Abstract
Studies focusing on biofilm assembly in deep-sea environments are rarely conducted. To examine the effects of substrate type on microbial community assembly, biofilms were developed on different substrates for different durations at two locations in the Red Sea: in a brine pool and in nearby bottom water (NBW) adjacent to the Thuwal cold seep II. The composition of the microbial communities in 51 biofilms and water samples were revealed by classification of pyrosequenced 16S rRNA gene amplicons. Together with the microscopic characteristics of the biofilms, the results indicate a stronger selection effect by the substrates on the microbial assembly in the brine pool compared with the NBW. Moreover, the selection effect by substrate type was stronger in the early stages compared with the later stages of the biofilm development. These results are consistent with the hypotheses proposed in the framework of species sorting theory, which states that the power of species sorting during microbial community assembly is dictated by habitat conditions, duration and the structure of the source community. Therefore, the results of this study shed light on the control strategy underlying biofilm-associated marine fouling and provide supporting evidence for ecological theories important for understanding the formation of deep-sea biofilms.
The formation of biofilms, in which bacteria attach to a substrate and form a community, is a longstanding topic that remains under investigation1. In the marine environment, biofilms are often referred to as microfouling, resulting in an undesirable accumulation of microorganisms. Identifying key processes associated with microfouling, for instance, by understanding how biofilms form on different artificial materials, may provide useful information to improve control strategies. It has been shown that in the subtidal zone, the assembly of biofilms is influenced by the type of substrates to which they attached2. In addition, Bellou et al3 examined the effect of substrate type on the development of bacterial biofilm communities in the Eastern Mediterranean sea while Meier et al4 characterized biofilms that formed in the Cayman Trough. The above-mentioned studies indicated a strong influence of a variety of artificial materials on biofilm development. In contrast, we previously reported that the in situ environments might exert a strong selective effect on the microbes present in biofilms developed on artificial materials in locations near the Red Sea Thuwal cold seeps5, whereas the types of substrates had limited effects on biofilm development. However, further studies in cold seeps with an improved experimental design are required to verify previous conclusions and to reveal the principles governing the effects of substrate type on biofilm formation.
Understanding the processes that govern the composition of communities is an ecological question of great interest, and theories have been proposed to answer it6,7,8. In aquatic ecosystems, biofilm assembly is initiated by the microbial inhabitants of the source waters, and species sorting has been identified as a major underlying mechanism. Species sorting is a process that occurs when bacteria are selected from a pool of species to form a community due to selection by local abiotic and biotic environmental conditions9. A growing number of studies have examined species sorting for bacterial communities from subsurface environments, such as soil communities10 and stream biofilms9. It has been hypothesized that the effect of species sorting is stronger at the extremes of an environmental gradient11 and in the initial stages of community assembly12. In addition, species sorting has been suggested to be stronger for specialists rather than for generalists13. However, as nearly all of the existing tests were confined to the surface area of the earth11,12,14,15, whether this theory can explain the assembly of microbial communities in the deep sea remains unknown.
We have previously described microbial community profiles in a few locations in the Red Sea5,16,17,18. Several cold seeps formed in the Red Sea by the divergent movement of the Arabian and African continental plates resulting in the formation of new oceanic crust. Some of the seeps contain highly saline waters caused by the dissolution of Miocene evaporite deposits and form deep-sea brine pools19. The Thuwal cold seeps20 are newly discovered seeps located at the seafloor of the Red Sea at a depth of about 850 m, where hypersaline brine seeps out of the seabed and fills an adjacent bottom basin forming a brine pool. Lee et al5 showed that the brine pool, the seeping water and the NBW are dominated by sulfate-reducing, ammonia-oxidizing and sulfur-oxidizing microbes. In this study, we investigate the microbial communities in the same brine pool as well as in the NBW near the Thuwal cold seeps to reveal the effects of substrate type on biofilm formation in these environments. Six engineering materials were selected that are commonly used to build sensors for marine environments and which represent different surface properties. The biofilms were developed for three and six days, which were selected by referring to the durations used by Lee et al5.
