Abstract
We have previously identified a new centrosomal protein, centrosomal protein 4.1-associated protein (CPAP), which is associated with the γ-tubulin complex. Here, we report that CPAP carries a novel microtubule-destabilizing motif that not only inhibits microtubule nucleation from the centrosome but also depolymerizes taxol-stabilized microtubules. Deletion mapping and functional analyses have defined a 112-residue CPAP that is necessary and sufficient for microtubule destabilization. This 112-residue CPAP directly recognizes the plus end of a microtubule and inhibits microtubule nucleation from the centrosome. Biochemical and functional analyses revealed that this 112-residue CPAP also binds to tubulin dimers, resulting in the destabilization of microtubules. Using the tetracycline-controlled system (tet-off), we observed that overexpression of this 112-residue CPAP inhibits cell proliferation and induces apoptosis after G2/M arrest. The possible mechanisms of how this 112-residue motif in CPAP that inhibits microtubule nucleation from the centrosome and disassembles preformed microtubules are discussed.
INTRODUCTION
Microtubules (MTs), which are composed of α/β tubulin heterodimers, are essential for a variety of cellular functions, including maintenance of cell shape, cell polarity, intracellular transport, cell mitosis, and meiosis. MT networks are intrinsically highly dynamic and undergo dramatic reorganization during the cell cycle (Desai and Mitchison, 1997). When cells enter mitosis, the interphase MT network is rapidly disassembled and then is followed by the reorganization of MTs into the mitotic spindle. The precise regulation of microtubule assembly and disassembly at both kinetochores and centrosomes is thought to be important for the maintenance of spindle structure (Waters et al., 1996) and chromosome segregation during mitosis (Desai and Mitchison, 1995).
Regulation of MT dynamics is commonly achieved by two groups of proteins, namely, MT stabilizers and destabilizers (Heald and Nogales, 2002). The former group is exemplified by microtubule-associated proteins, which stabilize MTs by binding along the sides of MTs (Cassimeris and Spittle, 2001). Members of the latter group include Op18/stathmin (Belmont and Mitchison, 1996; Cassimeris, 2002), katanin (McNally and Vale, 1993; Hartman et al., 1998), and Kin I kinesins (Walczak et al., 1996; Desai et al., 1999), which are MT destabilizers with the capability of disassembling MTs.
Protein 4.1R, originally identified as an 80-kDa protein (4.1R-80) in human erythrocytes, plays a crucial role in maintaining the specialized mechanical properties of the red cell plasma membrane. Multiple protein 4.1R isoforms generated by alternative RNA splicing have been identified in erythroid (Conboy et al., 1988; Tang et al., 1988) and nonerythroid cells (Tang et al., 1990; Conboy et al., 1991; Huang et al., 1993). In nucleated cells, protein 4.1R is not exclusively associated with the plasma membrane-associated cytoskeleton. Immunoreactive epitopes of protein 4.1R have been detected at several intracellular sites, including the nuclear matrix (De Carcer et al., 1995; Krauss et al., 1997b), microtubules (Perez-Ferreiro et al., 2001), and centrosomes (Krauss et al., 1997a; Hung et al., 2000), which are crucial to cell division. We previously isolated a nonerythroid 4.1R isoform (4.1R-135), which is generated by alternative mRNA splicing that results in the addition of a 209-amino acid domain to the N terminus of erythroid 4.1R-80 (Tang et al., 1990). Interestingly, this nonerythroid isoform has been reported to interact with the nuclear mitotic apparatus protein (NuMA) in the interphase nucleus and form a complex with several spindle pole organizing proteins, including NuMA, dynein, and dynactin during cell division (Mattagajasingh et al., 1999). This suggests that 4.1R-135 may play an essential role in organizing the nuclear architecture and mitotic spindle (Mattagajasingh et al., 1999).
In addition to NuMA, which binds to the carboxyl-terminal domain of 4.1R-135, we searched for proteins that interact with the head domain (residues 1–209) of 4.1R-135. Using a yeast two-hybrid screening system, we isolated a novel centrosomal protein, centrosomal P4.1-associated protein (CPAP), which is associated with the γ-tubulin complex (Hung et al., 2000). Functional analysis revealed that the MTs nucleated from the purified centrosomes were significantly inhibited by anti-CPAP antibody, suggesting that CPAP may play a role in the regulation of MT assembling and centrosome function (Hung et al., 2000). In this study, we characterize the function of CPAP that shows it to possess a novel microtubule-destabilizing motif. This motif contains only 112 amino acid residues that not only inhibit MT polymerization but also depolymerizes taxol-stabilized MTs. Thus, besides the known microtubule destabilizers (Op18/stathmin, katanin, and Kin I kinesins), CPAP may possibly represent a novel type of MT destabilizer.
MATERIALS AND METHODS
Purification of Glutathione S-Transferase (GST)-recombinant Proteins
The cDNAs encoding different portions of CPAP were fused in-frame to GST in pGEX-2T (Amersham Biosciences, Piscataway, NJ). Overexpression and affinity purification of GST recombinant proteins were performed as described previously (Hung et al., 2000).
In Vitro Microtubule Nucleation Assay
Centrosome isolation and microtubule nucleation tests were performed as described previously (Hung et al., 2000). To test the effect of different GST-CPAP truncated proteins on microtubule nucleation, the purified centrosomes (4 μl/each reaction) were preincubated with various GST-CPAP truncated proteins for 10 min at 4°C. The reaction mixture was then incubated in 65 μl of RG1 buffer (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgCl2, and 1 mM GTP) containing 125 μg of bovine brain tubulin (Cytoskeleton, Denver, CO) for 4 min at 37°C. Microtubules were fixed by adding 200 μl of 1% glutaraldehyde in RG1 buffer, sedimented onto acid-treated coverslips, and then examined by immunofluorescence microscopy by using anti-α-tubulin monoclonal antibodies N356 (Amersham) or fluorescein isothiocyanate (FITC)-conjugated anti-α-tubulin monoclonal antibodies (DM1A-FITC; Sigma-Aldrich, St. Louis, MO). The GST-CPAP truncated proteins were detected with anti-GST polyclonal antibody (Molecular Probes, Cincinnati, OH).
To test the effect of different GST-CPAP-truncated proteins on microtubule polymerization in absence of the centrosome, we followed the above-mentioned procedure except that the centrosomes were removed from the reaction mixture. The polymerized MTs were stained by FITC-conjugated anti-α-tubulin monoclonal antibodies (DM1A-FITC). Nocodazole (15 μM) was used as a positive control to evaluate the depolymerization of MTs.
