Abstract
Single CD34+ cells from adult human peripheral blood show mtDNA sequence heterogeneity. In this study, we compared mtDNA sequence variation in single CD34+ cells from peripheral blood (PB) mononuclear cells (MNCs) from the same donors but under different conditions of storage and transport: group I, MNCs from heparinized PB that inadvertently required six days to be transported to the testing laboratory; group II, MNCs which were isolated from PB within a day of phlebotomy and frozen prior to transportation and storage. We observed more cell death for MNCs of group I than group II. Concordantly, group I CD34+ cells had a very low potential for hematopoietic colony formation in vitro compared with group II cells. CD34+ cells of group II showed an unexpectedly higher level of mtDNA sequence heterogeneity than was present group I cells. These observations suggest that reduced mtDNA sequence heterogeneity in single CD34+ cells of group I was likely due to elimination of cells harboring mutations. CD34+ cells that survive stress ex vivo may be more enriched in quiescent primitive hematopoietic stem cells, with fewer mtDNA mutations than committed progenitors. Technically, attention is required for conditions of preparation of human blood samples for single cell mtDNA analysis.
Keywords: mtDNA, single cell analysis, mutation, hematopoietic stem cell, committed progenitors
1. Introduction
Human mtDNA is a circular double-stand molecule which encodes 2 rRNA, 22 tRNA, and 13 proteins essential for oxidative phosphorylation [1]. Because of the unique characteristics of this molecule, such as maternally inheritance, lack of recombination, a high mutation rate, and many copies per cell, mtDNA has been extensively used as a marker in the reconstruction of demographic history for ethnic populations and in forensic science identification [2, 3]. Germline and somatic mutations in mtDNA, which affect the transcription and translation of mtDNA genes or lead to defective protein production, cause mitochondrial dysfunction and contribute to a wide variety of disorders [4, 5]. The cumulative burden of mtDNA mutations is central to the “mitochondrial theory of aging” [6], which hypothesizes that aging is caused by a vicious cycle between reactive oxygen species generation and mtDNA mutation induction [7, 8]. However, this theory has been challenged by recent data, and further there are common misconceptions and criticisms concerning the theory itself [9, 10].
Over several years, we have systematically analyzed mtDNA mutation in single hematopoietic cells, with the intention to characterize potential roles of accumulated mtDNA mutations during the aging of human hematopoietic system and as they may relate to hematologic diseases [11–15]. We are also interested in whether somatic mtDNA mutations in single CD34+ cells are stable and can be utilized as markers to trace the hematopoietic stem cell (HSC) and its progeny [16]. We detected aging-dependent accumulation of mtDNA mutations in single colonies of CD34+ cells from cord blood, peripheral blood (PB), and bone marrow [12, 13]. CD34+ cells marked by certain mtDNA variant(s) are stable in vivo and repopulate recipient after HSC transplantation [16]. Moreover, some mtDNA variants in differentiated cells, such as T-cells, B-cells, and granulocytes, also are present in CD34+ cells from that individual, suggesting common origin of these mature cells from the same HSC clone [11].
In subsequent experiments, we aimed to discern mtDNA sequence variations in single CD34+ cells from multiple members of a large pedigree whose ages varied widely [15]. Due to unexpected delay in the arrival of blood samples from Europe to the testing laboratory and apparently anomalous results, we were forced to analyze mtDNA sequence heterogeneity in CD34+ cells isolated from peripheral blood (PB) mononuclear cells (MNCs) from the same donor twice. In the first round of sample collection (group I), PB was collected in heparin and shipped by express “overnight” service to the testing laboratory in Bethesda. However, the shipment was delayed, and MNCs were isolated upon receiving the samples six days after the initial phlebotomy (and subsequent isolation of cells and freezing in liquid nitrogen). Because of our concern for the integrity of the DNA ultimately extracted and anomalous results, we undertook a second round of sample collection from the same individuals (group II); MNCs were isolated from PB within 24 hours of blood drawing, frozen in a local laboratory by the same procedure, and then cells were transported to the testing laboratory in dry ice and transferred to liquid nitrogen upon arrival. We observed markedly different levels of mtDNA sequence heterogeneity in single CD34+ cells from the same donors’ MNCs after these different collection, storage and transportation protocols. Our observation suggested that mtDNA sequence heterogeneity in single CD34+ cells may unexpectedly alter in vitro, most likely due to changes in the composition of the CD34+ cell population.
