Abstract
The hallmarks of Lynch syndrome (LS) include a positive family history of colorectal cancer (CRC), germline mutations in the DNA mismatch repair (MMR) genes, tumours with high microsatellite instability (MSI-H) and loss of MMR protein expression. However, in ∼10–15% of clinically suspected LS cases, MMR mutation analyses cannot explain MSI-H and abnormal immunohistochemistry (IHC) results. The highly variable phenotype of MUTYH-associated polyposis (MAP) can overlap with the LS phenotype, but is inherited recessively. We analysed the MUTYH gene in 85 ‘unresolved' patients with tumours showing IHC MMR-deficiency without detectable germline mutation. Biallelic p.(Tyr179Cys) MUTYH germline mutations were found in one patient (frequency 1.18%) with CRC, urothelial carcinoma and a sebaceous gland carcinoma. LS was suspected due to a positive family history of CRC and because of MSI-H and MSH2–MSH6 deficiency on IHC in the sebaceous gland carcinoma. Sequencing of this tumour revealed two somatic MSH2 mutations, thus explaining MSI-H and IHC results, and mimicking LS-like histopathology. This is the first report of two somatic MSH2 mutations leading to an MSI-H tumour lacking MSH2–MSH6 protein expression in a patient with MAP. In addition to typical transversion mutations in KRAS and APC, MAP can also induce tumourigenesis via the MSI-pathway.
Keywords: MUTYH, base excision repair, somatic mutations, Lynch syndrome, immunohistochemistry, DNA mismatch repair
Introduction
Lynch syndrome (LS) is the most frequent autosomal dominant predisposition for early-onset colorectal cancer (CRC) (MIM# 114500) and associated tumours,1 and is diagnosed by the detection of a germline mutation in one of the mismatch repair (MMR) genes MLH1, MSH2, MSH6 or PMS2. In tumours, the typical clues for LS include high microsatellite instability (MSI-H) and the loss of MMR protein expression in immunohistochemistry (IHC). However, in 10–15% of our cases clinically suspected of LS, the MSI-H and abnormal IHC results in the tumours cannot be explained by MMR germline mutation analyses.2, 3
Germline mutations in the human Mut Y homolog (MUTYH; MIM# 604933) also lead to a predisposition to CRC and adenomas but with an autosomal recessive inheritance. The clinical picture of MUTYH-associated polyposis (MAP, MIM' 608456) is extremely variable and ranges from severe polyposis coli to attenuated forms with late age of onset or few adenomas or CRC, which creates phenotypic overlap with LS.4, 5, 6, 7, 8, 9
So far, three MAP patients have been described in the literature with an IHC loss of MLH1 protein staining and an MSI-H tumour: one male with a biallelic MUTYH germline mutation c.1227_28dupGG; p.(Glu410Glyfs*43) affecting the base excision repair (BER), and one somatic mutation in the MLH1 gene detected in his left-sided CRC at age of 49 years,10 the second with two heterozygous MUTYH mutations c.536A>G; p.(Tyr179Cys) and c.1187G>A; p.(Gly396Asp) and biallelic methylation of the MLH1 promoter in her caecal carcinoma at the age of 50 years11 and the third with homozygous MUTYH mutation c.536A>G; p.(Tyr179Cys) and no analyses for methylation or somatic mutation in MLH1 performed in the right-sided colon carcinoma diagnosed at age 50 years.12 In some publications CRC patients with MSI-H tumours were excluded for MUTYH analyses or MSI was not analysed.13, 14 Three further MAP patients were identified with MSI-H tumours but no information on IHC for MMR proteins was given,15, 16 whereas other studies detected no biallelic MUTYH mutation carrier in patients with MSI-H tumours.17, 18, 19, 20 To investigate the role of MAP as a possible cause of somatic mutations in MMR genes we analysed MUTYH in blood DNA of 85 ‘unresolved' patients suspected of having LS due to IHC loss of MMR protein expression.
