Abstract
Dysfunctional mucus barriers can result in important pulmonary and gastrointestinal conditions, but model systems to study the underlying causes are largely missing. We identified and characterized five mucin homologues in zebrafish, and demonstrated a strategy for fluorescence labeling of one selected mucin. These tools can be used for in vivo experiments and in pharmacological and genetic screens to study the dynamics and mechanisms of mucosal physiology.
The mucus hydrogel that lines the epithelia of the respiratory, gastrointestinal and urogenital tracts has critical, but poorly understood functions to protect the body from contact with pathogens and toxins, while enabling passage of nutrients, oxygen and sperm1,2,3,4,5,6,7,8. The main gel-forming constituents of the mucus are mucins: long and thread-like polymers which are densely O-glycosylated and secreted by specialized goblet cells. Changes in the amount and properties of secreted mucins occur naturally to regulate the passage of sperm, but they can also lead to an increased susceptibility to pathogenic infections, and contribute to disorders in the oral cavity, gastrointestinal tract, and cystic fibrosis lungs9,10,11,12,13,14. The mechanisms that regulate the properties, amount, and location of secreted mucus remain largely obscure, mainly due to the lack of animal models that allow for the live tracking of mucus function.
Results and Discussion
Zebrafish has emerged as a valuable model to study live cellular processes15,16 due to its sequenced genome, transparency through early adulthood, and amenability to high-throughput screens17,18. To determine if zebrafish is a useful in vivo model for the study of mucus physiology, we searched its genome for genes with homology to secreted mucins from other vertebrates. We focused on polymeric secreted mucins, the major gel-forming building blocks of the mucus barrier19,20,21. One characteristic of gel-forming mucins is the concurrence of two protein domains, the Proline, Threonine and Serine (PTS) domain, which is the main site of O-linked glycosylation on the protein, and the Von Willebrand Factor D (VWD) domain, which contributes to the polymerization of mucins. Using previously characterized computational tools22 we identified five putative mucin genes in zebrafish that contain coding regions for both PTS and VWD domains (Supplementary Fig. S1). Based on the subsequent analysis (see below) we named the five putative mucin genes muc5.1 (Ensembl ID ENSDARG00000070331), muc5.2 (ENSDARG00000058556), muc5.3 (ENSDARG00000089847), muc2.1 (ENSDARG00000074142) and muc2.2 (ENSDARG00000078994).
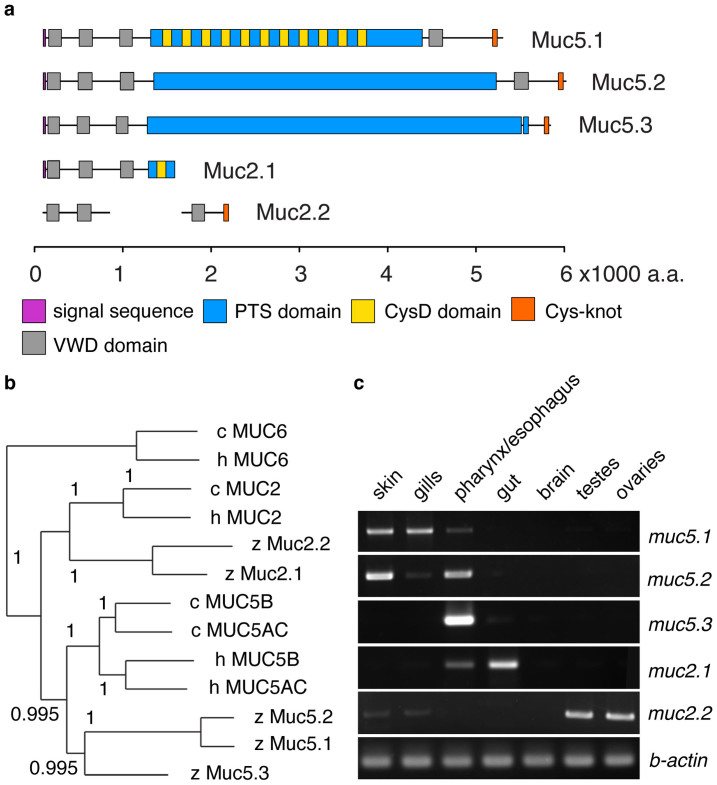
The organization of the protein domains in the identified zebrafish mucins is shown in Figure 1a. The better-studied mammalian polymerizing mucins MUC2, MUC5AC and MUC5B are characterized by an arrangement of four VWD domains with an extensive PTS domain between the third and fourth VWD domains. In addition, they contain CysD domains that are interspersed in the PTS domains, and a cystine knot at the C terminus; both CysD domains and the cystine knot are involved in polymerization. The mammalian MUC6 mucin is similar in architecture but lacks the CysD domains as well as a fourth VWD domain at its C terminus23. Our data show that two identified members of the zebrafish Muc5-family, Muc5.1 and Muc5.2, contain the characteristic arrangement of four VWD domains and a PTS domain localized between the third and fourth VWD domains (Figure 1a, Supplementary Figs. S1, S2, S3). Muc5.3 is different in that it appears to contain only the first three VWD domains. For the members of the Muc2-family (Muc2.1 and Muc2.2) we had less sequence information available and therefore, offer a more preliminary interpretation. For Muc2.1 we identified three VWD domains and a truncated PTS domain in the predicted N-terminal portion of the protein. For Muc2.2, only a