2. Materials and Methods
2.1. Sample collection
PB was drawn from the maternally related healthy donors living in Italy and the United States (Table S1); these donors were from Family B described in our recent publication [15]. In group I, whole blood (8 – 10 mL) from each of ten donors was collected in a heparinized tube in Europe and mailed to Bethesda. MNCs were separated by Ficoll density gradient centrifugation upon arrival, but after an unexpected six days after blood collection, followed by storage in liquid nitrogen. In group II, a similar amount of PB was obtained from ten donors (eight of them had been sampled in group I) in a heparinized tube and processed in a proximate laboratory in Italy within 24 hrs of phlebotomy by sedimentation of MNCs using the same standard protocol, freezing, and storage in liquid nitrogen. Freezing medium was composed of 10% dimethyl sulfoxide (DMSO) and 90% fetal bovine serum (FBS). The frozen MNCs were shipped to Bethesda on dry ice and then transferred to liquid nitrogen until they were thawed for sorting. MNCs of three healthy donors from the family living in the United States were processed within 24 hrs of blood collection and were stored promptly in liquid nitrogen. All donors gave informed consent according to a protocol for sample collection approved by the institutional review board of the National Heart, Lung, and Blood Institute.
2.2. Single CD34+ cell sorting and culture
Frozen MNCs were thawed and washed with phosphate buffered saline (PBS), as described [11, 15, 16]. Briefly, about 1 × 106 cells suspended in 100 µL of PBS containing 0.5% bovine serum albumin (BSA) were incubated with anti-CD34 phycoerythrin (PE)-conjugated monoclonal antibody (BD Bioscience, San Jose, CA) for 30 min at 4°C. Cells were then washed and resuspended in 600 µL of PBS supplemented with 0.5% BSA. We added 4 µL of 7-amino-actinomycin D (7-AAD) into the cell suspension and kept the cells on ice for approximately 20 min or until sorting on the MoFlo Cytometer (Dako-Cytomation, Ft Collins, CO). Cell sorting was performed using 100 milliwatts of the 488 nm line of an argon laser (I-90, Coherent Inc, Palo Alto, CA) for excitation. Forward scatter was the triggering parameter. Single cell deposition was accomplished using the CyClone automated cloner (Dako-Cytomation) in the 0.5 single drop mode with gating based on forward scatter and fluorescence, as described in our previous studies [11, 14, 16]. Single CD34+ cells from four donors, with ages of 51, 60, 87, and 89 years, respectively (Table 1), were collected in one or two 96-well plates (one cell/well) for mtDNA analysis, depending on the cell number. The remaining cells were then sorted into 96-well round-bottom cell culture plates (Corning Inc., Corning, NY) in the same mode as used for culture. Each well contained 50 µL of StemSpan® Serum-Free Expansion Medium supplemented with StemSpan® CC100 cytokine cocktail (StemCell Technologies Inc., Vancouver, Canada). Single CD34+ cells were cultured for a week in 5% CO2 at 37 °C. Colonies in wells of the culture plate were viewed using an inverted Olympus IX50 microscope. For the remaining six samples in group I and nine samples in group II (Table S1), we counted the ratio for CD34+ cells/live cells of each sample, to show the relative number of CD34 positive staining cells.
Table 1.
Number of mtDNA sequence haplotypes in the single CD34+ cell population
| Donor1 | Sample2 | Age/Sex | Live cells / Gated cells (%)3 |
CD34+ cells / Live cells (%)3 |
No. of cells for mtDNA analysis |
No. of haplotypes |
No. of haplotypes defined by nucleotide substitutions |
No. of haplotypes / 100 cells |
No. of haplotypes by substitutions / 100 cells |
|---|---|---|---|---|---|---|---|---|---|
| Donor #1 | Sample 5 | 51/M | 62.55 | 0.208 | 96 | 10 | 5 | 10.4 | 5.2 |
| B-8 | 91.92 | 0.371 | 94 | 35 | 28 | 37.2 | 29.8 | ||
| Donor #2 | Sample 10 | 89/F | 70.29 | 0.079 | 95 | 22 | 14 | 23.2 | 14.7 |
| B-3 | 96.41 | 0.026 | 86 | 48 | 42 | 55.8 | 48.8 | ||
| Donor #3 | Sample 6 | 60/F | 77.95 | 0.099 | 95 | 19 | 10 | 20.0 | 10.5 |
| B-6 | 89.71 | 0.025 | 95 | 47 | 38 | 49.5 | 40.0 | ||
| Donor #4 | Sample 9 | 87/F | 71.24 | 0.019 | 95 | 13 | 8 | 13.7 | 8.4 |
| B-5 | 87.69 | 0.025 | 95 | 49 | 40 | 51.6 | 42.1 |
These four donors were maternally related and belonged to family B described in our recent study [15]. Donors #1, #2, #3, and #4 refer to family members B-8, B-3, B-6 and B-5, respectively.