Microtubule Sedimentation Assay
Pure bovine brain α/β-tubulin (15 μM) obtained from Cytoskeleton was preincubated with various amounts of GST-CPAP truncated proteins in 50 μl of RG1 buffer for 30 min at 37°C. Tubulin polymerization was initiated by the addition of 50 μl of GMRG buffer (RG1 buffer containing 12 mM MgCl2 and 50% glycerol) and incubation at 37°C for 1.5 h. The reaction mixture was then centrifuged at 300,000 × g for 15 min at 37°C in a TL-100 ultracentrifuge (Beckman Coulter, Fullerton, CA). Supernatants and pellets were subjected to SDS-PAGE analysis followed by Coomassie Blue staining. In another experiment, MTs were prepolymerized by 25 μM paclitaxel (taxol) for 10 min at 37°C in RG1 buffer containing 4 mM MgCl2, 4 mM ATP, and 4 mM GTP. GST-PN2-2 or GST-PN2-3 recombinant proteins were then added to the polymerized MTs and incubated at 37°C for an additional 20 min. After incubation, the reaction mixture was centrifuged on a 50-μl glycerol cushion (50% glycerol, 10 μM taxol, and 2 mM GTP in RG1 buffer) at 100,000 × g for 30 min at 37°C in a Beckman TL-100 ultracentrifuge.
In Vitro Tubulin Dimer Binding Assay
GST- or GST-CPAP–truncated proteins were affinity purified and immobilized on glutathione-agarose beads (Sigma-Aldrich). The immobilized beads were then incubated with 7.5 μM α/β-tubulin (Cytoskeleton) in 50 μl of 1× RG1 buffer for 30 min at 4°C or at room temperature with nocodazole (15 μM). After incubation, the supernatants were collected and then the beads were washed three times with 1× RG1 buffer, followed by 1× RG1 buffer containing 150 mM NaCl, and finally 1× RG1 buffer. The supernatants and beads were then subjected to SDS-PAGE analysis and the protein bands were stained with Coomassie Blue.
To perform gel filtration chromatography of PN2-3-His6 and tubulin, the samples were injected into a Superdex 200 HR10/300 GL (Amersham Biosciences) packed in RG2 buffer (80 mM PIPES, 1 mM MgCl2, and 1 mM EGTA, pH 6.8). The column was run with RG2 buffer at 0.4 ml/min, and 0.5-ml fractions were collected with an AKTA purified 10-system (Amersham Biosciences). The elution profiles of proteins were analyzed for α-tubulin and PN2-3-His6 by immunoblotting with anti-α-tubulin antibody (Molecular Probes) or anti-His antibody (Serotec, Oxford, United Kingdom), followed by scanning densitometry. The column was calibrated with ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (68 kDa), and ovalbumin (45 kDa) as sizing standards (Amersham Biosciences).
Cell Culture and Transfection
HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. The cDNA clones encoding different portions of CPAP were subcloned in-frame into a cytomegalovirus promoter-driven enhanced green fluorescent protein (EGFP)-C1 expression vector (BD Biosciences Clontech, Palo Alto, CA) and then were transiently transfected into cells by LipofectAMINE 2000 as suggested by the manufacturer (Invitrogen, Carlsbad, CA).
Cold Treatment, Immunofluorescence, and Confocal Microscopy
Cultured cells were grown on coverslips for >24 h and then incubated at 4°C for 1 h. After cold treatment, the cold medium was replaced with warm medium and further incubated for 2 min at 37°C. Cells were then fixed with 3.7% formaldehyde at room temperature for 10 min. The fixed cells were probed with anti-α-tubulin monoclonal antibodies (N356; Amersham Biosciences) and anti-γ-tubulin polyclonal antibodies (Sigma-Aldrich). DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). The anti-α-tubulin monoclonal antibodies (N356) were detected with either Alexa 568, a Texas Red-conjugated goat anti-mouse IgG, or Alexa 647, a far-red fluorophore-conjugated goat anti-mouse IgG. The anti-γ-tubulin polyclonal antibodies were detected with Alexa 568 (Molecular Probes). Coverslips were mounted and observed with a laser scanning confocal system (MRC 1000; Bio-Rad, Hercules, CA).
Generation of Green Fluorescent Protein (GFP)-PN2-3–inducible Cell Lines
The cDNAs encoding GFP-PN2-3 (pEGFP-PN2-3) and GFP (pEGFP-C1) were subcloned into a pTRE2hgy vector (BD Biosciences Clontech) downstream of the tetracycline-responsive promoter. HeLa Tet-Off cells (BD Biosciences Clontech) were transfected with the above-mentioned constructs by using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's protocol. One day after transfection, the cells were subjected to selection with hygromycin B (200 μg/ml) in the presence of 0.5 μg/ml doxycycline (BD Biosciences Clontech). Hygromycin-resistant cell lines were screened for induction of GFP-PN2-3, or GFP expression upon removal of doxycycline by immunoblotting with anti-GFP antibodies (Molecular Probes).
FACScan Analysis
GFP- and GFP-PN2-3–inducible HeLa Tet-Off cell lines were cultured in medium without doxycycline for the indicated time. Total cells were harvested and fixed with 1% formaldehyde for 30 min at 4°C and then treated with 70% ethanol for another hour at -20°C. After fixation, the cells were incubated in 1 ml of phosphate-buffered saline solution containing RNase (100 U/ml) and propidium iodide (50 μg/ml) for 1 h in the dark place. GFP and DNA content analyses were performed using FACSCalibur (BD Biosciences, San Jose, CA) with CellQuest or ModFit LT software (BD Biosciences).
Cell Proliferation Assay
Both adherent and nonadherent cells were harvested at indicated time points and were analyzed in 96-well assay plates by a nonradioactive 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTS) cell proliferation kit according to the manufacturer's protocol (Promega, Madison, WI). Cellular proliferation was measured by reduction of MTS (a tetrazolium compound) into the formazan product by the dehydrogenase enzymes found in metabolically active cells. The quantity of formazan product as measured by 490-nm absorbance is directly proportional to the number of living cells and metabolic activity. MTS assay were performed in triplicate, and the mean values ± SD at 490 nm were measured.
