:10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. doi:10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. doi:10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. doi:10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. doi:10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. doi:10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29(4):637–649. doi: 10.1016/j.immuni.2008.08.009. doi:10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337(6101):1553–1556. doi: 10.1126/science.1220961. doi:10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. doi:10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen W, Hutzler M, Abel S, Alter C, Stockmann C, Kliche S, Albert J, Sparwasser T, Sakaguchi S, Westendorf AM, Schadendorf D, Buer J, Helfrich I. Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. The Journal of experimental medicine. 2012;209(11):2001–2016. doi: 10.1084/jem.20111497. doi:10.1084/jem.20111497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Tanaka S, Motomura Y, Ohno S, Yanagi Y, Inoue H, Kubo M. The 3' enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity. 2012;36(2):188–200. doi: 10.1016/j.immuni.2012.02.002. doi:10.1016/j.immuni.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334(6057):825–829. doi: 10.1126/science.1208421. doi:10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 32(1):116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. doi:10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. doi:10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K, Vahedi G, Ghoreschi K, Yang XP, Nakayamada S, Kanno Y, O'Shea JJ, Laurence A. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology. 2011;134(3):235–245. doi: 10.1111/j.1365-2567.2011.03483.x. doi:10.1111/j.1365-2567.2011.03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature reviews Immunology. 2010;10(3):159–169. doi: 10.1038/nri2710. doi:10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Hori S. Regulatory T cell plasticity: beyond the controversies. Trends in immunology. 2011;32(7):295–300. doi: 10.1016/j.it.2011.04.004. doi:10.1016/j.it.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. doi:10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Horwitz DA, Zheng SG, Gray JD. Natural and TGF-beta-induced Foxp3(+)CD4(+) CD25(+) regulatory T cells are not mirror images of each other. Trends Immunol. 2008;29(9):429–435. doi: 10.1016/j.it.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nature immunology. 2013a;14(11):1190–1198. doi: 10.1038/ni.2712. doi:10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O'Connell RM. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. Journal of immunology. 2013b;190(12):5972–5980. doi: 10.4049/jimmunol.1300351. doi:10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Brustle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Low E, Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(52):20846–20851. doi: 10.1073/pnas.0809077106. doi:10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nature reviews Immunology. 2009;9(2):83–89. doi: 10.1038/nri2474. doi:10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O'Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2(6):1697–1709. doi: 10.1016/j.celrep.2012.10.025. doi:10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. The Journal of experimental medicine. 2005a;202(9):1289–1300. doi: 10.1084/jem.20051044. doi:10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005b;307(5708):430–433. doi: 10.1126/science.1103336. doi:10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. doi:10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. doi:10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. doi:10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang EJ, Park HR, Hong JH, Hwang ES. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. Journal of immunology. 2013;190(11):5764–5770. doi: 10.4049/jimmunol.1203403. doi:10.4049/jimmunol.1203403. [DOI] [PubMed] [Google Scholar]
- Janson PC, Winerdal ME, Marits P, Thorn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3(2):e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. doi:10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209(2):243–250. doi: 10.1084/jem.20111174. doi:10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. doi:10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten LA, Leoni F, Meghji S, Mascagni P. Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol Med. 2011;17(5-6):391–396. doi: 10.2119/molmed.2011.00058. doi:10.2119/molmed.2011.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A. Pathophysiology of translational regulation by microRNAs in multiple sclerosis. FEBS Lett. 2011;585(23):3738–3746. doi: 10.1016/j.febslet.2011.03.052. doi:10.1016/j.febslet.2011.03.052. [DOI] [PubMed] [Google Scholar]
- Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nature immunology. 2001;2(1):37–44. doi: 10.1038/83144. doi:10.1038/83144. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annual review of immunology. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. doi:10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. doi:10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. doi:10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei CL, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155(7):1507–1520. doi: 10.1016/j.cell.2013.11.039. doi:10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Sklarz T, Banks LB, Gohil M, Waickman AT, Skuli N, Krock BL, Luo CT, Hu W, Pollizzi KN, Li MO, Rathmell JC, Birnbaum MJ, Powell JD, Jordan MS, Koretzky GA. Natural and inducible TH17 cells are regulated differently by Akt and mTOR pathways. Nature immunology. 2013;14(6):611–618. doi: 10.1038/ni.2607. doi:10.1038/ni.2607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. doi:10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. The Journal of experimental medicine. 2009;206(5):1001–1007. doi: 10.1084/jem.20090313. doi:10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. doi:10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496(7446):518–522. doi: 10.1038/nature11868. doi:10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37(3):501–510. doi: 10.1016/j.immuni.2012.05.031. doi:10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-Hora M, Kodama T, Tanaka S, Bluestone JA, Takayanagi H. Pathogenic conversion of Foxp3(+) T cells into TH17 cells in autoimmune arthritis. Nature medicine. 2014;20(1):62–68. doi: 10.1038/nm.3432. doi:10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. doi:10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. doi:10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31(6):941–952. doi: 10.1016/j.immuni.2009.10.008. doi:10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont V, Astoul E, Laurence A, Liautard J, Cantrell D. The T cell antigen receptor activates phosphatidylinositol 3-kinase-regulated serine kinases protein kinase B and ribosomal S6 kinase 1. FEBS Lett. 2000;486(1):38–42. doi: 10.1016/s0014-5793(00)02235-3. [DOI] [PubMed] [Google Scholar]
- Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. Journal of immunology. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Amarnath S, Mariotti J, Kim YC, Foley J, Eckhaus M, O'Shea JJ, Fowler DH. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity. 2012;37(2):209–222. doi: 10.1016/j.immuni.2012.05.027. doi:10.1016/j.immuni.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nature immunology. 2012;13(10):991–999. doi: 10.1038/ni.2416. doi:10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21(3):274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, Modena D, Moras ML, Pozzi P, Reznikov LL, Siegmund B, Fantuzzi G, Dinarello CA, Mascagni P. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11(1-12):1–15. doi: 10.2119/2006-00005.Dinarello. doi:10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Dona G, Fossati G, Sozzani S, Azam T, Bufler P, Fantuzzi G, Goncharov I, Kim SH, Pomerantz BJ, Reznikov LL, Siegmund B, Dinarello CA, Mascagni P. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2995–3000. doi: 10.1073/pnas.052702999. doi:10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. doi:10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, Goronzy JJ. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nature medicine. 2012;18(10):1518–1524. doi: 10.1038/nm.2963. doi:10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134(3):392–404. doi: 10.1016/j.cell.2008.07.025. doi:10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25(3):455–471. doi: 10.1016/j.immuni.2006.07.011. doi:10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007a;26(5):579–591. doi: 10.1016/j.immuni.2007.03.014. doi:10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007b;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. doi:10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. doi:10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nature immunology. 2011;12(6):551–559. doi: 10.1038/ni.2030. doi:10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. doi:10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HS, Hu CY, Chan HY, Liew YY, Huang HP, Lepescheux L, Bastianelli E, Baron R, Rawadi G, Clement-Lacroix P. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br J Pharmacol. 2007;150(7):862–872. doi: 10.1038/sj.bjp.0707165. doi:10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. The Journal of experimental medicine. 2008;205(9):1993–2004. doi: 10.1084/jem.20081062. doi:10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205(6):1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42(12):1113–1117. doi: 10.1038/ng.710. doi:10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11808–11812. doi: 10.1073/pnas.182425099. doi:10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta}. The Journal of biological chemistry. 2010;285(53):41328–41336. doi: 10.1074/jbc.M110.146852. doi:10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, Anderson SM, Wei L, Sun H, O'Shea JJ, Schwartzberg PL. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35(4):622–632. doi: 10.1016/j.immuni.2011.07.015. doi:10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142(6):914–929. doi: 10.1016/j.cell.2010.08.012. doi:10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. doi:10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, Arkwright PD, Kreins AY, Averbuch D, Engelhard D, Magdorf K, Kilic SS, Minegishi Y, Nonoyama S, French MA, Choo S, Smart JM, Peake J, Wong M, Gray P, Cook MC, Fulcher DA, Casanova JL, Deenick EK, Tangye SG. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119(17):3997–4008. doi: 10.1182/blood-2011-11-392985. doi:10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, Bult CJ, Fletcher CF, Forrest AR, Furuno M, Hill D, Itoh M, Kanamori-Katayama M, Katayama S, Katoh M, Kawashima T, Quackenbush J, Ravasi T, Ring BZ, Shibata K, Sugiura K, Takenaka Y, Teasdale RD, Wells CA, Zhu Y, Kai C, Kawai J, Hume DA, Carninci P, Hayashizaki Y. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS genetics. 2006;2(4):e62. doi: 10.1371/journal.pgen.0020062. doi:10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani CF, Alberigo G, Bacchetta R, Serafini G, Andreani M, Roncarolo MG, Gregori S. Killing of myeloid APCs via HLA class I, CD2 and CD226 defines a novel mechanism of suppression by human Tr1 cells. European journal of immunology. 2011;41(6):1652–1662. doi: 10.1002/eji.201041120. doi:10.1002/eji.201041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nature immunology. 2003;4(12):1183–1190. doi: 10.1038/ni1004. doi:10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods in molecular biology. 2013;997:23–33. doi: 10.1007/978-1-62703-348-0_3. doi:10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & development. 1997;11(2):156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–935. doi: 10.1038/nature05478. doi:10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31(1):51–59. doi: 10.1002/bies.080099. doi:10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. doi:10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8(12):1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Cua DJ. Th17 cell differentiation: the long and winding road. Immunity. 2008;28(4):445–453. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. The Journal of experimental medicine. 2013;210(11):2181–2190. doi: 10.1084/jem.20130376. doi:10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. doi:10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. doi:10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36(2):262–275. doi: 10.1016/j.immuni.2011.12.012. doi:10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci. 2008;121(Pt 7):939–946. doi: 10.1242/jcs.024646. doi:10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. doi:10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, Attinger A, Shui JW, Kim G, Lena CJ, Sakaguchi S, Miyamoto C, Wang P, Atarashi K, Park Y, Nakayama T, Honda K, Ellmeier W, Kronenberg M, Taniuchi I, Cheroutre H. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nature immunology. 2013;14(3):281–289. doi: 10.1038/ni.2523. doi:10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–260. doi: 10.1126/science.1145697. doi:10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 32(5):616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. Journal of immunology. 2011;187(5):2213–2221. doi: 10.4049/jimmunol.1003952. doi:10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. doi:10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagome K, Imamura M, Kawahata K, Harada H, Okunishi K, Matsumoto T, Sasaki O, Tanaka R, Kano MR, Chang H, Hanawa H, Miyazaki J, Yamamoto K, Dohi M. High expression of IL-22 suppresses antigen-induced immune responses and eosinophilic airway inflammation via an IL-10-associated mechanism. Journal of immunology. 2011;187(10):5077–5089. doi: 10.4049/jimmunol.1001560. doi:10.4049/jimmunol.1001560. [DOI] [PubMed] [Google Scholar]
- Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis and rheumatism. 2008;58(5):1284–1292. doi: 10.1002/art.23429. doi:10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O'Shea JJ. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35(6):919–931. doi: 10.1016/j.immuni.2011.11.012. doi:10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu Y, Nishida K, Miyazawa S, Komiyama T, Kadota Y, Abe N, Yoshida A, Hirohata S, Ohtsuka A, Ozaki T. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthritis Cartilage. 2008;16(6):723–732. doi: 10.1016/j.joca.2007.10.014. doi:10.1016/j.joca.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33(1):12–24. doi: 10.1016/j.immuni.2010.07.006. doi:10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. doi:10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Komiyama T, Miyazawa S, Shen ZN, Furumatsu T, Doi H, Yoshida A, Yamana J, Yamamura M, Ninomiya Y, Inoue H, Asahara H. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis and rheumatism. 2004;50(10):3365–3376. doi: 10.1002/art.20709. doi:10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- Nistala K, Adams S, Cambrook H, Ursu S, Olivito B, de Jager W, Evans JG, Cimaz R, Bajaj-Elliott M, Wedderburn LR. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14751–14756. doi: 10.1073/pnas.1003852107. doi:10.1073/pnas.1003852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala K, Moncrieffe H, Newton KR, Varsani H, Hunter P, Wedderburn LR. Interleukin-17-producing T cells are enriched in the joints of children with arthritis, but have a reciprocal relationship to regulatory T cell numbers. Arthritis and rheumatism. 2008;58(3):875–887. doi: 10.1002/art.23291. doi:10.1002/art.23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. doi:10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. doi:10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325(5943):1001–1005. doi: 10.1126/science.1176676. doi:10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry. 2012;287(14):11234–11239. doi: 10.1074/jbc.M111.324046. doi:10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010a;33(4):607–619. doi: 10.1016/j.immuni.2010.09.009. doi:10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nature reviews Immunology. 