Results
Microscopic images and cell density
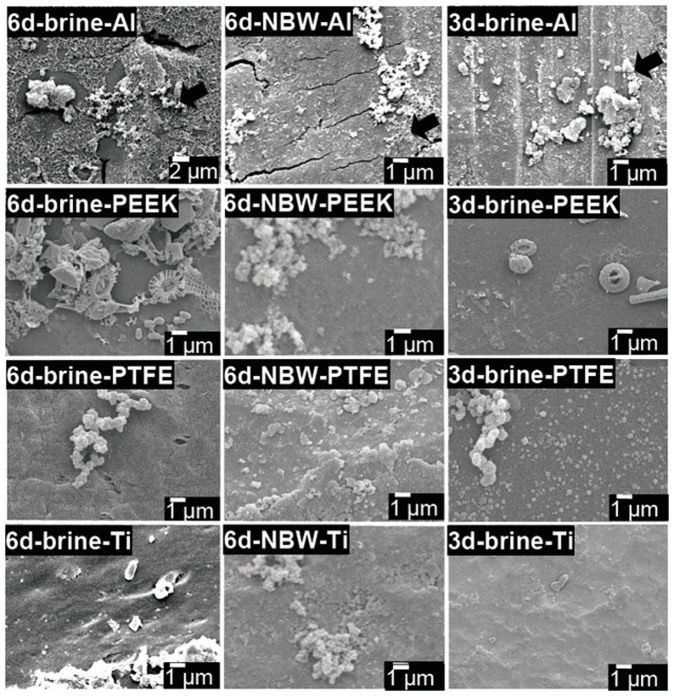
Deploying a Remotely Operated Vehicle (ROV) and special devices (Fig. 1), biofilms developed in the deep-sea on six different substrates: aluminium (Al), polyether ether ketone (PEEK), polyvinyl chloride (PVC), polytetrafluoroethene (PTFE), stainless steel (SS) and titanium (Ti), were obtained. Scanning electron microscopy (SEM) imaging revealed the structures of the 3- and 6-day biofilms that had developed on the surface of different materials, as highlighted by the micro-colonies and the microbes embedded in exopolymeric substance (EPS, as indicated in Fig. 2). In the brine pool, biofilms developed on Al and PEEK had the greatest biomass; whereas the PTFE substratum had the lowest biomass (Fig. 2). In addition, bacterial cells attached to the different materials displayed different phenotypes, indicating a selection process associated with the substrates. But in the NBW, biofilms developed on different materials showed less differences in biomass abundance based on the SEM images, compared with the brine pool biofilms. It has to be noted that the selected SEM images did not necessarily reflect the quantitative analyses, and thus cell density calculation was performed (Table 1). The average cell density tended to be higher for the 6-day biofilms in the brine pool than the 6-day biofilms in the NBW (one-way ANOVA, P < 0.01); and higher for the 6-day biofilms than the 3-day biofilms in both locations (one-way ANOVA, P < 0.01). The Levene test was used to test differences in homogeneity between different biofilm groups and the results suggested that the 6-day brine pool biofilms had higher variations in cell density compared with the 6-day NBW biofilms (p < 0.05), whereas the 3-day biofilms displayed higher variations than the 6-day biofilms (p < 0.01).
Figure 1. Set-up used for biofilm development in and adjacent to a brine pool near the Thuwal cold seeps.
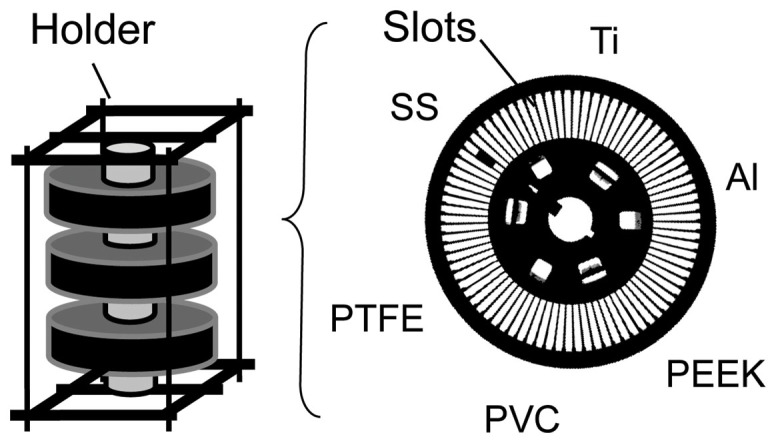
The samplers contained six types of substrate for biofilm development, including aluminum (Al), polyether ether ketone (PEEK), Polyvinyl chloride (PVC), polytetrafluoroethene (PTFE), stainless steel (SS) and titanium (Ti). Material pieces were embedded in the slots of the carousel. Three containers were fixed in the frame and then immersed in the water.
Figure 2. Selected scanning electron microscopy (SEM) images of material surfaces deployed in the brine pool and NBW for different durations (3 and 6 days).
Exopolymeric substance (EPS) can be observed (arrow) and scale bars are shown at the bottom of the images.
Table 1. Cell density of biofilm communities (×103 cell/cm2). Bacterial enumeration was performed using confocal laser scanning microscopy, and the cell density of two replicates was obtained for each substrate type.