Terminal Deoxynucleotidyl Transferase dUTP Nick-End Labeling (TUNEL) Assay
Apoptosis was measured by TUNEL and DAPI staining. HeLa Tet-Off-GFP-PN2-3 or GFP cells were cultured for 4 d in the absence of doxycycline. Both adherent and nonadherent cells were trypsinized and cytospined onto coverslips and then subjected to TUNEL assay by using an in situ cell death detection kit, TMR red (Roche Diagnostics, Mannheim, Germany). To confirm the TUNEL assay findings, apoptotic cells were also analyzed by DAPI staining. The cells were cytospined onto coverslips and fixed by 3.7% formaldehyde for 1 h at room temperature followed by treatment with 0.1% Triton X-100 for 5 min at 4°C. After washing, the cells were incubated with a TUNEL reaction mixture (Roche Diagnostics) and followed by DAPI staining. The cells were mounted on coverslips and observed under a fluorescence microscope (Axiovert 200 M; Carl Zeiss, Jena, Germany) coupled with a cool charge-coupled device system. The HeLa Tet-Off cells pretreated 0.1 μM okadaic acid (OA) were used as a positive control for TUNEL assay.
RESULTS
The PN2-3 Domain of CPAP Inhibits Microtubule Nucleation from the Centrosome
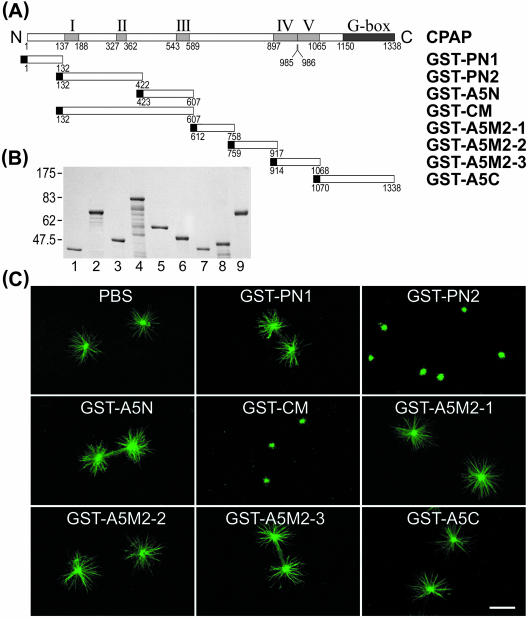
We previously identified a novel centrosomal protein CPAP, which may participate in centrosome function (Hung et al., 2000). To examine how CPAP regulates MT assembly from the centrosome, we have established an in vitro microtubule nucleation assay. We tested whether CPAP possesses a functional domain that affects MT assembly from the centrosome. A series of GST-tagged CPAP truncated clones (Figure 1A) were constructed, expressed in Escherichia coli and the chimeric proteins affinity purified (Figure 1B). The purified centrosomes were incubated with various GST-CPAP–truncated proteins and then tested for their capacity to nucleate MTs. As shown in Figure 1C, the formation of MT asters from the purified centrosome was specifically and significantly inhibited by GST-CM and GST-PN2 but not by other GST-CPAP–truncated proteins. A quantitative analysis of the inhibitory effect on the length as well as the number of growing MT is shown in Table 1. Both the number and the length of the microtubules were significantly affected by coincubation with GST-CM or GST-PN2 recombinant proteins. These results suggests that the PN2 (residues 132–422) domain in CM (residues 132–607) has an ability to obviate centrosome-dependent MT nucleation and assembly in vitro.
Figure 1.
Inhibition of microtubule nucleation from the purified centrosome. (A) cDNA constructs containing various portions of CPAP were subcloned into the pGEX-2T vector and expressed as GST-CPAP–truncated proteins in E. coli. (B) Coomassie Blue-stained gel with various affinity-purified GST-CPAP–truncated proteins used in microtubule nucleation assay. GST-PN1 (lane 1), GST-PN2 (lane 2), GST-A5N (lane 3), GST-CM (lane 4), GST-A5C (lane5), GST-A5M2-1 (lane 6). GST-A5M2-3 (lane7), GST-A5M2-2 (lane 8), and bovine serum albumin (BSA) (lane 9, 1 μg). (C) In vitro microtubule nucleation assay. The isolated centrosomes were preincubated with various GST-CPAP–truncated proteins (0.15 μM) for 10 min at 4°C, and then tubulin was added to initiate the microtubule nucleation. The microtubule asters were stained with FITC-conjugated anti-α-tubulin antibodies (DM1A-FITC). Bar, 10 μm. Five α-helical coiled-coil structures (I–V) and a G-box (glycine repeats) are indicated (Hung et al., 2000).
Table 1.
Effect of GST-CPAP-truncated proteins on microtubule nucleation in vitro
| No. of microtubules/centrosomea
|
||||
|---|---|---|---|---|
| GST-CPAP recombinant proteins (μM) | Mean of microtubule length (μm) ± SD | <10 | 10-20 | >20 |
| Control | 6.39 ± 0.47 | 0 | 0 | 56 |
| GST-PN1 (0.15) | 6.00 ± 0.36 | 0 | 0 | 57 |
| GST-PN2 (0.06) | 2.74 ± 0.75 | 10 | 8 | 32 |
| (0.15) | 0.87 ± 0.28 | 37 | 7 | 0 |
| GST-CM (0.06) | 3.25 ± 0.68 | 10 | 23 | 18 |
| (0.15) | 0.94 ± 0.19 | 46 | 2 | 0 |
The in vitro microtubule nucleation test was performed as described in the text. Photographs were taken, and the length and number of microtubules were calculated for ∼50 asters.
