Abstract
A series of sulfamide and triazole benzodiazepines were obtained with the principle of bioisosterism. The p53-murine double minute 2 (MDM2) inhibitory activity and in vitro antitumor activity were evaluated. Most of the novel benzodiazepines exhibited moderate protein binding inhibitory activity. Particularly, triazole benzodiazepines showed good inhibitory activity and antitumor potency. Compound 16 had promising antitumor activity against the U-2 OS human osteosarcoma cell line with an IC50 value of 4.17 μM, which was much better than that of nutlin-3. The molecular docking model also successfully predicted that this class of compounds mimicked the three critical residues of p53 binding to MDM2.
Keywords: p53-MDM2, small molecule inhibitors, sulfamidebenzodiazepine, triazolebenzodiazepine, antitumor activity
1. Introduction
The p53, a tumor suppressor, plays a critical role in DNA repair, differentiation, senescence, apoptosis, and cell-cycle arrest [1,2]. In approximately 50% of all human cancers, the p53 has been deactivated or deleted [3,4]. The p53 retains its wild-type form in the remaining human cancers and the activity is effectively inhibited through direct interaction with the human murine double minute 2 (MDM2) oncoprotein [5,6,7,8]. Thus, targeting p53-MDM2 protein-protein interaction has emerged as a novel strategy for development of anticancer drugs. Blocking this protein-protein interaction, releasing the p53 and recovering its function are the mechanism and new strategy of cancer therapy [9,10].
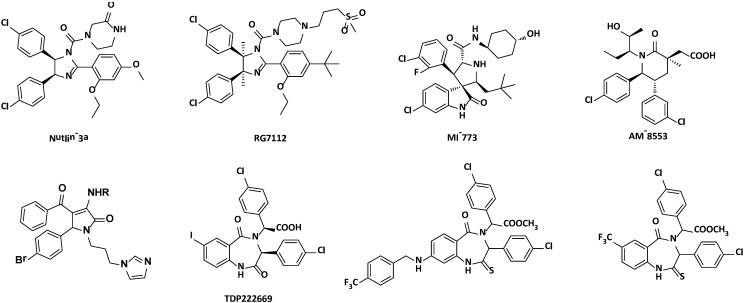
The binding cavities within MDM2, namely Phe19, Trp23, Leu26 pockets, are important hot spots for designing inhibitors that would release p53 [9,11]. Recently, a number of non-peptide small-molecule inhibitors based on the three hot spots have been reported (Figure 1). Inspiringly, six of them have been advanced into Phase I clinical trials [12], such as the nutlins (RG7112) [13,14,15], spiro-oxindoles (MI-773) [16]. Scientists from Amgen have reported another class of potent MDM2 inhibitors featuring a 1,3,5,6-tetrasubstituted piperidinone scaffold, AM-8553 [17]. Very recently, a pyrrolidone scaffold was identified by structure-based design in our group [18]. And a follow-up research was performed to develop a novel pyrrolo[3,4-c]pyrazole scaffold as dual inhibitors of p53-MDM2 interaction and the NF-κB pathway [19]. TDP222669, a benzodiazepine-based inhibitor of p53-MDM2 was reported by Grasberger et al. in 2005 [20]. However, this compound suffered from limited bioavailability and rapid in vivo clearance [21,22,23]. Thus, a series of thio-benzodiazepines were then designed based on the principle of bioisosterism, possessing both p53-MDM2 inhibitory activity and in vitro antitumor activity (Figure 1) [24,25].
Figure 1.
Representative p53-murine double minute 2 (MDM2) inhibitors.
In this study, we designed a series of sulfamide and triazole benzodiazepines based on the principle of bioisosterism. The triazole benzodiazepines showed good biological activity and could be used as promising lead structures for further optimization.
2. Results and Discussion
2.1. Chemistry
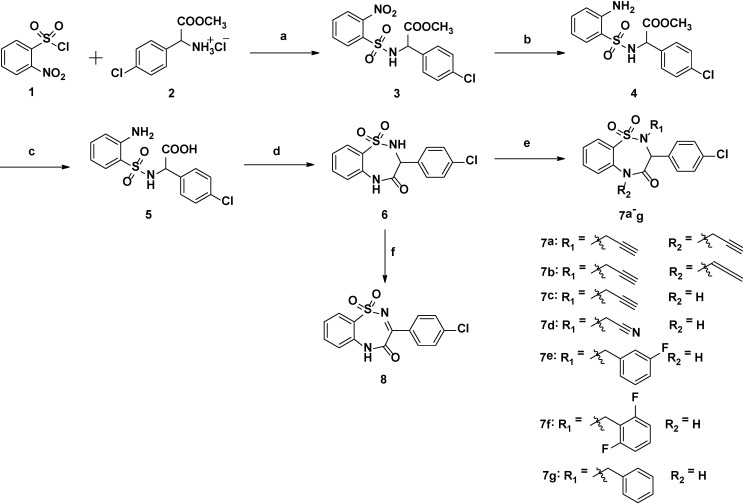
In this study, O-nitrobenzenesulfonyl chloride (1) and methyl amino(4-chlorophenyl)acetate hydrochloride (2) were used to perform the condensation reaction to get intermediate 3. Then, the intermediate 6 was made by reduction, hydrolysis and intramolecular condensation. The target compounds (7a–7g) were obtained by introducing aromatic and aliphatic groups on compound 6 using well-established conditions for substitution reaction. The compound 8 could be obtained without adding haloalkane using the similar condition (see Scheme 1).
Scheme 1.
Synthetic route of sulfamide-benzodiazepines.
