Abstract
In Saccharomyces cerevisiae, the nuclear-encoded protein Cbp1 promotes stability and translation of mitochondrial cytochrome b transcripts through interaction with the 5′ untranslated region. Fusion of a biotin binding peptide tag to the C terminus of Cbp1 has now allowed detection in mitochondrial extracts by using peroxidase-coupled avidin. Cbp1 is associated with the mitochondrial membranes when high ionic strength extraction conditions are used. However, the protein is easily solubilized by omitting salt from the extraction buffer, which suggests Cbp1 is loosely associated with the membrane through weak hydrophobic interactions. Gel filtration analysis and blue native PAGE showed that Cbp1 is part of a single 900,000-Da complex. The complex was purified using the biotin tag and a sequence-specific protease cleavage site. In addition to Cbp1, the complex contains several polypeptides of molecular weights between 113 and 40 kDa. Among these, we identified another message-specific factor, Pet309, which promotes the stability and translation of mitochondrial cytochrome oxidase subunit I mRNA. A hypothesis is presented in which the Cbp1–Pet309 complex contains several message-specific RNA binding proteins and links transcription to translation of the mRNAs at the membrane.
INTRODUCTION
Mitochondria are the power plants of most eukaryotic cells, producing ATP through oxidative phosphorylation. In Saccharomyces cerevisiae, the mitochondrial respiratory chain and ATP synthase consist of four large multisubunit complexes. The majority of subunits are encoded by nuclear genes (Tzagoloff and Dieckmann, 1990), whereas a smaller number are encoded by the mitochondrial genome (Foury et al., 1998). For example, of the many subunits of complex III, only cytochrome b is encoded by a mitochondrial gene (the COB gene) and synthesized within the organelle. Expression of COB requires several nuclear-encoded, message-specific factors that are imported into the organelle where they regulate processing, stability, and translation of the RNA (Dieckmann and Mittelmeier, 1987; Rödel and Fox, 1987; Michaelis et al., 1991; Dieckmann and Staples, 1994; Chen and Dieckmann, 1994, 1997; Grivell et al., 1999; Islas-Osuna et al., 2002). One of these is the Cbp1 protein, which is specifically required for the stability and translation of COB transcripts (Dieckmann et al., 1982, 1984b; Islas-Osuna et al., 2002). In cbp1 mutant cells, COB transcripts are degraded, cytochrome b is not synthesized, and the cells are unable to grow on nonfermentable carbon sources such as glycerol.
Cbp1 protects and promotes translation of COB mRNA through interaction with the 5′ untranslated region (UTR) of the message (Chen and Dieckmann, 1997; Islas-Osuna et al., 2002). The COB gene is cotranscribed with tRNAglu (Christianson et al., 1983), the 3′ end of which is 1100 base pairs upstream of the COB start codon. The mature 5′ end of COB mRNA maps 145 nucleotides downstream of the tRNA. Analyses of yeast strains with mutations in the AU-rich COB 5′ UTR have shown that a CCG trinucleotide 11 nucleotides downstream of the 5′ end of the mature mRNA plays a key role in the interaction with Cbp1; mutations that change this sequence to ACG, CCU, or CAG result in decreased stability of the COB mRNA (Chen and Dieckmann, 1997). Previous studies have identified suppressor mutations in the Cbp1 protein that rescue the defect caused by the ACG or CCU point mutations (Chen and Dieckmann, 1997; Chen et al., 1999; Islas-Osuna et al., 2003). Although these data provide evidence for a functional interaction between Cbp1 and COB transcripts, the question of how Cbp1 physically protects the mRNA from degradation and promotes translation remains unanswered.
Biochemical characterization of Cbp1 has been difficult. Cbp1 is present at very low levels, and overexpression of Cbp1 in either yeast cells or in a heterologous bacterial system leads to aggregation of the protein in insoluble inclusion bodies (Weber and Dieckmann, 1990). In several cases, scarce mitochondrial proteins have been detected and characterized using epitope tags (Ackerman et al., 1992; Wiesenberger and Fox, 1997; Green-Willms et al., 2001; Nowakowski et al., 2001). Therefore, we fused the Cbp1 protein to the biotinylation site of bacterial transcarboxylase (Cronan, 1990). Here, we describe the use of this “Bio” tag to purify Cbp1 as part of a soluble 900,000-Da complex from mitochondria sonicated in buffer lacking salt. This large complex contains at least one other nuclear-encoded, message-specific factor, Pet309, which protects mitochondrial COX1 mRNA and promotes its translation. Additional data suggest that Cbp1 is associated with the membrane in vivo. We propose the hypothesis that the Cbp1–Pet309 complex contains several message-specific RNA binding proteins and links transcription to translation of the mRNAs at the membrane.
MATERIALS AND METHODS
Strains and Media
The strains used in this study are listed in Table 1. Construction of the strains containing the tagged Cbp1 protein is described below. Media used were YPD (2% glucose, 2% peptone, 1% yeast extract), YEPG (3% glycerol, 2% peptone, 1% yeast extract), and WO (2% glucose, 0.67% yeast nitrogen base without amino acids). Amino acid supplements were added to WO at final concentrations of 20 μg/ml. Solid media contained 2% agar. For purification of Cbp1, cells were grown in liquid YPD.
Table 1.
Names and genotypes of yeast strains used in this study
| Strain | Genotype or description | Reference |
|---|---|---|
| S150-2B | a, [ρ+], ura3-52, his3, leu2-3,2-112 | Mayer and Dieckmann (1991) |
| S150-CP1L | LEU2 insertion at the PstI site of CBP1 in S150-2B | Sparks et al. (1997) |
| Cbp1-Bio1 | LEU2 replacement by CBP1/Factor Xa/Bio-tag fusion, HIS3 is inserted between CBP1 and NUC1 | This study |
| Cbp1-Bio2 | LEU2 replacement by CBP1/Bio-tag-fusion, HIS3 is inserted between CBP1 and NUC1 | This study |
Construction of Cbp1-Bio and Pet309-HA
The plasmids used to add the Bio tags to the carboxy terminus of the Cbp1 coding sequence, with and without the protease Factor Xa site, were prepared in several steps. Plasmid 3′GT10 was constructed by ligating the BglII fragment from plasmid T10 (Dieckmann et al., 1984a), which contains the 3′ half of CBP1 and all of the neighboring NUC1 gene, to pBS(-). The PstI-BamH1 fragment of 3′GT10 was ligated to pKS(+) to yield pFISH1. The stop codon of the Cbp1 coding sequence in pFISH1 was changed to a HindIII site by site-directed mutagenesis to yield plasmid 4888#5. The ClaI-PstI fragment of 3′GT10 was added to p4888#5 to yield pF48GCP. The EcoR1-HindIII fragment from YEp352-BIO6 (Ackerman et al., 1992) was ligated with HindIII-EcoR1 linkers to the HindIII site at the C terminus of Cbp1 in pF48GCP. This ligation put the biotinylation peptide in-frame with the end of the Cbp1 coding sequence, forming pF48BIO. The ClaI-BglI fragment of pF48BIO replaced the ClaI-BglI fragment of 3′GT10 to yield 3′GT10BIO2. HIS3+, as a BamHI fragment, was ligated to the BclI site between CBP1 and NUC1 to form 3′GT10BIO2H::B. An EcoRV fragment of 3′GT10BIO2H::B was used to transform strain S150-CP1L, which had been engineered to contain a LEU2 marker in the PstI site of CBP1 (Sparks et al., 1997). His+ transformants were selected on plates lacking histidine and screened for leucine auxotrophy. Integration of the correct construct was verified by Southern blotting, and sequencing of polymerase chain reaction (PCR)-amplified products by using DNA extracted from His+ Leu- transformants as template. This “Cbp1-Bio2” construct lacks the Factor Xa protease recognition site. A similar procedure was used to make the “Cbp1-Bio1” construct, which includes the Xa site between Cbp1 and the Bio tag.