Data indicated the number of centrosomes with <10, 10-20, >20 microtubules per centrosome
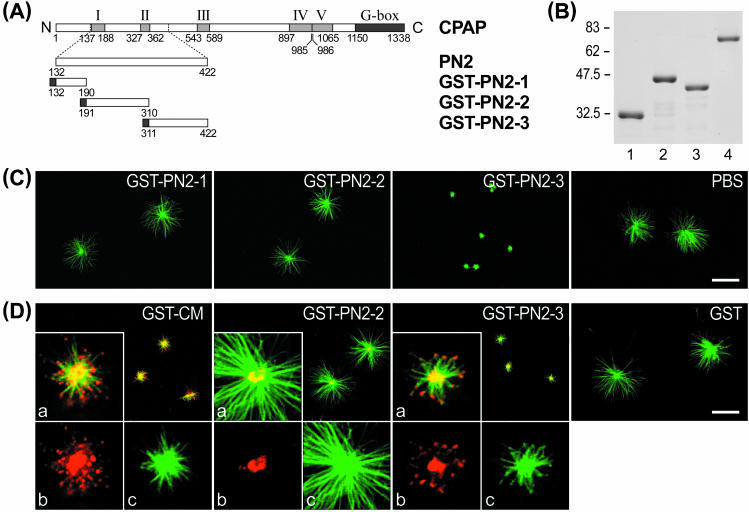
To define more precisely the region that exhibits the inhibitory activity in PN2, we further divided the PN2 into three subregions (PN2-1, PN2-2, and PN2-3) (Figure 2A), and GST recombinant proteins were affinity purified (Figure 2B). Figure 2C shows that only GST-PN2-3 (residues 311–422) has the ability to inhibit MT assembly from the centrosome. Furthermore, using anti-GST antibody, we detected that all three GST fusion proteins (GST-CM, GST-PN2-2, and GST-PN2-3) targeted the centrosome (see bright yellow spots in Figure 2D). However, the inhibitory effect was observed in GST-CM and GST-PN2-3 only, and not in GST-PN2-2 (Figure 2D) or GST-PN2–1 (our unpublished data), suggesting that PN2-3 located within the CM domain carries MT-destabilizing activity. Interestingly, confocal microscopy analysis showed that GST-CM and GST-PN2-3 seem to target and accumulate on the plus end of newly assembled MTs (see red spots in the inserted inlet in Figure 2D, a). GST-CM and GST-PN2-3 were present at the tip end in 81.44 and 81.25% of the MTs (n = 300 MTs), respectively.
Figure 2.
Mapping the microtubule-destabilizing activity of CPAP to the PN2-3 motif. (A) Three PN2 subregions, PN2-1, PN2-2, and PN2-3, were subcloned into a pGEX-2T vector. (B) Various GST-recombinant proteins, GST-PN2-1 (lane 1), GST-PN2-2 (lane 2), and GST-PN2-3 (lane 3), were affinity purified and stained with Coomassie Blue. Lane 4 represents 1 μg of bovine serum albumin (BSA). (C and D) In vitro microtubule nucleation assay. Various GST-CPAP–truncated proteins (0.45 μM) were preincubated with purified centrosomes as described in Figure 1C. The microtubule asters were stained with FITC-conjugated anti-α-tubulin antibodies (green color) (C), or doubly stained with both anti-α-tubulin monoclonal antibodies (green) and anti-GST polyclonal antibodies (red) (D). The inserted inlet (a) in Figure 2D is a merged photograph derived from two monochrome photos, b (anti-GST antibody, red) and c (anti-α-tubulin antibody, green). Bar, 10 μm.
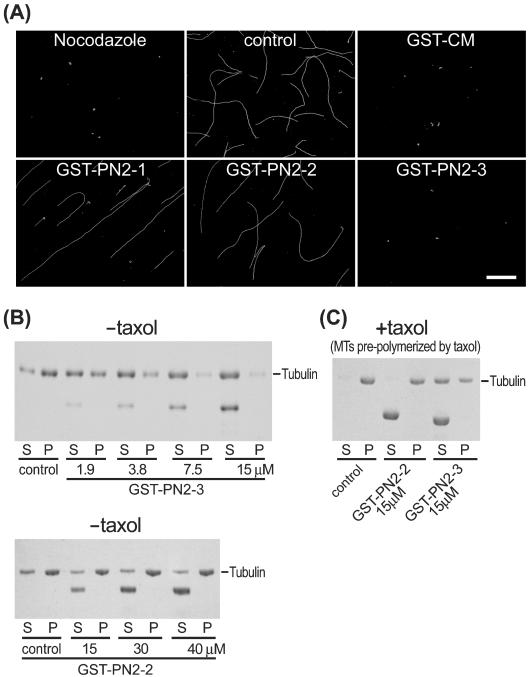
The PN2-3 Domain of CPAP Inhibits Tubulin Polymerization and Depolymerizes Taxol-stabilized Microtubules
We then tested how PN2-3 functions in the absence of the centrosome. We first examined the effects of PN2-3 polypeptide on microtubule polymerization by fluorescence microscopy. Figure 3A showed that microtubule polymerization is inhibited in the presence of GST-CM, GST-PN2-3, or nocodazole. However, no such effect was observed in GST-PN2-1– or GST-PN2-2–treated microtubules. This inhibition effect was further confirmed in a quantitative way by using an MT sedimentation assay. As shown in Figure 3B, GST-PN2-3 reveals a dose-dependent inhibition of tubulin polymerization. At equimolar concentrations, GST-PN2-3 almost completely inhibited the polymerization of a 15 μM tubulin solution. In contrast, no such effect was observed in GST-PN2-2 (even at 40 μM). Interestingly, GST-PN2-3 can depolymerize taxol-stabilized MTs (Figure 3C). In the presence of taxol, GST-PN2-3 but not GST-PN2-2 significantly disassembled taxol-stabilized MTs and prevented the sedimentation of MTs into the pellet (Figure 3C). Together, these results indicate that PN2-3, which contains amino acids 311–422 of CPAP, has the ability to inhibit tubulin polymerization and to depolymerize preformed MTs.
Figure 3.
(A) Testing the effects of PN2-3 polypeptide on microtubule polymerization by fluorescence microscopy. Tubulin (3 mg/ml) was incubated with different GST-CPAP–truncated proteins (0.45 μM) or nocodazole (15 μM). After incubation, the polymerized MTs were fixed and sedimented onto acid-treated coverslips as described in MATERIALS AND METHODS. MTs were analyzed by immunofluorescence assay using FITC-conjugated anti-α-tubulin antibodies (DM1A-FITC). Bar, 10 μm. Microtubules sedimentation assay (B and C). (B) Purified tubulins (15 μM) were incubated without (control) or with various amounts of GST-PN2-3 (top) or GST-PN2-2 (bottom) recombinant proteins. After centrifugation, the supernatants (S) and the pellets (P) were analyzed by SDS-PAGE, and the separated proteins were stained by Coomassie Blue. (C) Taxol-stabilized MTs (15 μM) were incubated with or without (control) indicated GST recombinant proteins and centrifuged through a glycerol cushion. After centrifugation, the supernatants (S) and the pellets (P) were analyzed by SDS-PAGE.