Samples from the same donors that were included in group I and group II were represented marked by “Sample” and “B-”, respectively. The mtDNA sequence data for samples of group II (B-8, B-3, B-6, and B-5) were taken from Yao et al. [15].
The gating for living cells and CD34+ cells was defined in Figure 1.
2.3. Single cell PCR amplification and sequencing
For mtDNA analysis, we followed our previously described procedure [11, 14–16]. In brief, single CD34+ cells were sorted into each well of an optical 96-well reaction plate (MicroAmp; Applied Biosystems, Foster City, CA) containing 50 µL of lysis buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl, 100 µg/mL Proteinase K, 1% Triton X-100). The plate was incubated at 56°C for 30 min, followed by incubation at 96°C for 8 min to release the mtDNA molecules. Two-step nested PCR was used to amplify the entire mtDNA control region in single CD34+ cells [14, 16]. The first PCR was performed in 30 µL of reaction mixture, which containing 5 µL of cell lysate, 400 µM of each dNTP, 1×LA PCRTM Buffer II (Mg2+ plus), 1 unit of TaKaRa LA TaqTM that with proof-reading activity (Takara Bio. Inc.), and 0.5 µM of each primer (L15594: 5’ - CGCCTACACAATTCTCCGATC -3’ and H901: 5’- ACTTGGGTTAATCGTGTGACC -3’). PCR amplification was performed on a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA) with the following cycles: one cycle of 94°C for 3 min; then 35 cycles of 94°C for 30 sec, 50°C for 40 sec and 72°C for 1 min with a 5 sec increase per cycle; and ending with a full extension cycle of 72°C for 10 min. The second PCR was performed in 50 µL of reaction mixture containing 400 µM of each dNTP, 1×LA PCR™ Buffer II (Mg2+ plus), 2 units of TaKaRa LA Taq™, 0.5 µM of each (L15990: 5’ – TTAACTCCACCATTAGCACC -3’ and H650: 5’ – GAAAGGCTAGGACCAAACCTA -3’), and 5 µL of each first PCR product. Amplification cycles for the second PCR consisted of one cycle of 94°C for 3 min; 35 cycles of 94°C for 30 sec, 52°C for 40 sec and 72°C for 90 sec; and ending with a full extension cycle of 72°C for 10 min.
Second PCR products were purified using the ExcelaPure 96-Well UF PCR Purification Kit (EdgeBiosystems, Gaithersburg, MD) and sequenced by using the BigDye Terminator v3.1 Cycle Sequencing Kit on a 3100 DNA sequencer (Applied Biosystems), according to the manufacturer’s manual. We used the second PCR primers and the following primers that were described in our previous studies [14–16] to cover the entire mtDNA control region: L15996, 5’- CTCCACCATTAGCACCCAAAGC -3’; L16209, 5’- CCCCATGCTTACAAGCAAGT-3’; L16517, 5’ - CATCTGGTTCCTACTTCAGG -3’; H26, 5’-GCATGGAGAGCTCCCGTGAGTGG-3’; L29, 5’ - GGTCTATCACCCTATTAACCAC-3’; L332, 5’- CCCGCTTCTGGCCACAGCAC-3’.
2.4. Scoring mtDNA sequence variation and statistical analysis
Our approach to define sequence variations in single CD34+ cells has been published [14–16]. In brief, sequences were aligned by SeqMan program in DNAstar package (DNASTAR Inc.) and were proof-read by eyes. Sequence variants were scored relative to the Cambridge Reference Sequence (rCRS) [1]. Cells with contamination were identified using the approach described before [17] and were excluded from the analysis. Due to the limits of sequencing, heteroplasmy (co-existence of wild-type and the mutant allele) of certain mtDNA variant was scored when a mutant allele was present at >10% level chromatography [14]. Because the four donors selected for mtDNA analysis were maternally related, their mtDNA sequences were essentially the same, and they contained a 16189T>C polymorphism that triggers heteroplasmy in multiple poly-C tracts in region 16184–16193; these length polymorphisms of poly-C tracts could be not reliably counted based on the sequencing electropherograms and were not analyzed in the current study. We scored the length variations of the C-tract in region 303–309 by direct counting of the base shift of T at site 310. The length mutation of the AC repeat in region 515–524 in the third hypervariable segment (HVS-III) was also based on sequencing electropherograms.
We counted the number of haplotypes in a population of cells from each sample and used it as an index to compare the level of mtDNA sequence heterogeneity between different samples. This index reflects the total number of mutations that have occurred or retained within a given number of cells, which in turn reflects the time over which the subclone has developed and the net mutation rate per unit time [14].