Materials and methods
The study cohort includes 85 patients suspected of having LS based on the MSI-H and pathological IHC results in their tumours: 23 had IHC loss of MLH1 and PMS2, 35 had loss of MSH2 and MSH6, 19 showed loss of MSH6 only, and 8 lacked expression of PMS2 only. All patients met at least one of the Bethesda criteria21 and were tested negative for germline mutations and deletions or duplications in the MLH1, MSH2, MSH6, EPCAM and PMS2 genes before this study. MLH1-deficient cases were included if MLH1 promoter methylation was absent in tumour DNA. Sixty nine patients were from Germany and sixteen from the USA. All patients provided their informed consent for cancer genetics research.
Control cohort: 403 CRC patients with either confirmed LS by pathogenic MMR germline mutation (N=326) or sporadic tumourigenesis by BRAF mutation and MLH1 promoter methylation in tumour DNA (N=77).
DNA extraction from EDTA blood, standard PCR and direct sequencing with ABI PRISM 3100 Avant (Big Dye v1.1) was performed according to standard protocols. For computer analysis of the sequences the software Mutation surveyor 3.1 (SoftGenetics, State College, PA, USA) was used.
Mutation pre-screening of MUTYH was performed by direct sequencing of exon 6–7 and 12–13 including the hotspot mutations p.(Tyr179Cys) and p.(Gly396Asp) after PCR amplification using Ampli-Taq Gold (Life Technologies, Carlsbad, CA, USA) and digestion with Exo-SAP kit (Affymetrix, Santa Clara, CA, USA). At least one of these two hotspot mutations is found in 82–98% of all MAP cases in Caucasian, Canadian, American and Australian populations12, 22 and in our own unpublished cohort. In a further step, sequencing has been completed for all 16 coding exons and flanking regions of MUTYH (GenBank: NM_001128425.1; NG_008189.1) for 73 patients, 12 patients could not be analysed completely due to lack of DNA. In addition, deletion/duplication screening of all MUTYH exons was carried out by multiplex ligation-dependent probe amplification (MLPA) with MLPA kit P378-A1 Lot0910 (MRC Holland, Amsterdam, The Netherlands) and analysed with GeneScan 3.7 software (Life Technologies).
Owing to the accepted nomenclature, which uses the longest MUTYH-transcript (NM_001128425.1) as a reference, nucleotide and amino acid numbering after nucleotide position 157 (amino acid 53) may differ from other publications by up to 42 nucleotides (14 amino acids). The frequent mutations p.(Tyr179Cys) in exon 7 and p.(Gly396Asp) in exon 13, previously reported as p.(Tyr165Cys) and p.(Gly382Asp) correspond to p.(Tyr176Cys) and p.(Gly393Asp) using other transcripts of MUTYH.
Germline MUTYH mutations were submitted to the LOVD database (http://chromium.liacs.nl/LOVD2/colon_cancer/home.php?select_db=MUTYH).
Tumour DNAs from formalin-fixed paraffin-embedded tissues available from the biallelic mutation carrier were sequenced for somatic MAP-specific G:C>T:A transversions in MSH2 (NM_000251.2; NG_007110.2), KRAS exon 2 and 3 (NM_004985.3; NG_007524.1), and BRAF exon 15 (NM_004333.3).
Results
Biallelic MUTYH germline mutations lead to biallelic somatic transversions in the MMR gene MSH2
In one patient displaying loss of MSH2 and MSH6 on IHC of his sebaceous gland carcinoma, we detected homozygosity for the pathogenic MUTYH germline mutation c.536A>G; p.(Tyr179Cys) in exon 7. A deletion of the second allele was excluded by MLPA. The patient was diagnosed with two synchronous rectal adenocarcinomas T2 N0 G2 and several tubulo-villous adenomas at age 55, an urothelial bladder carcinoma at age 65, a sebaceous gland carcinoma and a sebaceous gland hypertrophy at age 66 years. The rectal cancer and the urothelial bladder cancer were microsatellite stable and had normal MMR protein staining by IHC.