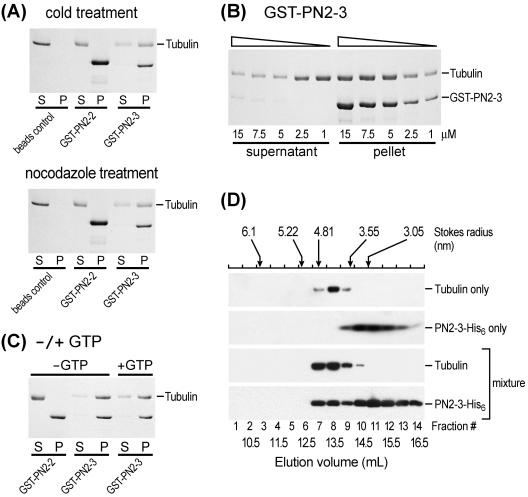
The PN2-3 Domain of CPAP Binds to a Tubulin Heterodimer
The molecular mechanism of how PN2-3 functions is not clear. One possible mechanism is that the PN2-3 polypeptide may bind to tubulin dimers, and thus inhibit tubulin polymerization. To test this hypothesis, we developed an in vitro tubulin dimer binding assay (low concentration of tubulin, incubation at 4°C, or in the presence of nocodazole) that favors the formation of tubulin dimers instead of polymers in the reaction solution. As shown in Figure 4A in cold-treated experiments, we observed that GST-PN2-3, but not GST-PN2-2, binds to tubulin dimers. Interestingly, the binding of PN2-3 to tubulin dimers revealed a dose-dependent (Figure 4B) and GTP-independent (Figure 4C) pattern. The stoichiometry of the GST-PN2-3 and a tubulin dimer was ∼1.0 (Figure 4B). Similar results were also observed in nocodazole-treated experiments (Figure 4A).
Figure 4.
GST-PN2-3 binds to a tubulin dimer. (A) Various GST-fusion proteins that had been preimmobilized on glutathione-agarose beads were incubated with purified tubulin (7.5 μM) in 1× RG1 buffer at 4°C for 30 min (cold treatment) or in the presence of nocodazole (15 μM). After centrifugation, the supernatants (S) and the pellets (P) containing the beads were analyzed on 12% SDS-PAGE, and the protein bands were stained by Coomassie Blue. (B) A dosage-dependent binding test. Various amounts of GST-PN2-3 fusion proteins were incubated with a fixed amount (7.5 μM) of purified tubulin and analyzed as described in A. (C) GTP requirement test. Various GST-fusion proteins were incubated with purified tubulins in 1× RG1 buffer with (+) or without (-) the addition of GTP. The reaction mixtures were analyzed as described in A. (D) Gel filtration chromatography of PN2-3-His6 and tubulin. The elution profiles of proteins on a Superdex 200 HR10/300 GL (Amersham Biosciences) gel filtration column were assayed for α-tubulin and PN2-3-His6 by immunoblotting analysis. According to the prerun protein size makers, the Stokes radius for a tubulin dimer and PN2-3-His6, which eluted at ∼13.2 and 15 ml, are 4.18 and 1.45 nm, respectively. The addition of tubulin to the PN2-3-His6 induced a shift of the PN2-3-His6 peak (∼13 ml) that may represent one PN-2-3-His6 molecule and one tubulin dimer.
The direct interaction of PN2-3 with a tubulin dimer was further confirmed by gel filtration chromatography (Figure 4D). To eliminate GST's possible steric effect and to create a better size distinction in the gel filtration profile, the GST-PN2-3 recombinant protein was replaced by PN2-3-His6. The protein elution profiles were examined by immunoblotting. As shown in Figure 4D, tubulin alone was eluted at ∼13.2 ml as a major peak in a dimer form in Stokes radius of 4.18 nm. This finding is consistent with previous reports (Fukata et al., 2002), indicating that tubulin is present largely as a heterodimer under these conditions. The addition of tubulin to the PN2-3-His6 induced a shift of the PN2-3-His6 peak (Figure 4D). Immunoblotting analysis indicated that this shift peak was composed of PN2-3-His6 and tubulin. The increased Stoke's radius of PN2-3 in the presence of tubulin is consistent with formation of a 1:1 complex with one PN2-3-His6 and one α/β tubulin heterodimer (Figure 4D).
Overexpression of Full-Length CPAP or PN2-3 Inhibits Microtubule Reassembly in Vivo
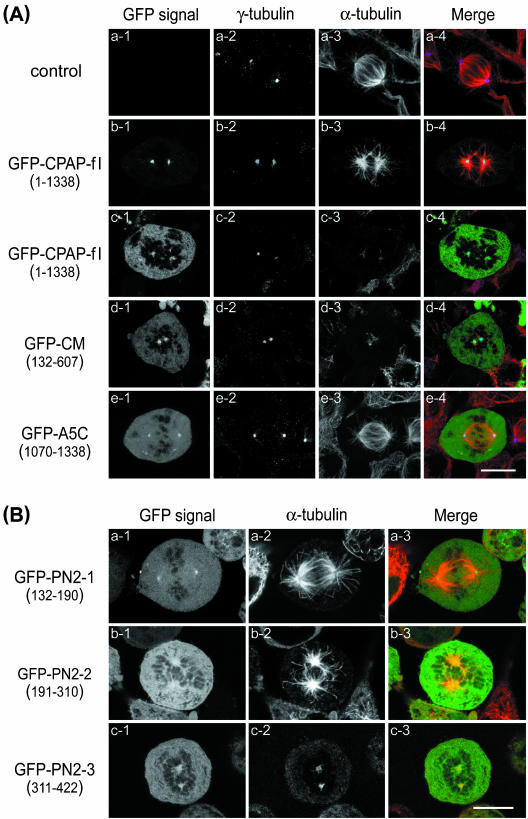
To examine how the CPAP molecule destabilizes MT in vivo, we prepared a series of GFP-tagged deletion constructs and expressed both full-length and truncated CPAP proteins in HeLa cells by transient transfection (Figure 5). The inhibitory effect of MT reassembly was observed in cold-treated cells. It has long been recognized that cold temperature (4°C) treatment can depolymerize microtubules into α/β tubulin dimers; however, microtubules will soon reassemble if the temperature returns to 37°C. Using this approach, we found that overexpression of GFP-CPAP-full-length (Figure 5A, c3; GFP-CPAP-fl, residues 1–1338), GFP-CM (Figure 5A, d3; residues 132–607), and GFP-PN2-3 (Figure 5B, c2; residues 311–422) significantly inhibit MTs reassembly in cold-treated mitotic cells. In those cells, GFP-CPAP-full length (Figure 5A, c1), GFP-CM–truncated proteins (Figure 5A, d1), and GFP-PN2-3 (Figure 5B, c1) were not only targeted the mitotic centrosomes but also expressed in a significant amount in the cytosol.