short Ensembl transcript was available, which was the basis for the depicted C-terminal VWD domain (Fig. 1a). In addition, muc2.1 mRNA was used as a guide to predict the exon/intron structure of the muc2.2 gene from the available genomic sequence. From the resulting muc2.2 gene model two further VWD domains were identified at the N-terminus of Muc2.2. In the genomic prediction we also found evidence for a PTS region (Supplementary Fig. S1). The PTS region was not included in the protein domain illustration because it was absent from the Ensembl transcript model at the time (Fig. 1a). However, should future updated transcript models include the sequence for the PTS, Muc2.2 would have the standard architecture of a MUC2 type mucin. For further details regarding the genomic organization and protein homology of the zebrafish mucins, as well as information on additional mucin-like transcripts, the reader is referred to Supplementary Figs. S1 and S3, and Supplementary Table S1.
Figure 1. Identification of five polymeric secreted mucins in zebrafish.
(a) – Illustration of predicted mucin protein domain architectures. Two members of the Muc5 family (Muc5.1 and Muc5.2) are composed of three successive VWD domains, followed by a PTS domain and a fourth VWD domain at the C-terminus. This architecture is typical for mammalian gel-forming secreted mucins. The third Muc5 family member, Muc5.3, has a similar predicted domain composition but lacks the fourth VWD domain at the C-terminus. For the Muc2 family members, Muc2.1 and Muc2.2, regions of the protein sequence are missing as the current genome assembly is incomplete. The Muc2.2 protein domain lacks a PTS domain because it was absent from the Ensembl transcript prediction, though such domain was found at the genomic level (Supplementary Fig. S1). VWD: Von Willebrand Factor type D domain; PTS: Proline, Threonine and Serine domain; CysD and Cys-knot are cysteine rich domains. All domain structures except the N-terminal portion of Muc2.2 were identified based on Ensembl transcripts. The sequences used for the construction of the depicted protein models are listed in Supplementary Add. S1. (b) - Phylogenetic tree comparing zebrafish mucins with chicken and human polymeric secreted mucins from N-terminal portions of the mucins containing the three first VWD domains. The numbers at the branches represent posterior probabilities. The tree shows that the identified mucins group with MUC5 and MUC2, but not with MUC6, from chicken and human. (c) - Tissue distribution of the mucin transcripts as detected by RT-PCR. The muc5 family of mucins is expressed in respiratory organs (skin, gills, pharynx and esophagus). muc2.1 expression is detected in the digestive system, predominantly in the gut which is typical for MUC2 mucins in mammals. muc2.2 expression is detected in reproductive organs.
Based on the N-terminal portions of the mucin proteins that contain the first three VWD domains we constructed phylogenetic trees (Fig. 1b: MrBayes, Supplementary Fig. S2: neighbor-joining tree). The results show that Muc5.1, Muc5.2 and Muc5.3 group with the human and chicken MUC5AC and MUC5B. Muc5.1 and Muc5.2 appear closely related to each other. Muc2.1 and Muc2.2 group with the human and chicken MUC2 mucins. Our data also suggest that none of the studied zebrafish genes are related to the vertebrate MUC6 mucin.
From the genomic structure we derived that the genes muc5.1, muc5.2 and muc2.2 are localized in a cluster on chromosome 25 (Supplementary Fig. S1). muc2.1 currently has an unassigned position in the genome, Zv9_NA774, with approximately two thirds of its length unknown. The muc5.3 gene, which is located on chromosome 7, has the gene tollip as its immediate neighbor. The occurrence of zebrafish mucin genes in a cluster, as well as the genomic synteny with the tollip gene, is reminiscent of the mucin gene organization in other vertebrates, including humans24.