| Brine pool | NBW | |||
|---|---|---|---|---|
| Substrate type | 3 days | 6 days | 3 days | 6 days |
| Al-1 | 33.3 ± 3.2 | 70.2 ± 2.1 | 13.6 ± 1.5 | 24.2 ± 6.4 |
| Al-2 | 35.3 ± 6.3 | 66.3 ± 6.4 | 15.7 ± 0.1 | 25.4 ± 1.6 |
| PEEK-1 | 22.6 ± 1.6 | 38.9 ± 0.7 | 12.1 ± 2.6 | 18.3 ± 5.3 |
| PEEK-2 | 23.7 ± 0.5 | 35.9 ± 5.6 | 13.2 ± 1.3 | 17.6 ± 1.4 |
| PTFE-1 | 4.2 ± 0.5 | 6.5 ± 3.1 | 9.5 ± 3.7 | 19.6 ± 0.1 |
| PTFE-2 | 2.2 ± 0.5 | 4.5 ± 2.8 | 7.8 ± 2.3 | 13.9 ± 0.7 |
| PVC-1 | 10.5 ± 3.4 | 28 ± 6.8 | 14.3 ± 0.2 | 11.2 ± 2.1 |
| PVC-2 | 12.5 ± 4.1 | 25.3 ± 5.4 | 14.7 ± 0.3 | 12.8 ± 0.4 |
| SS-1 | 14.3 ± 2.3 | 45.5 ± 7.6 | 11.2 ± 3.7 | 21.2 ± 1.6 |
| SS-2 | 42.6 ± 7.1 | 10.9 ± 2.0 | 26.5 ± 4.1 | |
| Ti-1 | 5.3 ± 1.6 | 18.6 ± 4.1 | 14.6 ± 5.4 | 13.5 ± 1.0 |
| Ti-2 | 4.7 ± 0.3 | 13.5 ± 1.3 | 12.3 ± 2.5 | 16.7 ± 0.5 |
Taxonomic profile
The barcoded 454 pyrosequenced 16S rRNA amplicons for the 47 biofilms and the four water samples are summarized in Table S1, in addition to the number of qualified 16S rRNA reads and bacterial and archaeal OTUs. Assignment of the reads to the phylum level revealed different bacterial compositions among the samples (Fig. S2). First, different taxonomic patterns were observed between the biofilm and the planktonic communities in both environments. The bacterial reads were assigned largely to Proteobacteria, which accounted for 50% to 95% of all samples. However, the proportions of several other phyla differed between the biofilm and the water samples. For example, Thaumarchaeota and Deferribacteres were prevalent in the brine pool water but diminished in both the 3-day and the 6-day brine pool biofilms. Similarly, Actinobacteria accounted for 20% of the NBW water community but was less common in the NBW biofilms. In addition, biofilms developed on different substrates displayed varied compositions, especially 3-day biofilms. For example, the relative abundance of Firmicutes tended to be higher in the 3-day Al (2.4%) and PTFE (9.1%) biofilms than in the 3-day biofilms developed on other materials in the brine pool. In the NBW, the Euryarchaeota could only be detected in 3-day Al (2.1%) and PTFE (4.9%) biofilms.
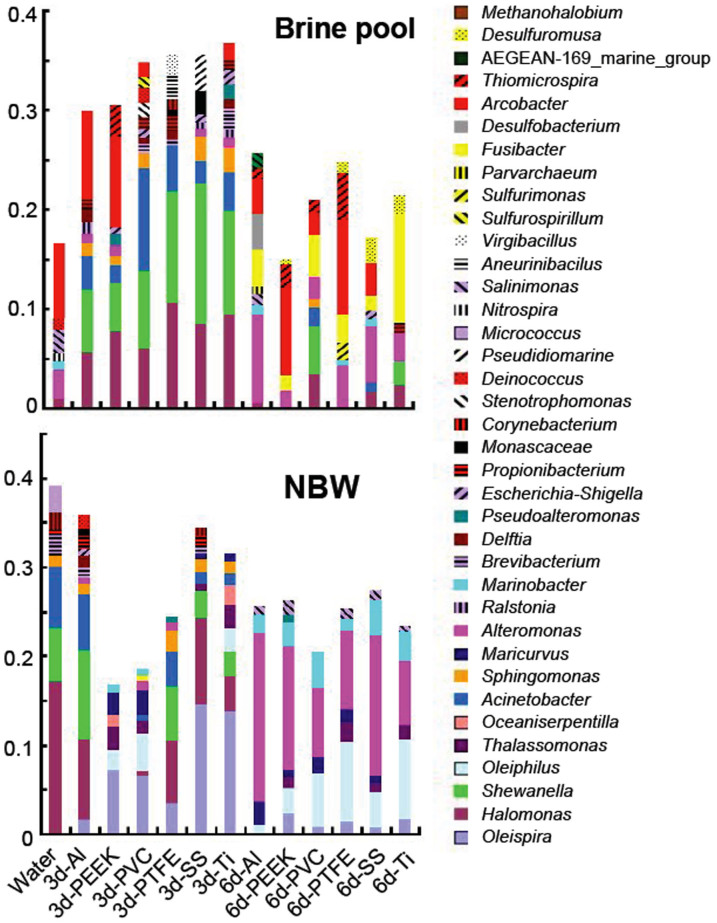
Reads that passed the quality filtering were further classified to the genus level, resulting in more than 800 genera. Genera with a relative abundance > 1.0% are summarized in Figure 3. Consistent with the results obtained at the phylum level, the water communities displayed different patterns at the genus level compared with the biofilms, especially for the brine pool samples. For instance, Shewanella, Halomonas, Sulfurimonas and Desulfuromusa were enriched in the brine pool biofilms (Shewanella and Halomonas showed high abundance in all of the 3-day brine pool biofilms, whereas Desulfuromusa were specific in the 6-day PEEK, PTFE, SS and Ti brine pool biofilms) compared with the water samples. However, several genera differed in relative abundance between the 3-day and the 6-day biofilms. For example, Acinetobacter and Shewanella dominated the 3-day brine pool biofilms, but they could not be detected in the 6-day Al, PEEK and PEFE biofilms). A significant difference between the biofilm community structures was confirmed using a two-way PERMANOVA test, which showed that both the type of substrate and the locations had a significant effect (p < 0.05) on the microbial community composition. In summary, biofilms from different substrates, stages and locations differed in both phenotype and structure. These results suggest 1) species sorting occurred during biofilm assembly from the water species; 2) the effects of substrate type might differ between the two time points and the two habitats.