Reagents and conditions: (a) N,N-diisopropylethylamine, DCM, 0–25 °C, overnight; (b) Fe/HOAc, 70 °C, 2.5 h; (c) LiOH, THF:MeOH:H2O = 4:2:1, overnight; (d) EDC·HCl, DMAP, overnight; (e) acetonitrile, K2CO3, RX, 60 °C, 3–6 h; (f) acetonitrile, K2CO3, 60 °C, 4 h.
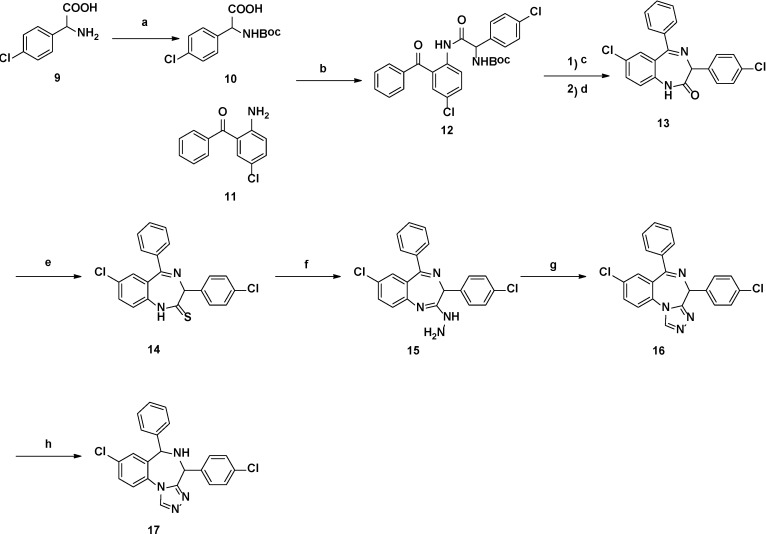
For the triazole benzodiazepines, imine 13 was intramolecular cyclized from compound 12 in the weak acid condition, following the HBTU condensation of compounds 10 and 11. Then, the triazole target compound 16 was synthesized by Lawesson’s reagent, hydrazine hydrate, and cyclization using triethylorthoformate. Compound 16 was then reduced by NaBH3CN to get compound 17 (see Scheme 2).
Scheme 2.
Synthetic route of triazole-benzodiazepines.
Reagents and conditions: (a) (Boc)2O, 1 M NaOH, dioxane, r.t., overnight; (b) HBTU, DMAP, r.t., overnight; (c) TFA, DCM, r.t., overnight; (d) HOAc, DCM, reflux, 5 h; (e) toluene, Lawesson’s Reagent, 70 °C, 4 h; (f) THF, NH2NH2.H2O, r.t., overnight; (g) Triethylorthoformate, toluene, reflux, 4 h; (h) NaBH3CN, MeOH, HOAc.
2.2. Disrupting the Binding of p53-MDM2
A well-established fluorescence polarization (FP) binding assay was performed to measure the inhibitory activity (Ki) of designed compounds. Nutlin-3, a commercial available p53-MDM2 inhibitor, was used as the reference compound. As presented in Table 1, most of this series of benzodiazepines exhibited moderate to good inhibitory activity. In order to explore whether the aramatic groups could be substituted by nonaramatic groups, aliphatic substituted compounds 7a–7c were synthesized based on the principle of scaffold hopping, which showed the inhibitory activity with Ki values in nanomolar range. Particularly, compound 7c had a Ki value of 0.26 μM, comparable to the positive compound nutlin-3.The further insight of compound 7c with MDM2 protein was re-examined by molecular modeling (Figure 2A). The two phenyl rings of (R)-7c were located in the Phe19 and Trp23 pockets respectively and the propargyl group was partly inserted into the Leu26 pocket. However, the disubstituted analogues (7a and 7b) were less active, probably due to limited space for binding. For similar reason, the aromatic substituted compounds (7e and 7f) were totally inactive. Only compound with unsubstituted benzyl group (7g) showed moderate p53-MDM2 inhibitory activity (Ki = 8.14 μM). Compound 8, reduced from compound 6, was observed dramatic decrease of the inhibitory activity, highlighting the importance of the conformational restriction.
Table 1.
In vitro activity of the designed compounds.
| Compounds | Ki (μM ) | IC50 (μM) | |||
|---|---|---|---|---|---|
| Saos-2 | U-2 OS | A549 | NCI-H1299 | ||
| (p53 null) | (wt-p53) | (wt-p53) | (p53 null) | ||
| 7a | 0.43 | >100 | >100 | >100 | >100 |
| 7b | 0.36 | >100 | >100 | >100 | >100 |
| 7c | 0.26 | >100 | >100 | >100 | >100 |
| 7d | >100 | >100 | >100 | 92.3 | >100 |
| 7e | >100 | >100 | 90.5 | >100 | >100 |
| 7f | 78.0 | >100 | >100 | >100 | >100 |
| 7g | 8.14 | >100 | >100 | >100 | >100 |
| 8 | 11.9 | >100 | >100 | >100 | >100 |
| 13 | 0.20 | 3.01 | 3.12 | 9.16 | 6.11 |
| 14 | 8.76 | 3.67 | 5.31 | 24.05 | 15.56 |
| 16 | 1.22 | 3.55 | 4.17 | 7.62 | 5.50 |
| 17 | 2.80 | 24.81 | 33.98 | 26.78 | 34.59 |
| Nutlin-3 | 0.09 | 20.8 | 16.3 | 12.7 | 4.15 |
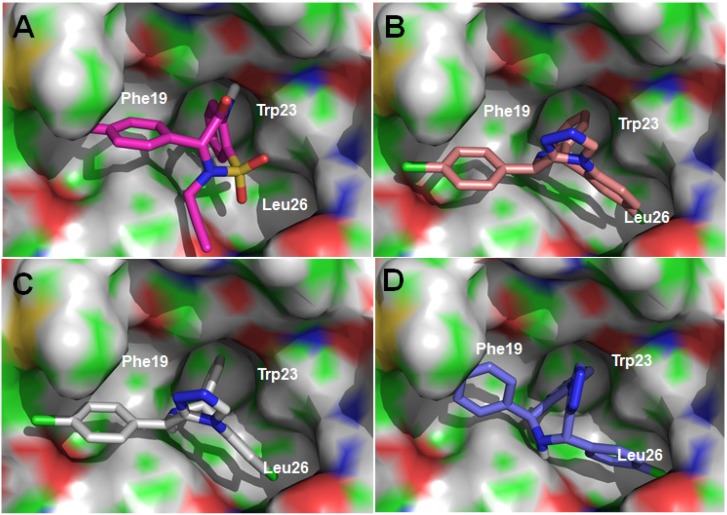
Figure 2.