To determine the tissue distribution of the putative zebrafish mucin gene transcripts we performed RT-PCR from tissue isolated from adult zebrafish (Fig. 1c). Our data show that muc5.1 and muc5.2 are both expressed in the skin, the gills and the pharynx/esophagus, while muc5.3 expression appears to be restricted to the pharynx/esophagus. By in situ hybridization on adult zebrafish sections we were able to further specify the distribution of muc5.1 and muc5.2 to the pharynx and muc5.3 to the esophagus (Supplementary Fig. S4). In addition, by separating the gill lamellae from the gill arches we saw that only muc5.1 is expressed in the lamellar part (Supplementary Fig. S5) and hence, appears to represent a bona fide respiratory mucin. muc2.1 is predominantly found in the gut, while muc2.2 is expressed in testes and ovaries (Fig. 1c). Together, the expression pattern of the zebrafish muc5 family appears reminiscent of the human MUC5 family, which is found in the respiratory and the upper digestive tracts25. Moreover, muc2.1 in zebrafish shares its tissue localization with human MUC2, the major mucin in the gut. The outlier is muc2.2, which is expressed in the reproductive organs in zebrafish; in humans, MUC5AC, MUC5B and MUC6 are found in the male urogenital tract26 and female endocervix. We also observed that expression of the mucin genes during zebrafish development correlates with the initiation of development of the respective organs, in which mucins are found in the adult fish27,28 (Supplementary Fig. S6).
The above sequence information on mucin genes was used to create a fluorescent reporter of mucin activity in zebrafish. The complete open reading frames of secreted mucins is difficult to tag due to its large size. As a consequence, thus far only one full length mucin, MUC5AC from the mouse, has been successfully fluorescently tagged and expressed under a constitutive promoter29. Our goal here was to generate a reporter in zebrafish that is expressed under the endogenous mucin promoter, which would enable the real time tracking of mucin production in response to physiological changes. To achieve this, we excised from the BAC CH211-19808 9.8 kb of genomic sequence of muc5.1 that comprises 4.6 kb upstream and 5.2 kb downstream of the mucin start codon ATG, and cloned it into the pBSII KS(+) vector. Our goal was to express the Red Fluorescent Protein (RFP) in frame with the ATG and the muc5.1 secretory signaling sequence to enable secretion of the mucin reporter. We inserted a Tag-RFP targeting cassette at 93 bp downstream from the mucin ATG, using Lambda-Red homologous recombination (Supplementary Fig. S7). In this construct, the RFP is terminated with the stop codon and hence, does not include the mucin coding sequence beyond the signaling sequence.
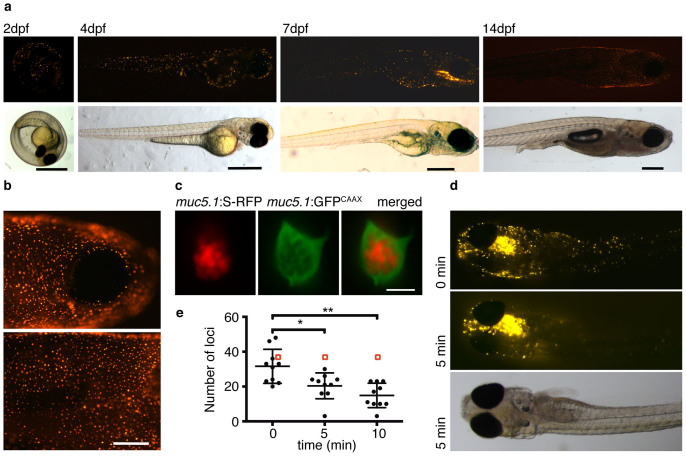
To test for the functionality of the mucin reporter, the construct was linearized, injected into one-cell stage zebrafish embryos, and the animals were raised to adulthood and screened for germ line transmission. In germ line transgenics of the muc5.1:S-RFP reporter, fluorescence became detectable at two days post-fertilization (dpf) as dots on the skin (Fig. 2a). At day four, the fluorescent signal was evident along the body axis including in the mouth of the larva (Fig. 2a). At two weeks post fertilization, the fluorescent signal was scattered throughout the skin at an average of 2500 dots/mm2 (Fig. 2b). Such dotted distribution has been reported for mucus-secreting cells in the zebrafish intestine and in the human respiratory epithelium30,31. A closer inspection of the fluorescent loci by confocal microscopy shows that the mucin reporter is stored in relatively large granules inside the cells, which resemble secretory vesicles characteristically produced by mucus-secreting cells (Fig. 2c)32. The cellular membranes in Fig. 2c were visualized with GFP fused to a CAAX motif, which targets the GFP to the membrane and which is expressed under the regulation of the muc5.1 promoter. The GFP-CAAX reporter delineates the cellular and vesicular membranes of the muc5.1 producing cells (Fig. 2c). Together these data suggest that the mucin reporter is produced in secretory cells and compartmentalized in granules, as is expected for secreted mucins.