Figure 3. Taxonomic classification of qualified bacterial reads retrieved from water and biofilm samples in the brine pool and NBW.
OTUs with 97% identity were classified down to the genus level using the RDP classifier in the QIIME pipeline. Genera with a relative abundance of >1% in at least one replicate and mean values calculated from two replicates are shown.
Community similarity
To further study substrate type-coupled species sorting in biofilm assembly, the similarity between the microbial composition of the water (NBW and the brine pool) and the biofilm communities (including the 3- and the 6-day biofilms) was examined. In the Principal component analysis (PCoA) with all the samples, the brine pool and NBW samples formed distinct groups, indicating different community compositions (Fig. S3). Moreover, PCoA of brine pool samples showed that PC1 (explaining 49.1% of the variance) clearly separated the water samples from the biofilm communities. PC2 (which explains 28.7% of the variance) clearly separated the water samples, the 3-day biofilms and the 6-day biofilms (Fig. 4). However, the water and the biofilm samples from the NBW could not be clearly separated by the PC1 (which explains 71.4% of the variance) or PC2 (which explains 12.9% of the variance) (Fig. 4). Moreover, the distance between the brine pool water and biofilm communities in the PCoA results was larger than that in the NBW communities. Another interesting finding was that the extent of dissimilarity (indicated by how far away the samples were from each other on the PCoA plots) between the 3-day biofilms in the brine pool and in the NBW on different substrates was larger than that of the 6-day biofilms, despite the drastic environmental differences in the habitats where they were developed.
Figure 4. Similarity of microbial communities from water and biofilm samples, as illustrated by the OTU composition and abundance-based PCoA plot.
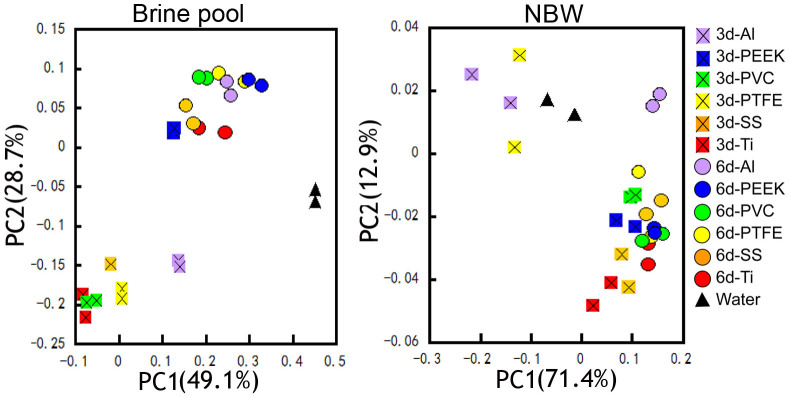
Community similarities in the brine pool and NBW are shown. The eigenvalues (percentage variance for the first two principal components) are indicated in the figure.
The results of the PCoA analysis described above were supported by the UPGMA hierarchical clustering, which revealed two large clusters with high bootstrap support for samples from the brine pool water and biofilms, The NBW water and biofilm samples have no clear separation (Fig. S4). Furthermore, the distance (distance matrix indicating the dissimilarity among all the tested samples) between the brine samples was significantly larger than between the NBW samples (0.2 versus 0.08), indicating a more important role of species sorting in the brine pool compared with the NBW.
Discussion
The effect of substrate type on marine biofilm formation has industrial implications, as the substrate type can affect biofilm metabolism and cause microfouling. Extending on our previous work5, we investigated biofilms developed in two habitats: a brine pool and NBW in the Thuwal cold seep system. Although sulfate-reducing and sulfur-oxidizing species were found in the brine pool biofilms in this study, their relative abundance and diversity in the current biofilms were less compared with the study by Lee et al. In contrast, the relative abundance of Halomonas, Shewanella and Arcobacter increased. This change was probably caused by seasonal changes in water seepage that varied with respect to the concentration of hydrogen sulfide. A greater concentration of hydrogen sulfide was recorded during the 2011 cruise by Lee et al., which resulted in sulfate accumulation. This may explain the increased number of sulfate-reducing and sulfur-oxidizing bacteria in the pool. This result is also supported by a comparison of the microbial communities present in the brine waters from the two cruises (unpublished results). Furthermore, Lee et al. developed the biofilms on a seeping vent not in the NBW. In addition, the primer pairs (U515F and U1390R) used to amplify the 16S rRNA fragments in the present study differed from those (U905F and U1492R) used by Lee et al. Although, the former primer pair targeted a longer region and overlapped with the latter primer pair21,22, both have been reported as suitable to amplify 16S rRNA fragments from uncultured samples. Lee et al. showed that the substrate effect was overarched by the in situ conditions of the deployment sites. To the contrary, our results revealed the selection effects of different substrates during the development of biofilms. It is interesting that high-biomass biofilms formed on Al, contrary to an earlier study that reported that no attached bacteria was found on Al samples from a deep-sea location outside the hydrothermal fluids of the Snake Pit site in the Mid-Atlantic Ridge23. Biofilm formation was lowest on PTFE and Ti, which indicates that these two materials may be less suitable for in situ microbial attachment, likely due to the smooth surface of PTFE and titanium toxicity. Several previous studies have pointed out the relationship between smoothness and toxicity of surfaces with bacterial EPS production and biofilm formation24,25.