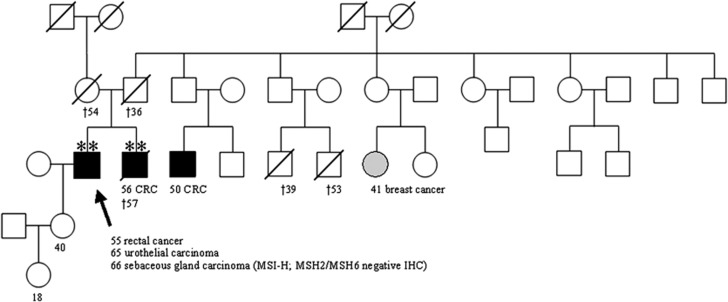
LS was suspected originally due to the patient's personal history in addition to his family history of a brother with caecal CRC T3 N2 G3 at age 56 years and two affected cousins (CRC at age 50; breast cancer at age 41 years) (Figure 1). Biallelic copies of the MUTYH mutation c.536A>G; p.(Tyr179Cys) were also found in DNA extracted from the paraffin-embedded normal tissue of the deceased brother. DNA of further family members was not available for genetic testing to assess the MUTYH mutation status.
Figure 1.
Pedigree of the family with the affected individual III:2 and his affected brother III:3 both bearing a homozygous MUTYH p.(Tyr179Cys) mutation (**). The sebaceous gland carcinoma revealed an MSI-H phenotype with loss of MSH2/MSH6 expression. Germline mutation analysis of MMR genes was negative. No genomic DNA was available from other affected family members with CRC as well as no detailed information concerning the parents.
Chronic lymphadenitis as a typical histological feature of LS was described in the CRC of the index patient and his brother. Information on Crohn's-like lymphocytic response and tumour-infiltrating lymphocytes reported in BER- and MMR-deficient tumours16, 23 was not given.
Tumour analyses
Tumour sequencing of the index patient's sebaceous gland carcinoma identified two somatic pathogenic transversion mutations in the MSH2 gene (c.2101G>T; p.(Glu701*) in exon 13 and c.2554G>T; p.(Glu852*) in exon 15, Supplementary Figure 1A) explaining the development of MSI-H in tumourigenes is and loss of the MSH2 and MSH6 proteins in this tumour. The phase of the MSH2 mutations in exon 13 and 15 in tumour DNA could not be determined, as we did not find heterozygous SNPs in the neighbouring genomic regions in blood DNA of the patient and the amplification of 4.6 kb from exon 13 to 15 in tumour DNA for subcloning was not successful due to the poor DNA quality. The other two tumours, a rectal carcinoma and an urothelial carcinoma that were microsatellite stable, both had a heterozygous somatic KRAS hotspot mutation c.34G>T; p.(Gly12Cys) transversion in exon 2 (Supplementary Figure 1B).
Monoallelic MUTYH germline mutations in LS suspected individuals and controls
None of the remaining patients analysed were found to have MUTYH mutations. The control cohort of 403 CRC patients revealed five patients with monoallelic heterozygous hotspot mutations (two in exon 7 and three in exon 13). No second MUTYH mutation was found in these patients.
Discussion
Biallelic MUTYH mutations impair the BER process and can result in somatic mutational inactivation of both MMR-alleles mimicking LS by displaying a MMR-deficient tumour. To our knowledge, in the literature six MAP patients with MSI-H tumours were described,15, 16 three of them with an IHC MLH1-deficiency in their MSI-H CRC.10, 11, 12 The loss of MLH1 staining in those three MAP tumours with MSI-H was caused by a somatic MAP-specific mutation in MLH1 together with a second undetectable defect,10 whereas biallelic methylation of the MLH1 promoter was causing the MLH1-deficiency in the other MAP patient.11
Here we present a MAP-patient, in which BER deficiency was associated with the development of two somatic MAP-specific G>T transversion mutations in the MSH2 gene in the tumour resulting in MMR deficiency, MSI-H and a loss of MSH2 and MSH6 expression in the patient's sebaceous gland carcinoma. This patient's CRC and urinary bladder carcinoma revealed the MAP-specific KRAS G>T transversion mutations.14 The somatic G>T transversions in MSH2 or KRAS, respectively, in three different tumours of the patient are very likely to be due to the germline BER defect.7, 8
Interestingly, three further cancer diagnoses were reported in this family including CRC and breast cancer, which are compatible with a recessive mode of inheritance and the clinical phenotype of MAP.23, 24, 25 Biallelic MUTYH mutations were detected in the brother and might also have induced the tumourigenesis in the cousins (not investigated). Heterozygous MUTYH mutations were not identified in the remaining 84 index patients, for which somatic mutations or further germline pathomechanisms, for example, large genomic rearrangements such as inversions in the MMR genes have to be considered.3, 26, 27, 28 In the control cohort the MUTYH mutation frequency of 1.24% was comparable to the frequencies of 0%–4.8% reported in the general population.12, 22, 29
As MAP can manifest in an extremely variable phenotype, MUTYH mutation analysis should also be considered in patients with a low number of adenomas, early-onset or synchronous or metachronous CRC and in patients with MSS tumours.30, 31 With an incidence of only 1.18% in MMR-deficient CRC patients with typical histological features of LS and several adenomas MAP can mimic LS. These patients need different surveillance and due to the different pattern of inheritance a different risk annotation for family members.32 We suggest to perform MUTYH genetic testing in these unresolved MSI-H cases with additional adenomas either in the two fragments harbouring the common mutations or in the course of next-generation sequencing cancer kits including the MMR genes, APC and MUTYH besides others.18
We reported for the first time a MAP patient initially suspected of LS, in which two MAP-specific somatic MSH2 mutations explained the IHC loss of MSH2/MSH6 in his MSI-H tumour. MAP might be a differential diagnosis for CRC patients with MSI-H tumours and several adenomas.