The Cbp1-Bio1 strain was further modified to include an HA tag on the Pet309 protein. Three copies of the HA tag were added in-frame to the carboxy terminus of Pet309 by using an HA-URA3-HA cassette amplified by PCR with the appropriate primer pair (Schneider et al., 1995). The primers and vector containing the cassette were kindly provided by Tom Fox (Cornell University, Ithaca, NY) (Naithani et al., 2003). The amplified sequence was flanked by 45 base pairs of sequence homologous to the PET309 gene and transformed into the Cbp1-Bio1 strain (ura3). Integration of the hemagglutinin (HA) cassette at the PET309 locus in Ura+ transformants was verified by sequencing PCR-amplified products.
Isolation and Fractionation of Mitochondria
Mitochondria were isolated following standard procedures (Faye et al., 1974). The protein concentration of the final mitochondrial suspension was determined, and mitochondria were frozen and stored at -80°C as pellets containing 5 mg of protein. The fractionation of isolated mitochondria from the Cbp1-Bio1 strain was done essentially as described by Siep et al. (2000). In brief, mitochondrial pellets were thawed on ice and resuspended in TE buffer (10 mM Tris, 1 mM EDTA) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 μg/ml pepstatin A). Five hundred microliters of this buffer was used per frozen pellet to make a final protein concentration of 10 mg/ml. After incubation on ice for 15 min, the suspension was subjected to sonication six times for8sat35% output. The sonicator was a Fisher model 300 equipped with a microtip. Care was taken to keep the suspension cold. After sonication, the soluble (matrix) and insoluble (membrane) fractions were separated by ultracentrifugation for 30 min at 100,000 × g in a TLA 100.3 rotor (Beckman Coulter, Fullerton, CA) at 4°C. In the case of the Cbp1-Bio1/Pet309-HA double tagged strain, sonication led to cleavage of Pet309-HA. Thus, the mitochondrial suspension was subjected to three consecutive cycles of freezing and thawing. After a 30-min centrifugation at 14,000 rpm (4°C), the supernatant containing the Cbp1–Bio1/Pet309–HA complex was analyzed using two-dimensional blue native PAGE (BN-PAGE) analysis.
For alkaline carbonate extraction, 0.5 ml of mitochondria at 2 mg/ml was mixed with an equal volume of 0.2 M sodium carbonate, 10 mM EDTA and incubated on ice for 30 min. After centrifugation at 100,000 × g for 30 min, the membrane pellet was washed and dissolved in sample buffer. The supernatant was precipitated with trichloroacetic acid and centrifuged. The protein pellet was rinsed with 70% ethanol, dried, and dissolved in sample buffer. Samples of each fraction, equivalent to 40 μg of starting mitochondrial protein, were separated by SDS-PAGE on a 10% polyacrylamide gel.
SDS-PAGE, Silver Staining, and Western Blot Analysis
Proteins were resolved on SDS-polyacrylamide gels following the protocol described by Laemmli (1970). Silver staining was performed according to Blum et al. (1987) except when proteins were analyzed by mass spectrometry (see below). For Western blot analysis, the transfer to ImmunoBlot polyvinylidene diflouride (PVDF) membrane (Bio-Rad, Hercules, CA) was done with a TransBlot semidry transfer cell (Bio-Rad) at 15 V for 15 min. Antisera were diluted as follows: 1:1000 for anti-Mss51 (A. Tzagoloff, Columbia University, New York, NY) and 1:500 for anti-Idh and 1:400 for anti-Mdh1 (L. McAlister-Henn, University of Texas Health Sciences Center, San Antonio, TX). Secondary antibody conjugated to horseradish peroxidase (HRP) was diluted 1:1000. For detection of Cbp1-Bio1, a 1:1500 dilution of Neutravidin conjugated to horseradish peroxidase (Neutravidin-HRP; Pierce Chemical, Rockford, IL) was used. Cox2 protein was detected using monoclonal antibodies from Molecular Probes (Eugene, OR) at 1 μg/ml as suggested by the supplier. Secondary antibody conjugated to horseradish peroxidase was diluted 1:2000. Proteins were visualized using the SuperSignal West Pico chemiluminescent substrate (Pierce Chemical). Biotinylated marker proteins (Bio-Rad) were used to estimate protein molecular weights.
Sucrose Gradients
Isolated mitochondria were sonicated in buffer containing 1 M KCl. After ultracentrifugation, the pellet fraction was suspended and dialyzed overnight at 4°C in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Protein (5 mg) in 900 μl of TE was loaded onto the top of a 40–70% (wt/vol) sucrose gradient prepared with 10 mM Tris, pH 7.5. The gradient was centrifuged for 15 h at 53,000 × g in a SW40Ti rotor (Beckman Coulter) at 4°C. Fractions (1 ml) were collected from the bottom of the gradient. Then, 15 μl of each fraction was analyzed on 7% SDS-polyacrylamide gels.
Gel Filtration
Solubilized mitochondrial extracts (950 μl + 50 μl of 100% glycerol) were fractionated on a 120-ml Sephacryl S300 column (Pharmacia, Peapack, NJ) equilibrated in TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). The flow rate was adjusted to 0.5 ml/min. The eluate was collected in fractions of 1 ml. Gel filtration fractions were concentrated by trichloroacetic acid precipitation and analyzed by SDS-PAGE and Western blots. For calibration of the column, bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), apoferritin (443 kDa), thyroglobulin (669 kDa), and dextran blue (for determination of the void volume) (Sigma-Aldrich, St. Louis, MO) were subjected to gel filtration in separate runs with the column adjusted to the same conditions as used for the mitochondrial extracts.
BN-PAGE of Soluble Mitochondrial Fractions
The compositions of blue native gels and gel buffers were as described by Jänsch et al. (1996). The resolving gel contained an acrylamide gradient from 4 to 16%. Sample (15 μl) containing ∼50 μg of protein was mixed with 5 μl of 4× ACA buffer (0.3 M aminocaproic acid, 2 mM EDTA, 200 mM Bis-Tris, pH 7.0), incubated for 20 min on ice, and centrifuged for 20 min at 14,000 rpm (4°C) in an Eppendorf centrifuge. Before loading onto the blue native gel, 6 μl of Serva Blue G solution (5% Serva Blue in 75 mM aminocaproic acid) was added to the supernatant. The protein complexes were separated at 4°C for a total of ∼5 h with increasing voltage (30 min at 50 V, 30 min at 75 V, 60 min at 100 V, 30 min at 125 V, 60 min at 150 V, 30 min at 175 V, 60 min at 200 V). When the Coomassie dye had migrated halfway through the gel, the cathode buffer was replaced by buffer with the same composition but lacking the dye. For Western blot analysis of the first dimension, the protein complexes were transferred to PVDF membrane by using standard protocols. For the second, denaturing gel dimension, a lane of the blue native gel containing the sample was cut into slices of 3-mm length starting at the top. The gel slices were equilibrated for 1 min at 95°C with 1× SDS-PAGE sample buffer containing 2% SDS and 10 mM dithiothreitol. After cooling on ice, they were transferred into the slots of a 7.5% SDS-PAGE gel that was electrophoresed at 20 V for 2 h.
Purification of Cbp1-Bio1 on Streptavidin-coated Magnetic Beads
Sonic supernatants or gel filtration fractions were fractionated by affinity chromatography on Streptavidin-coated magnetic beads (Roche Diagnostics, Indianapolis, IN). The beads were washed three times with 500 μl of 1× Tris-buffered saline/Tween 20 (TBST) (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20) by using a magnetic bead separator (Roche Diagnostics) to sediment the beads. Prewashed beads, equivalent to 12 μl of the original Roche suspension, were then added to 450 μl of soluble extract plus 50 μl of 10× TBST. The binding reaction was carried out at 4°C for at least 2 h on a rotating device. The unbound supernatant was discarded, whereas the beads containing the biotinylated proteins were washed six times with 500 μl of 1× TBST. For SDS-PAGE analysis, the bound fraction was eluted under denaturing conditions by incubating with protein sample buffer at 60°C for 3 min.