Docking modes of the sulfamide and triazole benzodiazepines with MDM2: (A) compound 7c (R); (B) compound 16 (R); (C) compound 17 (R, R); (D) compound 17 (S, S).
For the triazole derivatives, the target compound and intermediates were tested. Unsurprisingly, the benzodiazepine (13) showed good activity (Ki = 0.20 μM) on disrupting the p53-MDM2. The target triazole derivative (16) had a moderate Ki value of 1.22 μM, which is much better than compound 14 and 17. As shown in Figure 2, the R configuration of compound 16 was predicted to bind to MDM2 protein probably, which was similar to the compound 7c. Two 4-cholrophenyl groups and the benzene ring, mimicking the three key hydrophobic residues of p53, were inserted into Phe19, Leu26 and Trp23 pockets (Figure 2B). However, only two isomers of target compound 17 could bind well to MDM2 (Figure 2C,D), presumably the reason for the decrease of the activity compared to compound 16. Interestingly, these two isomers had different binding poses. The (R, R)-isomer could be inserted into the binding pockets as well as R-compound 16. For the (S, S)-isomer, the two 4-chlorophenyl groups were occupied the Trp23 and Leu26 pockets, while the benzene ring was located in Phe19 pocket.
2.3. In Vitro Antitumor Activity
To investigate the in vitro antiproliferative activity of the designed p53-MDM2 inhibitors, four human tumor cell lines, namely U-2 OS (wild-type p53), A549 (wild-type p53), Saos-2 (p53 null), and NCI-H1299 (p53 null), were chosen for assaying. Nutlin-3 was used as a reference compound. The obtained IC50 values for the p53-MDM2 inhibitors are summarized in Table 1. Unfortunately, all the sulfamide benzodiazepines showed poor in vitro antitumor activity, potentially due to their poor aqueous solubility. Comparing with the totally inactive sulfamide benzodiazepines, the triazole benzodiazepines showed promising antiproliferative activity. Notably, compound 16 showed better activity (IC50 = 7.62 μM) against the A549 cell line than thio-benzodiazepine 14 (IC50 = 24.05 μM) and nutlin-3 (IC50 = 12.7 μM). And its antiproliferative activity (IC50 = 4.17 μM) against the U-2 OS cell line was about 4-fold better than nutlin-3. Moreover, compound 17 exhibited poor activity against these four cell lines, which matched with the protein binding assay. Nevertheless, these compounds had poor selectivity over cancer cell lines with deleted p53, which inspired us to carry out further optimization to find more active analogues. Focused medicinal chemistry work is in progress to expand the SAR of this series of compounds.
3. Experimental Section
3.1. Chemistry
All reagents and solvents were purchased from commercial suppliers and used as received unless otherwise stated. 1H-NMR and 13C-NMR spectra were recorded on a BRUKER AVANCE 300 and 600 spectrometer (Bruker Company, Rheinstetten, Germany), using TMS as an internal standard and CDCl3 or DMSO-d6 as solvents. Chemical shifts (δ values) and coupling constants (J values) are given in ppm and Hz, respectively. TLC analysis was carried out on silica gel plates GF254 (QingdaoHaiyang Chemical, Qingdao, China). Flash column chromatography was carried out on silica gel 300–400 mesh. Anhydrous solvent and reagents were all analytical pure and dried through routine protocols.
Methyl 2-(4-chlorophenyl)-2-(2-nitrophenylsulfonamido)acetate (3). Methyl 2-amino-2-(4-chlorophenyl)acetate hydrochloride (2, 2.15 g, 9.1 mmol) and N,N-diisopropylethylamine (1.76 g, 13.6 mmol) was added into dry CH2Cl2 (80 mL). The solution was stirred at 0 °C then O-nitrobenzenesulfonyl chloride (1, 1.0 g, 4.5 mmol) was added slowly. After that, the mixture was stirred overnight at room temperature and concentrated under vacuum. The residue was purified by flash column chromatography to give compound 3, yield: 81%. 1H-NMR (DMSO-d6, 600 MHz): δ 9.39 (d, 1H, J = 4.3 Hz), 7.95–7.91 (m, 1H), 7.90–7.89 (m, 1H), 7.81–7.79 (m, 1H), 7.77–7.76 (m, 1H), 7.38–7.34 (m, 4H), 5.23 (d, 1H, J = 4.3 Hz), 3.54 (s, 3H); ESI-MS (m/z): 385.52 (M + H)+.