Figure 2. Expression of the fluorescent mucin reporter muc5.1:S-RFP in zebrafish.
(a) –Visualization of embryos at 2, 4, 7 and 14 days post fertilization (dpf) by fluorescence (top row) and bright field (bottom row) microscopy shows that the mucin reporter expresses in distinct loci distributed across the skin of the fish. Scale bars are 0.5 mm. (b) –Live visualization of the fluorescent mucin reporter in the head (top) and trunk (bottom) of 14 dpf fish. Scale bar is 200 μm. (c) – Confocal image of a muc5.1:S-RFP-positive locus shows that the mucin reporter is packed inside secretory vesicles in cells within the skin. The cell membranes are labeled with GFP that was expressed under the promoter of muc5.1 and targeted to the membrane via a CAAX motif. Scale bar is 5 μm. (d) – Exposure of fish to LPS results in the partial loss of muc5.1:S-RFP loci and a simultaneous appearance of fluorescence in the immediate surrounding of the fish, suggesting the secretion of the mucin reporter. The bright field image (bottom) shows that fish remain intact during this treatment. (e) – Quantification of fluorescent loci within a consistent region (approximately between the head and top of the trunk) in ten individual fish before and after exposure to LPS. The red square is the mean of 10 control fish (no LPS addition) at t = 0, 5, and 10 minutes. The mean value is 35.7 across all three time points. For the LPS addition, each point represents the number of fluorescent loci in the same fish at the various time points (10 fish total). The error bars indicate standard deviation. One * indicates p < 0.05 between 0 minutes and 5 minutes using paired two-tailed T-test. Two ** indicates p < 0.01 using the same test between 0 minutes and 10 minutes. In most fish, a substantial proportion of loci are lost on treatment with LPS, suggesting that the mucin-reporter is secreted on this stimulus.
To test if muc5.1:S-RFP can be expelled from the cells, hence allowing the observation of live secretion, we used lipopolysaccharide (LPS) from E. coli as a characterized stimulant33. Our data show that in 4 dpf fish, on exposure to 0.5 μg/μl LPS, the majority of muc5.1:S-RFP-producing cells lose fluorescence within minutes while the released RFP collects as a halo around the fish (Fig. 2d). A quantification of cells that expel muc5.1:S-RFP reveals that roughly 40% of the putative goblet cells per fish release the fluorescence within five minutes after induction with LPS, with further signal loss within the next five minutes (Fig. 2d, 2e). 10 minutes after initial addition of LPS the anesthetized fish displayed normal heartbeat and circulation, suggesting that the expulsion of the mucin-reporter is not due to toxicity.
In summary, we show evidence for five gel-forming secreted mucin genes in zebrafish with a high degree of homology to other vertebrate mucins in their genomic and protein domain organization, as well as their tissue specific expression. We developed a strategy to build a fluorescent mucin reporter expressed under native regulatory elements and show that its release can be triggered and quantified in the live fish. Together, our work offers a useful set of tools to study the dynamics of mucin secretion and expression in the unperturbed fish and upon pathogenic, pharmacological or genetic challenges. Our hope is that this experimental system may allow for screening of conditions that not just trigger mucus secretion, but cause long-term effects of goblet cell differentiation seen as metaplasia/hyperplasia in mucosal diseases.
Methods
Zebrafish maintenance
All fish were maintained according to standard procedures and on approval of the MIT Committee on Animal Care. Strain T/AB1434 was used for the RT-PCR, qRT-PCR and in situ hybridization experiments. The optically transparent casper strain35 was used as the genetic background for expression of the fluorescent mucin construct. Fish were exposed to 0.05% Tricaine in buffered fish water for anesthesia and to 0.1% of the same medium for euthanasia.