The unpaired t test was used to compare differences between the two groups of samples that had been processed differently. We used the Fisher exact test to quantify the difference of heterogeneity level in CD34+ cells from the same donor. A value of P < 0.05 was regarded as statistically significant.
3. Results and Discussion
3.1. Cell viability and colony formation
Upon receiving the heparinized blood shipped to Bethesda at day 6 after blood collection, we suspected hemolysis, as the serum was tinged red. After Ficoll density gradient centrifugation and washing in PBS, we discarded the cell clump and froze the remaining MNCs in freezing medium. Staining with trypan blue showed that 20–50% of suspended MNCs were dead cells among the samples.
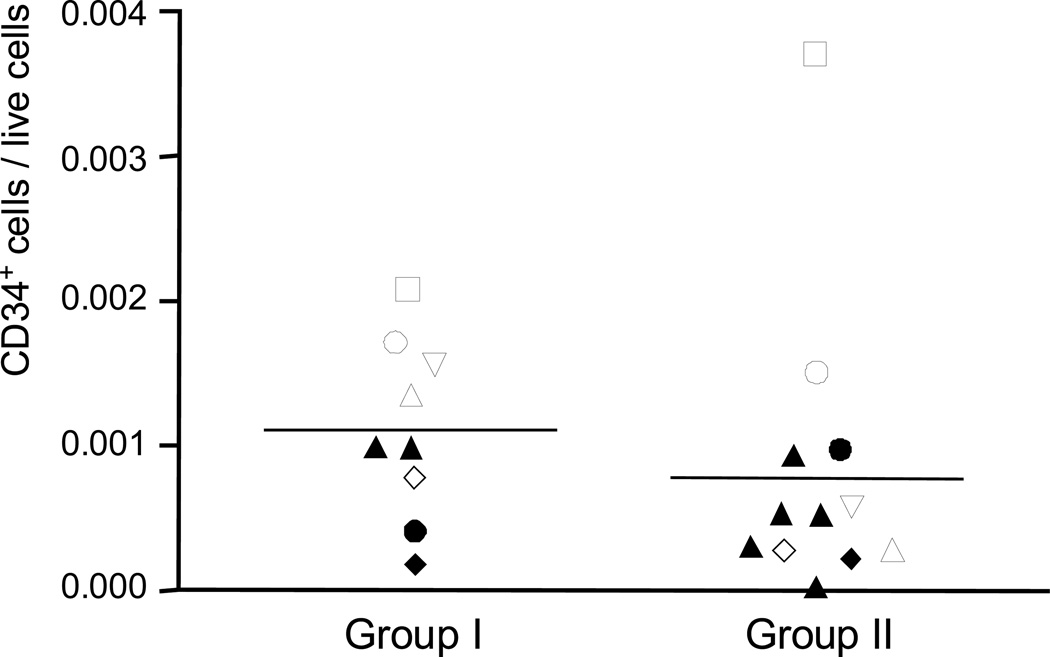
After thawing and washing of the frozen MNCs, we observed cell debris clumps for the samples of group I but not for samples of group II, suggesting that many of the MNCs in the former were dead. Staining with 7-AAD during the sorting for the MNCs confirmed that a high proportion (22%–58%) of MNCs of group I was dead cells, whereas of MNCs of group II, only 4%–15% cells were positive for 7-AAD staining (Fig. 1A). However, we observed a generally higher frequency of CD34+ cells in the gated area for MNCs in samples of group I compared to group II (except for donor #1, who showed an inverse pattern; Table 1, Table S1 and Fig. 2). Since we removed the dead cells by 7-AAD staining during the sorting, we do not think the enriched CD34+ cells in the MNCs of group I were artifacts of non-specific staining of dead cells. The enrichment of CD34+ cells in group I more likely and simply is explained by higher viability of CD34+ cells compared with more mature leucocytes, as others have shown that most hematopoietic progenitor cells, characterized by positive staining for CD34, were more resistant to cryopreservation injury than mature mononucleated cells, regardless of the overall post-thaw total nucleated cell viability [18, 19].