Methyl 2-(2-aminophenylsulfonamido)-2-(4-chlorophenyl)acetate (4). Compound 3 (1.0 g, 2.6 mmol) was stirred with AcOH (50 mL). The resulting solution was heated at 45 °C and treated under vigorous stirring with iron powder (2.0 g, 35.7 mmol) in one portion. When the exothermic reaction had subsided, the reaction mixture was heated at 65–70 °C for 2.5 h and then allowed to cool and stirred with CH2Cl2 (50 mL) and H2O (50 mL). The resulting suspension was filtered to remove the unreacted iron and the filtrate transferred to a separating funnel. The organic layer was washed with H2O (50 mL), NaHCO3 (aq, 2%; 50 mL), H2O (50 mL) and then separated, dried (Na2SO4), and evaporated to dryness to give product 4, 0.88 g, yield: 95%. 1H-NMR (DMSO-d6, 600 MHz): δ 8.79 (d, 1H, J = 4.1 Hz), 7.43–7.42 (m, 1H), 7.33–7.32 (m, 2H), 7.29–7.27 (m, 2H), 7.20–7.17 (m, 1H), 6.69–6.53 (m, 1H), 6.53–6.50 (m, 1H), 5.90 (s, 2H), 4.96 (d, 1H, J = 4.1 Hz), 3.48 (s, 3H); ESI-MS (m/z): 355.67 (M + H)+.
3-(4-Chlorophenyl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (6). Compound 4 (0.50 g, 1.4 mmol) was dissolved in a mixed solution (THF:MeOH:H2O = 4:2:1, 35 mL). Then, LiOH (0.10 g, 4.2 mmol) was added and the mixture was stirred overnight at room temperature. The resulting solution was evaporated to dryness and dissolved in H2O (30 mL), extracted by ether (20 mL × 2). The aqueous layer was acidified with HCl (1 mol/L) to reach pH 4. The resulting solid was filtered and then directly dissolved in CH2Cl2 (40 mL) with EDC·HCl (0.54 g, 2.8 mmol) and DMAP (0.05 g, 0.4 mmol), stirred overnight at room temperature and concentrated under vacuum. The residue was purified by flash column chromatography to give compound 6, yield: 71%. 1H-NMR (DMSO-d6, 600 MHz): δ 10.49 (s, 1H), 9.14 (d, 1H, J = 8.4 Hz), 7.81–7.79 (m, 1H), 7.63–7.60 (m, 1H), 7.43–7.42 (m, 2H), 7.39–7.37 (m, 2H), 7.31–7.29 (m, 2H), 5.29 (d, 1H, J = 8.0 Hz); ESI-MS (m/z): 323.58 (M + H)+.
3.2. General Procedure
3.2.1. Synthesis of 7a and 7b
Compound 6 (0.20 g, 0.6 mmol), haloalkane (0.35 g, 2.9 mmol) and potassium carbonate (0.09 g, 0.6 mmol) were mixed in acetonitrile (10 mL) and stirred at 60 °C for 5 h. The resulting precipitate was filtered off and the filtrate was then evaporated to dryness. The residue was purified by flash column chromatography to give two compounds. The compound with lower polarity was 7a (yield: 25%); compound 7b was the tautomer of compound 7a (yield: 25%).
3.2.2. Synthesis of 7c–7g
Compound 6 (0.20 g, 0.6 mmol), haloalkane (0.07 g, 0.58 mmol) and potassium carbonate (0.09 g, 0.6 mmol) were mixed in acetonitrile (10 mL) and stirred at 60 °C for 5 h. The resulting precipitate was filtered off and the filtrate was then evaporated to dryness. The residue was purified by flash column chromatography to give compounds 7c–7g, yield: 25%–49%.
3-(4-Chlorophenyl)-2,5-di(prop-2-yn-1-yl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7a). 1H-NMR (DMSO-d6, 600 MHz): δ 7.83 (dd, 1H, J1 = 1.44 Hz, J2 = 8.01 Hz), 7.49 (t, 1H, J = 7.65 Hz), 7.35 (t, 1H, J = 7.59 Hz), 7.15–7.09 (m, 3H), 6.89 (s, 2H), 5.96 (d, 1H, J = 6.10 Hz), 5.84 (s, 1H), 4.77 (dd, 1H, J = 2.38, 17.93 Hz), 4.66 (s, 1H), 4.50–4.47 (m, 1H), 4.19 (d, 1H, J = 14.05 Hz), 3.29 (s, 1H). 13C-NMR (150 MHz, DMSO-d6): δ 168.13, 138.78, 137.67, 135.04, 132.25, 131.89, 130.53, 128.09, 126.93, 126.52, 125.51, 124.27, 124.04. 78.91, 78.34, 75.31; ESI-MS (m/z): 399.82 (M + H)+.
3-(4-Chlorophenyl)-2-(prop-2-yn-1-yl)-5-(propa-1,2-dien-1-yl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7b). 1H-NMR (DMSO-d6, 600 MHz): δ 7.91–7.86 (m, 2H), 7.81 (s, 1H), 7.59 (t, 1H, J = 7.86 Hz), 7.44 (d, 2H, J = 7.98 Hz), 7.34 (s, 2H), 4.65–4.56 (m, 3H), 4.18 (d, 1H, J = 18.93 Hz), 3.24 (d, 1H, J = 18.26 Hz), 3.13 (t, 1H, J = 2.44 Hz), 3.01 (s, 1H); 13C-NMR (75 MHz, DMSO-d6): δ 165.68, 138.94, 135.68, 134.04, 133.36, 131.76, 130.23, 128.86, 128.23, 128.05, 125.84,78.98, 78.80,75.66; ESI-MS (m/z): 399.82 (M + H)+.