Bioinformatics for mucin gene identification
To perform a comprehensive search for mucin genes in zebrafish we applied two strategies. First, we searched for VWD and PTS domains in protein sequences derived from the genes and transcripts identified by Ensembl36. To identify VWD domains we used Hmmer, version 3 (http://hmmer.wustl.edu) and for the PTS domains we used an in-house Perl script PTSPRED22,23. Second, we analyzed zebrafish genomic sequences to identify regions encoding VWD and PTS domains. The latest zebrafish genome release (Zv9) was searched with GeneWise (Wise2), using a VWD HMM model as query37 and a version of PTSPRED adapted to nucleotide sequences. Proteins or genomic regions that contained both VWD and PTS domains were considered mucin candidates, and these were further screened against the H. sapiens genome in Ensembl. The N-terminus of Muc2.2 was predicted by GeneWise using a close homolog as the protein query sequence. For the phylogenetic analysis, regions of the protein sequences that contain the three N-terminal VWD domains were aligned with T-coffee and the resulting alignment was edited to remove gapped regions. Phylogenetic trees were produced with a neighbor-joining method as well as with MrBayes38 with the setting prset aamodelpr = mixed. Graphs of the protein domain structures were generated with R (http://www.r-project.org) and edited using Adobe Illustrator. Signal sequences were predicted with SignalP 4.039.
Verification of mucin transcripts
To verify the identity of the predicted mucin gene models, total RNA was isolated from three months old male and female fish. The fish were euthanized, snap-frozen in liquid nitrogen, and ground to powder with mortar and pestle. RNA was isolated using TRIzol (Life Technologies). cDNA was synthesized with an oligo dT and random hexamer primer mix using a long-range cDNA synthesis reaction. PCR was performed using a high-fidelity polymerase. Primers (listed in Table 1) were designed using Primer-BLAST.
Table 1. Primers used for mucin gene expression analysis.
| Mucin transcript RT-PCR | Tissue RT-PCR and in situ hybridization |
|---|---|
| muc5.1 | muc5.1 |
| F1- IJ1 – tgacatgggttgaaagcaaa | F – IJ7- tgttccaggcacacattgat |
| R1 – IJ4 – tggtggtgttgaaaatgtga | R – IJ28 - cgacaaatcgtgtgagaaca |
| F3 - IJ7 – catccctaggcacatccact | muc5.2 |
| R3 - IJ63 – tgttttgcgtcgcttaagaa | F – IJ68 - ctgagatgggtcatcctgct |
| muc5.2 | R – IJ31 - ggtactgctggcaaaccatt |
| F1 - IJ68 – ctgagatgggtcatcctgct | muc5.3 |
| R1 - IJ71 – gtctcgggtgatgaaggtgt | F – IJ80 - cgagcaatatgcacagcact |
| F2 - IJ50 – cagtgcatcgagaccaacag | R – IJ81 - gctcgagcagttgaaaaacc |
| R2 - IJ51 – tcatcattcgcctaattcca | muc2.1 |
| muc5.3 | F – IJ125 - tgctgcttctggcgctttctggt |
| F1 - IJ80 – cgagcaatatgcacagcact | R – IJ126 - ttgctgtcctccatgcgggtg |
| R1 - IJ81 – gctcgagcagttgaaaaacc | muc2.2 |
| F2 - IJ82 – agatgcctgctgtccagagt | F – IJ167 - accacaaccaaacccatgtt |
| R2 - IJ83 – aggttccacacaaccctgag | R – IJ168 - gttccacattggccttctgt |
| muc2.1 | |
| F1 – IJ125 – tgctgcttctggcgctttctggt | |
| R1 - GY2 – cataaccaatttccccatcg | Muc5.1 primers for in situ hybridization |
| muc2.2 | muc5.1 |
| F1 - IJ111 – actgtccgtgtcctcatggt | F – IJ1 - tgacatgggttgaaagcaaa |
| R1 - IJ112 – tctcctccagtgtcacatgc | R – IJ3 - aaccgtgaccgtttcttcac |
| F2 - IJ113 – accacaaccaaacccatgtt | |
| R2 - IJ114 – cctctcgaatgctggatctc | |
| qRT-PCR | |
| muc5.1 | muc2.1 |
| F – tggcaacttggctgatgata | F – GY3 - aatatgccttgcggaacaac |
| R – tcgtcacacggaccagtaga | R – GY4 - gtgctgaggttgcagaatga |
| muc5.2 | beta-actin2 |
| F – ggtgtctgttccgatcaatc | F - cgtgctgtcttcccatcca |
| R – tcatccttgtcgccattgta | R – tcaccaacgtagctgtctttctg |
| muc5.3 | |
| F – GY5 - ggggaaaactacaccagcaa | |
| R – GY6 - tgtgaattctgtgccagagc |
RT-PCR gene expression profiling in tissues and in situ hybridization
Selected tissues were dissected from five fish, washed in ice-cold PBS and immediately snap frozen in liquid nitrogen. For homogenization, the tissues were passed through a syringe with needle and centrifuged through a QIAShredder column. Total RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was synthesized using standard reagents and the transcript segments were amplified by PCR with Taq polymerase. Each amplified PCR product was ligated into pGEM-T Easy (Promega) and verified by sequencing.