The key findings are consistent with the hypotheses made in the framework of the species sorting theory. First, The effect of the substrate seems “stronger” (having a more divergent effect on community similarity) in the initial phase (3 days) than in the later stage (6 days). The type of substrate in the 3-day biofilms influenced the communities in terms of biomass and structure as compared with the 6-day biofilms. A similar finding was also obtained in our previous study of subtidal biofilms in which we hypothesized that the substrate type has strong influence on the biofilm composition during initial developmental stages and that this effect diminishes as the biofilm ages2. It has been suggested that when bacteria disperse into a new environment, environmental filtering is important during the initial step of the formation of a new community26, and it has been hypothesized that the effect of species sorting is stronger during initial rather than later stages of community assembly12. Thus, it could be a general principle shared by many tested microbial communities that the effect of local conditions become weaker as the community develops. When bacteria disperse into a local environment to build a new community, the underlying mechanism of biofilm formation might encompass the interaction between bacteria and abiotic factors initiating formation and then selecting corresponding bacterial traits. In the present study, bacterial attachment is likely affected by the properties of the substratum. For example, both the surface roughness and hydrophobicity of the stainless steel influenced bacterial adhesion, as only species with a greater adhesion ability could attach27. There is evidence that Acinetobacter, which dominated the 3-day brine pool biofilms (Fig. 3), are able to adhere to metals and form biofilms (such as Acinetobacter junii BB1A28). Also, some metals are toxic to the bacteria and only resistant species can attach to these metals and proliferate. In the present study, Shewanella was consistently one of the dominant genera in the 3-day biofilms (Fig. 3). Many species classified in this genus are metabolically versatile and capable of reducing metals, metalloids, and even radionuclides instead of oxygen during anaerobic growth29,30. The subsequent stage in the formation of a new community could be the interaction between the pioneer species and late arrivals from the regional (water) species pool. The pioneer species lay the foundation layer for subsequent species, hence reducing the constraints exerted by the environments on the new arrivals, as indicated by previous biofilm models31,32,33. Therefore, the effect of species sorting by substrate type becomes relatively weaker in shaping the community structure as the microbial community ages. There seems to be more bacteria with good abilities to adhere to the substrates in the NBW compared with the brine pool, based on the smaller differences among the 3-day NBW biofilms. However, in the brine pool, once the foundation layer has formed, a biofilm would be developed in a short time, as indicated by the high biomass of the 6-day brine pool Al, PEEK and SS biofilms. Thus, biofilm formation is probably an important strategy for some bacteria to adapt to the harsh environments in the brine pool.
Second, there was a clear separation in the PCoA analysis between the brine pool community and biofilms based on their composition; whereas the NBW water community clustered together with the NBW biofilms (Figs. 4 and S4). These results suggest that the effect of species sorting in structuring the biofilm communities was relatively stronger in the brine pool than in the NBW. A similar observation was also made by Lee et al5 for biofilms that had developed over the cold seep. In that study, greater variation was observed among brine pool biofilms compared with the seep vent biofilms. As suggested by the results of a previous study that evaluated the influence of deterministic environmental filtering on river community dynamics11, robust species sorting occurs in locations with extreme environmental variation. According to Pandit et al13, species sorting has a strong effect on specialists, whereas regional forces (patch or neutral dynamics) have a greater impact on generalists. In the present study, habitat specialists were found in the brine pool biofilms. For instance, Sulfurimonas was detected in the brine pool biofilms but rarely detected in the water and NBW samples (Fig. 3). Species of Sulfurimonas have been isolated from deep sea hydrothermal vents34. These organisms grow chemolithoautotrophically by using zero-valent sulfur, molecular hydrogen or reduced sulfur compounds as electron donors and nitrate, nitrite and oxygen as electron acceptors. The genera Desulfuromusa was enriched in the brine pool biofilms (Fig. 3), members of which are sulfate-reducing and Fe(III)-reducing bacteria, which can degrade organic compounds such as indolic compounds35. Members of Trichocomaceae, which could only be detected in the brine pool biofilms, have been shown to adapt to extreme environmental conditions36. These taxa may have a more restricted distribution and tend to be specific to extreme environments. In contrast, many genera in the NBW have a wide distribution, such as the Marinobacter, which are frequently isolated as hydrocarbon-degrading organisms in a wide variety of contaminated marine environments37, and Oleiphilus, which are frequently isolated from coastal sea waters38. Therefore, the presence of these habitat generalists in NBW biofilms may contribute to a low species sorting power. Notably, several bacterial groups that were found in the brine pool biofilms were not detected in the source community of the brine water. Thus, the prompt growth of these specialists in biofilms is consistent with the idea that biofilms consist of well-organized communities that can adapt and do not simply reflect the structure of the source communities9,39. This also explains why the effect of species sorting in structuring biofilm communities is stronger in the brine pool.