Elution of Streptavidin-bound Proteins by Factor Xa Protease
To elute Streptavidin-bound Cbp1-Bio1 under nondenaturing conditions, the magnetic beads with the bound protein fraction were washed and equilibrated with Factor Xa digestion buffer (20 mM Tris-HCl, 100 mM NaCl, 2 mM CaCl2, pH 8.0) (4 changes of 500 μl each) and were finally incubated with 30 μl of digestion buffer containing 1 μg of Factor Xa protease (Roche Diagnostics) for 2 h at room temperature plus 15 h at 4°C. The supernatant containing the eluate was collected and stored at -80°C. The protein fraction that remained on the beads and represented undigested proteins was eluted under denaturing conditions (see above).
Analysis of Proteins by Matrix-assisted Laser Desorption Ionization/Time of Flight (MALDI-TOF)
Proteins purified on Streptavidin beads and eluted with Factor Xa protease were separated by 7.5% SDS-PAGE. The gels were silver stained as described by Rabilloud (1992). The bands of interest were excised and stored at -20°C. Multiple bands of the same protein from parallel lanes of one gel were combined. For a background control, gel pieces from an empty lane of the same gel were excised. To enhance the sensitivity of mass spectrometric identification of peptides, silver-stained proteins were destained with chemical reducing agents as described previously (Gharahdaghi et al., 1999). Trypsin digestion was performed as described on the Web page of the mass spectrometry facility of the University of Arizona (www.chem.arizona.edu/facilities/msf) except that all volumes were increased fivefold to adapt for the large number of gel pieces. After extraction of the tryptic peptides from the gel pieces, the samples were vacuum-dried and resuspended in 10 μl of 0.1% trifluoroacetic acid (TFA). The samples were subsequently purified using ZipTipC18 pipet columns (Millipore, Billerica, MA). Briefly, the ZipTip columns were wetted with 10 μl of 0.1% TFA in 50% acetonitrile and then equilibrated with 0.1% TFA in H2O. The peptides were bound to the column by pipetting the sample slowly up and down 10 times. The column was washed with two changes of 0.1% TFA before the peptides were eluted in a two-step-process with 0.1% TFA in 50 and 95% acetonitrile, respectively. The combined eluate from both steps (20 μl) was reduced to 5 μl in a SpeedVac. This solution was subjected to MALDI-TOF on a Bruker Reflex-III MALDI-TOF instrument.
Monoisotopic peptide masses were compared against the Saccharomyces genome database with a mass tolerance of 0.1 Da by using ProFound (prowl.rockefeller.edu/cgi-bin/ProFound). The maximum number of missed cleavages was one.
RESULTS
Mitochondrial Function Is Not Affected by Fusion of the Bio Tag to the Carboxy Terminus
Two versions of the Cbp1-Bio protein were made: the first fusion, Bio1, contains the tetrapeptide recognition site for Factor Xa protease (Nagai and Thogersen, 1984) between Cbp1 and the Bio tag, whereas the second version, Bio2, lacks the Factor Xa site. The fusions were introduced into the CBP1 chromosomal location by gene replacement (see MATERIALS AND METHODS). Strains containing the tagged Cbp1 proteins grew on the glycerol-containing medium YEPG at rates similar to the untagged wild-type strain S150, indicating that mitochondrial function was not impaired in these strains (our unpublished data).
Extraction and Solubility of Cbp1
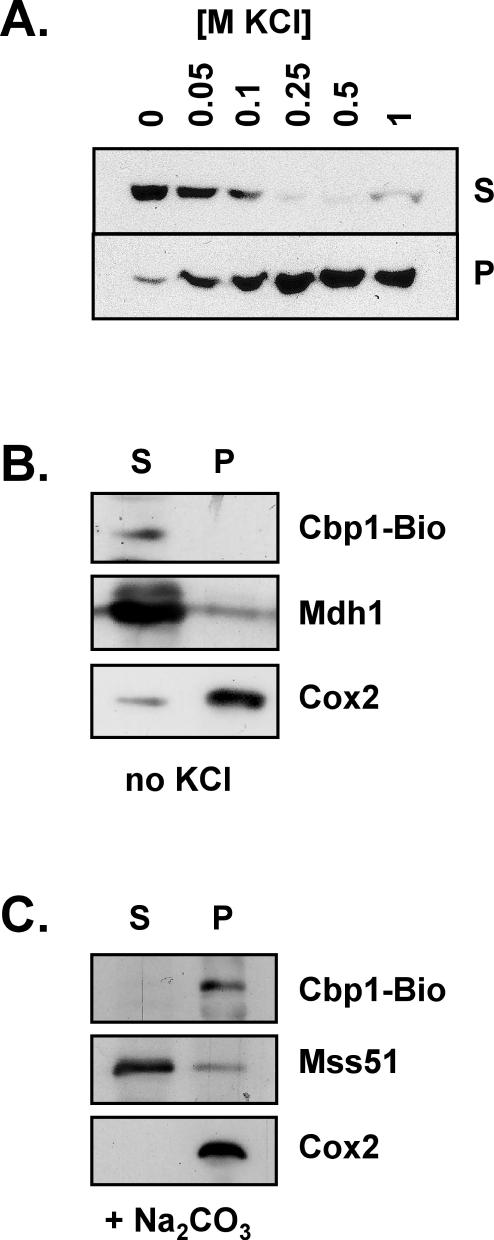
To determine the intramitochondrial localization of Cbp1, mitochondria from the Bio1 strain were disrupted by osmotic shock and sonication, and the soluble matrix fraction was separated from the insoluble membrane fraction by ultracentrifugation. Proteins in both the soluble supernatant and the insoluble pellet were analyzed by SDS-PAGE and Western blotting. Using standard sonication buffer containing 1 M KCl, Cbp1 was in the insoluble membrane fraction. However, when KCl was omitted from the sonication buffer, >90% of Cbp1 was in the soluble matrix fraction (Figure 1A). The solubility of Cbp1 was inversely related to the concentration of salt in the sonication buffer, suggesting hydrophobic interactions between Cbp1 monomers or between Cbp1 and other proteins.
Figure 1.
Extraction and solubility of Cbp1. (A) Effect of salt on extraction of Cbp1 from mitochondria. Mitochondria isolated from the Cbp1-Bio1 strain were disrupted by sonication in the presence of various concentrations of KCl. Soluble proteins (S) were separated from insoluble proteins (P) by ultracentrifugation (see MATERIALS AND METHODS). All fractions were analyzed by SDS-PAGE and Western blot by using HRP-coupled Neutravidin. The bands correspond to the Cbp1-Bio1 protein. (B) Effect of sonication in the absence of salt on soluble and membrane proteins. Mitochondria were disrupted in buffer lacking salt and centrifuged as described in A. Western blots were probed with Neutravidin to detect Cbp1-Bio, with antiserum to Mdh1, mitochondrial malate dehydrogenase, and with monoclonal antibodies to Cox2. (C) Effect of alkaline carbonate extraction on Cbp1. Mitochondria were treated with 100 mM sodium carbonate for 30 min on ice, and soluble proteins were separated from insoluble membrane proteins by ultracentrifugation (see MATERIALS AND METHODS). Western blots were probed with Neutravidin to detect Cbp1-Bio, with antiserum to Mss51, a peripheral membrane protein, and with monoclonal antibodies to Cox2, an integral membrane protein.
To investigate how the soluble enzyme malate dehydrogenase, Mdh1 (Thompson and McAlister-Henn, 1989), and the integral membrane protein cytochrome oxidase subunit II, Cox2 (Tsukihara et al., 1996), partitioned when mitochondria were sonicated in the absence of salt, Western blots were reacted with antiserum to Mdh1 or with monoclonal antibodies to Cox2 (Figure 1B). As expected, Mdh1 was primarily in the soluble fraction, and Cox2 was primarily in the membrane pellet fraction.
That Cbp1 can be solubilized by a no-salt sonication step suggests that the protein is peripherally associated with the membrane. A traditional method for extracting extrinsic membrane proteins is treatment of mitochondrial membranes with alkaline carbonate. After treatment with sodium carbonate, Cbp1 remained in the insoluble pellet fraction, whereas the extrinsic membrane protein Mss51 (Decoster et al., 1990; Siep et al., 2000) was found in the soluble fraction (Figure 1C). The integral membrane protein Cox2 remained in the pellet fraction as expected. Similar to the results with KCl (Figure 1A), carbonate treatment, which disrupts ionic interactions, strengthened the association between Cbp1 and the membrane, suggesting that this association is primarily through hydrophobic interactions.