In situ hybridization was performed on frozen sections of 5-week-old fish following a slight modification of the method described in40. Fish were euthanized and submersed three times (half an hour each) in fresh Tissue-Tek O.C.T. Compound (Sakura), then transferred to fresh O.C.T., frozen on dry ice, and stored at −80°C until cut into 10 μm thick sagittal cryosections. The cryosections were stored on glass slides at −80°C until needed. In situ hybridization probes were generated with the same primers as were used for the RT-PCR from tissues, with the exception of muc5.1 (see Table 1). In brief, total adult RNA was isolated and reverse transcribed into cDNA. The target cDNA sequence was amplified with the high-fidelity polymerase and cloned into pGEM-T Easy. Probes were generated to detect both sense and antisense RNA orientations. The plasmid was linearized with NdeI, phenol/chlorophorm extracted, and in vitro RNA transcribed using T7 polymerase with Dig-UTP nucleotide mix (Roche) in the presence of RNase inhibitor. The RNA products were subsequently purified through purification columns and tested on a gel for quality. Hybridization was detected with Anti-DIG-AP Fab fragments (Roche) and NBT/BCIP (Roche). In situ slides were visualized on Nikon Eclipse E600 and pictures were taken with a Spot RT Color camera.
For mucin gene expression profiling by qRT-PCR during zebrafish development, RNA was isolated from embryos on days 1, 2, 3, 4, 5 and 6 post-fertilization. 50, 30 and 20 embryos were used for days 1, 2, 3 and on, respectively, and kept frozen in liquid nitrogen at −80°C until used. Total RNA was extracted with the RNeasy Mini Kit. cDNA was synthesized from 200 ng total RNA and used for qRT-PCR. qRT-PCR was performed using the iQ™ SYBR® Green Supermix (Biorad) with a LightCycler 480 (Roche). The values were calculated from primer efficiencies according to the Pfaffl model41 where each mucin amplification efficiency value was compared to the values for β-actin. Individual mucin values were normalized to their corresponding day 6 expression values.
Generation and imaging of the fluorescent mucin reporter
muc5.1:S-RFP was designed following the protocols in42,43. BAC CH211-19808 was digested with KpnI and SpeI and the 9.88 kb fragment was cloned into the pBSII-KS(+) vector. Tag-RFP44 was inserted into the targeting cassette with high fidelity PCR using the primers listed in Table 2. The cassette was inserted in frame, by Lambda-Red homologous recombination, with the muc5.1 signaling sequence (93 bp from the ATG). The construct was linearized with NotI, column-purified and resuspended in 1 × Danieau buffer. The construct was injected into one-cell stage embryos at a concentration of 200 ng/μl. We obtained two fish with a stably inserted construct from the same mosaic parent.
Table 2. Primers used for generating the fluorescent mucin reporter and the membrane targeting GFP construct.
| muc5.1:S-RFP |
| F – atgggtcttcctactggccgggctgcagtccatacaagcaatggtgtctaagggcgaaga |
| R - cacactttccgcattaattaataatttttattttctaaccccatagagcccaccgcatcc |
| muc5.1: eGFPCAAX |
| F – IJ157 - tcctaatcctctcgaaagtggtc |
| R – IJ158 - cctcaagagctaatgtgagtcatct |
| F – memGFPF - aaaaaatctagaatggtgagcaagggcgagga |
| R – memGFPR - cgggatccaccggatcttgaccatggtttttt |
For the visualization of cellular membranes in mucin-expressing cells 2.2 kb of the muc5.1 promoter directly preceding the ATG was PCR amplified (primers listed in Table 2) and ligated into pCloneJet (ThermoScientific). GFPCAAX was amplified from a vector reported in45 using primers with XbaI/NcoI restriction sites (Table 2) and cloned downstream of the amplified muc5.1 promoter. The assembled construct was excised with NotI/NcoI, blunt-ended with Klenow Polymerase and ligated into the pXeX expression vector46. The final construct was linearized with BglI, column-purified and injected into muc5.1:S-RFP embryos for localization studies.
To visualize the expression of the fluorescent mucin reporter the fish were anesthetized and embedded in 3% methylcellulose. Pictures were taken with a Zeiss SteReo Discovery. V8 stereoscope equipped with an AxioCam MRc camera and AxioVision 4.8 software. For the visualization of individual goblet cells, whole 4 dpf fish were placed in a glass bottom culture dish (MatTek Corporation) filled with fish water containing anesthetics and covered with a glass coverslip. Cells were imaged with a spinning disc confocal microscope Nikon TE2000 with MetaMorph acquisition software and Hammamatsu Orca-ER camera.