Figure 5.
CPAP and its derived domains (CM, PN2-3) inhibit microtubule reassembly from cold-treated cells. (A) HeLa cells were transiently transfected with no vector (control, ∼a1–a4) or with GFP-CPAP-fl (full-length CPAP; ∼b1–b4 [low expression] and ∼c1–c4 [high expression]), GFP-CM (∼d1–d4), and GFP-A5C (∼e1–e4). Twenty-four hours subsequent to transfection, the cells were cold treated (4°C) for 1 h to disassemble the microtubules and then shifted to 37°C for 2 min to reassemble the microtubules. After treatment, the cells were fixed and doubly stained with anti-γ-tubulin (blue) and anti-α-tubulin (red) antibodies. The GFP-CPAP chimeric proteins (green) were directly observed with a confocal fluorescent microscope. Only the merged images of mitotic cells are displayed in color. Bar, 10 μm. (B) HeLa cells were transfected with GFP-PN2-1 (∼a1–a3), GFP-PN2-2 (∼b1–b3), or GFP-PN2-3 (∼c1–c3). After 24 h of transfection, cells were cold treated and analyzed by confocal microscopy as described above. Bar, 10 μm.
However, no significant inhibition on MT reassembly was observed in the transfected cells that overexpressed GFP-A5C (Figure 5A, e3; residues 1070–1338), other CPAP deletion constructs, such as GFP-PN2-1 (Figure 5B, a2; residues 132–190), GFP-PN2-2 (Figure 5B, b2; residues 191–310), GFP-PN1 (residues 1–132), GFP-A5N (residues 423–607), and GFP-A5M2 (residues 607-1070) (our unpublished data), or transfected cells with low-level expression of GFP-CPAP-fl (Figure 5A, b3; in these cells, the GFP-CPAP-fl green fluorescent signal was strongly detected on the mitotic centrosomes, but weakly detected in the cytosol; Figure 5A, b1). Similar results were observed in transfected 293 and SiHa cells (our unpublished data). Together, our results show that the PN2-3 motif of CPAP inhibits MT reassembling from the centrosome in cold-treated cells, suggesting that CPAP may act as a microtubule destabilizer in vivo.
Overexpression of the PN2-3 Polypeptide Inhibits Cell Proliferation and Induces G2/M Arrest as Well as Cell Apoptosis
To analyze the functions of PN2-3 within the cells in more detail, we established several HaLa-derived stable transfectants of GFP-PN2-3 and GFP in HeLa Tet-Off cells under the tetracycline control. Several stable cell lines of these two genes were expanded and screened for efficient gene induction by the removal of doxycycline. Representative transfectants were compared with parental cells for gene expression with or without doxycycline. Figure S1 (see supplementary data) shows an example of immunoblot analysis of several representative transfectants that were induced to express GFP-PN2-3 protein in the absence of doxycycline at different time intervals. The differential expression level of GFP-PN2-3 was observed in these cell lines. Clone #29 (GFP-PN2-3#29) with high level expression of PN2-3 was selected for the following analyses.
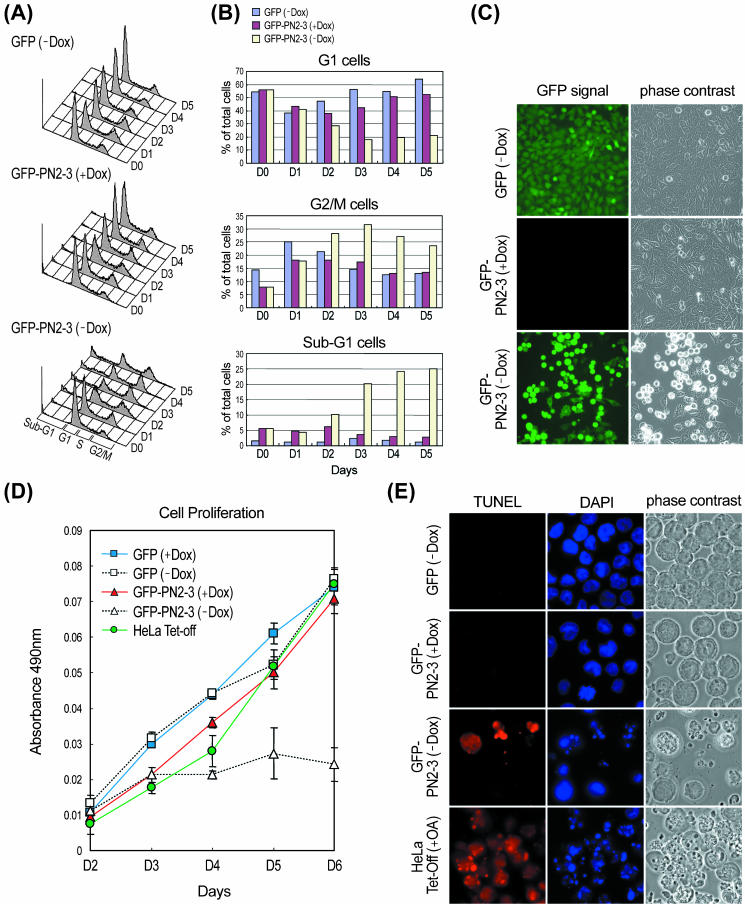
We first examined the effect of overexpression of PN2-3 on cell proliferation. HeLa Tet-Off-GFP-PN2-3, and -GFP cells were switched into the culture medium without doxycycline. At the indicated time interval after removal of doxycycline, the cell proliferation activity of each transfectant was analyzed by MTS assay. As shown in Figure 6D, induction of GFP-PN2-3 (-Dox) expression results in an inhibition of cell proliferation. In contrast, GFP alone has no effect on cell proliferation.
Figure 6.