In summary, the results of the current study provide evidence for substrate-type-coupled species sorting during biofilm formation in a cold seep system. Based on these findings, we hypothesize that in this system, the effect of substrate type on biofilm formation tends to be stronger 1) in extreme rather than in mild conditions; 2) in initial rather than later stages of biofilm development; 3) for specialists rather than generalists. Moreover, there are indications that the effect of species sorting is weak for autotrophic bacteria (e.g., the sulfur-oxidizing bacteria Thiomicrospira). The distribution of these bacteria tended to be random among brine pool biofilms (Fig. 3), warranting future investigation. These results should be informative in the selection of materials for equipment deployed in the deep-sea. In an environment with a strong influence of species sorting, there appears to be an increased necessity for the selection of anti-biofilm substrates in research facilities. The present findings also suggest that the meta-community processes known to predominantly structure bacterial communities in freshwater habitats and soils (species sorting), is also important in deep sea systems. Because the application of these theories towards an understanding of deep-sea microbial communities is still in its infancy and our ultimate objective is to manage marine fouling, additional studies are needed to elucidate several related issues, such as 1) the functional basis of species sorting, which may be revealed by analyses using metagenomic and metatranscriptomic approach; 2) general principles and specific hypotheses based on datasets from larger scale deep-sea environments; and 3) the gap between theoretical prediction and practical application.
Methods
Experimental design and sampling location
The experimental design and sampling locations are shown in Figure 1 and S1, respectively. Field sampling was conducted in May 2013 in the Thuwal cold Seep II (22°16N-38°53E) using an ROV Max Rover, DSSI, U.S.A. during the King Abdullah University of Science and Technology (KAUST) Red Sea exploration cruise. The brine pool was shallow, with a consistent depth of about 1.0 m. Water samples were collected from the brine pool and the NBW using CTD device. Water was filtered through a 1.6-μM GFA filter (Whatman International Ltd, Maidstone, UK) to remove large eukaryotes. Two separate environmental replicates were collected. Biofilms were developed on six different materials: Al, PEEK, PVC, PTFE, SS and Ti in the brine pool and in NBW (Fig. 1). Pieces of these materials (50 mm × 54 mm × 1 mm, with a total surface area of 5400 mm2) were embedded in special plastic carousers that were fixed in steel frames then placed in water at a depth of 850 m by the ROV. The set-up allowed the substrates i) to be completely immersed in water for biofilm development; ii) to be stable in the deep sea; iii) to be easily manipulated to reduce variations of the microbial community produced during launch and recovery by the ROV. The ROV, equipped with a built-in CCD color video system and a 5-function manipulator arm, was controlled remotely onboard. Two steel frames were placed in the brine pool and, in parallel, two were placed in the NBW to permit incubation for 3 days. Next, one frame from the brine pool and another from the NBW were recovered for DNA extraction. The remaining frames were further incubated in the two environments for three additional days (a total of six days) then recovered.
Microscopy and cell density analyses
The morphological properties of the biofilms on the different materials were examined using SEM (representative images are shown). Bacterial enumeration was performed according to a previously described protocol40. Briefly, bacterial cells were rinsed away from the substrate surface then re-suspended in 1 mL of PBS-buffered formaldehyde (final formaldehyde concentration of 2%). Polycarbonate membrane filters (0.2-μm pore size) were placed on the filter manifold. 5 mL of sterile, distilled water and 1 mL of sample were added to the filtration well. 25 μL of DAPI was subsequently added to the filtration well. Then, the filter was placed in the dark. After incubating for 20 minutes, the solution on top of the filter was gently vacuumed away, and the filter (sample side up) was placed on a microscope slide. Finally, a drop of non-fluorescent immersion oil was placed on top of the filter. The counting was performed using a confocal laser scanning microscopy (LSM7 DUO 710, Carl Zeiss, United States) at 40 × magnification. For bacterial enumeration, 10 fields were counted. Cell density (cell/cm2) was calculated using the following formula: bacteria/cm2 = (membrane conversion factor × number ratio)/area of substrate where the membrane conversion factor = filtration area/area of micrometer field; and the number ratio = total number of bacteria counted/number of micrometer fields counted. Variations in cell density among biofilms on the different substrates were calculated in the PAST41 software package. One-way ANOVA and Levene test were used to determine differences in the mean cell density value and variation between the brine pool and the NBW biofilm groups.
DNA extraction, PCR amplification and sequencing
Biofilms developed on different substrates in the brine pool and in the NBW were harvested (ten slides for each sample) using sterile cotton tips and re-suspended in Tris-HCl buffer. DNA extraction was performed according to methods described in previous studies42. Briefly, bacterial cells from the cotton tips (for biofilm samples) and filters (for water samples) were re-suspended in Tris-HCl buffer, pelleted by centrifugation at 4000 g for 10 min and then lysed with lysozyme, proteinase K and 10% SDS. Total nucleic acid was extracted and purified using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality and quantity of DNA were checked by agarose gel electrophoresis and with a Nanodrop device (ND-1000 spectrophotometer, DiaMed China Limited, Hong Kong) at 260 nm.