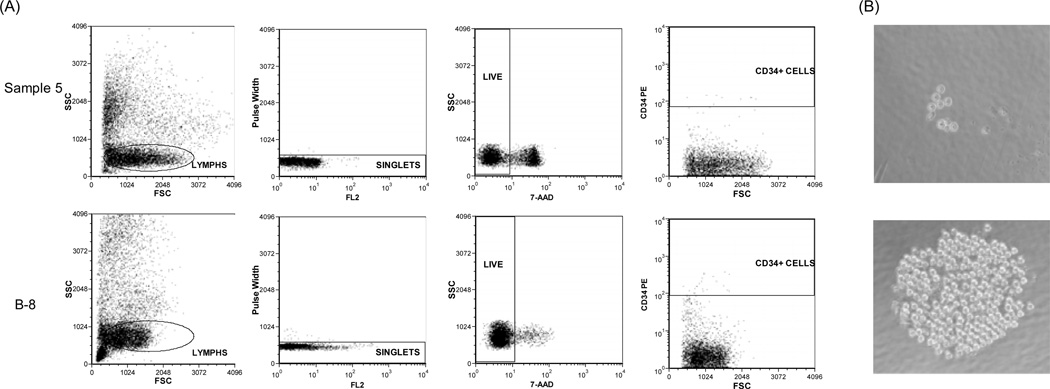
Figure 1.
Sorting for CD34+ cells using flow cytometry and culture assay for single CD34+ cells. (A) Dead cells in the mononuclear cells from donor #1 were demonstrated by staining with 7-AAD. Cells from Sample 5 (group I) had a lower level of viability than cells from B-8 (group II). (B) Single CD34+ cells from Sample 5 (group I) had less colony formation potential than those from B-5 (group II). A representative colony developed from individual CD34+ cell of each group was shown.
Figure 2.
Enriched CD34+ cells in mononuclear cells from group I but not in group II. The same donors that were sampled for both group I and group II studies were represented by the same icons. Donors sampled in either group I or group II study were demonstrated by filled triangles. The detailed information regarding samples was listed in Table S2.
In total, we collected 165 single CD34+ cells from the MNCs of group I for culture. After a week, we observed 7 colonies, all of which were relatively small (< 20 cells/colony). In contrast, we observed much more colony formation (727/1709) by single CD34+ cells of group II and a high frequency of large colonies (e.g. > 100 cells/colony; colony formation potential was statistically significant between the single CD34+ cells from the two groups by two tailed Fisher exact test, P < 1×10−6) (Fig. 1B). Thus, single CD34+ cells from samples of group II had a higher colony formation potential. As we only cultured CD34+ cells for a week, and this population was functionally heterogeneous, our conditions favored detection of growth of more differentiated lineage-committed progenitors rather than primitive HSCs that require more time in vitro to generate progeny. While we inferred that more quiescent cells with properties closer to a hematopoietic stem cell were preserved in group 1 specimens, as would be suggested by the literature [18, 19], we were not able to directly test for these cells in these samples.
3.2. Reduced level of mtDNA variation in group I cells
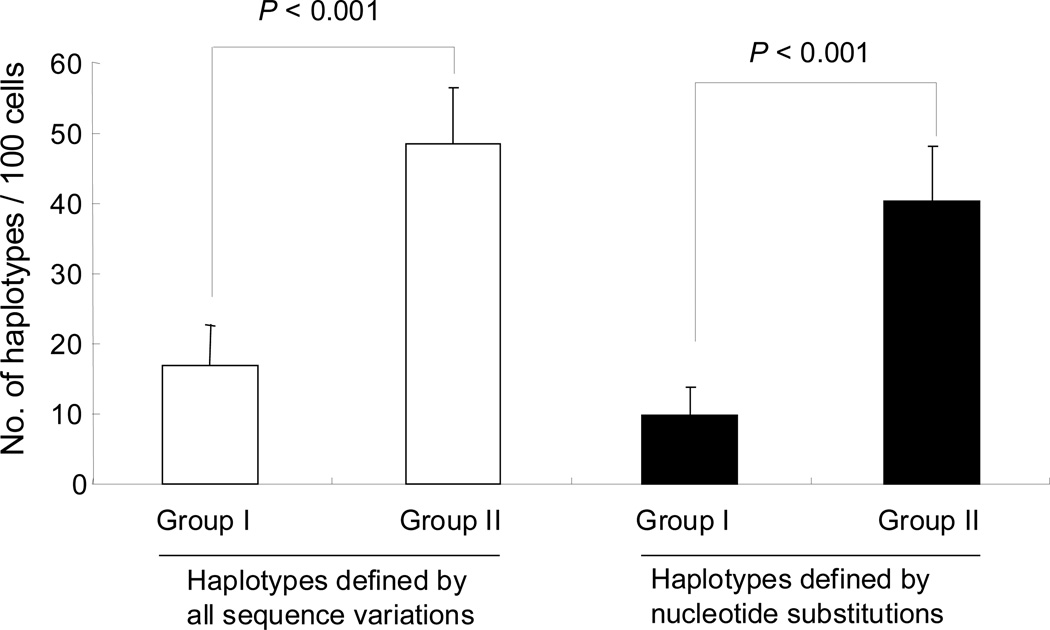