Effects of overexpression of GFP-PN2-3. HeLa Tet-Off-GFP and -GFP-PN2-3 cells were cultured with (+Dox) or without (-Dox) doxycycline for indicated days and harvested for fluorescence-activated cell sorting analysis (A). Three independent experiments produced similar results and a histogram of a representative experiment is shown. (B) Percentage of relative cell numbers of G1 (2N), G2/M (4N), and subG1 (<2N) cells were calculated based on the results shown in A. (C) Overexpression of GFP-PN2-3 (-Dox) (4 d after removal of doxycycline), but not GFP (-Dox), induced a change in cell morphology (round cells). (D) Effect of overexpression of GFP-PN2-3 or GFP on cell proliferation. After removing doxycycline at the indicated time, cell proliferation was monitored by MTS assay. (E) Apoptotic cell death induced by overexpression of GFP-PN2-3. HeLa Tet-Off-GFP-PN2-3 cells were cultured with or without doxycycline for 4 d and then subjected to TUNEL assay. Parental HeLa Tet-Off cells treated with OA (0.1 μM) for 18 h were included as a positive control. Condensed and fragmented nuclei were detected only in OA-treated and GFP-PN2-3–induced cells. Three independent experiments produced similar results.
To determine at which stage of cell cycle the cells were arrested, we performed cell cycle analysis. As shown in Figure 6A, overexpression of GFP-PN2-3 (-Dox) induced a progressive accumulation of cells in G2/M phase with a corresponding decrease of cells in G1 phase (also see Figure 6B). Hela-Tet-Off-GFP-PN2-3 (-Dox) cells also exhibited a change in cell morphology (Figure 6C, round cells). These cell cycle and morphological changes were not observed in cells that expressed only GFP (-Dox) or in noninduced GFP-PN2-3 (+Dox) cells that indicates that PN2-3 was responsible for the arrest in G2/M and the accompanying morphological changes.
The cell cycle analysis also revealed overexpression of PN2-3–induced cell apoptosis. As shown in Figure 6A, a progressive accumulation of subG1 phase cells with DNA content less than 2N was observed in GFP-PN2-3 (-Dox) cells. To confirm that this population was the result of apoptosis, cells were subjected to TUNEL assay. As shown in Figure 6E, an increase in TUNEL-positive cells was detectable in GFP-PN2-3 (-Dox) cells, but not in noninduced cells GFP-PN2-3 (+Dox). Induction of GFP alone revealed no or very few TUNEL-positive cells (GFP-Dox). Furthermore, we observed that the subG1 cells (apoptosis) were significantly increased right after an accumulation of the G2/M cells and an increase of subG1 cells is always correlated with a decrease of cells in G1 phase (Figure 6B). Similar results were also observed in clones #89 and #15, which showed similar GFP-PN2-3 expression level as clone #29 (our unpublished data). Our results suggested that overexpression of PN2-3 in transfected cells inhibits cell proliferation and induces apoptosis after G2/M arrest.
DISCUSSION
Perhaps the most dramatic change in MT reorganization occurs at the G2/M transition in the cell cycle. The interphase MT array is rapidly disassembled, followed by the reassembly of MTs into the mitotic spindle. MT assembly is initiated in vivo by γ-tubulin complexes at the centrosome (Schiebel, 2000). In this work, we identified a novel MT-destabilizing motif (PN2-3 with 112 amino acids) in CPAP, which possesses an ability to prevent tubulin polymerization and depolymerize taxol-stabilized MTs. To our surprise, no significant sequence homology was found between PN2-3 and other known MT destabilizers, including Op18/stathmin (Belmont and Mitchison, 1996; Cassimeris, 2002), katanin (McNally and Vale, 1993; Hartman et al., 1998), and Kin I kinesins (Walczak et al., 1996; Desai et al., 1999), suggesting that PN2-3 represents a novel type of MT-destabilizing motif.
Recently, Desai et al. (1999) proposed a model for the mechanism of MT destabilization by Kin I kinesins. Hartman and Vale (1999) suggested a mechanism of how katanin, a member of the AAA ATPase superfamily, uses nucleotide hydrolysis energy to sever and disassemble MTs. Furthermore, Op18/stathmin was proposed to destabilize MTs either by increasing the rate of catastrophes (Belmont and Mitchison, 1996) or by promoting tubulin-sequestering activity (Jourdain et al., 1997), or by using both mechanisms (Howell et al., 1999; Larsson et al., 1999). The sequence feature of the PN2-3 domain in CPAP differs from those of Kin I kinesins, katanin, and Op18/stathmin, suggesting that this 112-residue CPAP destabilizes MTs via a different mechanism.
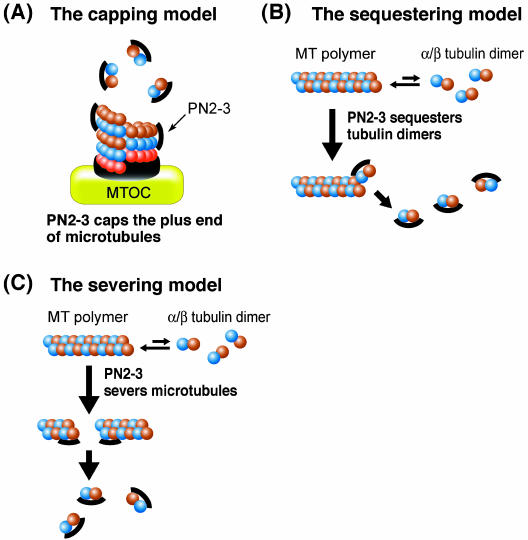
Based on the present study, we have proposed at least three possible mechanisms by which the PN2-3 domain in CPAP destabilizes MTs (Figure 7). One possible mechanism is that PN2-3 directly recognizes the plus end of a MT, which results in the inhibition of MT nucleation from the centrosome (Figure 7A). This view is supported by the observation that PN2-3 directly targets and accumulates to the plus end of newly assembled MTs from the centrosome (Figure 2D). However, in the absence of a centrosome, PN2-3 may destabilize MTs either by directly binding to a tubulin heterodimer, thereby depleting the pool of tubulin available for polymerization (tubulin-sequestering activity) (Figure 7B), or severing polymerized MTs (or taxol-stabilized MTs) via an unknown mechanism (tubulin-severing activity; Figure 7C). In the present study, we found that PN2-3 can depolymerize preformed MTs (or taxol-stabilized MTs) (Figure 3C), and PN2-3 directly binds to a tubulin heterodimer (Figure 4). These findings are consistent with the above hypotheses. Finally, the PN2-3 domain in CPAP may function in a combination of these mechanisms described above.
Figure 7.