3-(4-Chlorophenyl)-2-(prop-2-yn-1-yl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7c). 1H-NMR (DMSO-d6, 600 MHz): δ 10.62 (s, 1H), 7.79–7.77 (m, 1H), 7.71–7.68 (m, 1H), 7.43 (d, 2H, J = 8.12 Hz), 7.37–7.33 (m, 4H), 7.24 (d, 1H, J = 8.12 Hz), 4.94 (s, 1H), 4.17 (dd, 1H, J = 2.3, 18.54 Hz), 3.40 (dd, 1H, J = 2.3, 18.54 Hz); 13C-NMR (75 MHz, DMSO-d6): δ 167.89, 136.38, 135.18, 134.80, 133.61, 131.44, 129.72, 128.73, 127.75, 125.09, 123.52, 78.46, 75.35, 64.14; ESI-MS (m/z): 361.55 (M + H)+.
2-(3-(4-Chlorophenyl)-1,1-dioxido-4-oxo-4,5-dihydrobenzo[f][1,2,5]thiadiazepin-2(3H)-yl)acetonitrile (7d). 1H-NMR (DMSO-d6, 600 MHz): δ 10.75 (s, 1H), 7.82 (dd, 1H, J1 = 1.57 Hz, J2 = 8.04 Hz), 7.75–7.72 (m, 1H), 7.45 (d, 2H, J = 8.47 Hz), 7.41–7.38 (m, 3H), 7.28 (d, 1H, J = 8.07 Hz), 5.06 (s, 1H), 4.34 (d, 1H, J = 18.58 Hz), 3.96 (d, 1H, J = 18.58 Hz); ESI-MS (m/z): 362.29 (M + H)+.
3-(4-Chlorophenyl)-2-(3-fluorobenzyl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7e). 1H-NMR (DMSO-d6, 300 MHz): δ 10.63 (s, 1H), 7.79–7.76 (m, 1H), 7.68–7.62 (m, 1H), 7.32–7.17 (m, 7H), 6.97–6.76 (m, 3H), 5.28 (s, 1H), 4.24 (s, 2H); 13C-NMR (75 MHz, DMSO-d6): δ 169.06, 138.90, 138.80, 136.23, 135.99, 134.87, 132.99, 131.24, 130.55, 129.99, 128.25, 127.13, 124.73, 124.40, 122.92, 115.48, 115.19, 114.81, 114.54, 67.35, 53.29; ESI-MS (m/z): 431.66 (M + H)+.
3-(4-Chlorophenyl)-2-(2,6-difluorobenzyl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7f). 1H-NMR (DMSO-d6, 300 MHz): δ 10.56 (s, 1H), 7.86–7.61 (m, 3H), 7.56–7.34 (m, 7H), 6.85–6.82 (m, 1H), 5.11 (s, 1H), 4.40 (d, 1H, J = 14.4 Hz), 4.24 (d, 1H, J = 14.4 Hz); ESI-MS (m/z): 449.82 (M + H)+.
2-Benzyl-3-(4-chlorophenyl)-2,3-dihydrobenzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (7g). 1H-NMR (DMSO-d6, 600 MHz): δ 10.55 (s, 1H), 7.77 (dd, 1H, J1 = 1.41 Hz, J2 = 7.79 Hz,) 7.63–7.60 (m, 1H), 7.29–7.26 (m, 3H), 7.18–7.17 (m, 3H), 7.14–7.12 (m, 3H), 7.02–7.00 (m, 2H), 5.17 (s, 1H), 4.28 (d, 1H, J = 15.44 Hz), 4.16 (d, 1H, J = 15.44 Hz); 13C-NMR (75 MHz, DMSO-d6): δ 169.14, 135.96, 135.58, 131.09, 130.26, 128.84, 128.56, 128.29, 127.99, 127.04, 124.38, 122.83, 67.08, 53.64; ESI-MS (m/z): 413.61 (M + H)+.
3-(4-Chlorophenyl)benzo[f][1,2,5]thiadiazepin-4(5H)-one 1,1-dioxide (8). Compound 6 (0.10 g, 0.3 mmol), and potassium carbonate (0.05 g, 0.36 mmol) were mixed in acetonitrile (10 mL) and stirred at 60 °C for 4 h. The resulting precipitate was filtered off and the filtrate was then evaporated to dryness. The residue was purified by flash column chromatography to give compound 8, yield: 50%. 1H-NMR (DMSO-d6, 600 MHz): δ 8.14–8.12 (m, 2H), 7.88 (dd, 1H, J1 = 1.19 Hz, J2 = 8.12 Hz), 7.76–7.73 (m, 1H), 7.70–7.67 (m, 3H), 7.55–7.52 (m, 1H), 7.70–7.67 (m, 1H); 13C-NMR (75 MHz, DMSO-d6): δ 149.80, 140.28, 135.16, 134.06, 133.23, 132.09, 129.31, 127.91,124.01,119.37; ESI-MS (m/z): 321.33 (M + H)+.