In total, we analyzed 751 single CD34+ cells for mtDNA sequence variation from four healthy donors (Table 1). Among them, 381 cells were from MNCs of group I samples, and 370 cells were from MNCs prepared from samples in group II and were reported recently [15]. The values for the average number of haplotypes per 100 cells in each donor, defined by all sequence variations or by nucleotide substitutions only, were significantly lower (P<0.001) in single CD34+ cells of group I than in CD34+ cells from group II (Fig. 3 and Table 1). The reduced level of mtDNA heterogeneity in group I CD34+ cells was unexpected, as we had detected both global cell loss and deficiency of proliferating progenitor cells in these samples and anticipated DNA damage or stress might increase mtDNA mutations in these cells. Alternatively, these results are consistent with over representation of primitive and quiescent CD34+ cells in group I and a lower level of mtDNA mutation than in the committed progenitor cells. Clonal expansion of certain CD34+ cells in the heparinized blood during transport might also lead to a reduced level of mtDNA sequence heterogeneity, as we have observed for leukemic blasts [14], but would appear unlikely to occur under poor conditions for cell proliferation and further experiments should be carried out to solidify this speculation.
Figure 3.
A lower level of mtDNA sequence variations in single CD34+ cells from group I than group II.
3.3. Survival of CD34+ cells marked by specific mtDNA variants
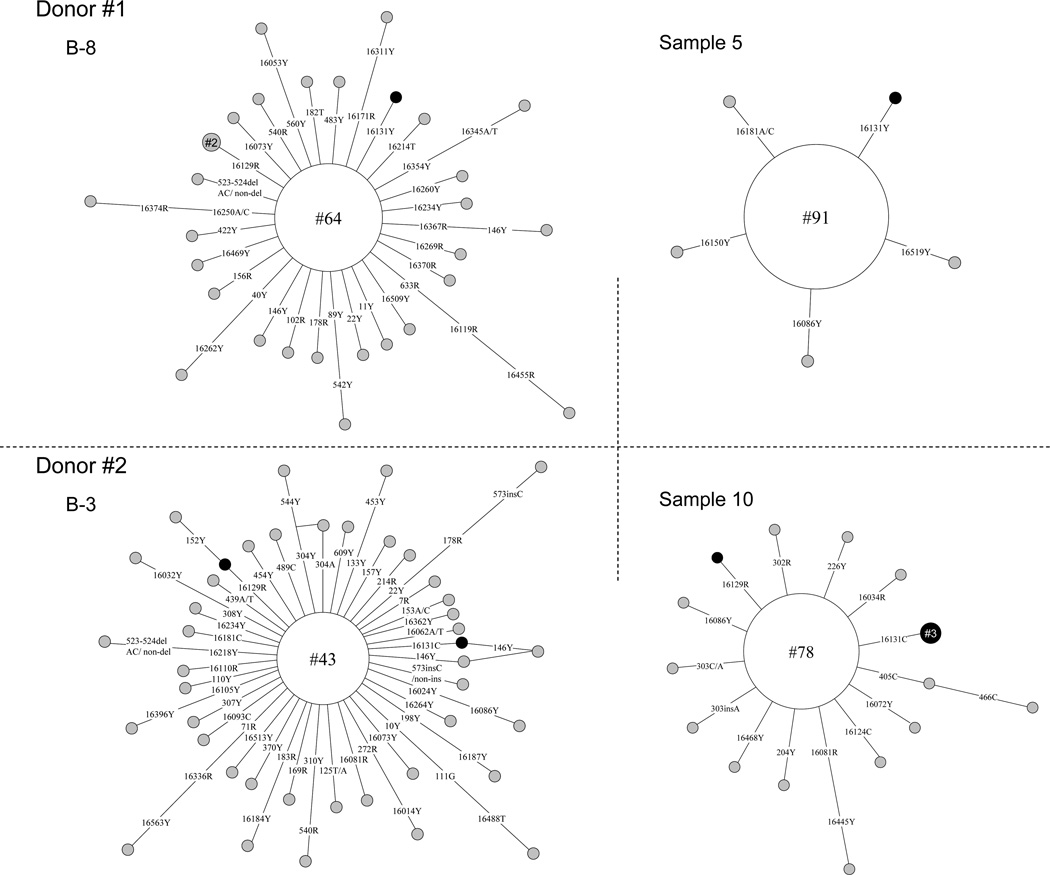
In our previous study, we found that mtDNA variants in CD34+ cells can exist in vivo for years and be transplanted to a recipient [16]. Disregarding the differences in sample storage and transport, the analyzed CD34+ cells from each donor represented two different samplings. It was of interest to compare the occurrence of clones marked by specific mtDNA variants in both batches of cells from the same donor to identify CD34+ cells capable of survival over the 6 days of transport. As the length mutations of the C-stretch in regions 16183–16193 (due to the 16189T>C variation) and 303–309 constitute hot mutational spots, we only considered the haplotypes defined by the nucleotide substitutions. For each donor, we identified at least one such haplotype in CD34+ cells, e.g. a cell with 16131T>Y was observed in two batches of CD34+ cells from donor #1; in donor #3, a cell clone with 182T>Y was observed in both groups (Fig. 4 and Supplementary Table 1). This unique pattern suggested that these CD34+ cells were presumably the primitive HSCs that better survived storage and transport.