Proposed models for microtubule destabilization by the PN2-3 polypeptide. (A) MT capping model: the PN2-3 polypeptide is proposed to bind to the plus end of a microtubule resulting in the inhibition of MT nucleation from the centrosome. (B) Tubulin-sequestering model: the PN2-3 polypeptide directly binds to a α/β tubulin heterodimer and thus depletes the pool of tubulin available for polymerization. (C) Tubulin-severing model: the prepolymerized or taxol-stabilized MTs can be severed by the PN2-3 polypeptide.
In this study, we report the identification of a novel microtubule-destabilizing motif in CPAP that possesses the ability to destabilize microtubules; however, the physiological function of the whole CPAP molecule is not entirely clear. It is possible that CPAP disassembles MTs via its PN2-3 motif and regulates MT dynamics at the centrosomes. Future experiments are clearly necessary to determine how an intact CPAP molecule destabilizes microtubules within the cells.
Supplementary Material
Acknowledgments
We are grateful for Drs. Yu-Li Wang (University of Massachusetts Medical School, Worcester, MA) and David Tu (The Pennsylvania State University, State College, PA) for helpful comments and discussion. We also thank Kuan-Yu Chou for assistance in confocal microscopy and Dr. C.V. Weaver for editing help. This work was supported by a grant from the National Science Council and Cancer Genomic Program project grant from Academia Sinica, Taipei, Taiwan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0121. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0121.
Online version of this article contains supporting material.
Online version is available at www.molbiolcell.org.
References
- Belmont, L.D., and Mitchison, T.J. (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623-631. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L. (2002). The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 14, 18-24. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., and Spittle, C. (2001). Regulation of microtubule-associated proteins. Int. Rev. Cytol. 210, 163-226. [DOI] [PubMed] [Google Scholar]
- Conboy, J.G., Chan, J., Mohandas, N., and Kan, Y.W. (1988). Multiple protein 4.1 isoforms produced by alternative splicing in human erythroid cells. Proc. Natl. Acad. Sci. USA 85, 9062-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy, J.G., Chan, J.Y., Chasis, J.A., Kan, Y.W., and Mohandas, N. (1991). Tissue- and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J. Biol. Chem. 266, 8273-8280. [PubMed] [Google Scholar]
- De Carcer, G., Lallena, M.J., and Correas, I. (1995). Protein 4.1 is a component of the nuclear matrix of mammalian cells. Biochem. J. 312, 871-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1995). A new role for motor proteins as couplers to depolymerizing microtubules. J. Cell Biol. 128, 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Desai, A., Verma, S., Mitchison, T.J., and Walczak, C.E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69-78. [DOI] [PubMed] [Google Scholar]
- Fukata, Y., et al. (2002). CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat. Cell Biol. 4, 583-591. [DOI] [PubMed] [Google Scholar]
- Hartman, J.J., Mahr, J., McNally, K., Okawa, K., Iwamatsu, A., Thomas, S., Cheesman, S., Heuser, J., Vale, R.D., and McNally, F.J. (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93, 277-287. [DOI] [PubMed] [Google Scholar]
- Hartman, J.J., and Vale, R.D. (1999). Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science 286, 782-785. [DOI] [PubMed] [Google Scholar]
- Heald, R., and Nogales, E. (2002). Microtubule dynamics. J. Cell Sci. 115, 3-4. [DOI] [PubMed] [Google Scholar]
- Howell, B., Larsson, N., Gullberg, M., and Cassimeris, L. (1999). Dissociation of the tubulin-sequestering and microtubule catastrophe-promoting activities of oncoprotein 18/stathmin. Mol. Biol. Cell 10, 105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.P., Tang, C.J., Kou, G.H., Marchesi, V.T., Benz, E.J., Jr., and Tang, T.K. (1993). Genomic structure of the locus encoding protein 4.1. Structural basis for complex combinational patterns of tissue-specific alternative RNA splicing. J. Biol. Chem. 268, 3758-3766. [PubMed] [Google Scholar]
- Hung, L.Y., Tang, C.J., and Tang, T.K. (2000). Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol. Cell. Biol. 20, 7813-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain, L., Curmi, P., Sobel, A., Pantaloni, D., and Carlier, M.F. (1997). Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry 36, 10817-10821. [DOI] [PubMed] [Google Scholar]
- Krauss, S.W., Chasis, J.A., Rogers, C., Mohandas, N., Krockmalnic, G., and Penman, S. (1997a). Structural protein 4.1 is located in mammalian centrosomes. Proc. Natl. Acad. Sci. USA 94, 7297-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss, S.W., Larabell, C.A., Lockett, S., Gascard, P., Penman, S., Mohandas, N., and Chasis, J.A. (1997b). Structural protein 4.1 in the nucleus of human cells: dynamic rearrangements during cell division. J. Cell Biol. 137, 275-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, N., Segerman, B., Howell, B., Fridell, K., Cassimeris, L., and Gullberg, M. (1999). Op18/stathmin mediates multiple region-specific tubulin and microtubule-regulating activities. J. Cell Biol. 146, 1289-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattagajasingh, S.N., Huang, S.C., Hartenstein, J.S., Snyder, M., Marchesi, V.T., and Benz, E.J. (1999). A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J. Cell Biol. 145, 29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and Vale, R.D. (1993). Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419-429. [DOI] [PubMed] [Google Scholar]
- Perez-Ferreiro, C.M., Luque, C.M., and Correas, I. (2001). 4.1R proteins associate with interphase microtubules in human T cells: a 4.1R constitutive region is involved in tubulin binding. J. Biol. Chem. 276, 44785-44791. [DOI] [PubMed] [Google Scholar]
- Schiebel, E. (2000). gamma-tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 12, 113-118. [DOI] [PubMed] [Google Scholar]
- Tang, T.K., Leto, T.L., Correas, I., Alonso, M.A., Marchesi, V.T., and Benz, E.J., Jr. (1988). Selective expression of an erythroid-specific isoform of protein 4.1. Proc. Natl. Acad. Sci. USA 85, 3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, T.K., Qin, Z., Leto, T., Marchesi, V.T., and Benz, E.J., Jr. (1990). Heterogeneity of mRNA and protein products arising from the protein 4.1 gene in erythroid and nonerythroid tissues. J. Cell Biol. 110, 617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., Mitchison, T.J., and Desai, A. (1996). XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37-47. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., Mitchison, T.J., Rieder, C.L., and Salmon, E.D. (1996). The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7, 1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.