tert-Butyl (2-((2-benzoyl-4-chlorophenyl)amino)-1-(4-chlorophenyl)-2-oxoethyl)carbamate (12). 4-chlorophenylglycine (9, 5.00 g, 48.6 mmol) was dissolved in 1,4-dioxane (60 mL) and NaOH (1 mol/L, 30 mL). Di-tert-butyl dicarbonate (9.2 g, 42.2 mmol) was then added into the mixture and stirred at room temperature for 12 h. After completion of reaction, the solution mixture was then acidified with HCl (1 mol/L) to reach pH 5, extracted by CH2Cl2 (100 mL × 3). The organic layer was then evaporated to dryness to get crude product 10 which was directly used in the next step. Compound 10, HBTU (11.2 g, 29.5 mmol), DMAP (0.66 g, 5.4 mmol) and compound 11 (6.24 g, 27.0 mmol) was dissolved in CH2Cl2 (200 mL) and stirred at room temperature for 12 h. The resulting mixture was evaporated to dryness. The residue was purified by flash column chromatography to give compound 12, 2.8 g yield: 21%. 1H-NMR (CDCl3, 300 MHz): δ 11.17 (s, 1H), 8.58 (d, 1H, J = 9.6 Hz), 7.67–7.61 (m, 3H), 7.53–7.51 (m, 3H), 7.41 (d, 2H, J = 8.2 Hz), 7.34 (d, 2H, J = 8.2 Hz), 7.26 (s, 1H), 5.76 (s, 1H), 5.27 (s, 1H), 1.4 (s, 9H); ESI-MS (m/z): 500.31 (M + H)+.
7-Chloro-3-(4-chlorophenyl)-5-phenyl-1H-benzo[e][1,4]diazepin-2(3H)-one (13). Compound 12 (0.4 g, 0.8 mmol) was added into CH2Cl2 (15 mL) with TFA (0.37 g, 3.2 mmol) and stirred overnight at room temperature. The mixture was then concentrated to dry and dissolved in CH2Cl2 (10 mL). Five drops of TFA was then added as the catalyst. The mixture was heated to reflux for 2 h. The resulting mixture was evaporated to dryness. The residue was purified by flash column chromatography to give compound 13, 0.25 g, yield: 83%. 1H-NMR (CDCl3, 600 MHz): δ 10.86 (s, 1H), 7.69 (dd, 1H, J1 = 2.46 Hz, J2 = 8.76 Hz), 7.60 (d, 2H, J = 8.4 Hz), 7.53–7.51 (m, 3H), 7.47–7.43 (m, 4H), 7.34 (d, 1H, J = 8.76 Hz), 7.30 (d, 1H, J = 2.46 Hz), 4.87 (s, 1H); 13C-NMR (75 MHz, DMSO-d6): δ 169.94, 167.32, 138.88, 138.84, 138.13, 132.43, 132.26, 131.47, 131.08, 130.05, 129.86, 128.94, 128.20, 127.16, 123.84, 67.23; ESI-MS (m/z): 381.05 (M + H)+.
7-Chloro-3-(4-chlorophenyl)-5-phenyl-1H-benzo[e][1,4]diazepine-2(3H)-thione (14). A dry toluene solution of compound 13 (0.20 g, 0.53 mmol) under an atmosphere of nitrogen was treated with Lawesson’s Reagent (2,4-bis(4-methoxyphenyl)-2,4-disulfide, 0.22 g, 0.54 mmol) under 70 °C for 3 h, cooled to room temperature, concentrated under vacuum and purified by column chromatography to give 14, 0.16 g, yield: 77%. 1H-NMR (CDCl3, 600 MHz): δ 12.67 (s, 1H), 7.74 (dd, 1H, J1 = 2.46 Hz, J2 = 8.82 Hz), 7.60 (d, 2H, J = 8.4 Hz), 7.55–7.52 (m, 3H), 7.49–7.46 (m, 3H), 7.41 (d, 2H, J = 8.4 Hz), 7.34 (d, 1H, J = 2.4 Hz), 5.15 (s, 1H); 13C-NMR (75 MHz, DMSO-d6): δ 201.57, 166.77,140.04, 139.25, 138.42, 132.36, 131.86, 131.28, 130.24, 129.95, 129.50, 129.00, 128.80, 127.88, 124.24, 71.22, 26.80; ESI-MS (m/z): 397.28 (M + H)+.
8-Chloro-4-(4-chlorophenyl)-6-phenyl-4H-benzo[f][1,2,4] (16). Compound 14 was added into THF (15 mL). The resulting solution was then treated with 85% hydrazine hydrate at 0 °C for 1 h, and then stirred at room temperature for 12 h. After completion of reaction, the mixture was evaporated to dryness to afford the crude product 15 which was directly added into a dry toluene (15 mL) solution of triethylorthoformate (10 mL). The mixture was then stirred under reflux for 2 h. The resulting mixture was then evaporated to dryness. The residue was purified by flash column chromatography to give compound 16, 0.14 g yield: 71%. 1H-NMR (CDCl3, 600 MHz): δ 9.20 (s, 1H), 7.99–7.95 (m, 2H), 7.77 (d, 2H, J = 8.46 Hz), 7.52–7.49 (m, 5H), 7.47 (d, 1H, J = 2.16 Hz), 7.45–7.43 (m, 2H), 5.61 (s, 1H); 13C-NMR (75 MHz, DMSO-d6): δ 166.26, 156.03, 143.31, 139.22, 137.37, 133.07, 132.86, 132.82, 131.81, 131.23, 129.87, 128.96, 128.86, 128.50, 125.80, 58.74; ESI-MS (m/z): 405.32 (M + H)+.
