Abstract
Newly synthesized secretory and transmembrane proteins are folded and assembled in the endoplasmic reticulum (ER) where an efficient quality control system operates so that only correctly folded molecules are allowed to move along the secretory pathway. The productive folding process in the ER has been thought to be supported by the unfolded protein response (UPR), which is activated by the accumulation of unfolded proteins in the ER. However, a dilemma has emerged; activation of ATF6, a key regulator of mammalian UPR, requires intracellular transport from the ER to the Golgi apparatus. This suggests that unfolded proteins might be leaked from the ER together with ATF6 in response to ER stress, exhibiting proteotoxicity in the secretory pathway. We show here that ATF6 and correctly folded proteins are transported to the Golgi apparatus via the same route and by the same mechanism under conditions of ER stress, whereas unfolded proteins are retained in the ER. Thus, activation of the UPR is compatible with the quality control in the ER and the ER possesses a remarkable ability to select proteins to be transported in mammalian cells in marked contrast to yeast cells, which actively utilize intracellular traffic to deal with unfolded proteins accumulated in the ER.
INTRODUCTION
Proteins must acquire correct secondary and tertiary structures to carry out their functions as assigned by the genetic code. Newly synthesized secretory and transmembrane proteins are folded and assembled in the endoplasmic reticulum (ER) where a number of molecular chaperones and folding enzymes (collectively termed ER chaperones hereafter) are abundantly expressed to assist or facilitate conformational maturation of these cargo proteins (Gething and Sambrook, 1992; Helenius et al., 1992). The protein-folding process in the ER, albeit productive under normal conditions, can be compromised when cells are exposed to a variety of environmental stress conditions. It also runs into trouble when amounts of newly synthesized secretory or transmembrane proteins exceed the folding capacity in the ER. In addition, proteins possessing genetic mutation(s) in their amino acid sequences may not be able to be folded properly. Eukaryotic cells from yeast to humans counteract such protein unfolding or misfolding in the ER by activating a homeostatic response, termed the unfolded protein response (UPR; Kaufman, 1999; Mori, 2000; Patil and Walter, 2001; Harding et al., 2002). The UPR consists of translational and transcriptional controls. The aim of translational control is to decrease the burden on the ER by inhibiting protein synthesis whereas that of transcriptional control is to augment the folding capacity in the ER by inducing ER chaperones.
Two basic leucine zipper (bZIP) proteins ATF6 and XBP1 have been identified as key regulators of transcriptional control during the mammalian UPR (Yoshida et al., 1998, 2001a; Haze et al., 1999; Calfon et al., 2002; Lee et al., 2002). ATF6 and XBP1 are specifically activated in response to ER stress by highly characteristic posttranslational and post-transcriptional mechanisms, designated regulated intramembrane proteolysis and frame switch splicing, respectively (Brown et al., 2000; Mori, 2003). We recently demonstrated that mammalian cells take advantage of differential properties between the two mechanisms to determine the fate of unfolded or misfolded proteins in the ER (Yoshida et al., 2003). Thus, mammalian cells execute a time-dependent phase transition from the ATF6-mediated unidirectional phase (refolding only) to the XBP1-mediated bidirectional phase (refolding plus degradation), depending on the quality or quantity or both of unfolded proteins accumulated in the ER.
ATF6, despite its regulatory role in transcription, is constitutively synthesized as a type II transmembrane protein embedded in the ER and subjected to proteolysis when unfolded proteins are accumulated in the ER (Haze et al., 1999). The cytoplasmic fragment of ATF6 thereby liberated from the membrane carries bZIP and other domains necessary for an active transcription factor, translocates into the nucleus, and activates transcription of its target genes (Yoshida et al., 2000). ER chaperones are major targets of the ATF6 pathway (Okada et al., 2002). ER stress-induced cleavage of ATF6 is carried out by the sequential actions of Site-1 protease (S1P) and Site-2 protease (S2P; Ye et al., 2000); S1P and S2P were identified originally as enzymes that process sterol regulatory element-binding proteins (SREBP), transcription factors responsible for cholesterol homeostasis in mammalian cells (Brown and Goldstein, 1999). As S1P and S2P are localized in the Golgi apparatus, ATF6 must be exported to the Golgi apparatus to be cleaved as in the case of SREBP. ER stress-induced relocation of ATF6 from the ER to the nucleus via the Golgi apparatus was indeed demonstrated by analyzing both transiently transfected ATF6 (Chen et al., 2002) and endogenous ATF6 (Okada et al., 2003). It should be noted that although mammalian cells express two closely related ATF6 proteins, namely ATF6α encoded by the ATF6 gene and ATF6β encoded by the G13/cAMP response element-binding protein-related protein gene, ATF6α can be used as a representative because no significant differences have been revealed between ATF6α and ATF6β so far (Haze et al., 2001; Yoshida et al., 2001b).
Previous studies seem to have established that an efficient quality control system operates in the ER whereby correctly folded molecules are allowed to exit the ER and reach their final destinations, whereas unfolded molecules are retained in the ER (Ellgaard and Helenius, 2003). The finding that ATF6 is transported from the ER to the Golgi apparatus in response to ER stress has therefore raised the question of whether ATF6 is exported to the Golgi apparatus via the same route and by the same mechanism as cargo proteins and if so whether cargo proteins unfolded by ER stress are discriminated from ATF6 and retained in the ER or can exit the ER together with ATF6. In other words, we intended to investigate the effects of ER stress on the quality control in the ER.
MATERIALS AND METHODS
Construction of Plasmids
Recombinant DNA techniques were performed according to standard procedures (Sambrook et al., 1989). The integrity of all constructed plasmids was confirmed by extensive sequencing analysis. To construct the green fluorescent protein (GFP)-ATF6α fusion gene, a cDNA fragment encoding amino acids 6–670 of ATF6α (Haze et al., 1999) was inserted between the BglII and SalI sites of pEGFP-C1 (CLONTECH, Palo Alto, CA) so that ATF6α was fused in-frame to the C-terminus of GFP. In the resulting plasmid designated pCMVfull-EGFP-ATF6α, ATF6α was preceded by 14 amino acids (RSIWNS-DPRGHEGP) derived from the multicloning site of pEGFP-C1. The CMV promoter region of pCMVfull-EGFP-ATF6α was shortened to create pCM-Vshort-EGFP-ATF6α by removing the nucleotide region from the AseI site to the AatII site in pEGFP-C1 (nucleotide positions 8 and 443, respectively). pCMVshort-EGFP-ATF6α(S1P-) and pCMVshort-EGFP-ATF6α(S2P-) were generated by mutagenizing pCMVshort-EGFP-ATF6α using the Exsite site-directed mutagenesis kit and QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), respectively. pCMVshort-EYFP-ATF6α was constructed by replacing the GFP-encoding the NheI and BspEI fragment of pCMVshort-EGFP-ATF6α with the YFP-encoding the NheI and BspEI fragment of pEYFP-C1 (CLONTECH).
pCMVfull-EGFP is identical to pEGFP-C1 and pCMVshort-EGFP was constructed by removing the nucleotide region from the AseI site to the AatII site in pEGFP-C1. The promoter regions of pCMVfull-EGFP and pCMVshort-EGFP were amplified by PCR and inserted into the XhoI-BglII sites of the pGL3-Basic vector (Promega, Madison, WI), which contains the firefly luciferase coding sequence but lacks eukaryotic promoter or enhancer elements, to create pGL3-CMVfull and pGL3-pCMVshort, respectively.
pCDM8-VSVGts045-GFP (Nehls et al., 2000) was provided by Dr. J. Lippincott-Schwartz (National Institute of Health). cDNA encoding VSVG-ts045 was amplified from pCDM8-VSVGts045-GFP and subcloned into pECFP-N1 (CLONTECH) to create pECFP-N1-tsVSVG in which VSVG-ts045 was fused in-frame to the N-terminus of CFP. pREP9-A1AT (Hosokawa et al., 2001) was provided by Dr. N. Hosokawa (Kyoto University). A1AT cDNA was amplified from pREP9-A1AT by PCR using 5′ and 3′ primers flanked by EcoRI and SmaI sites, respectively, and inserted between the EcoRI and SmaI sites of pECFP-N1 to create pECFP-N1-A1AT.
Cell Culture and Transfection
CHO cells were grown in a 1:1 mixture of Ham's F12 and DMEM supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin sulfate) in a 5% CO2, 95% air incubator at 37°C. Two days before transfection, cells were plated in 60-mm dishes (Falcon, Oxnard, CA) or 35-mm glass base dishes (Iwaki, Tokyo, Japan) so that they reached 70–80% confluency on the day of transfection. Two micrograms of plasmid DNA was added to each 60-mm dish for transfection after mixing with 10 μl of Superfect (Qiagen, Chatsworth, CA) in 100 μl of serum-free culture medium according to the manufacturer's instruction. After incubation for 3 h at 37°C, cells were washed with PBS and then cultured in 4 ml of fresh medium for 5–8 h at the temperature indicated in the text. For double transfection experiments, 0.3 μg of pCMVshort-EYFP-ATF6α and 0.7 μg of pECFP-N1-tsVSVG or pECFP-N1-A1AT (total 1 μg of plasmid DNA) were added to each 35-mm dish after mixing with 5 μl of Superfect in 60 μl of serum-free culture medium.
Immunoblotting
Immunoblotting analysis was carried out according to the standard procedure (Sambrook et al., 1989) as described previously (Okada et al., 2002) using an enhanced chemiluminescence Western blotting detection system kit (Amersham Biosciences, Piscataway, NJ). Chemiluminescence was detected using an LAS-1000plus LuminoImage analyzer (Fuji Film, Tokyo, Japan). Rabbit anti-ATF6α polyclonal antibody raised against the N-terminal region of ATF6α (residues 6–307) was prepared previously (Haze et al., 1999). Rabbit anti-GFP polyclonal antibody (Living Colors A.v. Peptide Antibody) was obtained from CLONTECH. Mouse anti-GM130 mAb and anti-KDEL mAb (clone 10C3) were purchased from BD Biosciences (San Diego, CA) and StressGene Biotechnologies, respectively. Anti-ERGIC-53 antibody was a gift from Dr. H.-P. Hauri (University of Basel).
Northern Blot Hybridization and Luciferase Assay
Total RNA was extracted by the acid guanidinium-phenol-chloroform method using ISOGEN (Nippon Gene) and analyzed by the standard Northern blotting using an Alkphos direct labeling kit (Amersham Biosciences). Chemiluminescence was visualized using an LAS-1000plus LuminoImage analyzer. The luciferase assay was performed according to our published procedures (Yoshida et al., 2000)
Immunoprecipitation
CHO cells cultured in 60-mm dishes were solubilized, after being washed with PBS, for 30 min on ice in 400 μl of lysis buffer (20 mM Tris-HCl, pH 7.5, containing 1% Triton X-100, 10% glycerol, 0.15 M NaCl, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride, 4 μg/ml aprotinin, and 2 μg/ml pepstatin A). Lysates clarified by centrifugation at 10,000 × g for 5 min were incubated with 5 μl of anti-GFP mAb (mixture of clone 7.1 and 13.1, Roche Molecular Biochemicals, Indianapolis, IN) for 30 min on ice and then with 20 μl of protein G-Sepharose (Amersham Biosciences) for 30 min on ice with occasional shaking. Sepharose beads were collected by brief centrifugation and washed with lysis buffer three times. Immunoprecipitated materials were eluted by boiling for 5 min in 1× Laemmli's sample buffer. Two microliters of anti-GFP antibody was used to immunoprecipitate A1AT-CFP secreted into the medium.
Indirect Immunofluorescence
CHO cells cultured in 35-mm glass-base dishes were fixed with PLP solution (2% p-formaldehyde, 10 mM sodium metaperiodate, and 75 mM l-lysine in PBS) for 20 min on ice, permeabilized with PBS containing 0.2% Triton X-100 for 10 min at room temperature, and then reacted with various primary antibodies for 1 h at 37°C. Rabbit anti-ATF6α antibody was diluted 33-fold, whereas mouse anti-ERGIC-53, anti-KDEL, and anti-GM130 antibodies were diluted 50-fold with PBS containing 1% BSA before use. Primary antibodies were visualized by incubation for 1 h at 37°C with fluorescein isothiocyanate–conjugated goat anti-rabbit IgG antibody, rhodamine-conjugated goat anti-rabbit IgG antibody, or rhodamine-conjugated goat anti-mouse IgG antibody (ICN Pharmaceuticals, Costa Mesa, CA), followed by confocal laser scanning fluorescence microscopy performed using an LSM510 (Carl Zeiss, Thornwood, NY).
Fluorescence Microscopy
GFP or one of its variants in CHO cells cultured in 35-mm glass-base dishes was visualized using an IX71 inverted fluorescent microscope (Olympus, Tokyo, Japan). Fluorescent images of GFP and YFP were obtained through a U-MNIBA2 filter (Olympus, excitation: 470- to 490-nm, emission: 510- to 550-nm), whereas those of CFP were obtained through a U-MCFPHQ filter (Olympus, excitation: 425- to 445-nm, emission: 460- to 510-nm).
Microinjection
cDNAs encoding the wild-type and dominant negative mutant form of Sar1 (Kuge et al., 1994) were provided by Dr. O. Kuge (Kyushu University) and subcloned into the mammalian expression vector pCI-neo (Promega) to create pCI-Sar1WT and pCI-Sar1DN(T39N), respectively. After mixing with 1 μg/μl TRITC-conjugated dextran (Molecular Probes, Eugene, OR) used as an injection marker, these plasmids (a final concentration of 0.5 μg/μl) were micro-injected into nuclei of CHO cells using a semiautomatic system consisting of a FemtoJet and Micromanipulator 5171(Eppendorf, Fremont, CA).
RESULTS
GFP-ATF6α Fusion Protein Expressed at a Low Level Responds to ER Stress Similarly to Endogenous ATF6
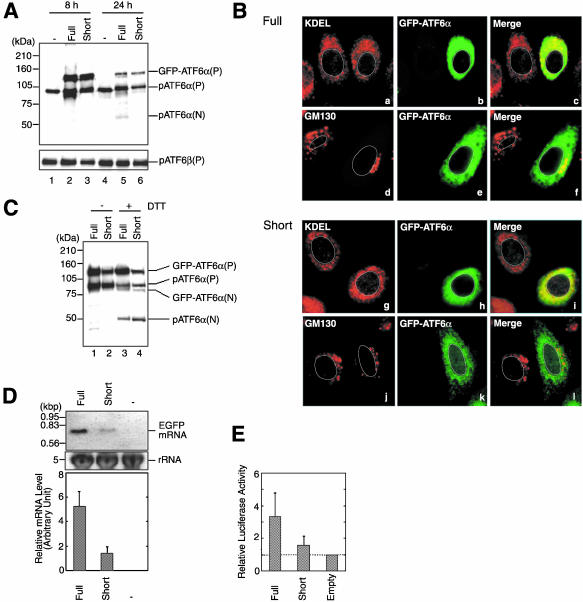
In an attempt to answer the questions described in the Introduction, we constructed GFP-ATF6α, which relocates from the ER to the nucleus via the Golgi apparatus in response to ER stress similarly to endogenous ATF6. ATF6α was fused to the C-terminus of GFP in the mammalian expression vector pEGFP-C1 in which expression of GFP is under the control of a strong CMV promoter, and the resulting plasmid pCMVfull-EGFP-ATF6α was used to transfect Chinese hamster ovary (CHO) cells. As a result, a band was detected with anti-ATF6α antibody 8 h after transfection at the molecular mass of 120 kDa, which corresponded to the sum of the size of GFP (30 kDa) and that of pATF6α(P), the ER membrane-bound precursor form of ATF6α (90 kDa; Figure 1A, lane 2). Because this band was recovered in the membrane fraction after fractionation and was immunoprecipitable using anti-GFP antibody (our unpublished data), this band was assigned to the full-length GFP-ATF6α bound to the membrane and thus designated GFP-ATF6α(P). In contrast to endogenous pATF6α(P), however, GFP-ATF6α(P) was localized in both the ER and the Golgi apparatus as the GFP fluorescence merged with the staining obtained not only with anti-KDEL antibody, which recognizes major ER chaperones such as BiP/GRP78 and GRP94, but also with anti-GM130 antibody, which recognizes the specific Golgi marker protein GM130 (Figure 1B, a–f). Furthermore, the level of pATF6α(P) was markedly enhanced in transfected cells as compared with untransfected cells (Figure 1A, compare lane 2 with lane 1). This was most likely due to partial degradation of GFP-ATF6α(P) because the level of the full-length ATF6β, pATF6β(P), remained unaffected by transfection with pCMVfull-EGFP-ATF6α (Figure 1A, bottom, compare lane 2 with lane 1). Critically, this enhancement of pATF6α(P) obscured the detection of ER stress-induced cleavage of GFP-ATF6α(P) as shown in Figure 1C. The molecular mass (90 kDa) of pATF6α(P) was very close to that (80 kDa) of the soluble and nuclear form of GFP-ATF6α, designated GFP-ATF6α(N), which was produced in response to dithiothreitol (DTT) treatment (Figure 1C, lane 3); DTT causes ER stress by disrupting disulfide bond formation (Kaufman, 1999). It should be also noted that this enhancement of pATF6α(P) eventually caused ER stress by itself in transfected cells because pATF6α(P) is a transmembrane protein in the ER; the active and nuclear form of ATF6α, pATF6α(N), was detected 24 h after transfection with pCMVfull-EGFP-ATF6α (Figure 1A, lane 5).
Figure 1.
Effects of using a full-length or truncated CMV promoter on the expression, localization and ER stress-induced processing of GFP-ATF6α. (A) CHO cells were transfected with pCMVfull-EGFP-ATF6α (Full) or pCMVshort-EGFP-ATF6α (Short). Eight or 24 h after transfection, cell lysates were prepared and analyzed by immunoblotting using anti-ATF6α or anti-ATF6β antibodies. The migration positions of GFP-ATF6α(P), endogenous pATF6α(P), endogenous pATF6α(N), and endogenous pATF6β(P) are marked. The positions of full-range rainbow molecular weight markers (Amersham Biosciences) are indicated on the left. (B) Eight hours after transfection with pCMVfull-EGFP-ATF6α (Full) or pCMVshort-EGFP-ATF6α (Short), CHO cells were fixed, stained with anti-KDEL or anti-GM130 antibodies, and analyzed by fluorescence microscopy. GFP-ATF6α was visualized by its own fluorescence (shown in green), whereas ER chaperones having the KDEL sequence at their C-termini and GM130 were visualized using rhodamine-conjugated secondary antibody (shown in red). An outline of the nucleus is indicated by a white line in each cell. (C) Eight hours after transfection with pCMVfull-EGFP-ATF6α (Full) or pCMVshort-EGFP-ATF6α (Short), CHO cells were untreated (-) or treated (+)with 1 mM DTT for 30 min before the preparation of cell lysates, which were then analyzed by immunoblotting using anti-ATF6α antibody. The migration positions of GFP-ATF6α(P), endogenous pATF6α(P), GFP-ATF6α(N), and endogenous pATF6α(N) are marked. (D) Twelve hours after mock transfection (-) or transfection of CHO cells with pCMVfull-EGFP (Full) or pCMVshort-EGFP (Short), total RNA was prepared and analyzed by Northern blot hybridization using a probe specific to GFP. The level of 28S rRNA in each lane is shown as a loading control. Similar experiments were carried out two more times. Chemiluminescence intensity of each band was determined using an LAS-1000plus Luminoimage analyzer and the averages are presented with standard deviations (error bars). (E) CHO cells were transfected with pGL3-Basic vector (Empty), pGL3-CMVfull (Full), or pGL3-CMVshort (Short) together with reference plasmid pRL-SV40. The relative luciferase activity was determined after overnight incubation of transfected cells. The averages from duplicate determination of three independent experiments are presented with standard deviations (error bars) after normalization to the value obtained for Empty.
To solve this problem, we shortened the CMV promoter by deleting ∼430 base pairs from the 5′ side because promoter strength usually correlates well with length if the 3′ region containing the TATA box is maintained; it is our experience that the analysis of a series of 5′ deletion mutants of a promoter almost always shows a gradual decrease in basal expression activity. To quantify the difference in the activity, we fused these “full” and “short” CMV promoters to the coding sequence of GFP or firefly luciferase and transfected CHO cells with the resulting plasmids. As shown in Figure 1D, the level of GFP mRNA expressed from the short promoter was about fourfold lower than that from the full promoter. Similarly, luciferase activity expressed from the short promoter was about fourfold lower than that from the full promoter when luciferase activity expressed from the vector alone (no promoter insertion) was subtracted (Figure 1E). Thus, the short promoter possessed considerably lower activity than the full promoter as we expected.
We expressed GFP-ATF6α from the short CMV promoter by transfecting CHO cells with the plasmid pCMVshort-EGFP-ATF6α. As a result, GFP-ATF6α(P) was detected at the expected molecular mass of 120 kDa 8 h after transfection (Figure 1A, lane 3), being recovered in the membrane fraction after fractionation and immunoprecipitable using anti-GFP antibody (our unpublished data). Importantly, GFP-ATF6α(P) expressed from the short CMV promoter was localized exclusively in the ER (Figure 1B, g–l). The level of pATF6α(P) in transfected cells was similar to that in untransfected cells (Figure 1A, compare lane 3 with lane 1), and pATF6α(N) was not produced even 24 h after transfection (Figure 1A, lane 6). Most importantly, conversion of GFP-ATF6α(P) to GFP-ATF6α(N) was clearly observed after DTT treatment of transfected cells (Figure 1C, lane 4).
To confirm that ER stress-induced cleavage of GFP-ATF6α(P) was carried out by the sequential actions of S1P and S2P as in the case of SREBP and ATF6, we mutated the sequences required for cleavage by S1P or S2P. The consensus sequence of the S1P cleavage site is RxxL/K (Brown and Goldstein, 1999). ATF6α contains two RxxL motifs in an overlapping manner (RRHLL) and it was previously shown that cleavage of ATF6α by S1P was abolished when both Arg415 and Arg416 were mutated to Ala (Ye et al., 2000). We thus constructed GFP-ATF6α carrying R415A and R416A mutations and designated it GFP-ATF6α(S1P-). On the other hand, cleavage of SREBP by S2P requires the Asn-Pro sequence present in the transmembrane domain (Brown and Goldstein, 1999). ATF6α contains an Asn and a Pro in its transmembrane domain although they are separated by two amino acids in contrast to the case of SREBP. It was previously shown that cleavage of ATF6α by S2P was completely blocked by simultaneous mutation of Asn391 and Pro394 to Phe and Leu, respectively (Ye et al., 2000). GFP-ATF6α carrying N391F and P394L mutations was thereby constructed and designated GFP-ATF6α(S2P-).
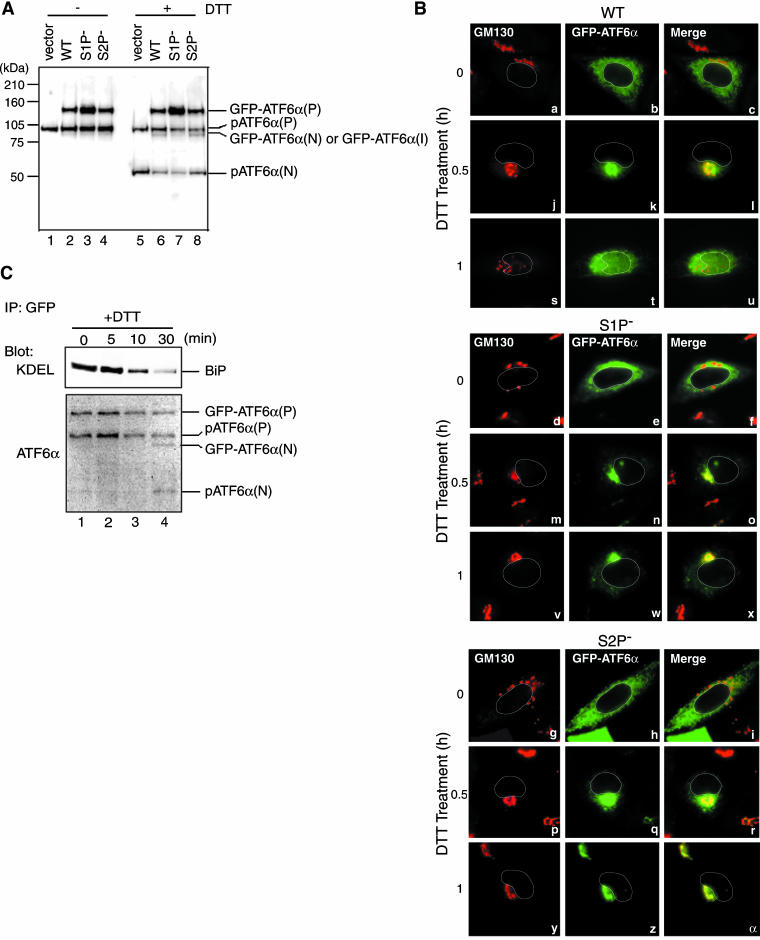
Both GFP-ATF6α(S1P-) and GFP-ATF6α(S2P-) were expressed in CHO cells by transfection similarly to the wild-type GFP-ATF6α using the short CMV promoter. All three fusion proteins were detected with anti-ATF6α antibody at the molecular mass of 120 kDa (Figure 2A, lanes 2–4) and were localized in the ER but not in the Golgi apparatus (Figure 2B, a–i) as expected. On treatment of transfected cells with 1 mM DTT, the wild-type GFP-ATF6α(P) was cleaved to produce GFP-ATF6α(N) (Figure 2A, lane 6), being consistent with the results shown in Figure 1C. Accordingly, the GFP portion of the wild-type GFP-ATF6α was translocated to the nucleus via the Golgi apparatus (Figure 2B, j–l and s–u). Thus, GFP-ATF6α expressed from the short CMV promoter at a low level exhibited the same behavior as endogenous ATF6 in ER-stressed cells. In marked contrast, no cleaved product was detected in cells expressing GFP-ATF6α(S1P-) upon DTT treatment (Figure 2A, lane 7), although GFP-ATF6α(S1P-) was transported to the Golgi apparatus similarly to the wild-type GFP-ATF6α (Figure 2B, m–o). Furthermore, GFP-ATF6α(S1P-) remained associated with the Golgi apparatus even 1 h after DTT treatment (Figure 2B, lanes v–x) in contrast to the wild-type GFP-ATF6α. These results indicated that the initial cleavage by S1P is critical for the processing of GFP-ATF6α and that only the cleaved product, GFP-ATF6α(N) can enter the nucleus.
Figure 2.
Effects of mutating the S1P or S2P cleavage site on ER stress-induced processing and relocation of GFP-ATF6α. (A) Eight hours after transfection with pCMVshort-EGFP-ATF6α (WT), pC-MVshort-EGFP-ATF6α(S1P-), pCMVshort-EGFP-ATF6α(S2P-), or vector alone, CHO cells were untreated (-) or treated (+) with 1 mM DTT for 30 min before the preparation of cell lysates, which were then analyzed by immunoblotting using anti-ATF6α antibody. The migration positions of GFP-ATF6α(P), endogenous pATF6α(P), GFP-ATF6α(N), or GFP-ATF6α(I), and endogenous pATF6α(N) are marked. (B) CHO cells transfected as in A were treated with 1 mM DTT for the indicated periods, fixed, stained with anti-GM130 antibody, and then analyzed by fluorescence microscopy as described in the legend to Figure 1B. An outline of the nucleus is indicated by a white line in each cell. (C) CHO cells transfected with pCMVshort-EGFP-ATF6α were treated with 1 mM DTT for the indicated periods. GFP-ATF6α was immunoprecipitated from cell lysates using anti-GFP antibody. Immunoprecipitates were analyzed by immunoblotting using anti-ATF6α or anti-KDEL antibodies.
In contrast, GFP-ATF6α(S2P-) was translocated to the Golgi apparatus in response to DTT treatment (Figure 2B, p–r) and was cleaved to produce a band migrating at the same position as that of GFP-ATF6α(N) (Figure 2A, lane 8). However, the GFP-ATF6α(N)-like molecule remained associated with the Golgi apparatus even 1 h after DTT treatment (Figure 2B, panels y–α). We therefore assigned the GFP-ATF6α(N)-like molecule produced from GFP-ATF6α(S2P-) as GFP-ATF6α(I), an intermediate fragment that had been cleaved by S1P but not yet by S2P and thereby remained associated with the membrane. It should be noted that endogenous ATF6α served as an internal control; pATF6α(P) was converted to pATF6α(N) in response to DTT treatment in all cases (Figure 2A, lanes 5–8).
Interestingly, when cell lysates expressing GFP-ATF6α were subjected to immunoprecipitation using anti-GFP antibody and the immunoprecipitates were analyzed by immunoblotting using anti-ATF6α antibody, both GFP-ATF6α(P) and endogenous pATF6α(P) were detected (Figure 2C, lane 1), suggesting that they formed a dimer. Furthermore, immunoblotting analysis of the immunoprecipitates using anti-KDEL antibody revealed an association with BiP (Figure 2C, lane 1), which was lost gradually after treatment with DTT (Figure 2C, lanes 2–4) as reported previously for transfected ATF6α (Shen et al., 2002). We concluded that GFP-ATF6α under the control of the truncated CMV promoter provides a powerful tool for studying the regulatory mechanism of ATF6 activation in conjunction with the quality control system operating in the ER.
ATF6 Is Transported from the ER to the Golgi Apparatus via the Same Route and by the Same Mechanism as Cargo Proteins
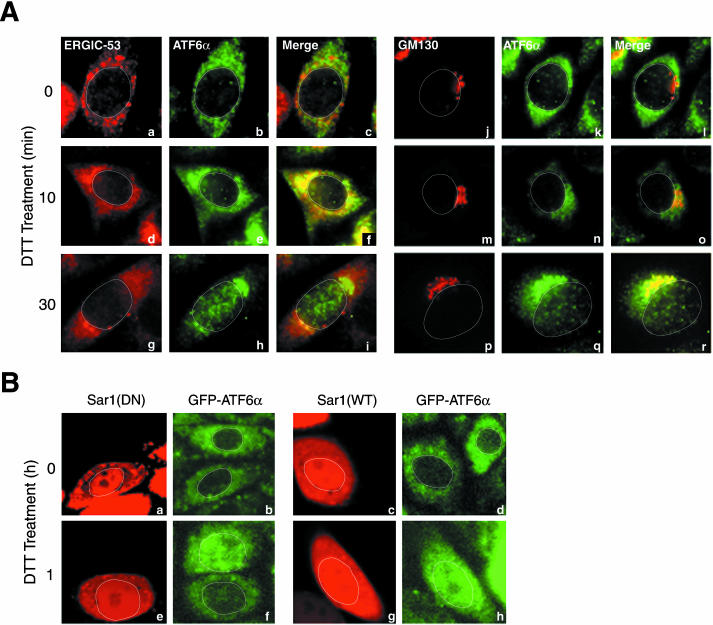
We investigated whether the route ATF6 takes to reach the Golgi apparatus is the same as that cargo proteins take during the course of secretion and whether the transport of ATF6 and cargo proteins is carried our by the same vesicles. It is well established that cargo proteins are transported from the ER to the Golgi apparatus through the ER-Golgi intermediate compartment (ERGIC, also called vesiculartubular clusters; Bannykh and Balch, 1997; Hauri et al., 2000). To determine whether ATF6 also transits ERGIC, we performed double immunostaining of CHO cells using anti-ATF6α and anti-ERGIC-53 antibodies; ERGIC-53 is a specific marker for ERGIC (Hauri et al., 2000). As shown in Figure 3A, anti-ATF6α antibody stained perinuclear structures of unstressed CHO cells (Figure 3A, b and k), which were distinct from punctate structures stained with anti-ER-GIC-53 (Figure 3Aa) or anti-GM130 (Figure 3Aj) antibody (at time 0). Ten and 30 min after the treatment of CHO cells with 1 mM DTT, ATF6α moved toward (Figure 3A, e and n) and reached (Figure 3A, h and q), respectively, a juxtanuclear region, which overlapped with GM130-positive punctate structures (Figure 3A, p–r). The nucleus also started to be stained with anti-ATF6α antibody 30 min after DTT treatment (Figure 3A, h and q). Importantly, ATF6α staining overlapped with ERGIC-53 staining at 10 min (Figure 3Af) but not at 30 min (Figure 3Ai) after DTT treatment, strongly suggesting that ATF6 transits ERGIC on the way from the ER to the Golgi apparatus as cargo proteins do.
Figure 3.
Analysis of the route and mechanism of ER stress-induced transport of ATF6. (A) CHO cells treated with 1 mM DTT for the indicated periods were fixed, double-stained with anti-ATF6α and anti-ERGIC-53 antibodies (a–i) or with anti-ATF6α and anti-GM130 antibodies (j–r), and then analyzed by fluorescence microscopy. ATF6α was visualized using FITC-conjugated secondary antibody (shown in green), whereas ERGIC-53 and GM130 were visualized using rhodamine-conjugated secondary antibody (shown in red). An outline of the nucleus is indicated by a white line in each cell. (B) Plasmid DNA encoding either the wild-type Sar1 [Sar1(WT)] or dominant negative mutant form of Sar1 [Sar1(DN)] was microinjected into nuclei of CHO cells expressing GFP-ATF6α (shown in green) together with TRITC-conjugated dextran (shown in red). Five hours later, cells were treated with 1 mM DTT for the indicated periods, and then analyzed by confocal laser scanning fluorescence microscopy. An outline of the nucleus is indicated by a white line in each cell.
It is also well established that cargo proteins are transported from the ER to the Golgi apparatus by vesicles coated with COP-II coat proteins (COP-II vesicles) and that Sar1, a cytoplasmic GTPase, plays a key role in the formation of COP-II vesicles (Antonny and Schekman, 2001). The exit of cargo proteins from the ER is blocked by a dominant negative mutant form of Sar1 [Sar1(DN)] such as the T39N mutant, which is no longer activated by the GDP/GTP exchange reaction (Barlowe et al., 1994; Kuge et al., 1994). This led us to examine whether the ER stress-induced exit of ATF6 from the ER is also blocked by Sar1(DN). Plasmid DNA encoding Sar1(DN) was microinjected into nuclei of CHO cells to overproduce Sar1(DN). However, we could not determine the effects of overproduced Sar1(DN) on the translocation of endogenous ATF6α because microinjected materials leaked from cells when they were fixed for immunofluorescence analysis. We therefore transfected CHO cells with pCMVshort-EGFP-ATF6α and isolated a stable transformant. As shown in Figure 3B, GFP-ATF6α exhibited a typical ER pattern under normal conditions regardless of the microinjection as expected (Figure 3B, a and b). Importantly, GFP-ATF6α relocated to the nucleus in uninjected cells in response to treatment with 1 mM DTT but remained associated with the ER even 1 h after DTT treatment of microinjected cells (Figure 3B, e and f). The wild-type Sar1 [Sar1(WT)] served as a negative control; DTT treatment-induced relocation of GFP-ATF6α was not affected by microinjection of plasmid DNA encoding Sar1(WT) (Figure 3B, c, d, g, and h). On the basis of these results, we concluded that the ER stress-induced transport of ATF6 is mediated by COP-II vesicles similarly to that of cargo proteins.
ATF6 Is Discriminated from Unfolded Cargo Proteins when Transported in Response to ER Stress
We next investigated whether the quality control system in the ER for cargo proteins is operating normally under ER stress conditions, in other words, whether unfolded cargo proteins are retained in the ER even though ATF6 is transported from the ER to the Golgi apparatus in response to ER stress. We used vesicular stomatitis virus G protein (VSVG) as a representative of transmembrane-type cargo proteins (Gallione and Rose, 1985). Extensive analyses conducted on the temperature-sensitive mutant (ts045) of VSVG have revealed a strict relationship between folding status and permitted export from the ER (deSilva et al., 1993; Nehls et al., 2000; Ellgaard and Helenius, 2003); ts045-VSVG cannot be folded properly because of a point mutation in its lumenal domain and thus is retained in the ER at the nonpermissive temperature 39°C, whereas ts045-VSVG attains native conformations and exits the ER to reach the plasma membrane at the permissive temperature 32°C. To compare the localization of ts045-VSVG and ATF6α in the same cell, ts045-VSVG was fused to the N-terminus of the cyan-emitting variant of GFP (the resulting fusion protein is referred to as VSVG-CFP), whereas ATF6α was fused to the C-terminus of the yellow-emitting variant of GFP. The resulting fusion protein designated YFP-ATF6α was expressed under the control of the short CMV promoter similarly to GFP-ATF6α.
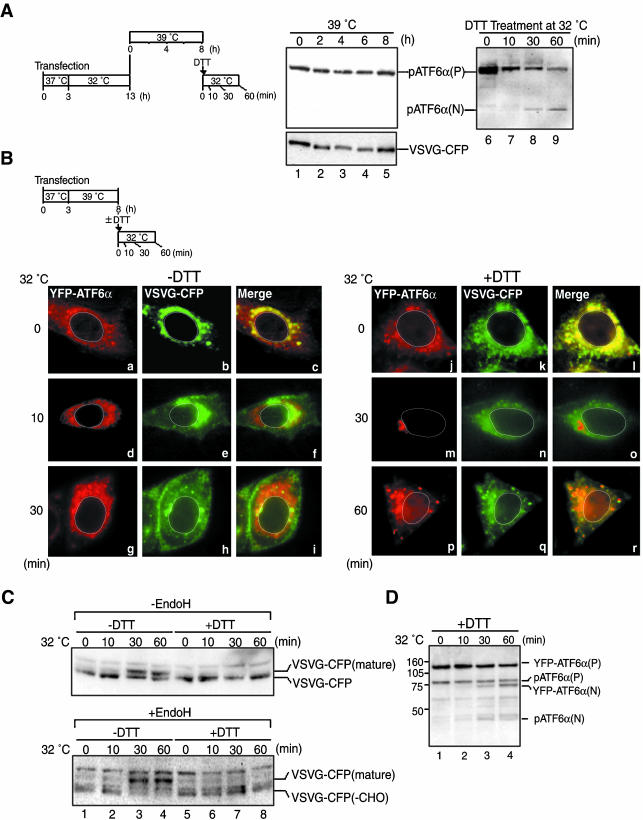
VSVG-CFP introduced by transfection was supposed to cause ER stress at 39°C; however, pATF6α(P) in transfected cells appeared to remain intact before (Figure 4A, lane 1) and after the temperature shift to 39°C (Figure 4A, lanes 2–5). The failure to detect pATF6α(N) was partly ascribed to rapid degradation by the proteasome; a small amount of pATF6α(N) was detected when the proteasome inhibitor MG132 was added to the culture before the temperature shift to 39°C (our unpublished data). However, as a great majority of ATF6α was present as pATF6α(P) even in the presence of MG132, misfolded VSVG-CFP might not be able to activate ATF6 efficiently (see DISCUSSION). In contrast, treatment of transfected cells with 1 mM DTT at 32°C caused rapid conversion of pATF6α(P) to pATF6α(N) (Figure 4A, lanes 6–9). We concluded that expression of VSVG-CFP by transfection did not significantly affect the behavior of endogenous ATF6α and that YFP-ATF6α should respond to ER stress properly in cells expressing VSVG-CFP.
Figure 4.