Two DNA replicates (biological replicates, namely two groups of separate environmental samples) were PCR-amplified using primers containing 6- or 8-nucleotide (nt) barcodes (Table S1) for multiplex pyrosequencing. Barcodes were added to the universal forward primer U515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and the reverse primer U1390R (5′-GACGGGCGGTGTGTRCAA-3′)22 to amplify the hypervariable V4–V8 region of the 16S rRNA genes in Bacteria and Archaea. A 20-μL PCR reaction mixture containing 0.4 U of Phusion High-Fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland), 1 × HF reaction buffer, 0.1 mM of each barcoded primer, 0.6 μL of DMSO, 0.2 mM of dNTPs (TaKaRa, Dalin, China) and 10 ng of purified DNA template. The PCR was performed in a thermal cycler (Bio-Rad, U.S.A.) under the following conditions: initial denaturation at 98°C for 30 s; 26 cycles of 98°C for 10 s, 60°C for 10 s and 72°C for 15 s; and a final extension at 72°C for 5 min. PCR products were purified using the TaKaRa Agarose Gel DNA Purification Kit (TaKaRa, Dalin, China) and quantified using the NanoDrop device. Pyrosequencing of the PCR products and two replicates was conducted on the ROCHE 454 FLX Titanium platform.
Taxonomic analysis
Bioinformatic analysis of the pyrosequencing data was performed using QIIME 1.7.043 as previously described44, and the following quality control criteria was used: i) removal of reads with ambiguous nucleotides, ii) removal of reads < 150 bp, iii) removal of reads containing homopolymers ≥ 6 bp. Reads were assigned to their corresponding samples according to their barcodes, clustered and then assigned to operational taxonomic units (OTUs) at 97% identity. The most abundant reads were selected as representatives from each OTU for de novo alignment using MUSCLE45 and reference-based alignment against the Silva108 database using PyNAST46. Chimeras in aligned reads were identified by ChimeraSlayer and then removed from the dataset. The significance of differences between biofilm communities were tested by a two-way PERMANOVA in the PAST41 software package using the habitat (brine versus NBW) and substrate type as input factors. Similarities among different microbial communities were determined by variance-covariance similarity matrices generated based on the composition of OTUs (relative abundance) and displayed using the principle coordinate analysis (PCoA) and weighted pair group method with arithmetic mean (UPGMA) clustering implemented in the PAST software package. Moreover, reads were assigned to different taxa using the RDP classifier version 2.2 against Silva108. The composition of samples at the phylum and genus levels are shown in Figure S2 and 3, respectively. A cut-off of 1% relative abundance for at least one replicate was used; that is, the phylum or genus which did not achieve an abundance greater than 1% in at least one out of two replicates were removed, and the mean value of the two replicates (biological replicates as mentioned above) is shown.
Author Contributions
W.P.Z., Y.W. and P.Y.Q. wrote the main manuscript. G.Z., W.P.Z., W.X., H.L.C., Y.H.W. and Z.B. performed the experiments. W.P.Z., R.M.T., S.B. and B.Y. analyzed the data. P.Y.Q., X.X.Z. and A.A.S. designed the experiment.
Supplementary Material
supplementary file
Acknowledgments
The authors are grateful to the suggestions provided by Dr. Y.X. Li and Dr. L.S. He as well as the crew of R/V Aegaeo for providing technical assistance during sample collection. We would also like to thank Mrs Soumaya Belkharchouche and Chan Colin for English editing. This study was supported by awards from the Sanya Institute of Deep Sea Science and Engineering, the Chinese Academy of Sciences (SIDSSE, CAS) (SIDSSE-201206), COMRA program of China (COMRRDA12SC02), the National Basic Research Program of China (973 Program, No. 2012CB417304), and the King Abdullah University of Science and Technology (SA-C0040/UK-C0016) to P.Y. Qian. This work was also supported by the Strategic Priority Research Program of CAS (XDB06010100 and XDB06010200) and SIDSSE-201305 from the SIDSSE to Y. Wang.