8-Chloro-4-(4-chlorophenyl)-6-phenyl-5,6-dihydro-4H-benzo[f][1,2,4]triazolo[4,3-a][1,4]diazepine (17). Glacial acetic acid (3 mL) and 16 (0.05 g, 0.12 mmol) were mixed in CH3OH (10 mL). Then, NaBH3CN (0.03 g, 0.48 mmol) was then added to the solvent, stirred for 12 h. Removal of the solvent under reduced pressure and the residue was purified by column chromatography to give 17, 0.04 g, yield: 80%. 1H-NMR (CDCl3, 600 MHz): δ 8.87 (s, 1H), 7.63 (d, 1H, J = 8.46 Hz), 7.56 (dd, 1H, J1 = 2.34 Hz, J2 = 8.46 Hz), 7.43 (d, 2H, J = 8.52 Hz), 7.33–7.29 (m, 7H), 7.01 (d, 1H, J = 2.42 Hz), 5.29 (d, 1H, J = 7.81 Hz), 5.19 (d, 1H, J = 7.81 Hz), 4.32 (t, 1H, J = 7.52 Hz); 13C-NMR (75 MHz, DMSO-d6): δ 153.29, 143.09, 141.91, 139.73, 138.26, 133.03, 132.49, 132.20, 130.53, 130.45, 129.85, 129.21, 128.58, 128.25, 127.34, 125.24, 59.74, 53.31; ESI-MS (m/z): 407.51 (M + H)+.
3.3. Computational Protocol
Molecular docking was used to predict the binding mode of the synthesized benzodiazepine derivatives. The crystal structure of MDM2 (PDB code: 1T4E) was prepared by removing the benzodiazepine and adding hydrogen atoms in GOLD 5.0. We used TDP222669 as a positive control. The active site was defined to encompass all MDM2 atoms within a 12 Å radius sphere from the center of 1T4E ligand. Other parameters were set by default.
3.4. p53-MDM2 Binding Assay
The dose-dependent binding experiments were carried out with serial dilution in DMSO of compounds. A 5 µL sample of the tested sample and preincubated (for 30 min) MDM2 binding domain (1–118) (10 nM) and PMDM6-F peptide (Anaspec, Fremont, CA, USA) (10 nM) in the assay buffer (100 mM potassium phosphate, pH 7.5; 100 µg/mL bovine gamma globulin; 0.02% sodium azide) were added into black 96-well microplates with F-bottom and chimney wells (Corning #3993) to produce a final volume of 115 µL. For each assay, the controls included the MDM2 binding domain and PMDM6-F. The polarization values were measured after 1 h of incubation at room temperature using Biotek Synergy H2 with a 480 nm excitation filter, a 528 nm static and polarized filter (BioTek, Winooski, VT, USA). The Ki values were determined from a plot using nonlinear least-squares analysis. And curve fitting was performed using GraphPad Prism software. Nutlin-3 (Sigma-Aldrich, Shanghai, China) was used as reference compound for validating the assay in each plate [18,19,24,25,26,27].
3.5. In Vitro Antitumor Activity
The cellular growth inhibitory activity was determined using four human cancer cell lines [U-2 OS (wild-type p53), A549 (wild-type p53), Saos-2 (p53 null), and NCI-H1299 (p53 null)]. 5–6 × 104 cells per well were plated in 96-well plates. After culturing for 24 h, test compounds were added onto triplicate wells with different concentrations and 0.1% DMSO for control. After 72 h of incubation, 20 μL of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide) solution (5 mg/mL) was added to each well, and after the samples were shaken for 1min the plate was incubated further for 4 h at 37 °C. Benzodiazepines were dissolved with 100 μL of DMSO. The absorbance (OD) was quantitated with microplate using Biotek Synergy H2 at 570 nm (BioTek, Winooski, VT, USA). Wells containing no drugs were used as blanks. Concentration of the compounds that inhibited cell growth by 50% (IC50) was calculated [28].
4. Conclusions
A series of novel sulfamide and triazole benzodiazepines inhibitors of p53-MDM2 protein-protein interaction were successfully obtained by the principle of bioisosterism. Most of the novel benzodiazepines exhibited moderate protein binding inhibitory activity. Particularly, triazole benzodiazepines showed good inhibitory activity and antitumor potency. The molecular docking model also successfully predicted that this class of compounds mimicked the three critical residues of p53 binding to MDM2. In conclusion, this class of benzodiazepines can be used as promising lead structures for further optimization.
Acknowledgments
Financial support for this research is provided by National Natural Science Foundation of China (grant 81373278, 81373331), the 863 Hi-Tech Program of China (grant 2012AA020302), Key Project of Science and Technology of Shanghai (grant 14431902300).
