Abstract
The Sec1/Munc18 (SM) family of proteins is thought to impart compartmental specificity to vesicle fusion reactions. Here we report characterization of Vps33p, an SM family member previously thought to act exclusively at the vacuolar membrane with the vacuolar syntaxin Vam3p. Vacuolar morphology of vps33Δ cells resembles that of cells lacking both Vam3p and the endosomal syntaxin Pep12p, suggesting that Vps33p may function with these syntaxins at the vacuole and the endosome. Consistent with this, vps33 mutants secrete the Golgi precursor form of the vacuolar hydrolase CPY into the medium. We also demonstrate that Vps33p acts at other steps, for vps33 mutants show severe defects in endocytosis at the late endosome. At the endosome, Vps33p and other class C members exist as a complex with Vps8p, a protein previously known to act in transport between the late Golgi and the endosome. Vps33p also interacts with Pep12p, a known interactor of the SM protein Vps45p. High copy PEP7/VAC1 suppresses vacuolar morphology defects of vps33 mutants. These findings demonstrate that Vps33p functions at multiple trafficking steps and is not limited to action at the vacuolar membrane. This is the first report demonstrating the involvement of a single syntaxin with two SM proteins at the same organelle.
INTRODUCTION
Active sorting of endosomal and vacuolar proteins from secreted and plasma membrane resident proteins occur in the late Golgi of Saccharomyces cerevisiae. A number of vacuolar protease precursors like the Golgi form of carboxypeptidase Y (p2 CPY) enter the vacuole from the late Golgi, transiting through the late endosome (Jones et al., 1997; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001). However, proteins like the alkaline phosphatase precursor (p2 ALP) and Vam3p bypass the endosome and go directly from the Golgi to the vacuole (Cowles et al., 1997; Stepp et al., 1997). Plasma membrane proteins are also targeted to the vacuole in a regulated manner (Jones et al., 1997; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001). All of these targeting steps involve formation of vesicles carrying the appropriate cargo at the donor membrane and targeting of these vesicles to the target organelle. Many protein players have been implicated in vesicle formation, docking, and fusion (Waters and Hughson, 2000). SNARE proteins (soluble N-ethylmaleimide sensitivity factor receptor protein) are present on both the vesicle (v-SNARE) and the target (t-SNARE) membranes (Rothman, 1994; Pelham, 2001). Although appropriate pairing of cognate SNAREs on the vesicle and target membranes is the key event that drives membrane fusion, they are promiscuous in nature (Gotte and von Mollard, 1998). Proteins of the Rab-GTPase family help in appropriate SNARE pairing, but are not solely responsible for this event (Pfeffer, 2001). Specificity of fusion might be a cooperative effect of multiple layers of protein interactions, or might be due to specific regulatory steps leading to specific activation of SNAREs.
Sec1/MUNC18 (SM) family proteins are involved in conferring specificity to these membrane events by binding t-SNAREs of the syntaxin family (Rizo and Sudhof, 2002; Gallwitz and Jahn, 2003; Toonen and Verhage, 2003). In yeast there are four SM family proteins: Sec1p, Sly1p, Vps45p, and Vps33p. Null mutants of each of these exhibit a block in protein traffic at the respective step(s), thus implying that these proteins have a positive role in yeast membrane fusion. Sec1p in yeast has been found to copurify not only with the cognate syntaxin, but also with its SNARE partners or the SNARE complex, implying that yeast SM proteins play a role during the fusion event (Carr et al., 1999). It has been demonstrated that although binding of an SM to its cognate syntaxin does not increase the rate of SNARE pairing, it prevents the syntaxin from promiscuous SNARE binding (Peng and Gallwitz, 2002), suggesting a role in specificity.
Although there are only four SM proteins, there are seven syntaxin homologues encoded in the yeast genome (Bock et al., 2001). All vesicle fusion events require the presence of at least one SM and one syntaxin; therefore, certain SM proteins must interact with more than one syntaxin. The SM protein Vps45p (Cowles et al., 1994) binds with the endosomal syntaxin Pep12p and the syntaxin Tlg2p, which is involved in early stages of cytoplasm-to-vacuole targeting (Cowles et al., 1994; Abeliovich et al., 1999; Bryant and James, 2001). Yeast Sly1p binds the Golgi syntaxin Sed5p and the ER syntaxin Ufe1p (Yamaguchi et al., 2002). However, the key point in support of SM proteins imparting compartmental specificity is that so far, only a single SM has been assigned to each organelle and each fusion step at a given organelle.
In yeast, Vps33p/Pep14p (Banta et al., 1990; Wada et al., 1990) is the SM protein assigned to the vacuole, where it interacts with the vacuolar syntaxin Vam3p (Sato et al., 2000). The mouse mutation buff has been recently localized to the mouse gene Vps33a (Suzuki et al., 2003), which has significant homology to the yeast gene, VPS33, and the Drosophila gene, carnation (Sevrioukov et al., 1999). buff mutants exhibit phenotypes similar to the human Hermansky-Pudlak syndrome (HPS), Chediak-Higashi syndrome, and Griscelli syndrome (Huizing et al., 2002).
If Vps33p were to act only at the vacuole with Vam3p, null mutants of VPS33 and of VAM3 should exhibit the same phenotype. However, vps33Δ cells have no discernable vacuole in the cell (Banta et al., 1990), whereas vam3Δ cells have fragmented vacuolar morphology (Srivastava and Jones, 1998), implying that Vps33p has more diverse functions in the cell than Vam3p. Cells bearing deletions in VPS33, PEP3/VPS18 (Preston et al., 1991), PEP5/VPS11 (Woolford et al., 1990), or VPS16 (Horazdovsky and Emr, 1993) exhibit a “no vacuole” phenotype. All of these genes are categorized as class C VPS genes, based on their mutant vacuolar morphology (Raymond et al., 1992). The protein products of these genes form a hetero-oligomeric complex (Rieder and Emr, 1997; Sato et al., 2000; Seals et al., 2000; Wurmser et al., 2000). It has been shown recently by our laboratory and others that the class C VPS proteins, Pep3p, Pep5p, and Vps16p, function at multiple steps of protein targeting to the yeast vacuole (Srivastava et al., 2000; Peterson and Emr, 2001; Whyte and Munro, 2002). These findings suggested the possibility that Vps33p function is not limited to the vacuolar membrane.
In this study we have investigated the role of Vps33p at different steps of targeting to the vacuole and have studied different protein interactions between Vps33p and proteins involved in trafficking at the yeast endosome. We find that Vps33p acts at the late endosome with the endosomal syntaxin, Pep12p, in addition to functioning with Vam3p at the vacuole. Our results indicate that two SM proteins, namely Vps45p and Vps33p, act with a common syntaxin, Pep12p, at the same organelle.
MATERIALS AND METHODS
Materials, Media, and Strains
Oligonucleotide primers, <30 nucleotides were purchased from One Trick Pony Oligos (Ramona, CA) and 60mer primers were purchased from Invitrogen Life Technologies (Carlsbad, CA). HA and TAP tags were added to genomic copies at the C terminus using 60-nucleotide-long primers as recommended by Longtine et al., (1998) and the Seraphin lab web page, www.embl-heidelberg.de/ExternalInfo/seraphin/TAP.html, respectively. α-factor antibody and Gap1p antibody were kind gifts from Howard Riezman and Bruno Andre, respectively. Pep12p monoclonal antibodies and FM4-64 were purchased from Molecular Probes (Eugene, OR). Pep5p antibody used was that described in Woolford et al. (1990). Cycloheximide and protease inhibitors were purchased from Sigma Chemicals (St. Louis, MO). Dithiobis[succinimidylpropionate] (DSP), was bought from Pierce Chemicals (Rockford, IL). Zymolase 20T was purchased from Seikagaku Corporation (Tokyo, Japan). All immunoblots were prepared using secondary antibodies conjugated to HRP followed by processing the blots for chemiluminescence using the Super Signal West Pico kit from Pierce Chemicals according to the manufacturer's instructions. IgG Sepharose was purchased from Amersham-Pharmacia (Piscataway, NJ); calmodulin beads were bought from Stratagene (La Jolla, CA), and TEV protease was purchased from GIBCO BRL (Gaithersburg, MD).
YEPD, YEPD + 5 mM ZnCl2, synthetic and LB media were prepared as described previously (Srivastava et al., 2000). All yeast strains were derived from X2180–1B as described in Srivastava et al. (2000) (Table 1). Standard genetic and molecular biological techniques were used (Hawthorne and Mortimer, 1960; Sambrook et al., 1989). All plasmids were propagated in LM1035 or DH5α Escherichia coli strain (Table 2).
Table 1.
List of strains
| Strain | Genotype | Source |
|---|---|---|
| BJ 5146 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2 kex2Δ::HIS3-5 | Robert Fuller |
| BJ 5147 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2 KEX2 | Robert Fuller |
| BJ 7962 | MATa pep5Δ::TRP1 trp1-Δ101 ura3-52 leu2-Δ1 his3-Δ200 | Lab strain |
| BJ 7965 | MATα pep5Δ::TRP1 trp1-Δ101 ura3-52 leu2-Δ1 lys2-801 | Lab strain |
| BJ 8920 | MATα his3-Δ200 ura3-52 leu2Δ1 trp1 | Lab strain |
| BJ 8921 | MATα his3-Δ200 ura3-52 leu2Δ1 lys2-801 | Lab strain |
| BJ 8927 | MATα his3-Δ200 ura3-52 leu2Δ1 pth1Δ::HIS3 | Lab strain |
| BJ 8928 | MATa trp1-1 his3-Δ200 ura3-52 leu2Δ1 lys2-801 pep12Δ::LEU2 | Lab strain |
| BJ 8929 | MATa his3-Δ200 leu2-Δ1 ura3-52 trp1 lys2-801 pep12Δ::LEU2 pth1Δ::HIS3 | Lab strain |
| BJ 9680 | MATa his3-Δ200 ura3-52 leu2-Δ1 trp1 vps33Δ::HIS3 | This study |
| BJ 9681 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33Δ::HIS3 | This study |
| BJ 9702 | MATa his3-Δ200 ura3-52 leu2 trp1 VPS8::TAP::URA3 | This study |
| BJ 9832 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-1 | This study |
| BJ 9833 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-3 | This study |
| BJ 9835 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-5 | This study |
| BJ 9863 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-8 | This study |
| BJ 9864 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-14 | This study |
| BJ 9975 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 VPS16::HA::HIS3MX6 | This study |
| BJ 9976 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-801 VPS33::HA::HIS3MX6 | This study |
| BJ 9985 | MATa his3-Δ200 ura3-52 leu2 trp1-1 VPS8::TAP::URA3 VPS33::HA::HIS3MX6 | This study |
| BJ 9986 | MATa his3-Δ200 ura3-52 leu2 trp1-1 VPS8::TAP::URA3 VPS16::HA::HIS3MX6 | This study |
| BJ 10166 | MATα vps27Δ::HIS3 his3-Δ200 ura3-52 leu2Δ1 lys2-801 | This study |
| BJ 10170 | MATα vps27Δ::HIS3 his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-5 | This study |
| BJ 10177 | MATα vps27Δ::HIS3 his3-Δ200 ura3-52 leu2-Δ1 lys2-801 vps33-14 | This study |
Table 2.
List of plasmids
| Plasmid | Insert | Vector |
|---|---|---|
| pBJ 2473 | YEp24 (High copy plasmid) | YEp24 |
| pBJ 4272 | 3.8-kb SalI PEP7 fragment | YEp24-SalI |
| pBJ 4896 | 4.6-kb BamHI/SphI PEP12 fragment | YEp24-BamHI/SphI |
| pBJ 7695 | 3.7-kb SalI/XbaI fragment of VPS33 | pRS315-SalI/XbaI |
| pBJ 8842 | 2.1-kb SmaI/SphI VPS45 fragment | YEp24-SmaI/SphI |
| pBJ 9794 | vps33ts-1-ORF with 280 bp upstream and 208 bp downstream | pRS316/SmaI |
| pBJ 9795 | vps33ts-3-ORF with 280 bp upstream and 208 bp downstream | pRS316/SmaI |
| pBJ 9796 | vps33ts-5-ORF with 280 bp upstream and 208 bp downstream | pRS316/SmaI |
| pBJ 9797 | vps33ts-8-ORF with 280 bp upstream and 208 bp downstream | pRS316/SmaI |
| pBJ 9798 | vps33ts-14-ORF with 280 bp upstream and 208 bp downstream | pRS316/SmaI |
| pBJ 9806 | 3.7-kb SalI/XbaI fragment of vps33-1 | pRS315-SalI/XbaI |
| pBJ 9807 | 3.7-kb SalI/XbaI fragment of vps33-14 | pRS315-SalI/XbaI |
| pBJ 9810 | 3.7-kb SalI/XbaI fragment of vps33-5 | pRS315-SalI/XbaI |
| pBJ 9811 | 3.7-kb SalI/XbaI fragment of vps33-8 | pRS315-SalI/XbaI |
| pBJ 9812 | 3.7-kb SalI/XbaI fragment of vps33-1 | pRS306-SalI/XbaI |
| pBJ 9813 | 3.7-kb SalI/XbaI fragment of vps33-5 | pRS306-SalI/XbaI |
| pBJ 9815 | 3.7-kb SalI/XbaI fragment of vps33-3 | pRS315-SalI/XbaI |
| pBJ 9816 | 3.7-kb SalI/XbaI fragment of vps33-3 | pRS306-SalI/XbaI |
| pBJ 9817 | 3.7-kb SalI/XbaI fragment of vps33-14 | pRS306-SalI/XbaI |
| pBJ 9820 | 3.7-kb SalI/XbaI fragment of vps33-8 | pRS306-SalI/XbaI |
Generation of Temperature-sensitive vps33 Mutants
The VPS33 ORF including 282 base pairs upstream and 206 downstream was PCR amplified (2323 base pairs PCR product) with primers that incorporated ends homologous to 40 bp on either side of the SmaI site in pRS316. PCR was carried out with skewed nucleotide concentrations: 1/10th concentration of either dATP, dCTP, dGTP, or dTTP (Muhlrad et al., 1992). This mutagenized PCR product was cotransformed with SmaI digested pRS316 into a vps33Δ (BJ9680) strain, and transformants were selected on C-ura plates, by virtue of in vivo gap repair. The colonies from the transformation plates were patched on C-ura plates and grown at 30°C for 48 h. At the end of 48 h these patches were replicated onto four YEPD plates that were incubated at 26, 30, 34, and 37°C, respectively, for 48 h and then screened for CPY activity using the APE overlay assay (Jones, 2002). Transformants (n = 2600) were screened in this manner and 5 plasmids were recovered as containing bona fide temperature-sensitive alleles, after shuttling through E. coli. All of the temperature-sensitive alleles were subcloned by gap repair into BJ7695 (wild-type VPS33 in pRS315) cut with SnaBI and SphI to include more upstream and downstream sequences in order to obtain unique enzymes sites for pop-in/pop-out integration (Rothstein, 1991). These alleles were checked for their ability to confer a temperature-sensitive CPY phenotype and further subcloned as SalI/XbaI fragments into pRS306. These plasmids were cut with either SnaBI, SphI, or BspMII, transformed into wild-type yeast (BJ8921), and were selected on C-ura plates for integrants (pop-in). These transformants were then plated onto 5-FOA plates (pop-outs). These pop-outs were screened for CPY activity by the APE assay, and the ones that retained the temperature-sensitive phenotype for CPY activity were used for further analysis of Vps33p function.
Electron Microscopy
Electron microscopy was performed as described in Webb et al. (1997a) and Srivastava et al. (2000).
Kinetic Assay of CPY Processing Using Cycloheximide
Twenty milliliters of wild-type or mutant cells were grown in YEPD to OD600 ∼1.5 at 25°C. Twenty ODs of cells were harvested and resuspended in 20 ml YEPD. Cycloheximide was added to 20 μg/ml from a 10 mg/ml stock solution. The cultures were incubated for 6–9 h at 25°C to allow turnover of internal CPY. Three milliliters of cells were removed for the 0-h time point. The remainder of the cells were washed two times with fresh YEPD, resuspended in 16 ml YEPD, and split into two aliquots. One sample was incubated at 25°C and the other at 37°C. Three milliliters from each was removed after 60- and 120-min incubation. The cells were pelleted and 50 μl of reducing sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 1% SDS, 0.1% bromophenol blue, 1% β-mercaptoethanol) was added to the cells, to represent the internal fraction. 300 μl of 100% TCA, and 30 μl of 10% Triton X-100 was added to the cell-free supernatant followed by incubation on ice for 20 min. After a 15-min spin at 4°C in the microfuge at 10,000 rpm, the pellet was washed two times with 100% ice-cold acetone and air-dried. This pellet representing the external fraction was resuspended in 50 μl RSB made in 4 M urea. After at least 1-h incubation on ice to allow solubilization of the pellet, the samples were boiled for 10 min, and 25-μl volumes of the internal and external fraction were subject to SDS-PAGE, followed by immunoblotting with anti-CPY antibody.
Alpha Factor Immunoblots
Strains were grown and pro-α factor from the external fraction was detected as described in Srivastava et al. (2000).
FM4-64 Staining
Cells were grown to OD600 0.5–0.9 at the permissive temperature of 25°C. Twenty ODs worth of cells for each sample were harvested and either retained at the permissive or shifted to the restrictive temperature of 37°C for 15 min. FM4-64 was added to a final concentration of 20 μM starting from a 16 mM stock, and the cells were incubated for 10 min at the appropriate temperature. Cells were spun at 700 × g for 3 min. A chase was performed with prewarmed YEPD added to make the cell concentration 10 OD/ml. Subsequently, 100 μl of cells were removed at different time points and spun at 700 × g for 3 min. The cells were resuspended in YEPD containing 20 mM NaN3 to ∼3 OD/ml. Seven microliters of cells was placed on a concanavalincoated slide. Cells were analyzed using a fluorescence microscope (Nikon, Melville, NY) equipped with a Hamamatsu black-and-white cooled chargecoupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) using a rhodamine filter at 546 nm. Digital images were acquired in the program Photoshop (Adobe Systems, Mountain View, CA). (Concanavalincoated slides were prepared by placing 5–7 μl of a 1 mg/ml solution of concanavalin A on a slide, spreading it with a cotton swab, and air drying for a few minutes. Results are best if slides are made fresh).
Endocytic Assay for Gap1p Turnover
Media, growth conditions, and processing of cells for assaying Gap1p turnover were as described in Srivastava et al. (2000).