Solubilized Cbp1 Aggregates in Salt
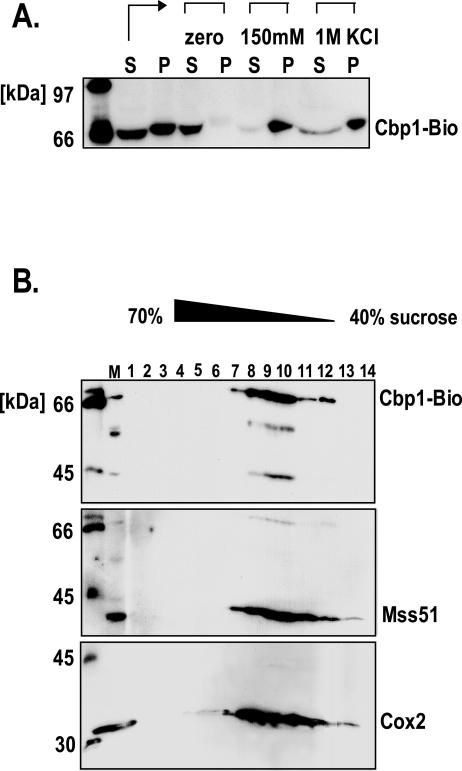
To examine the effect of salt on aggregation of Cbp1 after sonication, the Cbp1-containing soluble fraction obtained by sonication without salt was split three ways, and the aliquots were treated with no salt, or 150 mM or 1 M KCl for 1 h on ice. The samples were then centrifuged to pellet any aggregated proteins. In the salt-treated samples, Cbp1 pelleted with the insoluble fraction (Figure 2A). In the control experiment, where water was added instead of KCl, Cbp1 remained in the soluble supernatant. This result supports the hypothesis that solubilized Cbp1 and/or associated proteins have a hydrophobic patch, which causes the proteins to aggregate when salt is added.
Figure 2.
Solubility after sonication and membrane association in the presence of salt. (A) Solubility of Cbp1 when salt is added after sonication. Mitochondria were sonicated in buffer lacking KCl (S, P). After sonication and ultracentrifugation, aliquots of the soluble fraction (S) were incubated for 1 h on ice with either H2O or the indicated concentrations of KCl. The samples were then ultracentrifuged, and both the soluble supernatants (S) and the insoluble pellets (P) were analyzed by SDS-PAGE and Western blot by using HRP-coupled Neutravidin. (B) Sucrose gradient analysis of Cbp1 in the pellet fraction after sonication in buffer containing salt. Mitochondria were sonicated in buffer containing 1 M KCl, centrifuged, and the pellet fraction was resuspended and loaded onto a 40–70% sucrose gradient. Fractions 1–14 were collected and analyzed by Western blot. The blot was probed first with Neutravidin-HRP to detect Cbp1-Bio1. The blot was reprobed with anti-Mss51 antiserum, which detects the mitochondrial membrane protein Mss51 (Siep et al., 2000) and Cox2 antibodies, which detect the integral membrane protein Cox2 (Molecular Probes). Cbp1-Bio1, Mss51, and Cox2 were detected most heavily in fractions 8, 9, and 10.
Cbp1 Is Associated with Membranes When Mitochondria Are Sonicated in Buffer Containing Salt
Cbp1 is soluble when mitochondria are sonicated in the absence of salt, but the solubilized form aggregates when salt is added. This led us to question whether the Cbp1 protein found in the pellet fraction after sonication in the presence of salt is associated with the membranes or with an insoluble mass. To investigate this point, the mitochondrial pellet fraction obtained after sonication in buffer containing 1 M KCl was resuspended, dialyzed, and layered onto a 40–70% continuous sucrose gradient. After centrifugation, gradient fractions were analyzed by SDS-PAGE and Western blot. Cbp1-Bio banded at a position within the gradient that is typical for mitochondrial membranes, and Mss51 and Cox2 were detected in the same fractions that contained Cbp1-Bio (Figure 2B). Protein aggregates would be expected in the heavier sucrose fractions toward the bottom of the tube (Ackerman and Tzagoloff, 1990). Therefore, when mitochondria are sonicated in the presence of KCl, Cbp1-Bio is in the pellet fraction by virtue of its attachment to mitochondrial membranes.
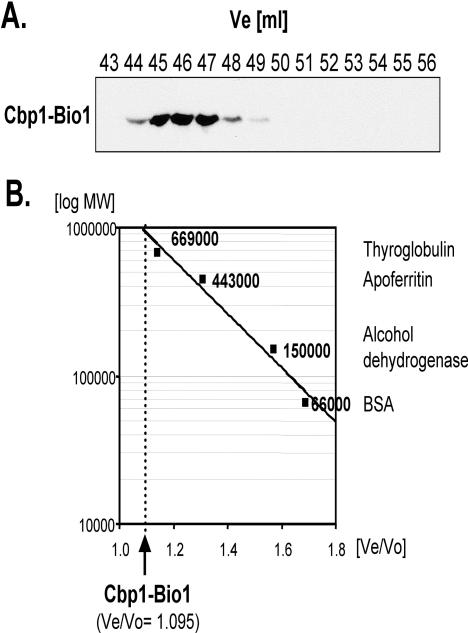
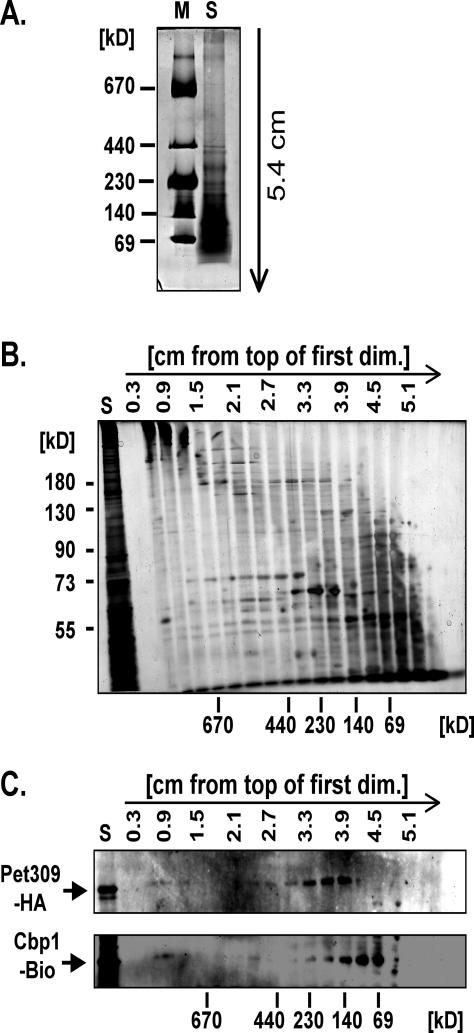
Cbp1 Elutes from a Size-Exclusion Column in the 900,000-Da Range
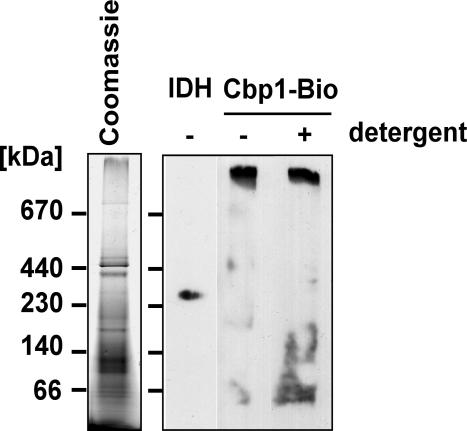
To analyze the soluble mitochondrial no-salt extract, sonic supernatant was fractionated on a Sephacryl S300 column. The deduced Ve/Vo value of Cbp1 was compared with Ve/Vo of several marker proteins that were used to calibrate the column. The peak of Cbp1 at fraction 46 corresponded to a molecular weight of ∼900 kDa (Figure 3, A and B). The high molecular weight of the gel filtration fractions containing Cbp1 suggested that Cbp1 associates with other mitochondrial proteins. To investigate whether Cbp1 is part of one or more complexes, the soluble fraction obtained by no-salt sonication of mitochondria was analyzed by blue native gel electrophoresis, which separates protein complexes based on size and the charge distribution on their surfaces (Schägger and von Jagow, 1991; Schägger et al., 1994). Immunoblot analysis of the first, native dimension showed that Cbp1 is detected in a high-molecular-weight band (Figure 4). The isocitrate dehydrogenase complex with a native molecular weight of 320 kDa migrated faster than the Cbp1 complex (Figure 4). When the nonionic detergent laurylmaltoside was added to a final concentration of 1%, the HRP-coupled Neutravidin reacted with several smaller molecular weight bands between 66 and 140 kDa in addition to the 900-kDa band, indicating that the Cbp1-containing complex is modestly sensitive to detergent treatment.
Figure 3.
Gel-filtration analysis of Cbp1-Bio1. (A) Fine mapping of the peak elution volume of Cbp1-containing fractions. Gel filtration fractions 43–56 (corresponding to the volume in milliliters, Ve) from strain Cbp1-Bio1 were electrophoresed, blotted, and probed for the presence of Cbp1-Bio1 protein with HRP-coupled Neutravidin. (B) Calculation of the molecular weight of the Cbp1-Bio1–containing complex. The graph shows the logarithm of molecular weights (y-axis) against Ve/Vo of the marker proteins used for calibration of the column (x-axis). Names and molecular weights of the proteins used for calibration are given on the right. The position of the peak Ve/Vo of the Cbp1–Bio1-containing complex is indicated by an arrow and a dotted line.
Figure 4.
Blue native PAGE analysis of Cbp1-Bio1. Soluble mitochondrial proteins from the Cbp1-Bio1 strain were subjected to blue native gel electrophoresis. One lane of the gel was restained with Coomassie Brilliant Blue R250 to visualize the protein complexes (left). The other lanes containing parallel samples were blotted and probed with an antibody directed against isocitrate dehydrogenase (IDH) or with Neutravidin-HRP to detect Cbp1-Bio1. Laurylmaltoside was added to one sample of the protein extract to a final concentration of 1% before electrophoresis (+ detergent).
Purification of Cbp1-Bio1 by Affinity Chromatography and Factor Xa Protease Elution
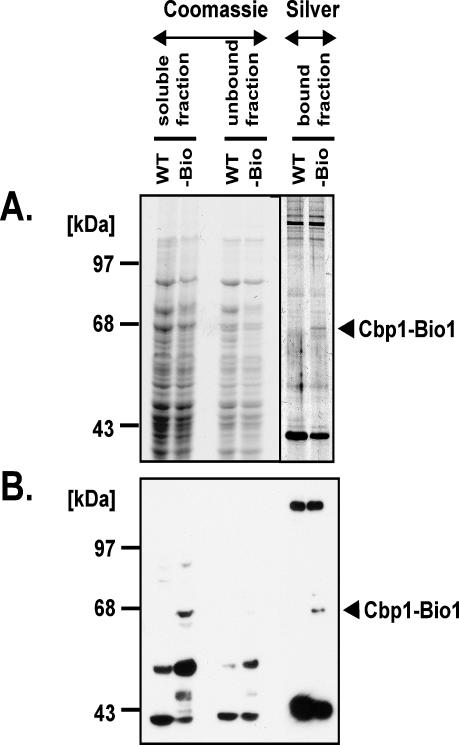
Cbp1-Bio1 was purified by adsorption of the soluble extract on Streptavidin-coated magnetic beads. The beads adsorb many different proteins from both the wild-type and the Cbp1-Bio1 extracts (Figure 5A, bound fraction). The protein patterns were identical except for a small number of weakly silver-stained bands, which were detected only in the Cbp1-Bio1 extract. One of these had a size expected for Cbp1-Bio1. In a Western blot of a duplicate gel, this band reacted with Neutravidin, confirming that it is Cbp1-Bio1. All of the Cbp1-Bio1 protein in the soluble fraction bound to the beads. Two other proteins of 120 and 45 kDa that either are biotinylated or have a protein motif with affinity for streptavidin also were adsorbed onto the affinity matrix (Figure 5B, bound fraction).
Figure 5.
Purification of Cbp1-Bio1 on Streptavidin-coated magnetic beads. (A) Visualization of protein patterns by staining. Soluble mitochondrial proteins from either the untagged strain (WT) or the Cbp1-Bio1 strain (-Bio) were allowed to adsorb onto Streptavidin-coated magnetic beads. Proteins that adsorbed were separated from those that did not by use of a magnet. The beads were washed and then boiled in gel-loading buffer. Bound and unbound fractions were separated on a 7.5% denaturing acrylamide gel. Samples containing the original soluble fraction (soluble fraction), or proteins that did not absorb to the beads (unbound fraction) were stained with Coomassie Blue. Samples of the material adhering to the streptavidin beads (bound fraction) were stained with silver nitrate. (B) Western blot analysis. A duplicate gel to that shown in A was analyzed by Western blot with HRP-coupled Neutravidin.
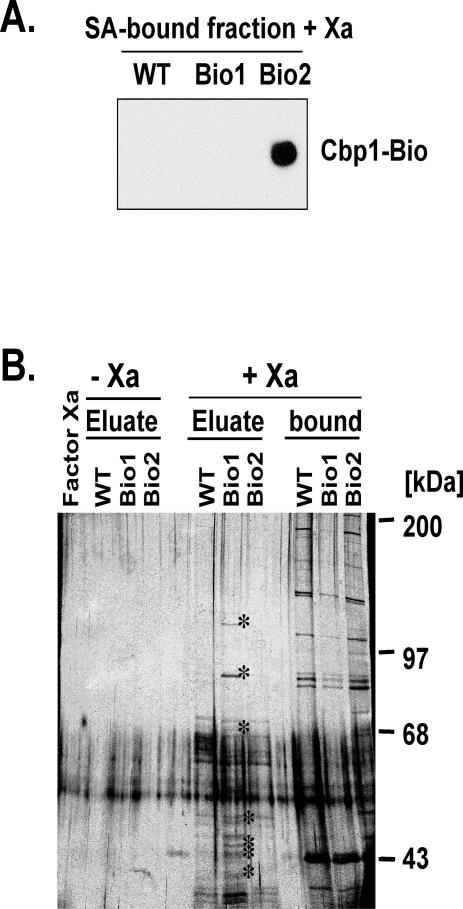
Cbp1-Bio1 was eluted from the beads by treatment with Factor Xa protease. As a control, extract containing Cbp1-Bio2, which lacks the Factor Xa site, was adsorbed on Neutravidin beads and treated with Factor Xa. Western blot and SDS-PAGE analysis showed that Cbp1-Bio2 remained bound to the beads after protease digestion, whereas Cbp1-Bio1 was efficiently released from the beads by treatment with protease (Figure 6A). A duplicate gel stained with silver revealed at least 10 protein bands that were present in the Factor Xa-eluted fraction from the Cbp1-Bio1 adsorbed beads but absent from the Cbp1-Bio2 and untagged wild-type Xa-eluted fractions (Figure 6B). The proteins unique to the protease-treated sample had estimated relative molecular weights of 110, 85, and 68 kDa, and several were in the 40- to 50-kDa range. The 68-kDa protein is of the correct size to be Cbp1.
Figure 6.
Elution of Cbp1-Bio1 from the Streptavidin beads with Factor Xa protease. (A) Western blot analysis of eluted proteins. The Streptavidin-bound fractions of mitochondrial extracts from WT, Cbp1-Bio1, and Cbp1-Bio2 were treated with Factor Xa protease. The proteins that remained on the Streptavidin beads were eluted under denaturing conditions. The proteins were analyzed by SDS-PAGE and Western blot by using HRP-coupled Neutravidin. (B) Silver stain analysis of eluted proteins. The silver-stained gel shows the protein composition of the eluate fractions containing the proteins that were cleaved from the beads with Factor Xa protease, the fractions that remained bound to the beads, and, as controls, the eluates obtained without addition of the protease. Protein bands unique to the eluate of the Cbp1-Bio1 strain are indicated by asterisks.
MALDI Analysis Identifies One of the Proteins as Pet309
The three proteins of highest molecular weight unique to the Cbp1-Bio1 + Xa eluate of Figure 6B (110, 85, and 68 kDa) could be excised from single dimension gels without the risk of including contaminating bands. These three bands were collected from the gels and prepared for MALDI analysis as described in MATERIALS AND METHODS. A sufficient number of peptide peaks to allow a general database search was obtained for only the 110-kDa band. Five of 10 peptides identified from this band matched the 113-kDa protein Pet309. Pet309 is a mitochondrial protein that specifically protects and promotes translation of the COX1 mRNA, analogous to the function of Cbp1 for COB mRNA (Manthey and McEwen, 1995). As expected, two of three experimental peaks obtained for the 68-kDa protein by MALDI matched theoretical peptide masses determined for the Cbp1 protein. The identity of the proteins in the other unique bands will require additional investigation.
Blue Native PAGE Identifies Pet309 and Cbp1 as Components of the Same High-Molecular-Weight Complex
Two-dimensional blue native PAGE analysis was used to verify the putative interaction between Cbp1 and Pet 309. To detect both proteins in the same extract, a strain containing HA-tagged Pet309 in addition to Cbp1-Bio1 was constructed (see MATERIALS AND METHODS). Disruption of mitochondria either by sonication or by use of detergents resulted in cleavage of the Pet309-HA protein into several smaller fragments. However, we found that repeated treatment of a mitochondrial suspension by alternate freezing and thawing periods released enough of the high-molecularweight complex containing Cbp1 (our unpublished data) to allow analysis by two-dimensional blue native PAGE. As shown in Figure 7, this extract contained at least two visible bands of molecular weights >670 kDa (Figure 7A). These bands gave rise to a complex protein pattern in the second dimension denaturing gel as visualized by silver staining (Figure 7B). Both Cbp1-Bio and Pet309-HA were detected in the second and third lanes (0.6–0.9 cm from the top of the first dimension gel) containing the high-molecular-weight bands, which suggests that they are part of a single complex (Figure 7C). In addition, a large amount of monomeric Pet309-HA protein was detected in lanes corresponding to an approximate molecular weight of ∼140 kDa (3.3–3.9 cm from the top of the first dimension gel). Likewise, monomeric Cbp1-Bio was mostly observed in the lanes corresponding to protein or protein complex sizes of 69–140 kDa (3.9–4.5 cm from the top of the first dimension gel).
Figure 7.
Two-dimensional analysis of a soluble mitochondrial extract of strain Cbp1-Bio1/Pet309-HA. (A) Soluble proteins were extracted from Cbp1-Bio1/Pet309-HA mitochondria in the absence of salt by repeated freezing and thawing and electrophoresed in the presence of Serva Blue as described in MATERIALS AND METHODS. An aliquot of the “HMW calibration kit for native electrophoresis” (Amersham Biosciences, Piscataway, NJ) was used as a molecular weight marker. The direction of electrophoresis is indicated by the arrow. (B) For subsequent analysis of the protein composition of the complexes by SDS-PAGE, the first-dimension gel was cut into slices of 3 mm starting at the top. The gel pieces were subjected to SDS-PAGE. For comparison, the soluble mitochondrial extract (S) also was electrophoresed. After separation, the proteins were stained with silver nitrate. On the left, the positions of molecular weight marker proteins of the denaturing second dimension are shown. Below, the relative positions of the first-dimension molecular weight markers are indicated. (C) A gel identical to the one in B was blotted onto PVDF membrane and probed consecutively with anti-HA-peroxidase (diluted to 50 mU/ml) (Roche Diagnostics) to detect HA-tagged Pet309 and Neutravidin-HRP (1:1500 dilution) (Pierce Chemical) to detect Cbp1-Bio.
DISCUSSION
Cbp1 Associates with the Mitochondrial Membrane Peripherally
Mitochondrial message-specific protectors and translational activators are of very low abundance and difficult to detect with highly specific antisera and monoclonal antibodies. When overexpressed in yeast, they are imported into mitochondria where they form large aggregates that can be solubilized only by strong denaturing detergents such as SDS (Weber and Dieckmann, 1990; Ackerman et al., 1992). Heterologous expression in Escherichia coli and other systems has not yielded soluble material (Weber and Dieckmann, unpublished observations).
In the current work, two new approaches were used to isolate a soluble form of Cbp1 that could be purified by affinity chromatography. First was the fusion of Cbp1 to the biotin-binding peptide tag. Detection of Cbp1-Bio with HRP-avidin proved much more sensitive than detection of Cbp1 with antibodies and allowed analysis of crude mitochondrial fractions. Second was the sonication of mitochondria in the absence of salt. Cbp1 partitions cleanly into the soluble supernatant after centrifugation of the sonicated mitochondria (Figure 1A). As long as salt is omitted from the sonication buffer and from the buffers of subsequent purification steps, Cbp1 remains soluble (Figure 2A). In contrast, analysis of the insoluble pellet obtained by the standard protocol with 1 M KCl in the sonication buffer, showed that Cbp1 primarily resides in the membrane fractions in the middle of a 40–70% continuous sucrose gradient (Figure 2B). These results suggest that in vivo Cbp1 is a peripheral inner membrane protein that associates with the membrane primarily through hydrophobic interactions. Cbp1 itself may contain a hydrophobic patch that is associated with the membrane, or there may be hydrophobic regions on one or more of the proteins associated with Cbp1.
Cbp1 Is Part of a Large Heterogeneous Complex
The high propensity of Cbp1 to aggregate in the presence of salt made traditional ion exchange chromatography impractical. Size exclusion chromatography and blue native gel electrophoresis yielded the result that Cbp1 is associated with a large, unique complex of ∼900,000 Da. The differential use of a Factor Xa protease site placed between the C terminus of Cbp1 and the Bio tag allowed visualization of several proteins specifically associated with Cbp1 in the large complex. From the silver-stained gel in Figure 6B, we estimate that the complex contains at least one 113-, one 85-, one 68- (Cbp1), and several 40- to 50-kDa proteins. These proteins account for at least 70% of the mass estimated for the entire complex, not taking into account differences in stoichiometry, which cannot be determined from the gel. The proteins must be soluble themselves, or they must have disruptable hydrophobic interactions with the membrane. It is possible that there are additional subunits of this complex that are more firmly bound to the membrane and are not released during the no-salt sonication step.
The COX1-specific 5′ UTR Protector and Translational Activator Pet309 Is in the Complex with Cbp1
The largest silver-stained band eluted with Factor Xa protease contains the 113-kDa protein Pet309. This protein is required for the stability and translational activation of the mitochondrial COX1 mRNA, which encodes the largest subunit of cytochrome c oxidase (Manthey and McEwen, 1995; Manthey et al., 1998). Blue native PAGE analysis provided additional evidence for the colocalization of Cbp1 and Pet309 in a large protein complex. In contrast to the sonication supernatant, in which at least 50% of Cbp1-Bio was found in the high-molecular-weight complex (Figure 4), the protein extract of the double-tagged strain used for this analysis showed only a low percentage of subunits bound in the complex (Figure 7C). The freeze-thaw procedure may not release all of a factor or factors necessary for complex integrity, and/or the freeze-thaw procedure may itself disrupt the complex. However, this treatment was necessary to avoid degradation of the Pet309-HA protein (Krause, Souza, and Dieckmann, unpublished data). Originally, Pet309 was characterized as an integral membrane protein when overexpressed (Manthey et al., 1998), whereas in the current studies the protein is extracted in buffer containing no salt. The difference in localization may be due to overexpression, leading to aggregation and cofractionation with the membranes, or the no-salt extraction procedure may sever the protein from a transmembrane domain. However, Pet309-HA seemed to be full-length or very close to the correct size when extracted with the freeze-thaw protocol (our unpublished data).
Pet309 is a member of the TPR/PPR family of proteins, which have 34/35 amino acid repeats. The TPR/PPR motif has been proposed to promote protein–protein and protein–RNA interactions (Small and Peeters, 2000). Several TPR/PPR proteins have been found to protect, process, and/or promote translation of organellar mRNAs and are found in large multiprotein complexes (Coffin et al., 1997; Fisk et al., 1999; Boudreau et al., 2000; Vaistij et al., 2000). Mutations in the human homolog of PET309, LRPPRC, cause the mitochondrial Leigh's type Saguenay-Lac St-Jean syndrome in a subpopulation of French Canadians (Mootha et al., 2003).
Transcription Is Linked to Translation at the Membrane
For many years, we have wondered why the expression of each of the seven mitochondrial mRNAs requires a unique set of splicing, protection, and translation initiation proteins. Through the present work and that of others, we are learning that these seemingly independent systems have a higher order of organization. Factors that affect individual mRNAs have been found to associate with factors that affect another mRNA and links of these protection/translational factors to the transcriptional apparatus have been made.
Here, we show that Cbp1, which protects and promotes translation of COB mRNA is physically associated with Pet309, which protects and promotes translation of COX1 mRNA. Naithani et al. (2003) reported interactions between translational activators for COX1, COX2, and COX3 mRNAs. They also showed interaction between Pet309 and Nam1, which protects and promotes translation of the ATP8/6 mRNA (Groudinsky et al., 1993; Wallis et al., 1994). Finally, Nam1 associates with the non-T7 like, N-terminal portion of the catalytic subunit, Rpo41, of mitochondrial RNA polymerase (Wang and Shadel, 1999; Rodeheffer et al., 2001).
Both the mitochondrial transcriptional apparatus and several message-specific translational activators have been found to associate with the inner mitochondrial membrane. Rpo41 is found in nucleoids, which contain mitochondrial DNA and many other proteins (Butow, personal communication; Rodeheffer and Shadel, 2003), and are firmly bound to the membrane (Albring et al., 1977). Rpo41 also has been linked to the membrane through the association between Nam1 and Sls1, which is a membrane-bound protein (Rouillard et al., 1996; Bryan et al., 2002). Here, we show that Cbp1 is likely associated with the membrane in vivo. Cbs1 and Cbs2, which promote translation of COB mRNA, also are associated with the membrane. Cbs1 is an integral membrane protein that requires detergent for extraction, whereas Cbs2 associates only loosely with the membrane (Michaelis et al., 1991) and may be a component of the Cbp1-Pet309 complex.
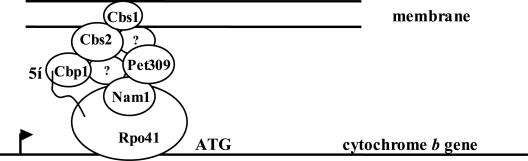
Based on our data and that of Rodeheffer et al. (2001), Rodeheffer and Shadel (2003), and Naithani et al. (2003), we propose a model that builds on that proposed by Bryan et al. (2002). In the model, message-specific polypeptides that protect the 5′ ends of newly synthesized transcripts are arranged in a large complex localized to the inner mitochondrial membrane. This complex interacts with the transcription machinery through Nam1 (Figure 8). As the 5′ end of any nascent mRNA emerges from the polymerase, its corresponding protection protein is poised to associate with the mRNA partner. In addition to providing protection, proteins such as Cbp1 and Pet309 promote translation through association with translational activators that are more firmly embedded in the membrane; the association may be direct, or via bridging activators that are also in the Cbp1–Pet309 complex. When the complex is associated with a particular mRNA docked at the membrane, ribosomes are attracted and translation commences. Localization at the membrane allows direct insertion of the nascent, highly hydrophobic mitochondrial gene products into the lipid bilayer for assembly into the large, multisubunit respiratory chain complexes and ATP synthase.
Figure 8.
Model linking transcription to mRNA protection and translation at the mitochondrial membrane. The mitochondrial RNA polymerase, labeled Rpo41, is shown transcribing mitochondrial DNA encoding cytochrome b (the COB gene) from left to right starting from the promoter (rightward pointing arrow). The wavy line represents the 5′ end of COB RNA emerging from the polymerase and associating with Cbp1, which is responsible for stabilizing the RNA and promoting its translation (Mittelmeier and Dieckmann, 1993; Islas-Osuna et al., 2002). Nam1 has been shown to interact with the N-terminal portion of the Rpo41 subunit of the polymerase (Rodeheffer et al., 2001) and also interacts with Pet309 (Naithani et al., 2003), which is required for the stability and translation of COX1 mRNA (Manthey and McEwen, 1995; Manthey et al., 1998). In the present work, Pet309 and Cbp1 were found in the same large complex of proteins. Although not yet proven to interact with the other proteins shown, Cbs1 and Cbs2 also are required for the translation of COB mRNA (Rödel, 1986). Cbs1 is an integral membrane protein, whereas Cbs2 is a peripheral membrane protein (Michaelis et al., 1991). Solubilized Cbs2 has been found in the same large mass fraction from size exclusion chromatography as Cbp1 and Pet309 (Krause, Souza, and Dieckmann, unpublished observation).
Acknowledgments
We thank Alex Tzagoloff for the Mss51 antiserum, Lee McAlister-Henn for the Idh and Mdh1 antisera, Ted Weinert for sharing the gel filtration column, and Tom Fox for providing the vector containing the HA-tag cassette. Alex Tzagoloff, Antoni Barrientos, John Little, Ted Weinert, Roy Parker, Telsa Mittelmeier, Tim Ellis, Michael Rice, and Melissa Dellos critiqued various versions of the manuscript. This work was supported by National Institutes of Health grant GM-34893 (to C.L.D.). K.K. received partial support from the Deutscher Akademischer Austauschdienst.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0126. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0126.
References
- Ackerman, S.H., Martin, J., and Tzagoloff, A. (1992). Characterization of ATP11 and detection of the encoded protein in mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 267, 7386-7394. [PubMed] [Google Scholar]
- Ackerman, S., and Tzagoloff, A. (1990). Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc. Natl. Acad. Sci. USA 87, 4986-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albring, M., Griffith, J., and Attardi, G. (1977). Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. USA 74, 1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, H., Beier, H., and Gross, H. (1987). Improved silver staining of plant proteins, RNA in polyacrylamide gels. Electrophoresis 8, 93-99. [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.D., Ossenbuhl, F., and Rochaix, J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19, 3366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, A.C., Rodeheffer, M.S., Wearn, C.M., and Shadel, G.S. (2002). Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics 160, 75-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., and Dieckmann, C.L. (1994). Cbp1p is required for message stability following 5′-processing of COB mRNA. J. Biol. Chem. 269, 16574-16578. [PubMed] [Google Scholar]
- Chen, W., and Dieckmann, C.L. (1997). Genetic evidence for interaction between Cbp1 and specific nucleotides in the 5′ untranslated region of mitochondrial cytochrome b mRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6203-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Islas-Osuna, M.A., and Dieckmann, C.L. (1999). Suppressor analysis of mutations in the 5′-untranslated region of COB mRNA identifies components of general pathways for mitochondrial mRNA processing and decay in Saccharomyces cerevisiae. Genetics 151, 1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T. Edwards, J. C. Mueller, D.M., and Rabinowitz, M. (1983). Identification of a single transcriptional initiation site for the glutamic tRNA and COB genes in yeast mitochondria. Proc. Natl. Acad. Sci. USA 80, 5564-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin, J.W., Dhillon, R., Ritzel, R.G., and Nargang, F.E. (1997). The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32, 273-280. [DOI] [PubMed] [Google Scholar]
- Cronan, J.E., Jr. (1990). Biotination of proteins in vivo: a post-translational modification to label, purify, and study proteins. J. Biol. Chem. 265, 10327-10333. [PubMed] [Google Scholar]
- Decoster, E., Simon, M., Hatat, D., and Faye, G. (1990). The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 224, 111-118. [DOI] [PubMed] [Google Scholar]
- Dieckmann, C.L., Homison, G., and Tzagoloff, A. (1984a). Assembly of the mitochondrial membrane system: nucleotide sequence of a yeast nuclear gene (CBP1) involved in 5′ end processing of cytochrome b pre-mRNA. J. Biol. Chem. 259, 4732-4738. [PubMed] [Google Scholar]
- Dieckmann, C.L., Koerner, T.J., and Tzagoloff, A. (1984b). Assembly of the mitochondrial membrane system: CBP1, a yeast nuclear gene involved in 5′ end processing of cytochrome b pre-mRNA. J. Biol. Chem. 259, 4722-4731. [PubMed] [Google Scholar]
- Dieckmann, C.L., and Mittelmeier, T.M. (1987). Nuclearly encoded Cbp1 interacts with the 5′ end of mitochondrial cytochrome b pre-mRNA. Curr. Genet. 12, 391-397. [DOI] [PubMed] [Google Scholar]
- Dieckmann, C.L., Pape, L.K., and Tzagoloff, A. (1982). Identification and cloning of a yeast nuclear gene (CBP1) involved in expression of mitochondrial cytochrome b. Proc. Natl. Acad. Sci. USA 79, 1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann, C.L. and Staples, R.R. (1994). Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. In: International Review of Cytology, ed. K.W. Jeon and J.W. Jarvik, San Diego, CA: Academic Press. [DOI] [PubMed]
- Faye, G.C., Kujawa, C., and Fukuhara, H. (1974). Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J. Mol. Biol. 88, 185-203. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury, F., Roganti, T., Lecrenier, N., and Prunelle, B. (1998)., The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440, 325-331. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C.R., Meagher, D.A., Imai, B.S., and Mishe, S.M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601-605. [DOI] [PubMed] [Google Scholar]
- Green-Willms, N.S., Butler, C.A., Dunstan, H.M., and Fox, T.D. (2001). Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 276, 6392-6397. [DOI] [PubMed] [Google Scholar]
- Grivell, L.A., Artel-Sanz, M., Hakkart, G., de Jong, L., Nijtmans, L.G., van Oosterum, K., Siep, M., and van der Spek, H. (1999). Mitochondrial assembly in yeast. FEBS Lett. 452, 57-60. [DOI] [PubMed] [Google Scholar]
- Groudinsky, O., Bousquet, I., Wallis, M.G., Slominsky, P.P., and Dujardin, G. (1993). The NAM1/MTF2 nuclear gene product is selectively required for the stability and/or processing of mitochondrial transcripts of the atp6 and of the mosaic, cox1 and cytb genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 240, 419-427. [DOI] [PubMed] [Google Scholar]
- Islas-Osuna, M.A., Ellis, T.P., Marnell, L.L., Mittelmeier, T.M., and Dieckmann, C.L. (2002). Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 277, 37987-37990. [DOI] [PubMed] [Google Scholar]
- Islas-Osuna, M.A., Ellis, T.P., Mittelmeier, T.M., and Dieckmann, C.L. (2003). Suppressor mutations define two regions in the Cbp1 protein important for mitochondrial cytochrome b mRNA stability in Saccharomyces cerevisiae. Curr. Genet. 43, 327-336. [DOI] [PubMed] [Google Scholar]
- Jänsch, L., Kruft, V., Schmitz, U.K., and Braun, H.-P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357-368. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Manthey, G.M., and McEwen, J.E. (1995). The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey, G.M., Przybyla-Zawislak, B.D., and McEwen, J.E. (1998). The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 255, 156-161. [DOI] [PubMed] [Google Scholar]
- Mayer, S.A., and Dieckmann, C.L. (1991). Yeast CBP1 mRNA 3′ end formation is regulated during the induction of mitochondrial function. Mol. Cel. Biol. 11, 813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, U., Körte, A., and Rödel, G. (1991). Association of cytochrome b translational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol. Gen. Genet. 230, 177-185. [DOI] [PubMed] [Google Scholar]
- Mittelmeier, T.M., and Dieckmann, C.L. (1993). In vivo analysis of sequences necessary for Cbp1-dependent accumulation of cytochrome b transcripts in yeast mitochondria. Mol. Cell. Biol. 13, 4203-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha, V.K., et al. (2003). Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA 100, 605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, K., and Thogersen, H.C. (1984). Generation of beta-globin by sequence-specific proteolysis of a hybrid protein produced in Escherichia coli. Nature 309, 810-812. [DOI] [PubMed] [Google Scholar]
- Naithani, S., Saracco, S.A., Butler, C.A., and Fox, T.D. (2003). Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 324-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski, D.W., Swayne, T.C., and Pon, L.A. (2001). Epitope tagging and visualization of nuclear-encoded mitochondrial proteins in yeast. Methods Cell Biol. 65, 257-276. [DOI] [PubMed] [Google Scholar]
- Rabilloud, T. (1992). A comparison between low background silver diammine and silver nitrate protein stains. Electrophoresis 13, 429-439. [DOI] [PubMed] [Google Scholar]
- Rodeheffer, M.S., Boone, B.E., Bryan, A.C., and Shadel, G.S. (2001). Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 276, 8616-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer, M.S., and Shadel, G.S. (2003). Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 278, 18695-18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel, G. (1986). Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5′-untranslated COB leader. Curr. Genet. 11, 41-45. [DOI] [PubMed] [Google Scholar]
- Rödel, G., and Fox, T.D. (1987). The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5′-untranslated leader. Mol. Gen. Genet. 206, 45-50. [DOI] [PubMed] [Google Scholar]
- Rouillard, J.M., Dufour, M.E., Theunissen, B., Mandart, E., and Dujardin, G. (1996). SLS1, a new Saccharomyces cerevisiae gene involved in mitochondrial metabolism, isolated as a synthetic lethal in association with an SSM4 deletion. Mol. Gen. Genet. 252, 700-708. [DOI] [PubMed] [Google Scholar]
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220-230. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223-231. [DOI] [PubMed] [Google Scholar]
- Schneider, B.L., Seufert, W., Steiner, B., Yang, Q.H., and Futcher, A.B. (1995). Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11, 1265-1274. [DOI] [PubMed] [Google Scholar]
- Siep, M., van Oosterum, K., Neufeglise, H., van der Spek, H., and Grivell, L.A. (2000). Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 37, 213-220. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 6-7. [DOI] [PubMed] [Google Scholar]
- Sparks, K.A., Mayer, S.A., and Dieckmann, C.L. (1997). Premature 3′-end formation of CBP1 mRNA results in the downregulation of cytochrome b mRNA during induction of respiration in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4199-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.M., and McAlister-Henn, L. (1989). Dispensable presequence for cellular localization and function of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 12091-12096. [PubMed] [Google Scholar]
- Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R., and Yoshikawa, S. (1996). The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272, 1136-1144. [DOI] [PubMed] [Google Scholar]
- Tzagoloff, A., and Dieckmann, C.L. (1990). PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Boudreau, E., Lemaire, S.D., Goldschmidt-Clermont, M., and Rochaix, J.D. (2000). Characterization of Mbb1, a nucleus-encoded tetratri-copeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97, 14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, M.G., Groudinsky, O., Slonimsky, P.P., and Dujardin, G. (1994). The NAM1 protein (NAM1p), which is selectively required for cox1, cytb and atp6 transcript processing/stabilization, is located in the yeast mitochondrial matrix. Eur. J. Biochem. 222, 27-32. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and Shadel, G.S. (1999). Stability of the mitochondrial genome requires an amino-terminal domain of yeast mitochondrial RNA polymerase. Proc. Natl. Acad. Sci. USA 96, 8046-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, E.R., and Dieckmann, C.L. (1990). Identification of the CBP1 polypeptide in mitochondrial extracts from Saccharomyces cerevisiae. J. Biol. Chem. 265, 1594-1600. [PubMed] [Google Scholar]
- Wiesenberger, G., and Fox, T.D. (1997). Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol. Cell. Biol. 17, 2816-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]