2010b;10(2):111–122. doi: 10.1038/nri2708. doi:10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. doi:10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, Yamato M, Vacanti CA. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014;505(7485):641–647. doi: 10.1038/nature12968. doi:10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. The Journal of experimental medicine. 2011;208(5):1001–1013. doi: 10.1084/jem.20102144. doi:10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nature immunology. 2012;13(4):405–411. doi: 10.1038/ni.2242. doi:10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Li MO. TGF-beta: guardian of T cell function. Journal of immunology. 2013;191(8):3973–3979. doi: 10.4049/jimmunol.1301843. doi:10.4049/jimmunol.1301843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, Huehn J, Fehling HJ, Sparwasser T, Nakai K, Sakaguchi S. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. doi:10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38(3):414–423. doi: 10.1016/j.immuni.2013.03.002. doi:10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature genetics. 1998;19(3):219–220. doi: 10.1038/890. doi:10.1038/890. [DOI] [PubMed] [Google Scholar]
- Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O'Brien S, Blank R, Lamb E, Natarajan S, Kastenmayer R, Hunter C, Grigg ME, Belkaid Y. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31(5):772–786. doi: 10.1016/j.immuni.2009.10.001. doi:10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin AC, Ramachandran V, Acharya S, Lewis DB. Human neonatal naive CD4+ T cells have enhanced activation-dependent signaling regulated by the microRNA miR-181a. Journal of immunology. 2013;190(6):2682–2691. doi: 10.4049/jimmunol.1202534. doi:10.4049/jimmunol.1202534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. Journal of immunology. 2009;182(12):7738–7748. doi: 10.4049/jimmunol.0900603. doi:10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101. doi: 10.1186/ar2493. doi:10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peine M, Rausch S, Helmstetter C, Frohlich A, Hegazy AN, Kuhl AA, Grevelding CG, Hofer T, Hartmann S, Lohning M. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS biology. 2013;11(8):e1001633. doi: 10.1371/journal.pbio.1001633. doi:10.1371/journal.pbio.1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295. doi: 10.1016/j.cell.2013.04.053. doi:10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. In vivo regulation of Bcl6 and T follicular helper cell development. Journal of immunology. 2010;185(1):313–326. doi: 10.4049/jimmunol.0904023. doi:10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. doi:10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217(2):133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. The Journal of clinical investigation. 1993;92(1):38–53. doi: 10.1172/JCI116576. doi:10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. doi:10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):3111–3116. doi: 10.1073/pnas.0910667107. doi:10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL, Sallusto F, Lanzavecchia A. Division of labor with a workforce of one: challenges in specifying effector and memory T cell fate. Science. 2007;317(5838):622–625. doi: 10.1126/science.1143775. [DOI] [PubMed] [Google Scholar]
- Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nature immunology. 2009;10(4):385–393. doi: 10.1038/ni.1715. doi:10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis and rheumatism. 1990;33(11):1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. doi:10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007;2(11):e1182. doi: 10.1371/journal.pone.0001182. doi:10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. doi:10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncador G, Brown PJ, Maestre L, Hue S, Martinez-Torrecuadrada JL, Ling KL, Pratap S, Toms C, Fox BC, Cerundolo V, Powrie F, Banham AH. Analysis of FOXP3 protein expression in human CD4+CD25+ regulatory T cells at the single-cell level. European journal of immunology. 2005;35(6):1681–1691. doi: 10.1002/eji.200526189. doi:10.1002/eji.200526189. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. doi:10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. doi:10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky AY. Regulatory T cells and Foxp3. Immunological reviews. 2011;241(1):260–268. doi: 10.1111/j.1600-065X.2011.01018.x. doi:10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. doi:10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. doi:10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166. doi: 10.1016/j.cell.2012.06.053. doi:10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo S, Cousins DJ, Winkelmann N, Triantaphyllopoulos K, Staynov DZ. Chromatin structure and DNA methylation of the IL-4 gene in human T(H)2 cells. Chromosome Res. 2009;17(4):485–496. doi: 10.1007/s10577-009-9040-3. doi:10.1007/s10577-009-9040-3. [DOI] [PubMed] [Google Scholar]
- Saouaf SJ, Li B, Zhang G, Shen Y, Furuuchi N, Hancock WW, Greene MI. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87(2):99–104. doi: 10.1016/j.yexmp.2009.06.003. doi:10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D'Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2(0):e01749. doi: 10.7554/eLife.01749. doi:10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzki B, Kingsley CI, Oliveira V, Karim M, Herber M, Wood KJ. IFN-gamma production by alloantigen-reactive regulatory T cells is important for their regulatory function in vivo. The Journal of experimental medicine. 2005;201(12):1925–1935. doi: 10.1084/jem.20050419. doi:10.1084/jem.20050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Hansmann L, Andreesen R, Edinger M, Hoffmann P, Rehli M. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naive Treg. European journal of immunology. 2011;41(5):1491–1498. doi: 10.1002/eji.201041067. doi:10.1002/eji.201041067. [DOI] [PubMed] [Google Scholar]
- Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson-Dupuis S, Hamlin F, Tran MV, Blankenship D, Pascual V, Savino DA, Banchereau J, Casanova JL, Ueno H. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121(17):3375–3385. doi: 10.1182/blood-2012-08-448902. doi:10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8(7):732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31(4):551–564. doi: 10.1016/j.immuni.2009.08.021. doi:10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MD, Huang L, Choi JH, Lee EJ, Wilson JM, Lemos H, Pan F, Blazar BR, Pardoll DM, Mellor AL, Shi H, Munn DH. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. 2013;38(5):998–1012. doi: 10.1016/j.immuni.2013.01.013. doi:10.1016/j.immuni.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11381–11386. doi: 10.1073/pnas.1019711108. doi:10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. Rna. 2010;16(2):324–337. doi: 10.1261/rna.1441510. doi:10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM. Regulatory T cells in autoimmmunity*. Annu Rev Immunol. 2000;18:423–449. doi: 10.1146/annurev.immunol.18.1.423. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in immunology. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. doi:10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. doi:10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, Stenvang J, Kauppinen S, Alenius H, Lauerma A, Homey B, Winqvist O, Stahle M, Pivarcsi A. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126(3):581–589. e581–520. doi: 10.1016/j.jaci.2010.05.045. doi:10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nature immunology. 2004;5(10):1017–1027. doi: 10.1038/ni1115. doi:10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. doi:10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33(2):192–202. doi: 10.1016/j.immuni.2010.07.014. doi:10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nature medicine. 2007;13(2):139–145. doi: 10.1038/nm1551. doi:10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- Stock P, Akbari O, Berry G, Freeman GJ, Dekruyff RH, Umetsu DT. Induction of T helper type 1-like regulatory cells that express Foxp3 and protect against airway hyper-reactivity. Nature immunology. 2004;5(11):1149–1156. doi: 10.1038/ni1122. doi:10.1038/ni1122. [DOI] [PubMed] [Google Scholar]
- Stockinger B, Veldhoen M, Martin B. Th17 T cells: linking innate and adaptive immunity. Seminars in immunology. 2007;19(6):353–361. doi: 10.1016/j.smim.2007.10.008. doi:10.1016/j.smim.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185(5):817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O'Shea JJ. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nature immunology. 2012;13(6):587–595. doi: 10.1038/ni.2286. doi:10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis and rheumatism. 2009;60(4):1065–1075. doi: 10.1002/art.24436. doi:10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- Thomas RM, Gamper CJ, Ladle BH, Powell JD, Wells AD. De novo DNA methylation is required to restrict T helper lineage plasticity. The Journal of biological chemistry. 2012;287(27):22900–22909. doi: 10.1074/jbc.M111.312785. doi:10.1074/jbc.M111.312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. Journal of immunology. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. doi:10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nature immunology. 2009;10(8):864–871. doi: 10.1038/ni.1770. doi:10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, Hosokawa H, Koseki H, Tokoyoda K, Suzuki Y, Motohashi S, Nakayama T. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39(5):819–832. doi: 10.1016/j.immuni.2013.09.012. doi:10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infection and immunity. 1999;67(7):3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18(3):415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, Meerding J, Berkers CR, Barbi J, Grone A, Sijts AJ, Maurice MM, Kalkhoven E, Prakken BJ, Ovaa H, Pan F, Zaiss DM, Coffer PJ. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 2013;39(2):259–271. doi: 10.1016/j.immuni.2013.05.018. doi:10.1016/j.immuni.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. The Journal of experimental medicine. 2009;206(1):43–49. doi: 10.1084/jem.20081438. doi:10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008a;453(7191):106–109. doi: 10.1038/nature06881. doi:10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Veldhoen Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008c;9(12):1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77(10):5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojinovic J, Damjanov N, D'Urzo C, Furlan A, Susic G, Pasic S, Iagaru N, Stefan M, Dinarello CA. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis and rheumatism. 2011;63(5):1452–1458. doi: 10.1002/art.30238. doi:10.1002/art.30238. [DOI] [PubMed] [Google Scholar]
- Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH, Arima K, Bover L, Hanabuchi S, Khalili J, Marinova E, Zheng B, Littman DR, Liu YJ. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106(12):4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Pan D, Lee YH, Martinez GJ, Feng XH, Dong C. Cutting edge: Smad2 and Smad4 regulate TGF-beta-mediated Il9 gene expression via EZH2 displacement. Journal of immunology. 2013;191(10):4908–4912. doi: 10.4049/jimmunol.1300433. doi:10.4049/jimmunol.1300433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011a;474(7351):390–394. doi: 10.1038/nature10006. doi:10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers. 2011b;30(4):171–179. doi: 10.3233/DMA-2011-0766. doi:10.3233/DMA-2011-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. European journal of immunology. 2007;37(1):129–138. doi: 10.1002/eji.200636435. doi:10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009a;8(12):969–981. doi: 10.1038/nrd3031. doi:10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011c;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. doi:10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011d;35(3):337–348. doi: 10.1016/j.immuni.2011.08.012. doi:10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009b;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. doi:10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annual review of immunology. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. doi:10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner HL, da Cunha AP, Quintana F, Wu H. Oral tolerance. Immunological reviews. 2011;241(1):241–259. doi: 10.1111/j.1600-065X.2011.01017.x. doi:10.1111/j.1600-065X.2011.01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. doi:10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- Winders BR, Schwartz RH, Bruniquel D. A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. Journal of immunology. 2004;173(12):7377–7384. doi: 10.4049/jimmunol.173.12.7377. [DOI] [PubMed] [Google Scholar]
- Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, Paul WE, Bosselut R, Wei G, Zhao K, Oukka M, Zhu J, Belkaid Y. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. The Journal of clinical investigation. 2011;121(11):4503–4515. doi: 10.1172/JCI57456. doi:10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496(7446):513–517. doi: 10.1038/nature11984. doi:10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33(3):313–325. doi: 10.1016/j.immuni.2010.09.001. doi:10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, Ruminski P, Weiss D, Von Schack D, Bluestone JA. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. The Journal of experimental medicine. 2012;209(10):1713–1722. S1711–1719. doi: 10.1084/jem.20120822. doi:10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465(7299):704–712. doi: 10.1038/nature09229. doi:10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Paul WE. Memory CD4+ T cells: fate determination, positive feedback and plasticity. Cell Mol Life Sci. 2012;69(10):1577–1583. doi: 10.1007/s00018-012-0966-9. doi:10.1007/s00018-012-0966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. miR-146a controls the resolution of T cell responses in mice. The Journal of experimental medicine. 2012;209(9):1655–1670. doi: 10.1084/jem.20112218. doi:10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, Feng XH, Jetten AM, Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008a;29(1):44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008b;28(1):29–39. doi: 10.1016/j.immuni.2007.11.016. doi:10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O'Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature immunology. 2011;12(3):247–254. doi: 10.1038/ni.1995. doi:10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496(7446):461–468. doi: 10.1038/nature11981. doi:10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR, Penix L, Wilson CB, Melvin AJ, McGurn ME, et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. Journal of immunology. 1994;153(8):3603–3610. [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–468. doi: 10.1016/j.immuni.2009.07.002. doi:10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272(34):21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. doi:10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–940. doi: 10.1038/nature05563. doi:10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8(5):457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009a;30(5):646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453(7192):236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature immunology. 2009b;10(9):1000–1007. doi: 10.1038/ni.1774. doi:10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunological reviews. 2010;238(1):247–262. doi: 10.1111/j.1600-065X.2010.00951.x. doi:10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annual review of immunology. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. doi:10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484(7395):514–518. doi: 10.1038/nature10957. doi:10.1038/nature10957. [DOI] [PubMed] [Google Scholar]