To visualize the effects of LPS on mucin secretion, larvae (4 dpf) were anesthetized and placed in a drop of fish water on a glass slide. Lipopolysaccharide (LPS) from E. coli (Sigma) was added to the water at a final concentration of 0.5 μg/μl, and pictures were taken with identical exposure times at 0, 5 and 10 minutes after addition of LPS. The number of mucin-expressing cells was counted manually before and after LPS exposure within the same region on the skin of the fish. As controls fish were incubated for the same amount of time in water with anesthetic but without LPS.
Author Contributions
I.J. and K.R. designed the experiments. T.S. performed the bioinformatics analysis. G.Y. performed the qRT-PCR experiment. I.J. performed RT-PCR, in situ hybridization, design and the analysis of the fluorescent mucin reporter. A.A. provided advice and support on the zebrafish experiments. I.J. and K.R. wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge financial support from the NIH-NIEHS Center Grant P30-ES002109 to K.R.; the Johnson & Johnson Corporate Office of Science and Technology; the Erik Philip-Sörensen Foundation to T.S.; NIH grant CA106416 and the Kathy and Curt Marble Cancer Research Fund; and the Koch Institute zebrafish core facility. We thank Tim Angelini for taking care of the fish, Isaac Han for generating the in situ construct of muc5.2, JrGang Cheng (BAC Facility, UNC Chapel Hill) for assembly of the fluorescent muc5.1:S-RFP construct, Wendy Salmon (Whitehead Institute) for assistance with confocal imaging, Isabel Brachmann (Whitehead Institute) for assistance with the fluorescent fish imaging, the Swanson Histology Facility (Koch Institute) for zebrafish cryosections, and Julio Amigo (MIT) for the GFPCAAX construct. Gene names were approved by the Zebrafish Nomenclature Committee. We thank Nicole Kavanaugh and Wesley Chen for help in preparing the figures and the entire Ribbeck lab for critical comments on the manuscript. We are grateful to Nancy Hopkins for supporting this project.
References
- Amerongen A. V., Bolscher J. G. & Veerman E. C. Salivary mucins: protective functions in relation to their diversity. Glycobiology 5, 733–740 (1995). [DOI] [PubMed] [Google Scholar]
- Becher N., Adams Waldorf K., Hein M. & Uldbjerg N. The cervical mucus plug: structured review of the literature. Acta Obstet Gynecol Scand 88, 502–513, 10.1080/00016340902852898 (2009). [DOI] [PubMed] [Google Scholar]
- Cone R. A. Barrier properties of mucus. Adv Drug Deliver Rev 61, 75–85, 10.1016/j.addr.2008.09.008 (2009). [DOI] [PubMed] [Google Scholar]
- Filipe M. I. Mucins in the human gastrointestinal epithelium: a review. Investigative & cell pathology 2, 195–216 (1979). [PubMed] [Google Scholar]
- Hollingsworth M. A. & Swanson B. J. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4, 45–60, 10.1038/nrc1251 (2004). [DOI] [PubMed] [Google Scholar]
- Linden S. K. et al. Mucins in the mucosal barrier to infection. Mucosal Immunol 1, 183–197, 10.1038/mi.2008.5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin M. A., Linden S. K., Sutton P. & Florin T. H. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9, 265–278, 10.1038/nrmicro2538 (2011). [DOI] [PubMed] [Google Scholar]
- Suarez S. S. & Pacey A. A. Sperm transport in the female reproductive tract. Hum Reprod Update 12, 23–37, 10.1093/humupd/dmi047 (2006). [DOI] [PubMed] [Google Scholar]
- Corfield A. P. et al. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut 47, 589–594 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood C. K. et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med 5, e54, 10.1371/journal.pmed.0050054 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell S. H. & Boucher R. C. Effective mucus clearance is essential for respiratory health. Am J Respir Cell Mol Biol 35, 20–28, 10.1165/rcmb.2006-0082SF (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. C. & Voynow J. A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 86, 245–278, 10.1152/physrev.00010.2005 (2006). [DOI] [PubMed] [Google Scholar]
- Tabak L. A., Levine M. J., Mandel I. D. & Ellison S. A. Role of salivary mucins in the protection of the oral cavity. J Oral Pathol 11, 1–17 (1982). [DOI] [PubMed] [Google Scholar]
- Venables C. W. Mucus, pepsin, and peptic ulcer. Gut 27, 233–238 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke G. J. & Currie P. D. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8, 353–367, 10.1038/nrg2091 (2007). [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Mahowald M. A., Ley R. E. & Gordon J. I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433, 10.1016/j.cell.2006.08.043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey R. D. & Zon L. I. Small molecule screening in the zebrafish. Methods 39, 255–261, 10.1016/j.ymeth.2005.09.019 (2006). [DOI] [PubMed] [Google Scholar]