Comparison of ER stress-induced transport of YFP-ATF6α with folding status-dependent transport of VSVG-CFP. (A) VSVG-CFP fusion protein was expressed in CHO cells by transfection. Transfected cells were maintained at the permissive temperature (32°C) for 10 h and then incubated at the nonpermissive temperature (39°C) for the indicated periods as schematically shown on the left. Aliquots of transfected cells cultured at 39°C for 8 h were further incubated at 32°C in the presence of 1 mM DTT for the indicated periods. Cell lysates were prepared and analyzed by immunoblotting using anti-ATF6α or anti-GFP antibodies. The positions of endogenous pATF6α(P), endogenous pATF6α(N), and VSVG-CFP are indicated. (B) YFP-ATF6α and VSVG-CFP were simultaneously expressed in CHO cells by transfection. Transfected cells were maintained at 39°C for 5 h and then incubated at 32°C for the indicated periods in the presence (+) or absence (-) of 1 mM DTT as schematically shown at the top. The localization of YFP-ATF6α (shown in red) and VSVG-CFP (shown in green) was determined using fluorescence microscopy. An outline of the nucleus is indicated by a white line in each cell. (C) CHO cells were transfected and cultured as in B. After incubation of the cells at 32°C in the presence (+) or absence (-) of 1 mM DTT for the indicated periods, cell lysates were prepared, treated with (+) or without (-) endoglycosidase H (Endo H), and analyzed by immunoblotting using anti-GFP antibody. VSVG-CFP represents the fusion protein translocated into the ER and yet having Endo H-sensitive carbohydrates, whereas VSVG-CFP(mature) represents the processed form of the fusion protein having Endo H-resistant carbohydrates. VSVG(-CHO) represents the fusion protein lacking a carbohydrate moiety. (D) CHO cells were transfected and cultured as in B. After incubation of the cells at 32°C in the presence of 1 mM DTT for the indicated periods, cell lysates were prepared and analyzed by immunoblotting using anti-ATF6α antibody. The migration positions of YFP-ATF6α(P), endogenous pATF6α(P), YFP-ATF6α(N), and endogenous pATF6α(N) are marked.
VSVG-CFP and YFP-ATF6α were then expressed in CHO cells simultaneously by cotransfection. When transfected cells were maintained at 39°C, misfolded VSVG-CFP was localized in the ER as expected (Figure 4B, b and k). Ten minutes after the temperature shift to 32°C, VSVG-CFP moved toward the Golgi apparatus (Figure 4Be), reaching the plasma membrane 30 min later (Figure 4Bh). In contrast, YFP-ATF6α remained associated with the ER (Figure 4B, a, d, and g). The visualized movement of VSVG-CFP was consistent with the results of biochemical analyses shown in Figure 4C. VSVG-CFP was detected as a single band when transfected cells were maintained at 39°C (Figure 4C, top, lane 1). From 30 min after the temperature shift to 32°C, a slower migrating band appeared (Figure 4C, top, lanes 3 and 4). Importantly, the slower migrating band but not faster migrating band was resistant to digestion with endoglycosidase H (Endo H; Figure 4C, bottom, lanes 1–4). The conversion of N-linked carbohydrates from an Endo H–sensitive high mannose-type to Endo H–resistant complex-type occurs in medial to trans regions of the Golgi apparatus (Helenius and Aebi, 2001). Thus, VSVG-CFP correctly folded at 32°C indeed moved along the secretory pathway.
Strikingly different results were obtained when DTT was added to the culture medium at a final concentration of 1 mM immediately before the temperature shift to 32°C. Because the folding of VSVG requires correct disulfide bond formation (de Silva et al., 1993), DTT treatment prevented VSVG-CFP from gaining native conformations. As a result, VSVG-CFP was retained in the ER (Figure 4B, k, n, and q) and remained sensitive to Endo H (Figure 4C, lanes 5–8) even at 60 min after the temperature shift. In marked contrast, both endogenous pATF6α(P) and YFP-ATF6α(P) were cleaved to produce pATF6α(N) and YFP-ATF6α(N), respectively, in response to DTT treatment (Figure 4D), and YFP-ATF6α relocated from the ER to the nucleus via the Golgi apparatus (Figure 4B, j, m, and p). Thus, the ER can precisely select transmembrane proteins to be transported to the Golgi apparatus.
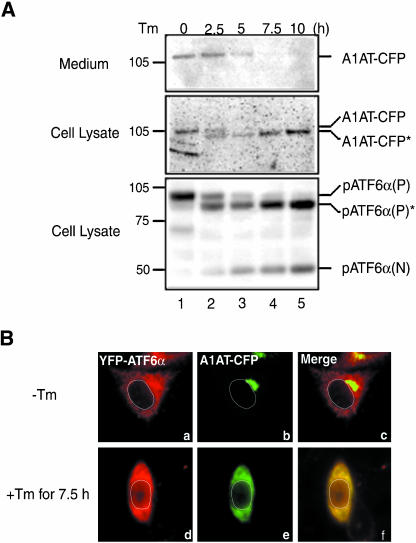
We next used α1-antitrypsin (A1AT), a serum glycoprotein, to examine the quality control system for soluble cargo proteins. A1AT was fused to the N-terminus of CFP and the resulting fusion protein designated A1AT-CFP was expressed in CHO cells by transfection. Under normal conditions, A1AT was synthesized and secreted into the medium as shown in Figure 5A (top and middle, lane 1). Microscopic analysis indicated that intracellular A1AT-CFP was accumulated in Golgi-like structures (Figure 5Bb), suggesting that export of A1AT-CFP from the Golgi apparatus is a rate-limiting step in the course of secretion of A1AT-CFP. On treatment of transfected cells with tunicamycin, which induces ER stress by inhibiting protein N-glycosylation (Kaufman, 1999), endogenous pATF6α(P) was converted to pATF6α(N) and, in addition, a nonglycosylated form of pATF6α(P), pATF6α(P)*, was produced (Figure 5A, bottom, lanes 2–5). Similarly, a nonglycosylated form of A1AT-CFP, A1AT-CFP*, was produced after tunicamycin treatment (Figure 5A, middle, lanes 2–5), leading to a gradual decrease in the level of A1AT-CFP secreted into the medium (Figure 5A, top, lanes 2–5). Microscopic analysis revealed that intra-cellular A1AT-CFP was present in perinuclear structures in cells treated with tunicamycin for 7.5 h (Figure 5Be), indicating that secretion of A1AT is blocked at the exit from the ER. Thus, unfolded cargo proteins of both transmembrane-type and soluble-type are retained in the ER even under ER stress. Importantly, however, some YFP-ATF6α was found in the nucleus (Figure 5B, d), demonstrating the ability of the ER to discriminate proteins to be transported from those to be retained under conditions of ER stress.
Figure 5.
Effects of tunicamycin treatment on the transport of α1-antitrypsin. (A) A1AT-CFP was expressed in CHO cells by transfection and subsequent culture at 37°C for 5 h. Then, cells were treated with 2 μg of tunicamycin (Tm) in 1 ml of culture medium for the indicated periods. Cell lysates were prepared and analyzed by immunoblotting using anti-GFP or anti-ATF6α antibodies. The level of secreted A1AT-CFP was also determined by immunoblotting using anti-GFP antibody after immunoprecipitation of A1AT from the culture medium with anti-GFP antibody. A1AT-CFP* and pATF6α(P)* denote deglycosylated forms of A1AT-CFP and endogenous pATF6α(P), respectively. (B) A1AT-CFP and YFP-ATF6α were expressed simultaneously in CHO cells by transfection and subsequent culture at 37°C for 5 h. Then, transfected cells were untreated (-) or treated (+) with 2 μg/ml tunicamycin (Tm) for 7.5 h. The localization of YFP-ATF6α (shown in red) and A1AT-CFP (shown in green) was determined using fluorescence microscopy. An outline of the nucleus is indicated by a white line in each cell.