TAP Tag Copurifications
One liter of each strain was grown overnight at 30°C in YEPD to OD600 ∼ 1.0. The cells were harvested and washed with 1 L of ice-cold water followed by a wash in 50 ml lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 1 mM DTT, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, 2.1 μM leupeptin). The cells were resuspended in 10 ml lysis buffer, and an equal volume of chilled glass beads was added. The cells were lysed by vortexing eight times for 30 s in ice. The lysate was centrifuged for 15 min at 6000 × g and the cell debris was discarded. Total protein was estimated using the protein assay from Bio-Rad (Richmond, CA) and an equal amount of protein from each sample was loaded to columns containing 250 μl of IgG-Sepharose beads that were preequilibrated with lysis buffer. After incubation with IgG beads overnight at 4°C, the unbound lysate was allowed to flow-through, followed by five 1-ml washes with lysis buffer. After this, five 2-ml washes were performed with TEV cleavage buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT). The bound beads were incubated with 1 ml TEV cleavage buffer and 150 U of TEV enzyme for 2 h at 4°C in the column. The TEV eluate was collected and loaded to columns containing 250 μl of calmodulin beads prewashed with calmodulin-binding buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 1 mM PMSF, 8.7 mM β-mercaptoethanol, 1 mM Mg-acetate, 1 mM imidazole, 2 mM CaCl2) along with 3 ml calmodulin-binding buffer and 3 μl of 1 M CaCl2 for every 1 ml of TEV eluate. After a 2-h incubation the bound beads were washed five times with 1 ml calmodulin binding buffer. Copuri-fied proteins were eluted first with 200 μl of calmodulin elution buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 1 mM PMSF, 8.7 mM β-mercaptoethanol, 1 mM Mg-acetate, 1 mM imidazole, 2 mM EGTA), followed by incubation with 500 μl of the same buffer. After this 30-min incubation, the 500 μl eluate was collected and a final elution was performed with 200 μl of the elution buffer. Proteins were precipitated with 10% TCA and 0.03% deoxycholate and analyzed by SDS-PAGE, followed by immunoblotting with appropriate antibodies.
Cross-linking and Coimmunoprecipitation
Wild-type and VPS33-HA tagged cells were grown in YEPD at 30°C to OD600 between 0.8 and 1.0. Eighteen ODs of cells were harvested for each sample and resuspended in 8 ml 0.1 M Tris-Cl, pH 9.0, 10 mM DTT and incubated at room temperature for 10 min. Cells were washed with 1 ml spheroplasting buffer (10 mM Tris-Cl, pH 7.0, 1.2 M sorbitol, 5% glucose, 0.5× YEPUAD [1× YEPUAD is YEPD with 40 mg/l each of uracil and adenine]) and then resuspended in 1 ml spheroplasting buffer + 1.2 mg zymolase 20T and incubated at 30°C for 45 min with gentle agitation. These cells were centrifuged at 7000 rpm in a microfuge for 7 min and the supernatant was discarded. The spheroplasts were lysed in 0.2 M sorbitol, pH 7.5, in the presence or absence of 200 μg/ml of the cross-linking agent DSP in DMSO at room temperature for 30 min. Excess DSP was quenched by adding Tris-Cl, pH 7.5, to a final concentration of 50 mM and further incubating for 15 min. NaCl and Triton X-100 were then added to final concentrations of 50 mM and 2%, respectively, and the cross-linked lysates were incubated on ice for 30 min. The lysates were further homogenized using a needle. One milliliter of 5 mg/ml cross-linked lysate was precleared using rabbit anti-mouse IgG-bound beads for 30 min at 4°C. This precleared lysate was bound to 20 μl protein A beads that were prebound to rabbit anti-mouse IgG and α-HA 12CA5 (monoclonal) antibody overnight at 4°C. The bound beads were washed five times with 1 ml wash buffer (50 mM NaCl, 50 mM Tris, pH 7.5, and 2% Triton X-100). The beads were resuspended in 2× USB (8 M urea, 4% SDS, 10% β-mercaptoethanol, 125 mM Tris, pH 6.8), which results in reversal of the cross-links, and subjected to immunoblotting with anti-HA and anti-Pep12p monoclonal antibodies.
RESULTS
vps33Δ Cells Exhibit Severe Morphological Defects Comparable to pep12Δ vam3Δ Cells
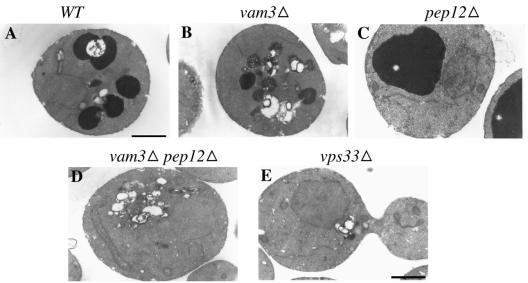
It has been shown previously that strains with deletions in any of the four class C VPS genes, namely PEP3, PEP5, VPS16, and VPS33, exhibit a “vestigial vacuole” or a “no vacuole” phenotype (Raymond et al., 1992). It was reported that pep5Δ (vps11Δ) cells have vacuolar morphologies comparable to pep12Δ vam3Δ cells (Peterson and Emr, 2001). This comparison was based on uptake of CMAC, a vital dye used to label the vacuolar lumen, and the observations were made using fluorescence microscopy. A punctate staining pattern was observed for both pep5Δ cells and pep12Δ vam3Δ cells. We wanted to determine the nature of these punctate dots and whether differences in vacuolar morphology are observed between pep12Δ vam3Δ cells and vps33Δ cells (a class C mutant) at the electron microscopic level. A similarity in the morphology of vps33Δ cells to pep12Δ vam3Δ cells would suggest that Vps33p might be functioning at the endosome with Pep12p, in addition to its function at the vacuolar membrane with Vam3p.
Wild-type, pep12Δ, vam3Δ, pep12Δ vam3Δ, and vps33Δ cells were grown and processed for electron microscopic observations of the vacuole as described in Srivastava et al. (2000). Using this procedure for vacuolar staining, vacuoles appear as darkly stained structures, presumably due to polyphosphates in the vacuolar lumen, when viewed under the electron microscope. As expected, wild-type cells showed normal vacuolar morphology (Figure 1A), pep12Δ cells exhibited a single large vacuole phenotype (class D phenotype; Figure 1C) and vam3Δ cells showed fragmented vacuoles (class B phenotype; Figure 1B). Neither pep12Δ vam3Δ cells nor vps33Δ cells displayed any discernible vacuolar structure in the cell (class C phenotype; Figure 1, D and E). However, both strains seemed to accumulate a number of unstained lipid-like structures as small clusters or large clusters (Figure 1, D and E). The number of these clusters, both small and large, was very similar in both strains. Both pep12Δ vam3Δ cells and vps33Δ cells showed a temperature-sensitive growth phenotype (unpublished data). The similarity in the phenotype of pep12Δ vam3Δ and vps33Δ cells suggested that Vps33p might function not only with Vam3p at the vacuolar membrane but also with Pep12p at the endosomal membrane.
Figure 1.
Vacuolar morphology of wild type and mutants. (A) Wild type (BJ 8921), (B) vam3Δ (BJ 8927), (C) pep12Δ (BJ 8928), (D) pep12Δ vam3Δ (BJ8929), and (E) vps33Δ (BJ9680) were grown to log phase at 30°C, fixed for 2 h, and spheroplasted. The spheroplasts were processed for vacuolar lumen staining and viewed under the electron microscope as described in Srivastava et al. (2000). Bar, 1 μM.
vps33 Mutants Secrete the Golgi Precursor Form of CPY
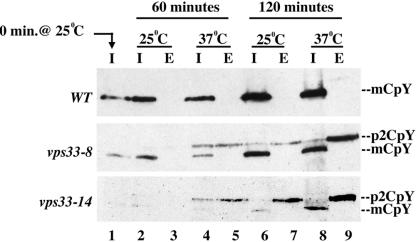
To study the role of Vps33p in different protein trafficking steps, we generated temperature-sensitive alleles of VPS33 by random PCR mutagenesis using skewed nucleotide concentrations (see MATERIALS AND METHODS). Mutant alleles were identified by assaying for temperature-sensitive production of the vacuolar hydrolase CPY by a plate assay (unpublished data). These alleles were integrated into the genome of wild-type yeast cells at the VPS33 locus. The mutant strains ranged from mild to very severe in their CPY activity phenotype. We used these mutants to assay whether the loss of CPY activity in these cells was due to the accumulation of p2 CPY inside the cell, indicative of a block only at the vacuolar membrane, or due to the complete loss of p2 CPY from the cell into the growth medium, a classic phenotype of cells blocked in transport between the Golgi and the endosome.
To investigate the fate of newly synthesized CPY in vps33 mutants, we added cycloheximide at 20 μg/ml to actively growing cells for 6–9 h, which resulted in a near complete loss of CPY in cells (lane 1 in Figure 2). At this point cells were collected and resuspended in fresh medium lacking cycloheximide to allow for new synthesis of CPY at 25 or 37°C for 60 or 120 min. Intracellular and extracellular levels of CPY were checked by Western blotting of cell extracts. Wild-type cell extracts showed increasing amounts of mature intracellular CPY with time, and no CPY was detected in the external fraction (Figure 2). However, the vps33ts mutant cells secreted a significant amount of p2 CPY into the extracellular fraction. Extracts from two representative mutant strains are shown in Figure 2. The vps33-8 strain represents a milder mutant (Figure 2), whereas the vps33-14 strain represents a stronger mutant that displays the CPY secretion phenotype at both permissive and restrictive temperatures (Figure 2). A similar p2 CPY secretion phenotype is exhibited by vps45 mutants when CPY maturation is assayed by metabolic labeling followed by immunoprecipitation with anti-CPY antibody (Cowles et al., 1994; Bryant et al., 1998). VPS45 encodes the SM homologue that acts with the endosomal syntaxin Pep12p. However, this is distinct from the intracellular accumulation of p2 CPY seen in vam3 cells (Srivastava and Jones, 1998). These results suggested that Vps33p function might be required at the late endosome. They also indicate that two SMs might be acting at the same organelle, presumably with the same syntaxin.
Figure 2.
Kinetic analysis of CPY processing. (A) Wild type (BJ 8921), (B) vps33-8 (BJ 9863), and (C) vps33-14 (BJ 9864) were grown to midlog phase at 25°C and incubated with cycloheximide for 6–9 h. After turnover of internal CPY, cells were collected and resuspended in fresh medium, and the samples were split in half and incubated at 25°C or 37°C for 2 h. Newly synthesized CPY in the internal (I) and External (E) fraction was extracted at 60- and 120-min time points and analyzed using SDS-PAGE and immunoblotting. p2 CPY is the Golgi form of CPY and mCPY is the mature vacuolar form of CPY
vps33 Cells Secrete Hyperglycosylated pro-α Factor into the Medium
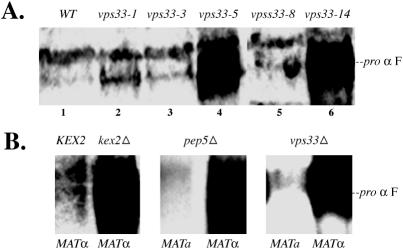
Kex2p is a protease required for the maturation of pro-α factor in the late Golgi. Kex2p constantly cycles between the late Golgi and the endosome (Fuller et al., 1988, 1989; Wilsbach and Payne, 1993). A defect in the retrieval of Kex2p back to the late Golgi results in the secretion of hyperglycosylated pro-α factor into the culture medium This defect in retrieval could be due to a block in transport from the endosome to the Golgi (Wilsbach and Payne, 1993) or due to mislocalization of Golgi resident proteins to the cell surface due to a block in transport between the Golgi and the endosome. Earlier studies in our laboratory have shown that pep3 and pep5 mutants exhibit severe defects in maturation of pro-α factor (Srivastava et al., 2000). We investigated the role of Vps33p in retrieval of Kex2p to the late Golgi.
MATα kex2Δ, pep5Δ, and vps33Δ cells secreted copious amounts of hyperglycosylated pro-α factor into the medium (Figure 3B). MATa pep5Δ and vps33Δ cells were used as controls, and these cells did not show any pro-α factor in the medium (Figure 3B). Only the two strong vps33ts mutants, namely vps33-5 and vps33-14, secreted hyperglycosylated pro-α factor into the medium, which appears as a large smear in immunoblots (Figure 3A, lanes 4 and 6), over the background levels seen as a single band in wild-type cells. This phenotype was present at the semipermissive temperature of 30°C, but there was no trace of pro-α factor outside the cells at the restrictive temperature of 35°C (unpublished data). None of the other temperature-sensitive vps33 mutants tested secreted hyperglycosylated pro-α factor into the medium at either the permissive (Figure 3A, lanes 2, 3, and 5), or the restrictive temperature (unpublished data) when compared with background levels in the wild-type cells. These results are compatible with the observation that at the semipermissive temperature the strong mutants have a defect in transport to the endosome from the Golgi and therefore Kex2p might be mislocalized to the cell surface. At the nonpermissive temperature, although there is a block in trafficking to the endosome from the Golgi, there might be sufficient retrieval of Kex2p to facilitate processing of pro-α factor due to higher rate of overall transport at elevated temperatures. We cannot rule out the possibility of a block in transport from the endosome to the Golgi at the permissive temperature, which results in lower levels of Kex2p in the Golgi and therefore a defect in processing of pro-α factor.
Figure 3.
pro-α factor secretion assay. (A) Lane 1: Wild-type (BJ 8921); lane 2, vps33-1 (BJ9832); lane 3, vps33-3 (BJ9833); lane 4, vps33-5 (BJ 9835); lane 5, vps33-8 (BJ 9863), and lane 6, vps33-14 (BJ 9864) were grown to late log phase (OD600 ∼3–6) at 30°C and total protein from the cell free supernatant was extracted as described elsewhere (Srivastava et al., 2000). (B) As controls, lanes 1, MATα KEX2 (BJ 5147) and 2, MATα kex2Δ (BJ 5146) were processed as described in A and assayed for their capacity to secrete pro-α factor into the medium. Null mutants, MATa pep5Δ (BJ 7962, lane 3) and MATα pep5Δ (BJ 7965, lane 4), and MATa vps33Δ (BJ 9680, lane 5) and MATα vps33Δ (BJ 9681, lane 6) were also assayed in a similar manner.
vps33 Cells Exhibit Severe Defects in Endocytosis
We wanted to determine if Vps33p function is required for fusion of vesicles carrying endocytic cargo with the late endosome. This was particularly interesting to us, because it is known that functions of the endosomal syntaxin Pep12p (Gerrard et al., 2000b), the rab GTPase Vps21p (Gerrard et al., 2000a), and the FYVE domain protein Pep7p are required for endocytic trafficking into the late endosome (Webb et al., 1997b). However, the function of the SM protein Vps45p is not required at this step (Bryant et al., 1998). vps45 mutants are able to degrade endocytic markers with wild-type kinetics (Bryant et al., 1998). Therefore Vps33p is a very plausible candidate to be acting between the early and late endosome because there are no other SM proteins known to be involved at this step.
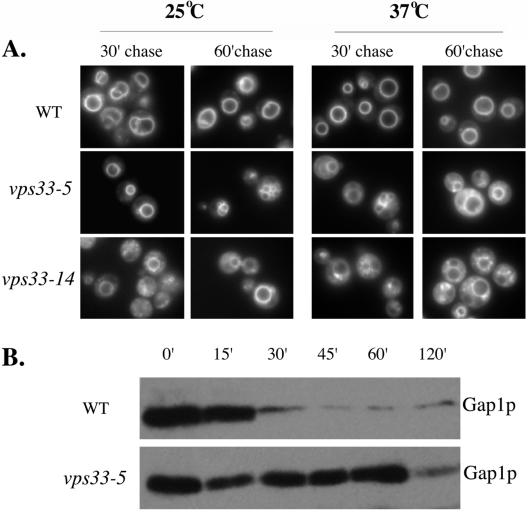
We assayed for endocytic trafficking using two independent assays. First, we assayed for the uptake of FM4-64 into wild-type and vps33 mutant cells. The advantage of using FM4-64 for assaying endocytic uptake is that it is a lipophilic dye that fluoresces only when inserted into a membrane (www.probes.com). Cells take up FM4-64 from the plasma membrane to the vacuolar membrane in a time-, temperature-, and energy-dependent manner via the endosomal compartments (Vida and Emr, 1995). We added FM4-64 to actively growing cells that were preincubated at either the permissive or the restrictive temperature and then performed the uptake assay. We observed in wild-type cells, at both temperatures tested, that the dye stained the vacuolar membrane within 30 min, with little or no extravacuolar diffuse staining (Figure 4A, top panel). vps33 mutants showed vacuolar membrane staining with some amount of extravacuolar diffuse staining after a 60-min chase period at the permissive temperature. However, at the nonpermissive temperature, there was a large amount of diffuse and punctate staining in all the mutants analyzed after 30 and 60 min of chase (Figure 4A). Because this dye is lipophilic and does not fluoresce in aqueous conditions, the extravacuolar diffuse staining is due to some lipid moiety. It is likely that this diffuse fluorescence is attributable to small vesicles presumably derived from the early endosome that are unable to fuse with the late endosomal compartment, and the punctate dots are late endosomal structures.
Figure 4.
Endocytosis assay. (A) Endocytic uptake of the lipophilic dye FM4-64. Wild-type (BJ 8921), vps33-5 (BJ 9835), vps33-8 (BJ 9863), and vps33-14 (BJ 9864) were grown to early log phase at 25°C and preincubated at 25°C or 37°C for 15 min. FM4-64 was added to 20 μM and incubated for 10 min. Excess FM4-64 was washed out and cells were incubated in fresh medium for 30 and 60 min. The cells were washed and placed on precoated slides and viewed under a fluorescence microscope with the rhodamine filter set. (B) Endocytic uptake of the plasma membrane marker, Gap1p. Cells were grown at the semipermissive temperature 30°C in minimal medium containing proline as the sole source of nitrogen. Cells were harvested and resuspended at 2 OD/ml and ammonium sulfate was added to the medium to a final concentration of 10 mM. One-milliliter aliquots were taken at the indicated time points and total protein was extracted as described in Srivastava et al. (2000). Samples were subject to SDS-PAGE followed by immunoblotting using anti-Gap1p antibody.
Second, we took advantage of the turnover of the plasma membrane marker, Gap1p, to look at endocytosis in vps33 mutants. Gap1p is synthesized and localized to the cell surface when cells are grown in a poor nitrogen source such as proline (Jauniaux and Grenson, 1990; Stanbrough and Magasanik, 1995; Hein and Andre, 1997). On shifting to a rich nitrogen source such as ammonium sulfate, Gap1p is ubiquitinated and reaches the vacuole via the endocytic pathway, where it is degraded (Springael and Andre, 1998). Any defect in the endocytic pathway leads to a loss of degradation of Gap1p upon addition of a rich source of nitrogen. Cells were grown in proline as the sole source of nitrogen at the semipermissive temperature. After addition of ammonium sulfate to the medium, total protein was extracted from equal numbers of cells and assayed for degradation of Gap1p by immunoblots. In wild-type cells we observed a large decrease in the amount of Gap1p within 30 min after ammonium sulfate addition (Figure 4B, top panel). In the vps33-5 mutant, however, the levels of Gap1p at the 0- and 60-min time points were comparable (Figure 4B, bottom panel). It was not until 120 min of chase that the mutant cells showed any decrease in the amount of Gap1p in the cells (Figure 4). The vps33-5 mutation resulted in stabilization of Gap1p, presumably by blocking transit of Gap1p to the late endosome.
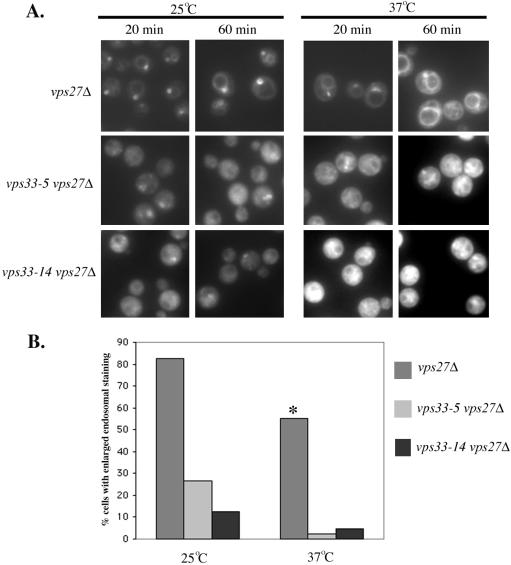
To address this issue more directly and to find out if the block in endocytic transport was occurring at the endosome, we deleted the wild-type copy of VPS27 in vps33 mutants. vps27Δ cells show a defect in traffic out of the late endosome, both in retrograde traffic to the late Golgi as well as forward transport to the vacuole (Piper et al., 1995). As a result, these cells exhibit an enlarged late endosome/PVC (prevacuolar compartment) phenotype. More than 80% of vps27Δ cells at 25°C accumulated FM4-64 in an enlarged endosomal compartment as early as 20 min into the chase (Figure 5, A and B) and a similar number of them showed staining after 60 min of chase (Figure 5A). At 37°C, presumably because of increased trafficking at higher temperature, some of the dye had moved from the endosomal membrane (20 min) to the vacuolar membrane (60 min). After 20 min of chase at 37°C only 55% of vps27Δ cells showed a distinct enlarged endosomal staining and a significant number of cells showed vacuolar membrane staining (Figure 5, A and B). However, we observed a different profile in vps27Δ vps33ts cells compared with vps27Δ cells. In vps27Δ vps33-5 and vps27Δ vps33-14 cells, we observed a more diffuse labeling pattern at both the permissive and nonpermissive temperatures at both 20 and 60 min of chase (Figure 5). After 20 min, 25% of vps27Δ vps33-5 cells and 13% of vps27Δ vps33-14 cells at 25°C showed signal in the enlarged endosome but <5% of these cells at 37°C showed any staining of an endosome-like compartment (or beyond) at either chase time point (Figure 5B). This indicates that even at the permissive temperature these cells are either not efficient in forming an enlarged endosome as seen in the vps27Δ cells or that trafficking to this endosomal compartment is compromised. As expected, this block in trafficking to the late endosome is more pronounced at the nonpermissive temperature. All these results imply that vps33 mutants are unable to deliver endocytic cargo to the endosomal compartment.
Figure 5.
Endocytosis in a vps27Δ background. (A) vps27Δ (BJ 10166), vps27Δ vps33-5 (BJ 10170), and vps27Δ vps33-14 (BJ 10177) cells were grown to early log phase at 25°C and preincubated at 25°C or 37°C for 15 min. FM4-64 was added to 20 μM and incubated for 10 min. Excess FM4-64 was washed out and cells were incubated in fresh medium for 20 and 60 min. The cells were washed and placed on precoated slides and viewed under a fluorescence microscope with the rhodamine filter set. (B) At 25°C, after 20 min of chase, vps27Δ cells (n = 55), vps27Δ vps33-5 cells (n = 43), and vps27Δ vps33-14 cells (n = 51) were scored for accumulation of FM4-64 in an enlarged endosomal compartment. At 37°C, after 20 min of chase, cells of vps27Δ (n = 48), vps27Δ vps33-5 (n = 44), and vps27Δ vps33-14 (n = 42) strain background were scored. From these counts, the percentage of cells showing the presence of the dye in an enlarged endosome was calculated and plotted as a bar graph. Asterisk on vps27Δ bar at 37°C indicates that although the percentage of cells showing endosomal staining is only 55%, many of the others showed vacuolar membrane staining as a result of the dye having been transported from the enlarged endosomal membrane (at an earlier time point) to the vacuolar membrane.
Vps33p and Other Class C Proteins Physically Interact with Vps8p
If Vps33p function is required for trafficking between the Golgi and the endosome, it might interact and function with proteins that are required for trafficking at this step. We chose to investigate Vps8p for interaction with Vps33p for two reasons. First, Vps8p function is required for anterograde and possibly retrograde trafficking between the Golgi and the endosome, and it has been shown to be required for the functionality of Vps21p, the Rab GTPase involved at this step (Chen and Stevens, 1996; Horazdovsky et al., 1996). Second, vps8-200 was isolated as a suppressor of the mutant class C PEP5 allele, pep5::TRP1 (Woolford et al., 1998).
We looked for interactions between Vps8p and the other class C VPS proteins Pep3p, Pep5p, and Vps16p as well, because the functions of these three proteins are required in trafficking between the late Golgi and the endosome in both anterograde and retrograde directions (Srivastava et al., 2000; Peterson and Emr, 2001). In this experiment we assayed for copurification of Vps33p and other class C proteins with Vps8p.
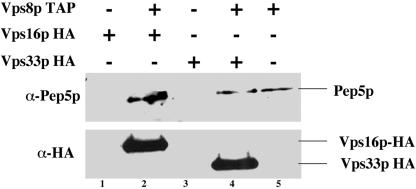
We tagged VPS8 with the TAP (tandem affinity purification) tag, which has a protein-A domain, a TEV protease cleavage site, and calmodulin-binding peptide (CBP domain; Rigaut et al., 1999), in strains that either had a VPS33-HA or a VPS16-HA epitope-tagged gene. All tags were inserted at the C terminus of the genomic copy so that the proteins are expressed under their normal promoters. We used VPS33-HA– or VPS16-HA–tagged strains without any TAP tag as our negative control. Using the two-step purification protocol with Vps8p-TAP tag, we pulled down Pep5p (Figure 6, lanes 2, 4, and 5), Vps33p-HA (Figure 6, lane 4), and Vps16p-HA (Figure 6, lane 2), as detected by immunoblots. In addition to Western blotting, mass spectrometric analysis of proteins copurifying with Vps8-TAP tag revealed its interaction only with the class C members namely, Pep3p, Pep5p, Vps16p, and Vps33p (unpublished data); no other protein was pulled down. More importantly, when Pep3p was TAP tagged and copurifications were performed, in addition to Vps8p and other class C proteins, we also pulled down Vps41p and Vam6p/Vps39p (detected by mass spectrometry; unpublished data). Vps41p and Vam6p/Vps39p are constituents of the HOPS complex involved exclusively in fusion at the vacuolar membrane (Price et al., 2000; Wurmser et al., 2000). These interactions are further corroborated by the interaction data available for the whole genome analysis (Gavin et al., 2002) at the Saccharomyces Genome Database. This indicates that Vps33p and other class C proteins might be present in at least two distinct complexes in the cell, one complex that includes Vps41p and Vam6p/Vps39p is required for vacuolar trafficking and the other complex that contains Vps8p functions in endosomal protein transport. However, we were unable to isolate such distinct protein complexes containing either Vps8p or Vam6p and Vps41p along with class C complex using sucrose density gradient sedimentation (unpublished data). This interaction of Vps8p with Vps33p implicates Vps33p in function at the late endosome in addition to its previous function at the vacuole.
Figure 6.
Vps33p, Vps16p, and Pep5p physically interact with Vps8p. One liter each Vps16p-HA (BJ9975), lane 1; Vps16p-HA Vps8p-TAP tag (BJ9986), lane 2; Vps33p-HA (BJ9976), lane 3; Vps33p-HA Vps8p-TAP tag (BJ 9985), lane 4; and Vps8p-TAP tag (BJ9702), lane 5 were grown to OD600 ∼1.0 at 30°C. The cells were harvested and lysed and lysates were adjusted to 6 mg/ml. An equal volume of lysates was first bound to IgG-Sepharose columns, followed by TEV protease cleavage. TEV eluates containing the copurifying complex were further purified using calmodulin columns. The eluates were TCA precipitated, subjected to SDS-PAGE, and immunoblotted with anti-Pep5p and anti-HA antibody.
Vps33p Physically Interacts with Endosomal Syntaxin, Pep12p
If Vps33p function is required at the late endosome, it is likely that it mediates its function by interacting with a syntaxin-like protein, because it is known that all SM proteins function with syntaxins on target membranes that receive donor vesicles (Gallwitz and Jahn, 2003; Toonen and Verhage, 2003). Because Pep12p is the only known syntaxin present in the late endosomal membrane of yeast (Becherer et al., 1996), we asked whether Vps33p interacts with Pep12p.
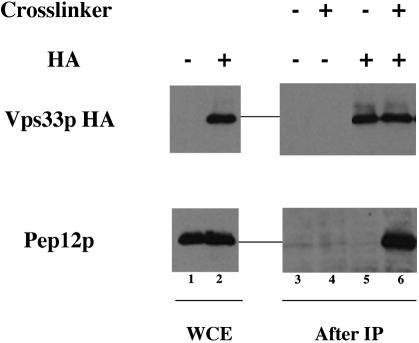
To look for a physical interaction between Vps33p and Pep12p, we lysed an untagged strain or a strain in which Vps33p-HA is the only copy of Vps33p in the presence or absence of 200 μg/ml the cross-linking agent DSP. These cross-linked lysates were subjected to immunoprecipitation with anti-HA mAb. The immunoprecipitates were analyzed by SDS-PAGE and immunoblotting after removal of the cross-links. Pep12p coprecipitation was observed only in the presence of both the HA tag on Vps33p and the cross-linker (Figure 7). This indicates that there is a transient or lowaffinity interaction between Pep12p and Vps33p in vivo. Quantitation of the immunoblots revealed that <10% of total cellular Pep12p is bound to Vps33p-HA under these experimental conditions.
Figure 7.
Syntaxin, Pep12p and SM, Vps33p physically interact. Untagged (BJ8921) and Vps33p-HA (BJ 9976) strains were grown at 30°C, spheroplasted, and lysed in the presence of 200 μg/ml cross-linking agent, DSP. The cross-linked lysates were subject to immunoprecipitation with monoclonal anti-HA antibody bound to rabbit anti-mouse IgG-coated protein A-Sepharose beads overnight at 4°C. After washing the beads thoroughly, Pep12p was detected with anti-Pep12p antibody by SDS-PAGE and immunoblots of reduced coimmunoprecipitates. Lanes 1 and 2 are whole cell extracts of BJ 8921 and BJ 9976, respectively. Lanes 3 and 4 are BJ 8921 without and with DSP, respectively after coprecipitation, and lanes 5 and 6 are BJ 9976 without and with DSP, respectively, after coprecipitation.
However, this result was unexpected because the wellcharacterized SM protein Vps45p has been shown to physically interact with Pep12p and is involved in targeting and fusion of Golgi derived vesicles on the late endosomal membrane (Burd et al., 1997). Also, to date, although there have been quite a few examples of one SM protein interacting with multiple syntaxins, there is no evidence for the same syntaxin interacting with more than one SM protein. At the same time, there is no instance in the literature where there are two SM proteins on the same organelle. The evidence is clear, however, that the SM proteins Vps45p and Vps33p both interact with Pep12p. Thus, this is a novel finding that two SMs can act with the same syntaxin on the same organelle.
PEP7 Genetically Interacts with vps33 and Alleviates Some Vacuolar Defects of vps33 Cells
Pep7p is a FYVE domain protein that mediates phosphatidyl inositol-3-phosphate (PI-3-P) signaling on the late endosomal membrane (Burd et al., 1997; Webb et al., 1997b; Burd and Emr, 1998). Pep7p genetically and physically interacts with the endosomal syntaxin Pep12p (Webb et al., 1997b; Burd and Emr, 1998), the GTP-bound form of the endosomal rab GTPase Vps21p (Tall et al., 1999) and the endosomal SM protein Vps45p (Webb et al., 1997b; Peterson et al., 1999). By means of these interactions, Pep7p integrates PI-3-P and GTPase regulatory signals on the endosomal membrane. pep7 mutants are blocked in transport of both biosynthetic and endocytic cargo to the endosomal membrane (Webb et al., 1997b). Because we have shown that vps33 mutants display a severe block in transport at the endosome, we wanted to determine if there was an interaction between Pep7p and Vps33p.
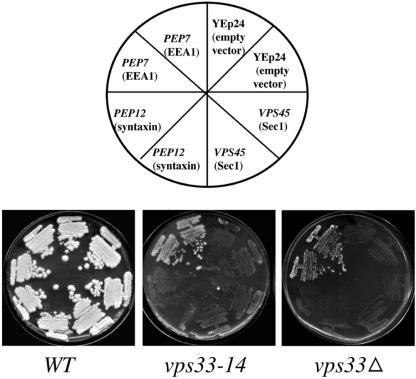
To address this issue we chose to take a genetic approach. Our laboratory has shown previously that the presence of high copy VPS45 overcomes some vacuolar defects of pep7 mutants (Webb et al., 1997a). It is known that cells that do not have a functional vacuole are unable to grow in medium containing divalent cations like Zn2+ or Sr2+. vps33 mutants fail to grow in medium containing 5 mM ZnCl2. We looked for rescue of this phenotype in vps33-14 and vps33Δ cells transformed with high copy PEP7, VPS45, or PEP12. High copy PEP7 restored growth of vps33-14 and vps33Δ strains on 5 mM ZnCl2 containing plates, but high copy VPS45 and PEP12 did not (Figure 8). The inability of Vps45p to replace the function of Vps33p suggests that these two SM proteins might be acting independently of each other. More importantly, we also found that presence of high copy VPS33 did not alleviate the mutant phenotypes of a vps45Δ strain, namely inability to produce mature CPY and lack of growth in medium containing Zn2+ or Sr2+ (unpublished data). Thus, the cell is unable to use the two SMs interchangeably and the functions of both the proteins are indispensable for proper protein trafficking to the vacuole via the late endosome.
Figure 8.
High copy PEP7 alleviates vacuolar defects of vps33Δ and vps33-14 cells. Wild type (8921), vps33-14 (BJ 9864), and vps33Δ (BJ 9680) were transformed with high copy empty vector, YEp24 (pBJ2473), PEP7 (pBJ4272), PEP12 (pBJ4896), or VPS45 (pBJ8842). Two transformants for each strain were streaked for singles onto c-ura plates. After growth for 3 days, they were replica-plated to YPD + 5 mM ZnCl2 plates and scored for growth at 30°C after incubation for 5–7 days.
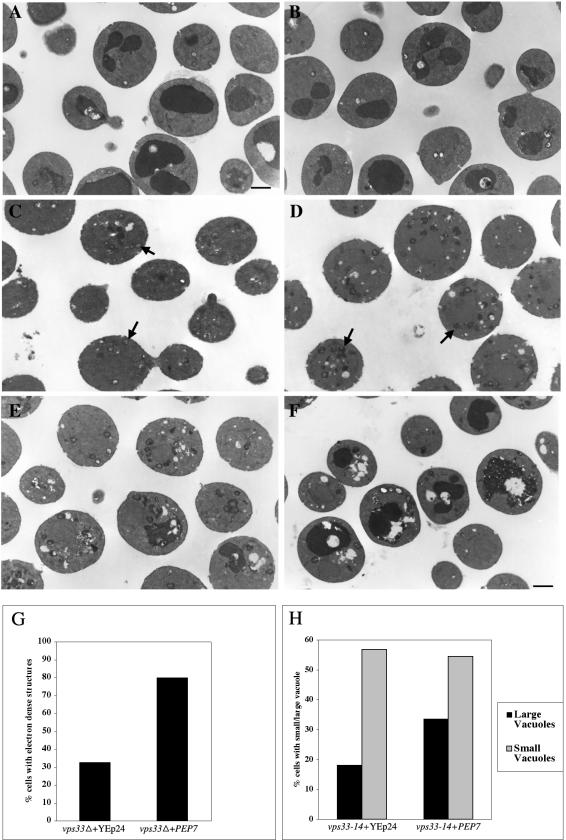
vps33Δ cells have a vestigial vacuole phenotype. Because high copy PEP7 restores the growth of vps33Δ on plates containing divalent cations, we suspected that these cells might have regained some vacuole-like structures, because cations are stored in the vacuole. To investigate this, we performed electron microscopic analysis on vps33Δ and vps33-14 cells transformed with a high copy empty vector or high copy PEP7. Wild-type cells showed identical vacuolar morphology with both YEp24 and high copy PEP7 plasmids (Figure 9, A and B). Strikingly 80% of vps33Δ cells transformed with PEP7 showed electron-dense structures (Figure 9, D and G). In contrast only 32% of vps33Δ cells transformed with the empty vector showed electron-dense structures and these were fewer in number per cell and also had much weaker staining (Figure 9, C and G). It is possible that the structures observed in vps33Δ cells transformed with YEp24 cells are not bona fide vacuoles or vestigial vacuoles. However, the electron-dense nature and abundance of these structures in vps33Δ cells transformed with PEP7 strongly argues that these cells have regained some fragmented vacuole like structures, reminiscent of the class B phenotype (Raymond et al., 1992).
Figure 9.
vps33Δ and vps33-14 cells regain vacuoles in the presence of high copy PEP7. (A) Wild type (BJ 8921)+YEp24, (B) wild type (BJ 8921)+high copy PEP7, (C) vps33Δ (BJ 9680) +YEp24, (D) vps33Δ (BJ 9680) + high copy PEP7, (E) vps33-14 (BJ 9864)+YEp24, and (F) vps33-14 (BJ 9864)+ high copy PEP7 were grown at 30°C and processed for analyzing vacuolar morphology as described elsewhere (Srivastava et al., 2000). Bar, 1 μM. The arrow in C points toward weakly stained “fragmented vacuoles.” The arrow in D points at more darkly stained electron-dense bona fide fragmented vacuoles. (G) The percentage of vps33Δ (BJ 9680) cells that carried YEp24 (n = 98) or high copy PEP7 (n = 135) that displayed electron-dense fragmented vacuoles were calculated and plotted. (H) The percentage of vps33-14 (BJ 9864) cells with either YEp24 (n = 171) or high copy PEP7 (n = 202) that displayed electron-dense smaller fragmented vacuoles and larger vacuoles were calculated and plotted.
Similar results were obtained when vps33-14 cells transformed with empty vector or high copy PEP7 were analyzed using electron microscopy. Eighteen percent of the cells with empty vector showed large electron-dense structures (Figure 9, E and H), whereas 33% of the cells with high copy PEP7 showed similar structures (Figure 9, F and H). However, the number of small electron-dense structures was almost the same in vps33-14 cells that were transformed with empty vector or high copy PEP7 (Figure 9, E, F, and H). Taken together, these results indicate that the cells with high copy PEP7 have reformed some vacuolar structures. The presence of high copy PEP7 mitigates the vacuolar defects of vps33 cells, possibly by means of routing more traffic from the endocytic pathway to the late endosome.
DISCUSSION
Our laboratory had previously shown that two class C Vps proteins, Pep3p and Pep5p, are not limited to functioning at the vacuolar membrane, but are also involved in trafficking from the Golgi to the late endosome and from the endocytotic pathway to the late endosome and might also be involved in retrograde traffic from the endosome to the late Golgi (Srivastava et al., 2000). Since then, it has been shown that the class C protein Vps16p is also involved in trafficking at the late endosome (Peterson and Emr, 2001). However, the role of the class C Vps protein Vps33p at steps other than the vacuolar membrane was unclear, because Vps33p belongs to the SM family of proteins, which has been shown to confer specificity in vesicle targeting and fusion. The results presented here clearly show that Vps33p function is required not only at the vacuolar membrane but also at the endosomal membrane. Our results indicate that two SM proteins, Vps45p and Vps33p, each act with the syntaxin Pep12p of the endosome and that there can be two SMs functioning on the same organelle. Our results therefore bring into question the role of SM proteins in determining compartmental specificity.
Secretion of p2 CPY in vps33 mutants (Figure 2) can result from a defect in trafficking of Golgi-derived vesicles to the late endosome. However this phenotype might also be a result of a defect in proper localization of the CPY receptor, Vps10p (Marcusson et al., 1994; Cooper and Stevens, 1996), due to a retrograde block in transport between the Golgi and the endosome. The hyperglycosylated pro-α factor secretion phenotype in the strong vps33 mutants suggests that the Golgi protease Kex2p is mislocalized, presumably to the cell surface, because of the inability of Golgi-derived vesicles to fuse with the endosomes (Figure 3). Again, this phenotype might also be a result of improper recycling of proteins, including Kex2p, from the endosome to the Golgi. The physical interaction of Vps8p with Vps33p (Figure 6) and other members of the class C family along with the phenotypes exhibited by vps33 mutants keeps the possibility open that Vps33p acts in both directions between the Golgi and the endosome.
Our results indicate that there are severe defects in endocytosis in vps33 mutants. Using FM4-64 as a marker for endocytosis, we were able to demonstrate both a kinetic delay in delivery of FM4-64 to the vacuole as well as accumulation of the dye both in punctate structures, which would include early endosomal structures, and small diffuse staining vesicles (Figure 4A). Using vps27Δ vps33 double mutants, we have demonstrated that a functional copy of VPS33 is required for trafficking to the late endosome/PVC from the plasma membrane (Figure 5, A and B). The vps33 mutants are also extremely delayed in endocytosis and turnover of Gap1p triggered by addition of ammonium sulfate to cells growing on proline (Figure 4B). Pep12p, Vps21p, and Pep7p have been shown to be involved in both the biosynthetic and endocytic pathways at the late endosome. Vps45p is exclusively involved in the biosynthetic route. This suggests that Vps33p might be the factor on the late endosome that distinguishes endocytic vesicles from Golgi-derived vesicles. Genetic interaction between PEP7 and mutant alleles of vps33 is another indication that Vps33p acts at the late endosome and links PI-3-P signaling to endocytic trafficking on the late endosome (Figures 8 and 9). Alleviation of vacuolar morphology phenotypes of vps33 mutants in the presence of high copy PEP7 suggests that in these suppressed cells there might be more trafficking through the endosome and this somehow leads to formation of vacuole-like structures. However, it is unclear what the molecular nature of this suppression might be. Action of Vps33p at the endosome is further indicated by its specific interaction with endosomal proteins Vps8p (Figure 6) and Pep12p (Figure 7).
Most importantly, addition of excess Vps45p in vps33 mutants does not mitigate vacuolar defects (Figure 8) and presence of excess Vps33p does not relieve the vps45Δ strain of its mutant phenotypes. This has also been shown in mammalian cells, where it was demonstrated that SM proteins could not be used interchangeably (Toonen and Verhage, 2003). This implies that Vps33p and Vps45p might be acting independently of each other and have indispensable roles in fusion at the Pep12p-containing compartment. This result argues that there is some specificity imparted by SM family proteins.
On the basis of our results, we propose that both Vps45p and Vps33p are involved in fusion of Golgi-derived vesicles on the late endosomal membrane and that both these proteins might be present in a single large complex that includes members of the class C complex (Pep3p, Pep5p, Vps16p, and Vps33p) in addition to Vps8p, Pep12p, Pep7p, and Vps21p. In the case of vesicles that originate from the endocytic route, the class C complex, Pep12p, Pep7p, and Vps21p, are involved. In yeast, no stable complex containing any of the class C members or Vps8p, and Vps45p could be isolated (Subramanian, S. and Jones, E.W., unpublished observations). However, in a recent article, studies with mammalian Pep3p/Vps18p revealed a physical interaction between Pep3p and Vps45p (Richardson et al., 2003). It is possible that the interaction between yeast class C protein(s) and Vps45p was not observed owing to its weak binding and/or transient nature. If two SM proteins are required to confer proper conformation of similar nature on the Pep12p molecule or in expediting trans-SNARE pairing, then overexpression of Vps45p should have partially rescued the vps33 mutant phenotype or vice versa. Because we did not observe such suppression, we postulate that these two SM proteins on the endosomal membrane are performing noninterchangeable functions and might be serving a dual purpose. First, these proteins might be involved in maintaining the integrity of the large protein complex that is involved in fusion of Golgi-derived vesicles with the late endosome/PVC and second, in maintaining the right conformation of Pep12p. We have also demonstrated biochemically that the class C complex displays strong physical interactions with Vps8p, a protein that directs traffic from the late Golgi to the late endosome. We also demonstrate a physical interaction between Vps33p and the syntaxin, Pep12p. On the basis of these observations, we propose that the reason why vps33Δ cells display a “no vacuole” phenotype is because they are defective for fusion of vesicles at the vacuolar membrane and endosomal membrane as well as for multiple steps in endocytosis and cytoplasm to vacuolar trafficking.
We do not know if the interaction between Vps33p and Pep12p is direct or indirect and whether the molecular nature of this interaction is similar to the one between Vps33p and Vam3p. It has been demonstrated previously that interaction of Vps33p with the vacuolar syntaxin Vam3p is abolished in pep3Δ cells (Sato et al., 2000). Because Pep3p function has also been implicated on the endosomal membrane, it will be interesting to test whether the Vps33p-Pep12p interaction also requires Pep3p and/or the class C complex (Vps16p, Pep3p, and Pep5p). It is not presently clear whether these large complexes are bridges between the SMs and syntaxins or are just involved in facilitating stronger interactions.
The results in this article raise some interesting questions: 1) Do both of the SM proteins interact with the same molecule of Pep12p or do they interact with two separate Pep12p molecules? If they do interact with the same Pep12p molecule, then is Pep12p different from the other members of the syntaxin family, in that it requires the presence of two SM proteins to prime it? 2) Is this phenomenon of two SM proteins at the same organelle unique to the endosome because the endosome receives vesicles from various origins? The vacuolar membrane in yeast also receives cargo from a variety of sources. However, only one SM protein, Vps33p, has been shown to be involved at this site. Further investigation needs to be done to answer these questions.
Acknowledgments
We are grateful to Bruno Andre for sending us the Gap1p antibody and Howard Riezman for providing us with the α-factor antibody. We acknowledge Tatyana Aleynikova's contribution in the initial screening for vps33ts alleles. We thank Joe Suhan for help with electron microscopy and Adam Linstedt for the fluorescence microscope. Finally, we thank Jeff Brodsky, Tina Lee, and Manojkumar Puthenveedu for valuable advice and critical reading of the manuscript. This research was supported by Grant GM29713 from the National Institutes of Health to E.W.J.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0767. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0767.
References
- Abeliovich, H., Darsow, T., and Emr, S.D. (1999). Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 18, 6005-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta, L.M., Vida, T.A., Herman, P.K., and Emr, S.D. (1990). Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol. Cell. Biol. 10, 4638-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer, K.A., Rieder, S.E., Emr, S.D., and Jones, E.W. (1996). Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7, 579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, J.B., Matern, H.T., Peden, A.A., and Scheller, R.H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839-841. [DOI] [PubMed] [Google Scholar]
- Bryant, N.J., and James, D.E. (2001). Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J 20, 3380-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, N.J., Piper, R.C., Gerrard, S.R., and Stevens, T.H. (1998). Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur J. Cell Biol. 76, 43-52. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., and Emr, S.D. (1998). Phosphatidylinositol(3)-Phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2, 157-162. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., Peterson, M., Cowles, C.R., and Emr, S.D. (1997). A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell 8, 1089-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, C.M., Grote, E., Munson, M., Hughson, F.M., and Novick, P.J. (1999). Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 146, 333-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.J., and Stevens, T.H. (1996). The VPS8 gene is required for localization and trafficking of the CPY sorting receptor in Saccharomyces cerevisiae. Eur J. Cell Biol. 70, 289-297. [PubMed] [Google Scholar]
- Conibear, E., and Stevens, T.H. (1998). Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta 1404, 211-230. [DOI] [PubMed] [Google Scholar]
- Cooper, A.A., and Stevens, T.H. (1996). Vps10p cycles between the late-Golgi and pre-vacuolar compartments in its function as the sorting receptor for multiple yeast hydrolases. J. Cell Biol. 133, 529-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C.R., Emr, S.D., and Horazdovsky, B.F. (1994). Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 107, 3449-3459. [DOI] [PubMed] [Google Scholar]
- Cowles, C.R., Snyder, W.B., Burd, C.G., and Emr, S.D. (1997). Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 16, 2768-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, R.S., Brake, A., and Thorner, J. (1989). Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 86, 1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, R.S., Sterne, R.E., and Thorner, J. (1988). Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 50, 345-362. [DOI] [PubMed] [Google Scholar]
- Gallwitz, D., and Jahn, R. (2003). The riddle of the Sec1/Munc-18 proteins— new twists added to their interactions with SNAREs. Trends Biochem. Sci. 28, 113-116. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C. et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- Gerrard, S.R., Bryant, N.J., and Stevens, T.H. (2000a). VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol. Biol. Cell 11, 613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard, S.R., Levi, B.P., and Stevens, T.H. (2000b). Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic 1, 259-269. [DOI] [PubMed] [Google Scholar]
- Gotte, M., and von Mollard, G.F. (1998). A new beat for the SNARE drum. Trends Cell Biol. 8, 215-218. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D., and Mortimer, R. (1960). Chromosome mapping in Saccharomyces: centromere-linked genes. Genetics 45, 1085-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, C., and Andre, B. (1997). A C-terminal di-leucine motif and nearby sequences are required for NH4(+)-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol. Microbiol. 24, 607-616. [DOI] [PubMed] [Google Scholar]
- Horazdovsky, B.F., Cowles, C., Mustol, P., Holmes, M., and Emr, S.D. (1996). A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J. Biol. Chem. 271, 33607-33615. [DOI] [PubMed] [Google Scholar]
- Horazdovsky, B.F., and Emr, S.D. (1993). The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268, 4953-4962. [PubMed] [Google Scholar]
- Huizing, M., Boissy, R.E., and Gahl, W.A. (2002). Hermansky-Pudlak syndrome: vesicle formation from yeast to man. Pigment Cell Res. 15, 405-419. [DOI] [PubMed] [Google Scholar]
- Jauniaux, J.C., and Grenson, M. (1990). GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190, 39-44. [DOI] [PubMed] [Google Scholar]
- Jones, E.W. (2002). Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351, 127-150. [DOI] [PubMed] [Google Scholar]
- Jones, E.W., Webb, G.C., and Hiller, M.A. (1997). Biogenesis and function of the yeast vacuole. In: Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology, Vol. III, ed. E.W. Jones, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 363-470. [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Marcusson, E.G., Horazdovsky, B.F., Cereghino, J.L., Gharakhanian, E., and Emr, S.D. (1994). The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 77, 579-586. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., and Parker, R. (1992). A rapid method for localized mutagenesis of yeast genes. Yeast 8, 79-82. [DOI] [PubMed] [Google Scholar]
- Mullins, C., and Bonifacino, J.S. (2001). The molecular machinery for lysosome biogenesis. Bioessays 23, 333-343. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R. (2001). SNAREs and the specificity of membrane fusion. Trends Cell Biol. 11, 99-101. [DOI] [PubMed] [Google Scholar]
- Peng, R., and Gallwitz, D. (2002). Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J. Cell Biol. 157, 645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.R., Burd, C.G., and Emr, S.D. (1999). Vac1p coordinates Rab and phosphatidylinositol 3-kinase signalling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr. Biol. 9, 159-162. [DOI] [PubMed] [Google Scholar]
- Peterson, M.R., and Emr, S.D. (2001). The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2, 476-486. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. (2001). Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487-491. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131, 603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, R.A., Manolson, M.F., Becherer, K., Weidenhammer, E., Kirkpatrick, D., Wright, R., and Jones, E.W. (1991). Isolation and characterization of PEP3, a gene required for vacuolar biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A., Wickner, W., and Ungermann, C. (2000). Proteins needed for vesicle budding from the Golgi complex are also required for the docking step of homotypic vacuole fusion. J. Cell Biol. 148, 1223-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C.K., Howald-Stevenson, I., Vater, C.A., and Stevens, T.H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, S.C.W., Winistorfer, S.C., Poupon, V., Luzio, P.J., and Piper, R.C. (2003). Mammalian late Vps orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell E-publication (E03-06-0358). [DOI] [PMC free article] [PubMed]
- Rieder, S.E., and Emr, S.D. (1997). A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 8, 2307-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- Rizo, J., and Sudhof, T.C. (2002). Snares and Munc18 in synaptic vesicle fusion. Nat. Rev. Neurosci. 3, 641-653. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E. (1994). Mechanisms of intracellular protein transport. Nature 372, 55-63. [DOI] [PubMed] [Google Scholar]
- Rothstein, R. (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194, 281-301. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., R., F.E., and T. (Editors), M. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sato, T.K., Rehling, P., Peterson, M.R., and Emr, S.D. (2000). Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6, 661-671. [DOI] [PubMed] [Google Scholar]
- Seals, D.F., Eitzen, G., Margolis, N., Wickner, W.T., and Price, A. (2000). A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 97, 9402-9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov, E.A., He, J.-P., Moghrabi, N., Sunio, A., and Kramer, H. (1999). A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell 4, 479-486. [DOI] [PubMed] [Google Scholar]
- Springael, J.-Y., and Andre, B. (1998). Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisae. Mol. Biol. Cell 9, 1253-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A., and Jones, E.W. (1998). Pth1/Vam3p is the syntaxin homolog at the vacuolar membrane of Saccharomyces cerevisae required for the delivery of vacuolar hydrolases. Genetics 148, 85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A., Woolford, C.A., and Jones, E.W. (2000). Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics 156, 105-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbrough, M., and Magasanik, B. (1995). Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 177, 94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp, J.D., Huang, K., and Lemmon, S.K. (1997). The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 139, 1761-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Oiso, N., Gautam, R., Novak, E.K., Panthier, J.J., Suprabha, P.G., Vida, T., Swank, R.T., and Spritz, R.A. (2003). The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc. Natl. Acad. Sci. USA 100, 1146-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall, G.G., Hama, H., DeWald, D.B., and Horazdovsky, B.F. (1999). The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol. Biol. Cell 10, 1873-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen, R.F., and Verhage, M. (2003). Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 13, 177-186. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, Y., Kitamoto, K., Kanbe, T., Tanaka, K., and Anraku, Y. (1990). The SLP1 gene of Saccharomyces cerevisiae is essential for vacuolar morphogenesis and function. Mol. Cell. Biol. 10, 2214-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M.G., and Hughson, F.M. (2000). Membrane tethering and fusion in the secretory and endocytic pathways. Traffic 1, 588-597. [DOI] [PubMed] [Google Scholar]
- Webb, G.C., Hoedt, M., Poole, L.J., and Jones, E.W. (1997a). Genetic interactions between a pep7 mutation and the PEP12 and VPS45 genes: evidence for a novel SNARE component in transport between the S. cerevisiae Golgi complex and endosome. Genetics 147, 467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, G.C., Zhang, J., Garlow, S.J., Wesp, A., Riezman, H., and Jones, E.W. (1997b). Pep7p provides a novel protein that functions in vesicle-mediated transport between the yeast Golgi and endosome. Mol. Biol. Cell 8, 871-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte, J.R., and Munro, S. (2002). Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115, 2627-2637. [DOI] [PubMed] [Google Scholar]
- Wilsbach, K., and Payne, G.S. (1993). Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 12, 3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford, C.A., Bounoutas, G.S., Frew, S.E., and Jones, E.W. (1998). Genetic interaction with vps8–200 allows partial suppression of the vestigial vacuole phenotype caused by a pep5 mutation in Saccharomyces cerevisiae. Genetics 148, 71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford, C.A., Dixon, C.K., Manolson, M.F., Wright, R., and Jones, E.W. (1990). Isolation and characterization of PEP5, a gene essential for vacuolar biogenesis in Saccharomyces cerevisiae. Genetics 125, 739-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser, A.E., Sato, T.K., and Emr, S.D. (2000). New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151, 551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Dulubova, I., Min, S.W., Chen, X., Rizo, J., and Sudhof, T.C. (2002). Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell 2, 295-305. [DOI] [PubMed] [Google Scholar]