- Rihel J. et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351, 10.1126/science.1183090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy J. V. & Dickey B. F. Airway mucus function and dysfunction. N Engl J Med 363, 2233–2247, 10.1056/NEJMra0910061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler S. J. & Spicer A. P. Epithelial mucin genes. Annu Rev Physiol 57, 607–634, 10.1146/annurev.ph.57.030195.003135 (1995). [DOI] [PubMed] [Google Scholar]
- Thornton D. J., Rousseau K. & McGuckin M. A. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol 70, 459–486, 10.1146/annurev.physiol.70.113006.100702 (2008). [DOI] [PubMed] [Google Scholar]
- Lang T., Hansson G. C. & Samuelsson T. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics 7, 197, 10.1186/1471-2164-7-197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Hansson G. C. & Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A 104, 16209–16214, 10.1073/pnas.0705984104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigny P. et al. Human mucin genes assigned to 11p15.5: identification and organization of a cluster of genes. Genomics 38, 340–352, 10.1006/geno.1996.0637 (1996). [DOI] [PubMed] [Google Scholar]
- Audie J. P. et al. Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem 41, 1479–1485 (1993). [DOI] [PubMed] [Google Scholar]
- Russo C. L. et al. Mucin gene expression in human male urogenital tract epithelia. Hum Reprod 21, 2783–2793, 10.1093/humrep/del164 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guellec D., Morvan-Dubois G. & Sire J. Y. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio). Int J Dev Biol 48, 217–231, 10.1387/ijdb.031768dg (2004). [DOI] [PubMed] [Google Scholar]
- Wallace K. N. & Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol 255, 12–29 (2003). [DOI] [PubMed] [Google Scholar]
- Ehre C. et al. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci U S A 109, 16528–16533, 10.1073/pnas.1206552109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G. et al. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet 4, e1000240, 10.1371/journal.pgen.1000240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes A. L. et al. Epithelial mucin stores are increased in the large airways of smokers with airflow obstruction. Chest 130, 1102–1108, 10.1378/chest.130.4.1102 (2006). [DOI] [PubMed] [Google Scholar]
- Perez-Vilar J. et al. Mucin granule intraluminal organization in living mucous/goblet cells - Roles of protein post-translational modifications and secretion. Journal of Biological Chemistry 281, 4844–4855, 10.1074/Jbc.M510520200 (2006). [DOI] [PubMed] [Google Scholar]
- Smirnova M. G., Guo L., Birchall J. P. & Pearson J. P. LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell Immunol 221, 42–49 (2003). [DOI] [PubMed] [Google Scholar]
- Amsterdam A. et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev 13, 2713–2724 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. M. et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189, 10.1016/j.stem.2007.11.002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. et al. Ensembl 2011. Nucleic Acids Res 39, D800–806, 10.1093/nar/gkq1064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M. & Durbin R. GeneWise and Genomewise. Genome Res 14, 988–995, 10.1101/gr.1865504 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology 61, 539–542, 10.1093/sysbio/sys029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8, 785–786, 10.1038/nmeth.1701 (2011). [DOI] [PubMed] [Google Scholar]
- Braissant O. & Wahli W. A Simplified In Situ Hybridization Protocol Using Non-radioactively Labeled Probes to Detect Abundant and Rare mRNAs on Tissue Sections. Biochemica 1, 10–16 (1998). [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier J. & Cheng J. G. Recombineering-based procedure for creating Cre/loxP conditional knockouts in the mouse. Curr Protoc Mol Biol Chapter 23, Unit 23 13, 10.1002/0471142727.mb2313s85 (2009). [DOI] [PubMed] [Google Scholar]
- Hollenback S. M., Lyman S. & Cheng J. Recombineering-based procedure for creating BAC transgene constructs for animals and cell lines. Curr Protoc Mol Biol Chapter 23, Unit 23 14, 10.1002/0471142727.mb2314s95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C. et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5, 545–551, 10.1038/nmeth.1209 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc J. A., Amigo J. & Lawson N. D. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn 236, 3077–3087, 10.1002/dvdy.21354 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D. & Krieg P. A. pXeX, a vector for efficient expression of cloned sequences in Xenopus embryos. Gene 147, 223–226 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information