Correctly Folded Cargo Proteins Exit the ER under ER Stress Conditions
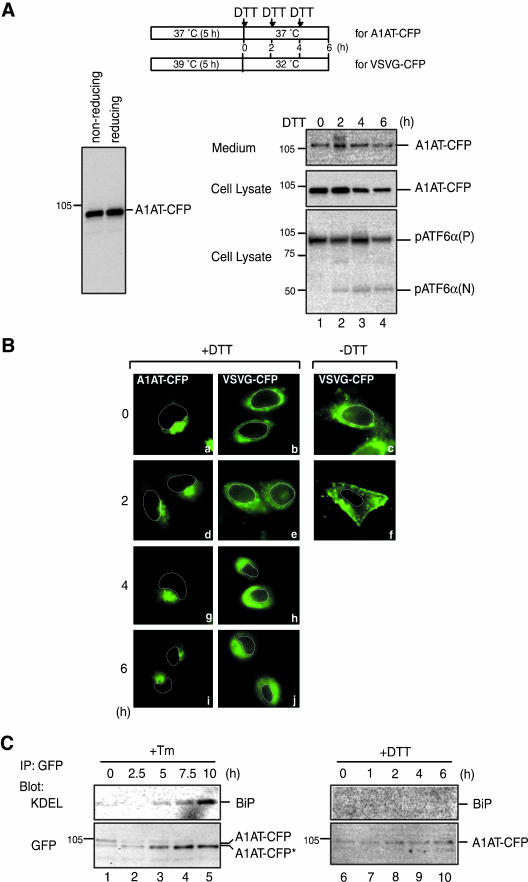
We next investigated whether cargo proteins are allowed to exit the ER even under ER stress conditions if they are correctly folded. In other words, we intended to determine whether only ATF6 is selectively transported to the Golgi apparatus or the secretion of folded molecules persists under ER stress. For this purpose, we determined the localization of A1AT-CFP and VSVG-CFP in cells treated with DTT. DTT treatment should have no effect on the folding of A1AT because A1AT contains only one cysteine residue and SDS-PAGE analysis of A1AT under both reducing and nonreducing conditions revealed no evidence of intermolecule disulfide bond formation (Figure 6A, left). On the other hands, DTT treatment causes malfolding of VSVG-CFP as already shown in Figure 4. Because preliminary experiments indicated that the DTT-induced retention of misfolded VSVG-CFP was effective for no more than 2 h, DTT was added to the culture medium of transfected cells at a final concentration of 0.5 mM every 2 h as depicted in Figure 6A. As a result, continuous conversion of endogenous pATF6α(P) to pATF6α(N) was observed (Figure 6A, bottom, lanes 1–4) and VSVG-CFP remained associated with the ER for 6 h (Figure 6B, b, e, h, and j). Under the conditions, the levels of intracellular and extracellular A1AT-CFP were not altered significantly (Figure 6A, top and middle, lanes 1–4) and intracellular A1AT-CFP remained associated with Golgi-like structures (Figure 6B, a, d, g, and i). Thus, A1AT-CFP continued to be synthesized and secreted in DTT-treated cells.
Figure 6.
Effects of dithiothreitol treatment on the transport of α1-antitrypsin. (A) A1AT-CFP was expressed in CHO cells by transfection and subsequent culture at 37°C for 5 h. Cell lysates were then prepared, subjected to SDS-PAGE under nonreducing and reducing conditions, and immunoblotted with anti-GFP antibody (left). Alternatively, DTT was added to the culture medium at a final concentration of 0.5 mM in a total volume of 1 ml every 2 h as schematically shown on the top. Cell lysates were prepared at the indicated time points and analyzed by immunoblotting using anti-GFP or anti-ATF6α antibodies (right). The level of secreted A1AT-CFP was also determined as in Figure 5A. (B) A1AT-CFP and VSVG-CFP were expressed in CHO cells separately by transfection. CHO cells expressing A1AT-CFP were treated with 0.5 mM DTT every 2 h as in A. CHO cells expressing VSVG-CFP were maintained at 39°C for 5 h after transfection and then treated with 0.5 mM DTT every 2 h after the temperature shift to 32°C. The localization of A1AT-CFP and VSVG-CFP (both in green) at the indicated time points was determined using fluorescence microscopy. An outline of the nucleus is indicated by a white line in each cell. (C) CHO cells expressing A1AT-CFP were prepared and treated with 2 μg/ml tunicamycin (Tm, left) and 0.5 mM DTT (right) for the indicated periods as in Figures 5A and 6A, respectively. A1AT-CFP was immunoprecipitated from cell lysates using anti-GFP antibody. Immunoprecipitates were analyzed by immunoblotting using anti-KDEL or anti-GFP antibodies.
To assess the folding status of A1AT-CFP, we examined whether A1AT in tunicamucin- or DTT-treated cells was bound to the ER chaperone BiP. A1AT-CFP was immunoprecipitated from cell lysates using anti-GFP antibody and then analyzed by immunoblotting using anti-GFP antibody or anti-KDEL antibody recognizing BiP. A1AT-CFP present in untreated cells was not associated with BiP (Figure 6C, lanes 1 and 6), indicating that it was correctly folded. In contrast, nonglycosylated A1AT-CFP* was associated with BiP (Figure 6C, lanes 2–5). Importantly, A1AT-CFP in DTT-treated cells was not bound to BiP (Figure 6C, lanes 7–10). These results are consistent with the notion that tunicamycin treatment caused retention of A1AT in the ER by preventing its folding, whereas DTT treatment had no effect on the folding of A1AT and thereby did not block its secretion. We concluded that the quality control system in the ER is not affected by ER stress so that unfolded cargo proteins are retained in the ER, whereas correctly folded cargo proteins are allowed to exit the ER. In other words, ATF6 is not an exceptional molecule to be transported from the ER to the Golgi apparatus under ER stress conditions.
DISCUSSION
In this report, we demonstrated that ATF6 is transported from the ER to the Golgi apparatus through ERGIC by COP-II vesicles similarly to cargo proteins. We also succeeded in expressing a GFP-ATF6α fusion protein that relocates from the ER to the nucleus via the Golgi apparatus in response to ER stress. The key was its expression level. As ATF6 is a transmembrane protein in the ER, its overexpression in itself causes ER stress. Indeed, we and others previously observed that overexpression of ATF6α driven by a strong CMV promoter resulted in constitutive cleavage, which was not enhanced further by ER stress treatment (Haze et al., 1999; Ye et al., 2000; Chen et al., 2002). Similarly, expression of GFP-ATF6α fusion protein from a strong CMV promoter was problematic as shown Figure 1; GFP-ATF6α was localized not only in the ER but also in the Golgi apparatus, partially degraded, and caused ER stress in CHO cells. To circumvent this technical problem, other investigators previously used weaker promoters such as the thymidine kinase promoter (Ye et al., 2000) and SV40 promoter (Chen et al., 2002) to drive the expression of ATF6α. Here, we found that shortening the CMV promoter by ∼430 base pairs from the 5′ side was sufficient to decrease the expression level of GFP-ATF6α. As a result, GFP-ATF6α expressed from the truncated promoter became localized exclusively in the ER and ER stress-induced cleavage was clearly evident on immunoblotting analysis (Figure 1). The GFP-ATF6α expressed from the short CMV promoter was transported to the Golgi apparatus in response to ER stress, where it was cleaved sequentially by S1P and S2P. Only a product cleaved by both enzymes could enter the nucleus as mutations at either the S1P cleavage site or S2P recognition site abolished the translocation of GFP-ATF6α from the Golgi apparatus to the nucleus (Figure 2). In addition, the constitutive association of GFP-ATF6α with BiP and ER stress-induced dissociation of the complex were observed (Figure 2) as reported for transfected ATF6α (Shen et al., 2002). We thus concluded that GFP-ATF6α expressed from the truncated CMV promoter behaved exactly like endogenous ATF6α.
It is noteworthy that misfolded VSVG-CFP did not activate ATF6 efficiently (Figure 4A) although the reason for the inefficiency is currently unclear. Ng et al. (1992) perviously analyzed the interaction of various mutant forms of the simian virus 5 hemagglutinin-neuraminidase glycoprotein (HN) with the ER chaperone BiP and found that there is a good correlation between the extent of association of an HN mutant with BiP and the extent of induction of BiP mRNA, a marker for activation status of the UPR; the more an HN mutant associates with BiP, the more BiP mRNA is induced in cells expressing the mutant HN (Ng et al., 1992). Therefore, it is interesting to determine the extent of association of VSVG-CFP with BiP in comparison with those of HN mutants as well as their ability to activate ATF6. It is possible that ATF6 is activated selectively in response to the accumulation of certain types of misfolded proteins but not to all of misfolded proteins accumulated in the ER.
Eukaryotic cells from yeast to humans have developed the UPR as a system to counteract protein unfolding or misfolding in the ER (Kaufman, 1999; Mori, 2000). Interestingly, however, several distinctions have been documented between the yeast and mammalian UPR. In the budding yeast Saccharomyces cerevisiae, the Ire1p-Hac1p pathway is solely responsible for transcriptional induction of all UPR target genes, whereas three signaling pathways function in mammals, namely the IRE1-XBP1, ATF6, and PERK-ATF4 pathways (Patil and Walter, 2001; Harding et al., 2002; Mori, 2003). The three mammalian pathways have distinct as well as overlapping target genes. The ATF6 pathway mainly regulates transcription of ER chaperone genes (Okada et al., 2002). The IRE1 pathway transactivates transcription of genes encoding ER chaperones and components required for ER-associated degradation of misfolded proteins (Yoshida et al., 2003). Activation of the PERK pathway results in the induction of genes involved in amino acid synthesis/metabolism/import, glutathione biosynthesis, and resistance to oxidative stress (Harding et al., 2003). These products are primarily induced to deal with various consequences of the accumulation of unfolded proteins in the ER. It is also postulated that the ATF6 and IRE1 pathways and the PERK and IRE1 pathways cooperate to elicit a maximal response to ER stress (Harding et al., 2002; Mori, 2003). It should be noted that the UPR in metazoan cells consists of not only transcriptional but also translational control (Harding et al., 2002). PERK in mammals and its homolog pek-1 in Caenorhabditis elegans are responsible for the translational attenuation that occurs immediately in response to ER stress. As a result, no more proteins enter the ER for a while, allowing metazoan cells to focus on dispatching preexisting unfolded proteins in the ER by refolding them or degrading them.
In contrast, yeast UPR consists of only transcriptional control because yeast cells do not express PERK-like proteins. Thus, proteins continue to be synthesized and delivered into the ER, which is in trouble under ER stress conditions. This might be the reason why yeast UPR target genes account for nearly 400 of the ∼6000 genes present in the yeast genome and include those encoding not only ER chaperones but also numerous proteins working at various stages in the secretory pathway (Travers et al., 2000). Organelles other than the ER must be involved to decrease the amount, concentration, or both of unfolded proteins accumulated in the ER. Recent studies further indicated that intracellular transport from the ER to the Golgi apparatus is actively utilized for ER-associated degradation of several substrate proteins (Vashist et al., 2001; Haynes et al., 2002; Taxis et al., 2002). It is unlikely that a novel trafficking pathway was activated to deal with specific unfolded proteins examined because conventional secretion genes, such as sec12 and sec18, were required for the degradation and unfolded proteins examined were indeed contained in COP-II vesicles. These results indicated that unfolded or misfolded proteins exit the ER in yeast cells in marked contrast to the results shown in this report; in mammalian cells, unfolded proteins are retained in the ER regardless of the presence or absence of ER stress. Yeast cells may have developed a system to counteract proteotoxicity exhibited by unfolded proteins that traverse in the secretory pathway for a while after the exit from the ER.
On the basis of the distinctions between yeast and mammals, we speculate that eukaryotic cells have established the ER-restricted management of unfolded proteins later in evolution with the advent of a PERK-like protein, which is responsible for ER stress-induced translational attenuation as well as with the mechanistic development to select proteins to be exported from the ER. The ER chaperone system may not be sufficient for such selectivity as they are well conserved from yeast to human. In any event, the GFP-ATF6α fusion gene constructed in this study should become a powerful tool for the biochemical and cell biological dissection of molecular events that lead to the activation of ATF6 as well as for understanding the molecular basis of the remarkable ability of the ER to distinguish proteins to be transported from those to be retained in the ER.
Acknowledgments
We are grateful to Drs. J. Lippincott-Schwartz, N. Hosokawa, H.-P. Hauri, and O. Kuge for providing materials. We thank Maki Shibata and Kaoru Miyagawa for technical and secretarial assistance. This work was supported, in part, by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (14037233 and 15GS0310 to K.M.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-09-0693. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-09-0693.
References
- Antonny, B., and Schekman, R. (2001). ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13, 438-443. [DOI] [PubMed] [Google Scholar]
- Bannykh, S.I., and Balch, W.E. (1997). Membrane dynamics at the endoplasmic reticulum-Golgi interface. J. Cell Biol. 138, 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., Rexach, M.F., Ravazzola, M., Amherdt, M., and Schekman, R. (1994). COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895-907. [DOI] [PubMed] [Google Scholar]
- Brown, M.S., Ye, J., Rawson, R.B., and Goldstein, J.L. (2000). Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391-398. [DOI] [PubMed] [Google Scholar]
- Brown, M.S., and Goldstein, J.L. (1999). A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96, 11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon, M., Zeng, H., Urano, F., Till, J.H., Hubbard, S.R., Harding, H.P., Clark, S.G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- Chen, X., Shen, J., and Prywes, R. (2002). The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277, 13045-13052. [DOI] [PubMed] [Google Scholar]
- de Silva, A., Braakman, I., and Helenius, A. (1993). Posttranslational folding of vesicular stomatitis virus G protein in the ER: involvement of noncovalent and covalent complexes. J. Cell Biol. 120, 647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- Gallione, C.J., and Rose, J.K. (1985). A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 54, 374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething, M.J., and Sambrook, J. (1992). Protein folding in the cell. Nature 355, 33-45. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Calfon, M., Urano, F., Novoa, I., and Ron, D. (2002). Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell. Dev. Biol. 18, 575-599. [DOI] [PubMed] [Google Scholar]
- Harding, H.P. et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619-633. [DOI] [PubMed] [Google Scholar]
- Hauri, H.P., Kappeler, F., Andersson, H., and Appenzeller, C. (2000). ER-GIC-53 and traffic in the secretory pathway. J. Cell Sci. 113, 587-596. [DOI] [PubMed] [Google Scholar]
- Haynes, C.M., Caldwell, S., and Cooper, A.A. (2002). An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158, 91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze, K., Okada, T., Yoshida, H., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001). Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355, 19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2001). Intracellular functions of N-linked glycans. Science 291, 2364-2369. [DOI] [PubMed] [Google Scholar]
- Helenius, A., Marquardt, T., and Braakman, I. (1992). The endoplasmic reticulum as a protein folding compartment. Trends Cell Biol. 2, 227-231. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., Wada, I., Hasegawa, K., Yorihuzi, T., Tremblay, L.O., Herscovics, A., and Nagata, K. (2001). A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R.J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211-1233. [DOI] [PubMed] [Google Scholar]
- Kuge, O. et al. (1994). Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 125, 51-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K., and Kaufman, R.J. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K. (2000). Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101, 451-454. [DOI] [PubMed] [Google Scholar]
- Mori, K. (2003). Frame switch splicing and regulated intramembrane proteolysis: key words to understand the unfolded protein response. Traffic 4, 519-528. [DOI] [PubMed] [Google Scholar]
- Nehls, S. et al. (2000). Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2, 288-295. [DOI] [PubMed] [Google Scholar]
- Ng, D.T.W., Watowich, S.S., and Lamb, R.A. (1992). Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol. Biol. Cell 3, 143-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T., Haze, K., Nadanaka, S., Yoshida, H., Seidah, N.G., Hirano, Y., Sato, R., Negishi, M., and Mori, K. (2003). A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 278, 31024-31032. [DOI] [PubMed] [Google Scholar]
- Okada, T., Yoshida, H., Akazawa, R., Negishi, M., and Mori, K. (2002). Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, C., and Walter, P. (2001). Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349-356. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99-111. [DOI] [PubMed] [Google Scholar]
- Taxis, C., Vogel, F., and Wolf, D.H. (2002). ER-Golgi traffic is a prerequisite for efficient ER degradation. Mol. Biol. Cell 13, 1806-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- Vashist, S., Kim, W., Belden, W.J., Spear, E.D., Barlowe, C., and Ng, D.T. (2001). Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 155, 355-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., Rawson, R.B., Komuro, R., Chen, X., Dave, U.P., Prywes, R., Brown, M.S., and Goldstein, J.L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355-1364. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J. Biol. Chem. 273, 33741-33749. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Hosokawa, N., Kaufman, R.J., Nagata, K., and Mori, K. (2003). A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4, 265-271. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001a). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., and Mori, K. (2000). ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20, 6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001b). Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21, 1239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]