Acknowledgments
We would like to thank the German Cancer Aid (Deutsche Krebshilfe grant 110780) and the Wilhelm Sander-Stiftung grant 2012.081.1 as well as grants P30 16058 and P01 CA124570 from the National Cancer Institute, USA, for support of this work. We also thank all patients for their participation in this study, as well as their respective doctors for contributing clinical information.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- Koehler U, Grabowski M, Bacher U, Holinski-Feder E. A new interphase fluorescence in situ hybridization approach for genomic rearrangements involving MLH1 and MSH6 in hereditary nonpolyposis colorectal cancer-suspected mutation-negative patients. Cancer Genet Cytogenet. 2007;175:81–84. doi: 10.1016/j.cancergencyto.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Morak M, Koehler U, Schackert HK, et al. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J Med Genet. 2011;48:513–519. doi: 10.1136/jmedgenet-2011-100050. [DOI] [PubMed] [Google Scholar]
- Aceto G, Cristina Curia M, Veschi S, et al. Mutations of APC and MYH in unrelated Italian patients with adenomatous polyposis coli. Hum Mutat. 2005;26:394. doi: 10.1002/humu.9370. [DOI] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Goergens H, et al. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C—>T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C—>T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- Morak M, Laner A, Bacher U, Keiling C, Holinski-Feder E. MUTYH-associated polyposis - variability of the clinical phenotype in patients with biallelic and monoallelic MUTYH mutations and report on novel mutations. Clin Genet. 2010;78:353–363. doi: 10.1111/j.1399-0004.2010.01478.x. [DOI] [PubMed] [Google Scholar]
- Lefevre JH, Colas C, Coulet F, et al. MYH biallelic mutation can inactivate the two genetic pathways of colorectal cancer by APC or MLH1 transversions. Fam Cancer. 2010;9:589–594. doi: 10.1007/s10689-010-9367-0. [DOI] [PubMed] [Google Scholar]
- Colebatch A, Hitchins M, Williams R, Meagher A, Hawkins NJ, Ward RL. The role of MYH and microsatellite instability in the development of sporadic colorectal cancer. Br J Cancer. 2006;95:1239–1243. doi: 10.1038/sj.bjc.6603421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SP, Cotterchio M, Jenkins MA, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136:1251–1260. doi: 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopperts AP, Nielsen M, Niessen RC, et al. Contribution of bi-allelic germline MUTYH mutations to early-onset and familial colorectal cancer and to low number of adenomatous polyps: case-series and literature review. Fam Cancer. 2013;12:43–50. doi: 10.1007/s10689-012-9570-2. [DOI] [PubMed] [Google Scholar]
- van Puijenbroek M, Nielsen M, Tops CM, et al. Identification of patients with (atypical) MUTYH-associated polyposis by KRAS2 c.34G>T prescreening followed by MUTYH hotspot analysis in formalin-fixed paraffin-embedded tissue. Clin Cancer Res. 2008;14:139–142. doi: 10.1158/1078-0432.CCR-07-1705. [DOI] [PubMed] [Google Scholar]
- Kambara T, Whitehall VL, Spring KJ, et al. Role of inherited defects of MYH in the development of sporadic colorectal cancer. Genes Chromosomes Cancer. 2004;40:1–9. doi: 10.1002/gcc.20011. [DOI] [PubMed] [Google Scholar]