Figure 4.
Network profiles of mtDNA haplotypes (CD34+ cell clones) observed in single CD34+ cell populations of donors #1 and #2 that were stored and transported by using two different approaches. The length mutation of C-tract in regions 16183–16192 and 303–309 in the mtDNA control region was not considered. The order of mutations on the branch is arbitrary. Each circle represents an mtDNA haplotype identified in a population of CD34+ cells, with its area being proportional to the frequency of the haplotype. We specified the number of cells sharing certain haplotype within the circle, for instance, “#64” means this haplotype was found in 64 single cells. The haplotype in the center of the network constitutes the consensus or aggregate sequence of the single cells.
3.4. Implications for primitive CD34+ cells and limitations of this study
For clinical reasons, the storage and transport of CD34+ cells have been extensively studied [18–21], mainly because hematopoietic reconstitution following transplantation depends upon the number of infused CD34+ cells [22]. In this study, we compared colony formation and the level of mtDNA sequence variations in CD34+ cells from the same donors that were stored and transported differently. We speculate that surviving CD34+ cells in group I were more primitive based on two lines of evidence: (1) lower colony formation potential during the short-term culture, and (2) low frequency of mtDNA sequence alterations. The persistence of certain CD34+ cell clones marked by specific mtDNA mutations in two different samplings of the same donor further suggested that these clones were stable [16] and had a strong survival potential in vitro. Further study will be required to characterize functionally the primitive status of enriched CD34+ cells as observed during the sorting for samples of group I and to elucidate why more primitive hematopoietic stem cells are more likely to survive stress. Regardless the mechanism, caution in interpreting mtDNA mutation data in human blood samples is warranted, with attention to the details of collection, processing, and storage, especially in increasingly global studies of the human genomes and its variants.
The current study has two limitations. First, reduced mtDNA sequence heterogeneity in single CD34+ cells of group I was based essentially on a single experiment and we did not repeat this experiment to confirm that there is a consistent finding. Nonetheless, we examined four pairs of samples in the analysis and we obtained similar pattern in all individuals, suggesting that this is unlikely to be a random bias. Second, except for the colony formation culture assay, we did not explore further to characterize the CD34+ cells of group I and to confirm that these cells are more primitive compared with the CD34+ cells of group II. Staining with more markers to distinguish the hematopoietic stem cells and progenitors will provide further evidence to clarify this issue.
Supplementary Material
Highlights.
mtDNA sequence heterogeneity in HSCs is markedly affected by storage and transport
CD34+ cells that survive stress ex vivo may be enriched in quiescent primitive HSCs
Primitive HSCs have fewer mtDNA mutations than committed progenitors
Acknowledgements
This study was supported by NIH intramural research grant. Y.-G.Y. was supported by the National Natural Science Foundation of China (30925021 and 31171225), Yunnan Province (2009CI119) and the Chinese Academy of Sciences. Y.-G.Y. and N.S.Y. designed the research, Y.-G.Y., L.S., S.K. and J.P.M. performed the research, Y.-G.Y. and L.S. analyzed the data. Y.-G.Y., S.K. and N.S.Y. wrote the paper, G.T. contributed essential samples.
Abbreviations
- mtDNA
mitochondrial DNA
- HSC
hematopoietic stem cell
- PB
peripheral blood
- MNC
mononuclear cell
- PBS
phosphate buffered saline
- BSA
bovine serum albumin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Conflicts of Interest
The authors declared no conflict of interest.