Author Contributions
Ideas and experiment design: C. Zhuang, Z. Miao, W. Zhang; Computational development: Z. Yu, C. Zhuang, C. Sheng; Chemistry and Biology: Z. Yu, C. Zhuang, Z. Guo, Y. Wu, J. Li, G. Dong, J. Yao; Analysis and data interpretation: Z. Yu, C. Zhuang, Z. Miao; Writing and review of the manuscript: All the authors; Study supervision: Z. Miao, W. Zhang.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Cochran A.G. Antagonists of protein-protein interactions. Chem. Biol. 2000;7:R85–R94. doi: 10.1016/S1074-5521(00)00106-X. [DOI] [PubMed] [Google Scholar]
- 4.Hainaut P., Hollstein M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000;77:81–137. doi: 10.1016/S0065-230X(08)60785-X. [DOI] [PubMed] [Google Scholar]
- 5.Oliner J.D., Kinzler K.W., Meltzer P.S., George D.L., Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 6.Oliner J.D., Pietenpol J.A., Thiagalingam S., Gyuris J., Kinzler K.W., Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 7.Picksley S.M., Lane D.P. The p53-mdm2 autoregulatory feedback loop: A paradigm for the regulation of growth control by p53? BioEssays. 1993;15:689–690. doi: 10.1002/bies.950151008. [DOI] [PubMed] [Google Scholar]
- 8.Wu X., Bayle J.H., Olson D., Levine A.J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 9.Kussie P.H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A.J., Pavletich N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 10.Fry D.C. Protein-protein interactions as targets for small molecule drug discovery. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Shangary S., Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: A novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray-Coquard I., Blay J.Y., Italiano A., Le Cesne A., Penel N., Zhi J., Heil F., Rueger R., Graves B., Ding M., et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 14.Tovar C., Graves B., Packman K., Filipovic Z., Higgins B., Xia M., Tardell C., Garrido R., Lee E., Kolinsky K., et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 15.Vu B., Wovkulich P., Pizzolato G., Lovey A., Ding Q.J., Jiang N., Liu J.J., Zhao C.L., Glenn K., Wen Y., et al. Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development. ACS Med. Chem. Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Yu S., Sun W., Kumar S., Sun D., Zou P., McEachern D., Zhao Y. Spiro-oxindole mdm2 antagonists. 8518984 B2. U.S. Patent. 2013 Aug 27;
- 17.Rew Y., Sun D., Gonzalez-Lopez De Turiso F., Bartberger M.D., Beck H.P., Canon J., Chen A., Chow D., Deignan J., Fox B.M., et al. Structure-based design of novel inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2012;55:4936–4954. doi: 10.1021/jm300354j. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang C., Miao Z., Zhu L., Dong G., Guo Z., Wang S., Zhang Y., Wu Y., Yao J., Sheng C., et al. Discovery, synthesis, and biological evaluation of orally active pyrrolidone derivatives as novel inhibitors of p53-MDM2 protein-protein interaction. J. Med. Chem. 2012;55:9630–9642. doi: 10.1021/jm300969t. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang C., Miao Z., Wu Y., Guo Z., Li J., Yao J., Xing C., Sheng C., Zhang W. Double-edged Swords as Cancer Therapeutics: Novel Orally Active Small Molecules Simultaneously Inhibit p53-MDM2 Interaction and the NF-κB Pathway. J. Med. Chem. 2014;57:567–577. doi: 10.1021/jm401800k. [DOI] [PubMed] [Google Scholar]
- 20.Grasberger B.L., Lu T., Schubert C., Parks D.J., Carver T.E., Koblish H.K., Cummings M.D., LaFrance L.V., Milkiewicz K.L., Calvo R.R., et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J. Med. Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 21.Marugan J.J., Leonard K., Raboisson P., Gushue J.M., Calvo R., Koblish H.K., Lattanze J., Zhao S., Cummings M.D., Player M.R., et al. Enantiomerically pure 1,4-benzodiazepine-2,5-diones as Hdm2 antagonists. Bioorg. Med. Chem. Lett. 2006;16:3115–3120. doi: 10.1016/j.bmcl.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 22.Parks D.J., LaFrance L.V., Calvo R.R., Milkiewicz K.L., Marugan J.J., Raboisson P., Schubert C., Koblish H.K., Zhao S., Franks C.F., et al. Enhanced pharmacokinetic properties of 1,4-benzodiazepine-2,5-dione antagonists of the HDM2-p53 protein-protein interaction through structure-based drug design. Bioorg. Med. Chem. Lett. 2006;16:3310–3314. doi: 10.1016/j.bmcl.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 23.Pevarello P., Fancelli D., Vulpetti A., Amici R., Villa M., Pittala V., Vianello P., Cameron A., Ciomei M., Mercurio C., et al. 3-Amino-1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: A new class of CDK2 inhibitors. Bioorg. Med. Chem. Lett. 2006;16:1084–1090. doi: 10.1016/j.bmcl.2005.10.071. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang C., Miao Z., Zhu L., Zhang Y., Guo Z., Yao J., Dong G., Wang S., Liu Y., Chen H., et al. Synthesis and biological evaluation of thio-benzodiazepines as novel small molecule inhibitors of the p53-MDM2 protein-protein interaction. Eur. J. Med. Chem. 2011;46:5654–5661. doi: 10.1016/j.ejmech.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z., Zhuang C., Zhu L., Zhang Y., Yao J., Dong G., Wang S., Liu Y., Chen H., Sheng C., et al. Structure-activity relationship and antitumor activity of thio-benzodiazepines as p53-MDM2 protein-protein interaction inhibitors. Eur. J. Med. Chem. 2012;56:10–16. doi: 10.1016/j.ejmech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Wu Y., Guo Z., Zhuang C., Yao J., Dong G., Yu Z., Min X., Wang S., Liu Y., et al. Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg. Med. Chem. Lett. 2014;24:2648–2650. doi: 10.1016/j.bmcl.2014.04.063. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang C., Narayanapillai S., Zhang W., Sham Y.Y., Xing C. Rapid identification of keap1-Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med. Chem. 2014;57:1121–1126. doi: 10.1021/jm4017174. [DOI] [PubMed] [Google Scholar]
- 28.Miao Z., Zhu L., Dong G., Zhuang C., Wu Y., Wang S., Guo Z., Liu Y., Wu S., Zhu S., et al. A new strategy to improve the metabolic stability of lactone: Discovery of (20S,21S)-21-fluorocamptothecins as novel, hydrolytically stable topoisomerase I inhibitors. J. Med. Chem. 2013;56:7902–7910. doi: 10.1021/jm400906z. [DOI] [PubMed] [Google Scholar]