- O'Shea AM, Cleary SP, Croitoru MA, et al. Pathological features of colorectal carcinomas in MYH-associated polyposis. Histopathology. 2008;53:184–194. doi: 10.1111/j.1365-2559.2008.03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez MD, Balaguer F, Bujanda L, et al. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin Cancer Res. 2010;16:5402–5413. doi: 10.1158/1078-0432.CCR-10-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanskanen T, Gylfe AE, Katainen R, et al. Exome sequencing in diagnostic evaluation of colorectal cancer predisposition in young patients. Scand J Gastroenterol. 2013;48:672–678. doi: 10.3109/00365521.2013.783102. [DOI] [PubMed] [Google Scholar]
- Wang L, Baudhuin LM, Boardman LA, et al. MYH mutations in patients with attenuated and classic polyposis and with young-onset colorectal cancer without polyps. Gastroenterology. 2004;127:9–16. doi: 10.1053/j.gastro.2004.03.070. [DOI] [PubMed] [Google Scholar]
- Urso E, Agostini M, Pucciarelli S, et al. Clinical and molecular detection of inherited colorectal cancers in northeast Italy: a first prospective study of incidence of Lynch syndrome and MUTYH-related colorectal cancer in Italy. Tumour Biol. 2012;33:857–864. doi: 10.1007/s13277-011-0312-0. [DOI] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aretz S, Tricarico R, Papi L, et al. MUTYH-associated polyposis (MAP): evidence for the origin of the common European mutations p.Tyr179Cys and p.Gly396Asp by founder events Eur J Hum Genet 2013. e-pub ahead of print 30 January 2013 doi: 10.1038/ejhg.2012.309 [DOI] [PMC free article] [PubMed]
- Nielsen M, de Miranda NF, van Puijenbroek M, et al. Colorectal carcinomas in MUTYH-associated polyposis display histopathological similarities to microsatellite unstable carcinomas. BMC Cancer. 2009;9:184. doi: 10.1186/1471-2407-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S, Jones N, Christian D, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis Gastroenterology 20091371976–1985.e1–10. [DOI] [PubMed] [Google Scholar]
- Wasielewski M, Out AA, Vermeulen J, et al. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat. 2010;124:635–641. doi: 10.1007/s10549-010-0801-7. [DOI] [PubMed] [Google Scholar]
- Chen JM. The 10-Mb paracentric inversion of chromosome arm 2p in activating MSH2 and causing hereditary nonpolyposis colorectal cancer: re-annotation and mutational mechanisms. Genes Chromosomes Cancer. 2008;47:543–545. doi: 10.1002/gcc.20556. [DOI] [PubMed] [Google Scholar]
- Wagner A, van der Klift H, Franken P, et al. A 10-Mb paracentric inversion of chromosome arm 2p inactivates MSH2 and is responsible for hereditary nonpolyposis colorectal cancer in a North-American kindred. Genes Chromosomes Cancer. 2002;35:49–57. doi: 10.1002/gcc.10094. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Chen JM, Ball EV, et al. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum Mutat. 2010;31:631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Houlston RS.Colorectal cancer risk in monoallelic carriers of MYH variants Am J Hum Genet 200679768–771.author reply 771-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper M, Plotz G, Juengling B, Trojan J, Lammert F, Raedle J. Adenoma development in a patient with MUTYH-associated polyposis (MAP): new insights into the natural course of polyp development. Dig Dis Sci. 2010;55:1711–1715. doi: 10.1007/s10620-009-0916-z. [DOI] [PubMed] [Google Scholar]
- Strate LL, Syngal S. Hereditary colorectal cancer syndromes. Cancer Causes Control. 2005;16:201–213. doi: 10.1007/s10552-004-3488-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Soler M, Perez-Carbonell L, Guarinos C, et al. Risk of cancer in cases of suspected lynch syndrome without germline mutation Gastroenterology 2013144926–932.e921; quiz e913-924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.