References
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salas A, Bandelt HJ, Macaulay V, Richards MB. Phylogeographic investigations: the role of trees in forensic genetics. Forensic Sci. Int. 2007;168:1–13. doi: 10.1016/j.forsciint.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 7.Trifunovic A. Mitochondrial DNA and ageing, Biochim. Biophys. Acta. 2006;1757:611–617. doi: 10.1016/j.bbabio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Harman D. Free radical theory of aging: an update: increasing the functional life span. Ann. N. Y. Acad. Sci. 2006;1067:10–21. doi: 10.1196/annals.1354.003. [DOI] [PubMed] [Google Scholar]
- 9.Howes RM. The free radical fantasy: a panoply of paradoxes. Ann. N. Y. Acad. Sci. 2006;1067:22–26. doi: 10.1196/annals.1354.004. [DOI] [PubMed] [Google Scholar]
- 10.Barja G. Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts. Antioxid. Redox. Signal. 2013 doi: 10.1089/ars.2012.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogasawara Y, Nakayama K, Tarnowka M, McCoy JP, Jr, Kajigaya S, Levin BC, Young NS. Mitochondrial DNA spectra of single human CD34+ cells, T cells, B cells, and granulocytes. Blood. 2005;106:3271–3284. doi: 10.1182/blood-2005-01-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin MG, Kajigaya S, McCoy JP, Jr, Levin BC, Young NS. Marked mitochondrial DNA sequence heterogeneity in single CD34+ cell clones from normal adult bone marrow. Blood. 2004;103:553–561. doi: 10.1182/blood-2003-05-1724. [DOI] [PubMed] [Google Scholar]
- 13.Shin MG, Kajigaya S, Tarnowka M, McCoy JP, Jr, Levin BC, Young NS. Mitochondrial DNA sequence heterogeneity in circulating normal human CD34 cells and granulocytes. Blood. 2004;103:4466–4477. doi: 10.1182/blood-2003-11-3949. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y-G, Ogasawara Y, Kajigaya S, Molldrem JJ, Falcão RP, Pintão M-C, McCoy JP, Jr, Rizzatti EG, Young NS. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007;109:756–762. doi: 10.1182/blood-2006-01-011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Y-G, Kajigaya S, Feng X, Samsel L, McCoy JP, Jr, Torelli G, Young NS. Accumulation of mtDNA variations in human single CD34+ cells from maternally related individuals: effects of aging and family genetic background. Stem Cell Res. 2013;10:361–370. doi: 10.1016/j.scr.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y-G, Childs RW, Kajigaya S, McCoy JP, Jr, Young NS. Mitochondrial DNA sequence heterogeneity of single CD34+ cells after nonmyeloablative allogeneic stem cell transplantation. Stem Cells. 2007;25:2670–2676. doi: 10.1634/stemcells.2007-0269. [DOI] [PubMed] [Google Scholar]
- 17.Yao Y-G, Bandelt H-J, Young NS. External contamination in single cell mtDNA analysis. PLoS ONE. 2007;2:e681. doi: 10.1371/journal.pone.0000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humpe A, Beck C, Schoch R, Kneba M, Horst HA. Establishment and optimization of a flow cytometric method for evaluation of viability of CD34+ cells after cryopreservation and comparison with trypan blue exclusion staining. Transfusion. 2005;45:1208–1213. doi: 10.1111/j.1537-2995.2005.00174.x. [DOI] [PubMed] [Google Scholar]
- 19.Reich-Slotky R, Colovai AI, Semidei-Pomales M, Patel N, Cairo M, Jhang J, Schwartz J. Determining post-thaw CD34+ cell dose of cryopreserved haematopoietic progenitor cells demonstrates high recovery and confirms their integrity. Vox Sang. 2008;94:351–357. doi: 10.1111/j.1423-0410.2007.001028.x. [DOI] [PubMed] [Google Scholar]
- 20.Antonenas V, Garvin F, Webb M, Sartor M, Bradstock KF, Gottlieb D. Fresh PBSC harvests, but not BM, show temperature-related loss of CD34 viability during storage and transport. Cytotherapy. 2006;8:158–165. doi: 10.1080/14653240600620994. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Acker JP, Cabuhat M, McGann LE. Effects of incubation temperature and time after thawing on viability assessment of peripheral hematopoietic progenitor cells cryopreserved for transplantation. Bone Marrow Transplant. 2003;32:1021–1026. doi: 10.1038/sj.bmt.1704247. [DOI] [PubMed] [Google Scholar]
- 22.Shpall EJ, Champlin R, Glaspy JA. Effect of CD34+ peripheral blood progenitor cell dose on hematopoietic recovery. Biol. Blood Marrow Transplant. 1998;4:84–92. doi: 10.1053/bbmt.1998.v4.pm9763111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.