References
- López D., Vlamakis H. & Kolter R. Biofilms. Cold Spring Harb Perspect Biol 2, a000398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. C. et al. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J 4, 817–828 (2010). [DOI] [PubMed] [Google Scholar]
- Bellou N., Papathanassiou E., Dobretsov S., Lykousis V. & Colijn F. The effect of substratum type, orientation and depth on the development of bacterial deep-sea. Biofouling 28, 199–213 (2012). [DOI] [PubMed] [Google Scholar]
- Meier A. et al. Analysis of hyper-baric biofilms on engineering surfaces formed in the Deep Sea. Biofouling 29, 1029–42 (2013). [DOI] [PubMed] [Google Scholar]
- Lee O. O. et al. In situ environment rather than substrate type dictates microbial community structure of biofilms in a cold seep system. Sci Rep 4, 3587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold M. A. et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 7, 601–613 (2004). [Google Scholar]
- Cottenie K. Integrating environmental and spatial processes in ecological community dynamics. Ecol Lett 8, 1175–1182 (2005). [DOI] [PubMed] [Google Scholar]
- Costello E. K. et al. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beseme K. et al. Unraveling assembly of stream biofilm communities. ISME J 6, 1459–1468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Bradford M. A. & Jackson R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007). [DOI] [PubMed] [Google Scholar]
- Stegen J. C. et al. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6, 653–1664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S. & Székely A. J. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5, 1086–1094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S. N., Kolasa J. & Cottenie K. Contrasts between habitat generalists and specialists: an empirical extension to the basic metacommunity framework. Ecology 90, 2253–2262 (2009). [DOI] [PubMed] [Google Scholar]
- Van der Gucht K. et al. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci U S A 104, 20404–20409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely A. J. & Langenheder S. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol Ecol 87, 102–112 (2014). [DOI] [PubMed] [Google Scholar]
- Qian P. Y. et al. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5, 507–518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools. Sci Rep 3, 1748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougouffa S. et al. Distinctive microbial community structure in highly stratified deep-sea brine water columns. Appl Environ Microbiol 79, 3425–3437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W. et al. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine–seawater interface of the Shaban Deep, Red Sea. Environ Microbiol 4, 758–763 (2002). [DOI] [PubMed] [Google Scholar]
- Batang Z. B. et al. First discovery of a cold seep on the continental margin of the central Red Sea. J Mar Syst 94, 247–253 (2012). [Google Scholar]
- Wang Y. et al. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4, e7401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Optimal eukaryotic 18S and universal 16S/18S ribosomal RNA primers and their application in a study of symbiosis. PLoS One 9, e90053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guezennec J., Ortega-Morales O., Raguenes G. & Geesey G. Bacterial colonization of artificial substrate in the vicinity of deep-sea hydrothermal vents. FEMS Microbiol Ecol 26, 89–99 (1998). [Google Scholar]
- Fang H. H. P., Chan K. Y. & Xu L. C. Quantification of bacterial adhesion forces using atomic force microscopy (AFM). J Microbiol Methods 40, 89–97 (2000). [DOI] [PubMed] [Google Scholar]
- Kerr A. et al. The biofouling resistant properties of six transparent polymers with and without pre-treatment by two antimicrobial solutions. Mater Design 22, 383–392 (2001). [Google Scholar]
- Székely A. J., Berga M. & Langenheder S. Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J 7, 61–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes P. C. et al. Work of adhesion of dairy products on stainless steel surface. Braz J Microbiol 43, 1261–1268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. & Chakraborty R. Quorum sensing in metal tolerance of Acinetobacter junii BB1A is associated with biofilm production. FEMS Microb Lett 282, 160–165 (2008). [DOI] [PubMed] [Google Scholar]
- Wu B. et al. Viability and metal reduction of Shewanella oneidensis MR-1 under CO2 stress: implications for ecological effects of CO2 leakage from geologic CO2 sequestration. Environ Sci Technol 44, 9213–9218 (2010). [DOI] [PubMed] [Google Scholar]
- Cheng Y. Y. et al. Promotion of iron oxide reduction and extracellular electron transfer in Shewanella oneidensis by DMSO. PloS One 8, e78466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E. Oral microbial communities: biofilms, interactions, and genetic systems 1. Annu Rev Microbiol 54, 413–437 (2000). [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Egland P. G., Diaz P. I. & Palmer R. J. Jr Genome–genome interactions: bacterial communities in initial dental plaque. Trends Microbiol 13, 11–15 (2005). [DOI] [PubMed] [Google Scholar]
- Donlan R. M. Biofilms: microbial life on surfaces. Emerg Infect Dis 8, 881–90 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K. et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen-and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 56, 1725–1733 (2006). [DOI] [PubMed] [Google Scholar]
- Bak F. & Widdel F. Anaerobic degradation of indolic compounds by sulfate-reducing enrichment cultures, and description of Desulfobacterium indolicum gen. nov., sp. nov. Arch Microbiol 146, 170–176 (1986). [Google Scholar]
- Pitt J. L., Samson R. A. & Frisvad J. C. in Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification (Amsterdam) (eds Robert, A. & Samson, J. I. P.) 9–49 (CRC press, 2000). [Google Scholar]
- Duran R. in Handbook of Hydrocarbon and Lipid Microbiology (eds Timmis, K. N.) 1725–1735 (Springer, 2010). [Google Scholar]
- Yakimov M. M. et al. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int J Syst Evol Microbiol 53, 779–785 (2003). [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W. & Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- Weinbauer M. G., Beckman C. & Höfle M. G. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microbiol 64, 5000–5003 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T. & Ryan P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4, 1–9 (2001). [Google Scholar]
- Zhang W. et al. Adaptation of intertidal biofilm communities is driven by metal ion and oxidative stresses. Sci Rep 3, 3180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K. & Bushman F. D. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. Toward Understanding the Dynamics of Microbial Communities in an Estuarine System. PloS one 9, e94449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary file