Abstract
Of the uncloned ODA genes required for outer dynein arm assembly in Chlamydomonas, ODA5 and ODA10 are of particular interest because they do not encode known subunits of the outer arm or the outer dynein arm-docking complex (ODA-DC), and because genetic studies suggest their products interact. Beginning with a tagged oda5 allele, we isolated genomic and cDNA clones of the wild-type gene. ODA5 predicts a novel, 66-kDa coiled-coil protein. Immunoblotting indicates Oda5p is an axonemal component that assembles onto the axoneme independently of the outer arm and ODA-DC and is uniquely missing in oda5 and oda10 axonemes. Oda5p is released from the axoneme by extraction with 0.6 M KCl, but the soluble Oda5p does not cosediment with the outer dynein arm/ODA-DC in sucrose gradients. Quantitative mass spectrometry by using isotope coded affinity tagging revealed that a previously unidentified adenylate kinase is reduced 35–50% in oda5 flagella. Direct enzymatic assays demonstrated a comparable reduction in adenylate kinase activity in oda5 flagella, and also in oda10 flagella, but not in flagella of other oda mutants. We propose that Oda5p is part of a novel axonemal complex that is required for outer arm assembly and anchors adenylate kinase in proximity to the arm.
INTRODUCTION
Dyneins are large, multisubunit microtubule motors that are involved in many types of cellular movements, including vesicle transport, nuclear migration, spindle formation and orientation, chromosome movements, and beating of cilia and flagella. Eukaryotic flagella contain three major classes of dynein: dynein 1b/2, the retrograde motor for intraflagellar transport (Pazour et al., 1999a; Porter et al., 1999); the heterogeneous inner arm dynein system, containing up to seven isoforms (Porter and Sale, 2000); and the outer arm dynein (Witman et al., 1994), of which there is only one known isoform. The outer dynein arms provide up to four-fifths of the power for flagellar movement (Brokaw, 1994) and are required for normal flagellar beat frequency and swimming speed. Failure to assemble the outer arms leads to poorly motile cilia and flagella in humans, and is a common cause of the inherited disease primary ciliary dyskinesia (Afzelius and Mossberg, 1995). Hence, the identification of outer dynein arm components and the mechanisms that mediate their assembly are of considerable interest.
The outer and inner dynein arms assemble independently and are located at specific sites within the axoneme. Inherent to this assembly process is the requirement for unique structural or biochemical landmarks that ensure the proper targeting of each dynein isoform to its correct site both around and along the length of the axoneme. The most extensively investigated paradigm for axonemal dynein assembly and targeting is the Chlamydomonas reinhardtii outer dynein arm (Pazour and Witman, 2000). This arm consists of at least 13 polypeptides, which include three heavy chains (HCs), two intermediate chains (ICs), and several light chains (LC1–8) (DiBella and King, 2001), and repeats at 24-nm intervals along the A-tubules of the flagellar outer doublet microtubules. The correct positioning of the outer arm is due at least in part to its association with the outer dynein arm-docking complex (ODA-DC), which is required for attachment of the outer arm onto the A-tubule. The ODA-DC, a heterotrimeric complex comprised of subunits DC1 (Koutoulis et al., 1997), DC2 (Takada et al., 2002), and DC3 (Casey et al., 2003), assembles onto the site normally occupied by the outer arm even in the absence of the latter structure.
Loss of the outer arm in Chlamydomonas results in a characteristic slow, jerky swimming phenotype, and this has been used to isolate “oda” mutants unable to assemble the outer dynein arm (Kamiya, 1988). To date, 16 ODA genes have been identified (Table 1), most of which have been cloned and determined to encode proteins of the outer dynein arm or ODA-DC. Of the uncloned ODA genes, ODA5, ODA8, and ODA10 are of particular interest for two reasons. First, although they are required for outer arm assembly, they do not encode known subunits of the outer arm or the ODA-DC (King, Pazour, and Witman, unpublished data). Second, complementation assays in temporary dikaryons suggest that the products of these three genes interact to form a complex (Kamiya, 1988). Thus, these genes may encode subunits of an unidentified component important for outer arm assembly.
Table 1.
Chlamydomonas ODA genes
| Gene | Proteina | Reference |
|---|---|---|
| ODA1 | DC2 | Kamiya, 1988; Takada et al., 2002 |
| ODA2 | γDHC | Kamiya, 1988; Mitchell and Rosenbaum, 1985; Wilkerson et al., 1994 |
| ODA3 | DC1 | Kamiya, 1988; Koutoulis et al., 1997 |
| ODA4 | βDHC | Kamiya, 1988; Mitchell and Brown, 1994; Sakakibara et al., 1993 |
| ODA5 | This study | Kamiya, 1988 |
| ODA6 | IC2 | Kamiya, 1988; Mitchell and Kang, 1991 |
| ODA7 | ? | Kamiya, 1988 |
| ODA8 | ? | Kamiya, 1988 |
| ODA9 | IC1 | Kamiya, 1988; Wilkerson et al., 1995 |
| ODA10 | ? | Kamiya, 1988 |
| ODA11 | αDHC | Sakakibara et al., 1991 |
| ODA12 | LC2 (Tctex2) | Koutoulis et al., 1997; Pazour et al., 1999b |
| ODA13 | LC6 (LC8 homolog) | King and Patel-King, 1995; Pazour and Witman, 2000 |
| ODA14 | DC3 | Casey et al., 2003 |
| ODA15 | LC7 | Bowman et al., 1999; Pazour and Witman, 2000 |
| ODA16 | ? | Ahmed and Mitchell, 2003 |
Proteins whose sequences are unknown or unpublished are indicated by ?
Key steps to understanding such a complex will be to clone the ODA5, ODA8, and ODA10 genes and characterize their products. To that end, we used insertional mutagenesis to generate a tagged oda5 allele, which was then used to isolate the wild-type gene. ODA5 encodes a novel axonemal protein that assembles independently of the outer arm and ODA-DC and is missing in oda5 and oda10 axonemes but not in axonemes of other oda mutants. The absence of Oda5p is correlated with a reduction in the level of a previously uncharacterized flagellar adenylate kinase (AK). The results suggest that Oda5p is part of a complex that includes the flagellar AK. Therefore, the Oda5p-containing complex is not only essential for outer arm assembly but also probably anchors AK in proximity to the arm, ensuring that both high-energy phosphate bonds of ATP can be efficiently utilized at the axoneme's major site of power production.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii strains used in this study include the following: CC-2454 (cw15, nit1-305, mt-), CC-48 (arg2, mt+), CC124 (nit1-137, nit2–137, mt-), CC-2229 (oda1, mt+), CC2233 (oda3-1, nit1, nit2, AC17, mt-), and 137C (nit1–137, nit2-137, mt+), all from the Chlamydomonas Genetics Center (Department of Biology (Duke University, Durham, NC); oda5+ (oda5-1, mt+), oda3+ (oda3-1, mt+), oda8+ (oda8, mt+), oda9+ (oda9, mt+), oda10+ (oda10-1, mt+) (Kamiya, 1988), V87.2 (oda10-2, nit1::NIT1, NIT2, agg1, mt-) (Koutoulis et al., 1997), oda9-V5 (oda9, mt+) (Wilkerson et al., 1995), and 45BO3 (oda5-2, cw15, nit1-305::NIT1, mt-), an insertional allele of oda5.
45BO3 was crossed to 137C to create strain 88b (oda5-2, NIT1, mt-). 88b was crossed to CC-48 to create strain 112b (oda5-2, arg2, mt-). 112b.76, 112b.150, 112b.219, and 112b.220 (oda5-2::ODA5, arg2, mt-) were created by transformation of the ODA5 gene into strain 112b. Strain 112b.221.4 (oda5-2::HA-ODA5, arg2, mt-) was created by transformation of a hemagglutinin (HA)-tagged ODA5 gene into strain 112b.
Chlamydomonas cells were grown in a 14:10 light:dark cycle in the following media: medium I of Sager and Granick (1953) modified to contain three times the original amount of phosphate (Witman, 1986); R-medium (medium I supplemented with 0.1% sodium acetate); R+Arg (R-medium supplemented with 50 μg/ml arginine); M-N (medium I without nitrogen); TAP (Harris, 1989); TAP+Arg (TAP-medium supplemented with 50 μg/ml arginine); and SGII/NO3 [medium II of Sager and Granick (1953) modified to contain 0.003 M KNO3 as the nitrogen source].
Transformations and Insertional Mutagenesis
All transformations were done using the glass bead method as described previously (Kindle et al., 1989; Koutoulis et al., 1997). The insertional mutant 45BO3 was generated by transforming CC-2454 cells with plasmid pMN24 (Fernandez et al., 1989) containing the Chlamydomonas NIT1 gene. Transformants positive for NIT1 were selected on SGII/NO3 media. Motility mutants were identified by growing positive transformants in liquid culture and screening by light microscopy. Cotransformations were performed using ODA5 genomic constructs and pARG7.8 plasmid (Debuchy et al., 1989); transformants were selected on TAP plates.
Electron Microscopy
Whole cells and isolated axonemes were processed as described previously (Hoops and Witman, 1983). Samples were embedded in a mixture of LX112/Araldite 502 epoxy resin and sectioned at 50–70 nm.
Isolation and Blotting of Nucleic Acids
Chlamydomonas RNA was obtained before deflagellation and 30–45 min after deflagellation by pH shock (Witman et al., 1972). Total RNA was isolated by LiCl precipitation (Wilkerson et al., 1994) and polyA+ mRNA selected using Oligo dT cellulose (Ambion, Austin, TX). Polyadenylated RNA was separated on 1% formaldehyde agarose gels and transferred to Duralon-UV (Stratagene, La Jolla, CA). RNA was cross-linked to the membranes by using a Stratalinker (Stratagene). Chlamydomonas genomic DNA isolations were performed as described previously (Koutoulis et al., 1997). Genomic DNA was separated on 0.8% agarose gels. Transfer to Duralon-UV and cross-linking were the same as for RNA. Hybridization probes were generated by random prime labeling by using the Prime-It II kit (Stratagene).
Genetic Analysis
Matings and temporary dikaryon, stable diploid, and tetrad analyses were performed according to standard procedures (Kamiya, 1988; Harris, 1989; Dutcher, 1995). The ODA5 allele was confirmed by crossing 45B03 with the oda5-1 strain. Progeny were scored for motility (Oda +/-) by light microscopy and for Arg+/- by comparing growth on TAP and TAP+Arg plates.
Motion Analysis
Swimming speed was determined as described in Kamiya (1988). The swimming velocity of 30 cells was used to determine the average swimming speed for each strain analyzed. Flagellar beat frequency was determined as described previously (Kamiya, 2000).
Cloning and Sequencing of the ODA5 Gene and cDNA
A probe, 36.1, to unknown sequence adjacent to the NIT1 insertion in oda5-2 was used to identify wild-type BAC clones (Clemson University Genome Institute, Clemson, SC) containing the unknown sequence. The BAC clones were tested for their ability to rescue the Oda5- phenotype by cotransforming strain 112b with the BAC clones and plasmid pARG7.8 (Debuchy et al., 1989). Transformants were scored for Oda+/- phenotype by light microscopy. The smallest rescuing fragment, 50.1, from one BAC clone was sequenced.
Primers to predicted exons were generated (Integrated DNA Technologies, Coralville, IA) and polymerase chain reaction (PCR) used to amplify fragments from a cDNA library constructed from polyA+ mRNA isolated 30′ postdeflagellation (Wilkerson et al., 1994) or a gametic cDNA library (gift of William Snell, University of Texas Southwestern Medical Center, Dallas, TX). cDNA clones were sequenced to confirm intron-exon boundaries. Primers designed within ODA5 introns amplified the exons from oda5-1 genomic DNA. Sequencing of three independent clones identified the oda5-1 mutation. All sequencing was performed by either the Iowa State DNA Sequencing Facility or the University of Massachusetts Medical School Nucleic Acid Facility. See online supplemental materials regarding the cloning of sequence flanking the oda5-2 insertion site and for construction of an HA-tagged ODA5 gene construct.
Computational Analysis
Sequence assemblies were performed using LaserGene SeqMan (DNAstar, Madison, WI). The LaserGene EditSeq module was used for translations and to obtain the theoretical isoelectric point and mass of the predicted Oda5 protein. Primer design was performed using PRIMER3 at www.genome.wi.mit.edu/cgi-bin/primer/primer3 (Rozen and Skaletsky, 2000). The BLAST server at www.ncbi.nlm.nih.gov/BLAST (Altschul et al., 1990) was used to search for homologous sequences. The COILS (www.ch.embnet.org/software/COILS_form.html) and PairCoil (http://paircoil.lcs.mit.edu/cgi-bin/paircoil) servers were used to predict coiled-coil regions in the Oda5 protein (Lupas et al., 1991; Berger et al., 1995). To predict regions of coding potential, the ODA5 and AK genomic sequences were analyzed using the GreenGenie gene prediction program (http://www.cse.ucsc.edu/%7Edkulp/cgi-bin/greenGenie) (Li et al., 2003). The AK protein was examined using the Joint Genome Institute (Walnut Creek, CA; JGI) Chlamydomonas version 2.0 genome database (http://shake.jgi-psf.org/chlre2/chlre2.home.html), and the PROSITE protein families and domains database (http://us.expasy.org/prosite/). The TreeTop-phylogenetic tree prediction program (http://www.genebee.msu.su/services/phtree_reduced.html) was used for comparisons of the C. reinhardtii flagellar AK versus Homo sapiens AK sequences.
Polyclonal Antibody Production
A cDNA encoding an NH2-terminal fragment of Oda5p was subcloned into the pMAL vector (Invitrogen, Carlsbad, CA) to create a construct containing the first 154 amino acids of Oda5p fused to the maltose binding protein. This fusion protein was bacterially expressed, purified, and used to immunize rabbits for polyclonal antibody production (Invitrogen). Affinity purification was performed using an Oda5-GST fusion protein containing the same NH2-terminal fragment of Oda5.
Isolation of Flagella and Flagellar Fractionations
Flagella were isolated by the method of Witman (1986) and extracted with 1% Nonidet P-40 (Calbiochem, La Jolla, CA) or 1% Tergitol Type NP-40 (Sigma-Aldrich, St. Louis, MO) in HMDEKP (30 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EGTA, 25 mM KCl, and 1 mM phenylmethylsulfonyl fluoride) as indicated. Demembranated axonemes were subsequently extracted with 0.6 M KCl in HMDEKP buffer. High-salt extracts were fractionated on 5–20% sucrose gradients under conditions that maintain the association of the outer arm and the ODA-DC (Takada et al., 2002). A mixture of bovine thyroglobulin, catalase, and bovine serum albumin (Sigma-Aldrich) was run in a parallel gradient for S-value determination.
Western Blots
SDS-PAGE and Western blots were performed according to standard methods. Protein extracts from intact cells were prepared by centrifuging cells and resuspending the cell pellets in sample buffer (10 mM Tris, pH 8.0, 32 mM dithiothreitol, 1 mM EDTA, 10% sucrose, and 1% SDS). Samples were heated for 10 min and sheared with a 22-gauge needle. Axonemal fractions were prepared as described above and dissolved in sample buffer. Proteins were separated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Oda5p was localized using the anti-Oda5p antibody diluted 1:1000 in 5% horse serum in 1× Tris-buffered saline/0.5% Tween 20. IC2, the γDHC, DC2, and IC140 were revealed using monoclonal antibodies 1869A and 12γB (King et al., 1985), an anti-DC62 polyclonal antibody (Wakabayashi et al., 2001), and an anti-IC140 polyclonal antibody (Yang and Sale, 1998), respectively. Horseradish peroxidase-conjugated secondary antibodies (Pierce Chemical, Rockford, IL; Sigma-Aldrich) were used at 1:2000.
Isotope-Coded Affinity Tagging (ICAT)
ICAT was performed at the University of Victoria GenomeBC Proteomics Centre (Vancouver, BC, Canada). Briefly, pellets of Tergitol-treated flagella from wild-type and oda5-1 were resuspended in 6 M urea, 0.1% SDS and then labeled using the ICAT reagent kit (Applied Biosystems, Foster City, CA). The samples were combined, digested with trypsin, and the resulting peptide mixture fractionated into four or 10 fractions. The peptides were then affinity purified using a streptavidin column. The results are the pooled analyses of both the four- and 10-fraction experiments. Analysis was performed on an Applied Biosystems/MDS QStar hybrid liquid chromatography/tandem mass spectrometry (MS/MS) quadripole time of flight system and quantitation performed using Applied Biosystems ProICAT software.
AK Assay
AK activity was determined by the method of Watanabe and Flavin (1976) with slight modifications. AK was assayed by coupling the formation of ATP from ADP to NADP+ reduction in the presence of hexokinase and glucose-6-phosphate dehydrogenase. The reaction mixture consisted of 55 mM Tris, pH 7.9, 40 mM glucose, 2 mM MgCl2, 1 mM ADP, 0.18 mM NADP+, 1 U each of hexokinase and glucose-6-phosphate dehydrogenase, and 1 mM sodium-(meta)vanadate to inhibit dynein ATPases. The reaction mixture was preincubated for 10 min to consume any ATP contaminating the ADP. Flagellar fractions were added to the reaction and the adenylate kinase activity measured by monitoring the change in absorbance at 340 nm that accompanied the production of NADPH. Data points were collected every 30 s for 10 min. Assays were performed in triplicate on three independently isolated flagellar samples.
RESULTS
The oda5-2 Insertional Mutant
To obtain insertional mutants with defects in motility, CC-2454 cells were transformed with plasmid pMN24 (Fernandez et al., 1989). Oda- phenotypes were distinguished from other motility mutants by their slow, jerky swimming motion that is characteristic of loss of the outer dynein arm. The new oda mutants were then identified by crossing them to known oda mutants. When transformant 45BO3 was crossed with the original oda5-1 mutant (Kamiya, 1988) and tetrads dissected, no progeny showed wild-type motility (PD: NPD:T, 37:0:0). Moreover, temporary dikaryons between oda5-1 and 45B03 did not undergo any increase in motility during 2 h of mating. Finally, oda5-1 and 45B03 did not complement in stable diploids. Thus, it is concluded that 45B03 is an insertional allele of the ODA5 gene. The 45B03 mutation hereafter will be referred to as oda5-2.
Isolation of the ODA5 Gene and Rescue of the oda5-2 Mutant
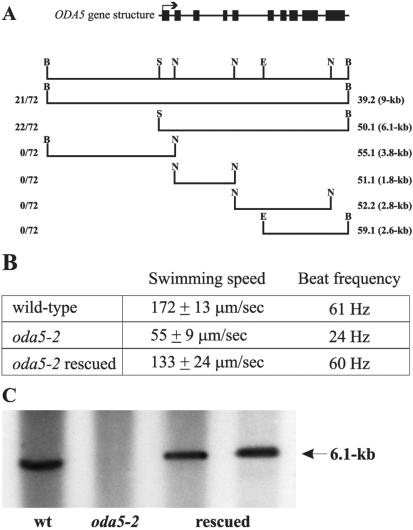
DNA flanking the integrated selectable marker in the oda5-2 strain (Figure S1) was cloned and used as a probe to identify four BACs containing that DNA. Restriction mapping indicated these BAC clones had overlaping inserts (our unpublished data). To test whether these BAC clones contained the ODA5 gene, we transformed oda5-2 with the BACs and screened for rescue of the Oda- phenotype. One BAC clone rescued the motility defect in oda5-2, suggesting it contained the ODA5 gene. This BAC contained an insert of ∼40 kb. To delimit the ODA5 gene within this BAC, we subcloned smaller fragments from this insert and tested them for rescue. The smallest rescuing genomic fragment, 50.1, is 6.1 kb (Figure 1A) and rescued 22 of 72 colonies that had been cotransformed with the ARG7 selectable marker.
Figure 1.
The ODA5 gene rescues the Oda5- motility phenotype and the transforming DNA is recovered in the oda5-2 rescued strains. (A) The upper line illustrates the intron-exon structure of the ODA5 gene. Rectangles indicate exons and solid lines indicate introns. The initial exon is marked by an arrow indicating the direction of transcription. The second line is the restriction map for the relevant portion of the rescuing BAC (N, Nco1; B, BamH1; E, EcoR1; S, Sal1). Subclones from the rescuing BAC were tested for their ability to rescue the Oda5- motility phenotype. The left column indicates the number of rescued transformants/total number of cotransformants screened. The right column indicates the construct name and the size of the genomic fragment. The smallest rescuing fragment was a 6.1-kb Sal1-BamH1 fragment (50.1). (B) Motility assays were performed on wild type, oda5-2, and one of the rescued strains (oda5-2 rescued). Both swimming speed and flagellar beat frequency were rescued to near wild-type levels in the rescued strain (n = 30 for each strain). (C) Southern blots of SacI/BamH1-digested DNA from wild type, oda5-2, and two strains rescued by transformation of oda5-2 were probed with the transforming DNA. Wild-type DNA contains hybridizing sequences from the ODA5 region. Oda5-2 does not contain hybridizing sequences as these regions are deleted. The two rescued strains contain hybridizing sequences, demonstrating that these sequences have been recovered.
Motility assays of wild-type, oda5-2, and a strain rescued with the 50.1 genomic fragment revealed that both swimming speed and flagellar beat frequency were restored to near wild-type levels in the rescued strain (Figure 1B). The swimming speed of the rescued strain (133 ± 24 μm/s) was slightly slower than that of wild-type (172 ± 13 μm/s), but clearly rescued compared with the oda5-2 mutant (55 ± 9 μm/s). The flagellar beat frequency was restored completely (61 Hz for wild-type vs. 60 Hz for the rescued strain). To further verify the rescued strains, we performed Southern blot analysis using a probe to the rescuing fragment. In wild-type, the probe hybridized to a band of the expected size, whereas in the oda5-2 strain, the probe did not hybridize to any band, indicating this region is deleted in oda5-2 (Figure 1C). The probe also hybridized to bands in the rescued strains, demonstrating that the deleted sequences have been restored (Figure 1C). Probes to either end of the 9-kb rescuing fragment (Figure 1A) indicate this entire region is deleted in the mutant (our unpublished data).
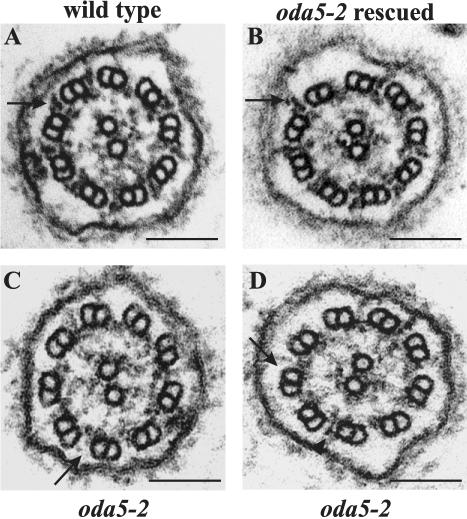
Electron microscopy demonstrated that the outer arms are missing in oda5-2 and restored in the rescued strains. Figure 2 shows cross sections through the flagella of wild-type, the oda5-2 insertional mutant, and one of the rescued strains. Wild-type and the rescued strain show the full complement of outer arms (Figure 2, A and B). The oda5-2 strain specifically lacks these structures (Figure 2, C and D), although residual material was occasionally observed at the site normally occupied by the outer arm (Figure 2D). These data demonstrate that the 50.1 rescuing fragment contains the ODA5 gene.
Figure 2.
The oda5-2 insertional mutant lacks outer dynein arms and the rescued strains have restored outer dynein arms. Electron micrographs of axonemal cross sections from (A) wild type, (B) an oda5-2 insertional mutant rescued with the 6.1-kb fragment containing the ODA5 gene (oda5–2 rescued), and (C and D) the oda5-2 insertional mutant. Arrows indicate the outer dynein arms in the wild-type and rescued flagella, but the absence of outer arms in the oda5-2 flagella. In occasional micrographs, some outer arms, or partial arms, were observed in oda5-2 (arrowhead in D). Bar, 100 nm.
Cloning the ODA5 cDNA
The rescuing genomic fragment was completely sequenced. To identify potential coding regions, we analyzed the sequence using the GreenGenie gene prediction program (Li et al., 2003) (Figure 1A). PCR primers designed from the predicted exons were used to amplify the ODA5 cDNA from wild-type Chlamydomonas cDNA libraries. Analysis of the cDNA revealed a large open reading frame (Figure 3A). The cDNA sequence spans the entire rescuing fragment, and no other genes are found in this region (Figure 1A). Note that fragments 51.1, 52.2, 55.1, and 59.1, which contain only portions of the ODA5 gene, do not rescue oda5-2, indicating that the gene is disrupted in these smaller fragments (Figure 1A).
Figure 3.
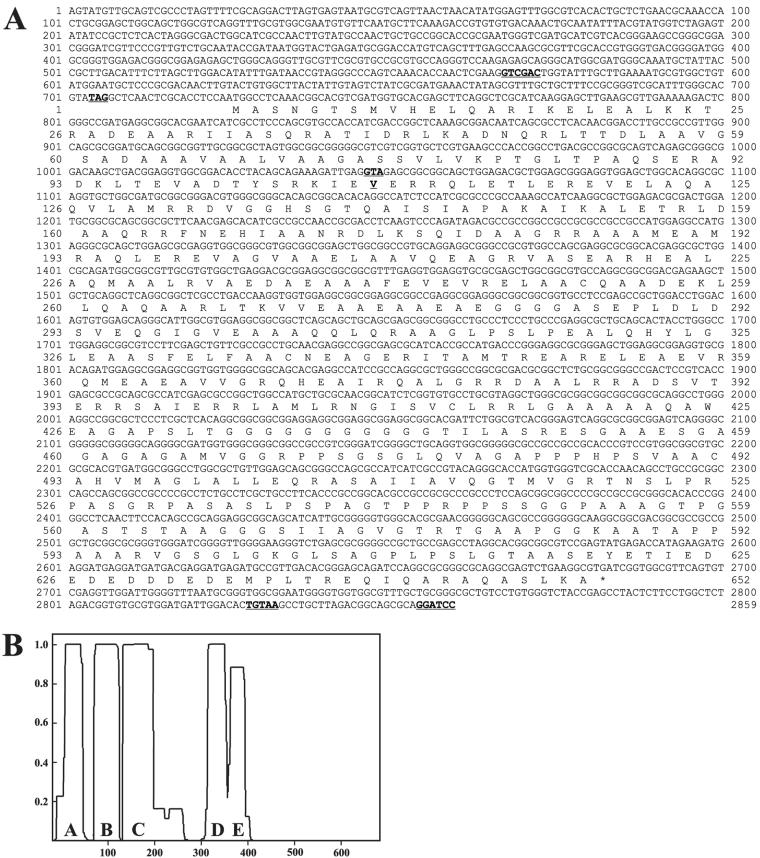
Sequence and predicted structure of the ODA5 gene and its product. (A) ODA5 cDNA sequence and its predicted amino acid sequence. In bold and underlined are a Sal1 restriction enzyme site (567–572), which denotes the 5′ end of the 50.1 rescuing fragment; an in-frame stop codon (704–706) that is upstream of the predicted start codon; a GTA codon (1046–1048), encoding valine 108, that is converted to a TAA stop codon in the oda5-1 mutant strain; a polyadenylation signal sequence (2828–2832); and a BamH1 restriction site (2854–2859), which denotes the 3′ end of the 50.1 rescuing genomic fragment. The sequence from the polyadenylation site to the BamH1 site is derived from the ODA5 genomic sequence. Additional 5′ untranslated region (UTR) sequences were identified by PCR amplification of cDNA and EST database searches. The additional 5′ UTR sequence matches genomic sequence 5′ to the Sal1 site. This indicates that the 50.1 rescuing fragment does not contain the entire 5′ UTR of ODA5, but it does contain the entire coding region and 3′ polyadenylation signal. These sequences have been deposited in GenBank/EMBL/DDBJ with accession no. AY452532. (B) Graphical representation of the predicted coiled-coil regions in the Oda5 protein as determined using the COILS program (MTIDK matrix, with a 2.5 weighting of hydrophobic positions a and d) (Lupas et al., 1991). The x-axis is amino acid number, and the y-axis is the probability that the sequence will form coiled-coil secondary structure.
The ODA5 cDNA predicts a 652-amino acid protein (Figure 3A) with a mass of ∼66-kDa and a pI of 5.2. Oda5p is predicted to contain several coiled-coil domains and a COOH-terminal noncoiled-coil region (Figure 3B). BLAST reports do not reveal significantly related sequences, suggesting that Oda5p is a novel protein.
The oda5-1 Mutant Has a Defect in the ODA5 Gene
To ascertain whether the oda5-1 mutant contains a defect in the ODA5 gene, the exons from oda5-1 genomic DNA were sequenced. A double-base pair replacement was identified 321 base pairs downstream of the initiating ATG. This mutation converts a GTA codon specifying a valine to the stop codon TAA (Figure 3A), which would result in translation of only 108 of the 652 amino acids encoded by the wild-type ODA5 gene. This result further verifies that the gene we have identified is the ODA5 gene and that the insertional mutant is an allele of oda5-1.
The ODA5 mRNA Is Up-Regulated by Deflagellation
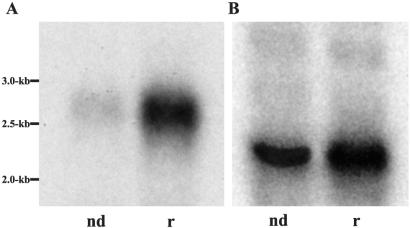
Transcription of genes encoding flagellar proteins is up-regulated in response to deflagellation (Silflow et al., 1982). To determine whether this is the case for ODA5, we performed Northern blot analysis on RNA isolated from wild-type cells either before deflagellation, or 30–45′ postdeflagellation. Using a probe to the antisense strand of the ODA5 cDNA, we found that the ODA5 transcript is a ∼2.7-kb mRNA that is up-regulated during flagellar regeneration (Figure 4A). This result suggests that ODA5 encodes a flagellar protein.
Figure 4.
ODA5 gene expression is up-regulated by deflagellation. PolyA+ mRNA was isolated from wild-type non-deflagellated (nd) cells and cells that were deflagellated and actively regenerating their flagella (r). (A) An ODA5 cDNA probe was hybridized to Northern blots of the isolated mRNA. The probe identifies an induced message at ∼2.7 kb, in good agreement with the 2.8-kb size of the ODA5 cDNA. (B) A cDNA probe to fructose-biphosphate aldolase recognizes an ∼2.0-kb mRNA that serves as a loading control; transcription of this gene is not up-regulated by deflagellation.
Oda5p Is an Axonemal Protein
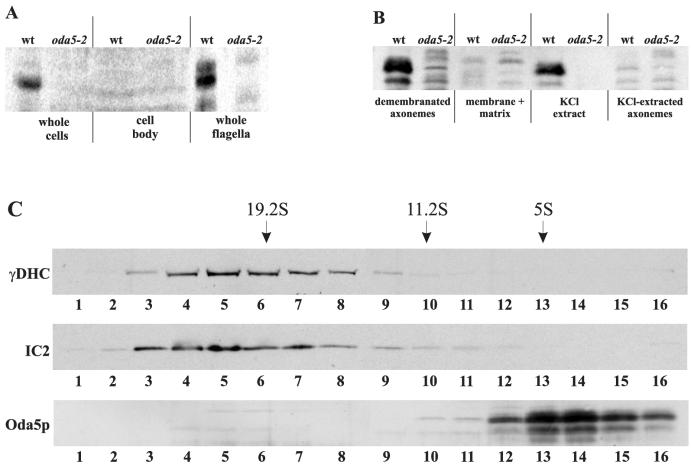
To facilitate the in vivo localization of the Oda5 protein, we generated a polyclonal antibody to an Oda5 fusion protein. In wild-type whole cells, the Oda5p antibody detected a band having a Mr of 76,000 on SDS-PAGE (Figure 5A). This band was not detected in the null strain, confirming our antibody recognized the correct protein. Although the antibody slightly cross-reacted with other proteins, we were readily able to follow the distribution of Oda5p by comparing fractions from wild type and the oda5-2 null strain. When cells were deflagellated and cell bodies analyzed, we detected little to no Oda5p in the cell body fraction. Oda5p was highly enriched in isolated whole flagella, demonstrating that Oda5p is a bona fide flagellar protein (Figure 5A).
Figure 5.
Western blot analysis indicates Oda5p is a salt-extractable, Mr 76,000 axonemal protein that sediments at ∼5S in sucrose density gradients. (A) The Oda5 antibody recognizes an Mr 76,000 band in wild-type whole cells that is absent from the oda5-2 whole cells, confirming the antibody recognizes the correct protein. This band is not detected in cell bodies lacking flagella (middle), yet it is readily detected in whole flagella (right). (B) Oda5p remains associated with the axoneme after Nonidet P-40 detergent extraction (demembranated axonemes) and is not detected in the Nonidet P-40 detergent-soluble membrane + matrix fraction (membrane + matrix). Extraction of demembranated axonemes with 0.6 M KCl releases Oda5p into the KCl extract; none remains in the KCl-extracted axonemes. (C) Sucrose gradient fractions were probed with antibodies to outer dynein arm components IC2 and γ-DHC, and with the Oda5p-antibody. The outer dynein arm/ODA-DC complex sediments at ∼23S as expected; however, Oda5p sediments at ∼5S.
To further localize Oda5p within the flagellum, we isolated whole flagella and extracted them with Nonidet P-40 detergent followed by 0.6 M KCl (Figure 5B). Oda5p is not present in the detergent-soluble membrane + matrix fraction, but it remains associated with the demembranated axoneme, demonstrating that Oda5p is an axonemal component and not a soluble or membrane-associated protein. When we subsequently extracted the demembranated axonemes with 0.6 M KCl, Oda5p was released from the axoneme.
Previous data have shown that the outer arm and the ODA-DC also are released from the axoneme by 0.6 M KCl (Pfister et al., 1982; Takada et al., 1992; Takada et al., 2002). To determine whether Oda5p associates with the outer arm or the ODA-DC, we subjected the high-salt extract to sucrose gradient sedimentation, by using conditions designed to maintain the outer arm/ODA-DC association (Takada et al., 1992). Western blots of sucrose gradient fractions were probed with antibodies to IC2 and the γDHC, which confirmed the migration of the outer arm/ODA-DC at the expected position of ∼23S (Figure 5C). In contrast, Oda5p migrated at ∼5S in these gradients. These data demonstrate that under conditions that remove the outer arm and the ODA-DC as an intact complex, Oda5p is not associated with these two components.
Oda5p Assembles onto the Axoneme Independently of the Outer Dynein Arm and the ODA-DC
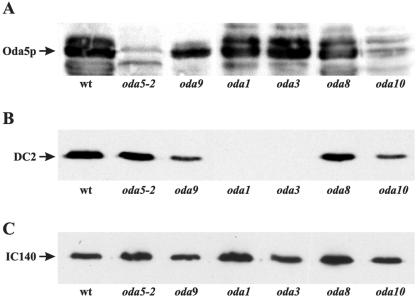
Inasmuch as Oda5p behaved independently of the outer arm and ODA-DC in sucrose gradients, we investigated whether Oda5p can assemble onto the axoneme in the absence of the latter structures. Western blots (Figure 6A) revealed that Oda5p is present in axonemes of an oda9 mutant, which is defective in the IC1 gene and fails to assemble an outer arm (Wilkerson et al., 1995), and of oda1 and oda3 mutants, which are defective in the DC2 and DC1 components of the docking complex, respectively (Koutoulis et al., 1997; Takada et al., 2002) and fail to assemble the ODA-DC and the outer arm. These results show that Oda5p can assemble onto the axoneme independently of the outer arm and the ODA-DC.
Figure 6.
Oda5p assembles onto the axoneme independently of the outer dynein arm and the ODA-DC. (A) Western blot of axonemes isolated from wild type (wt) and oda5-2, oda9, oda1, oda3, oda8, and oda10 and probed with the Oda5p-antibody. Oda5p is detected on axonemes from wild type and all the oda mutants except oda5-2 and oda10. (B) Western blot of axonemes from the same strains was probed with an antibody to the DC2 component of the ODA-DC. DC2 assembles onto axonemes in all strains except oda1 and oda3, which are defective in DC2 and DC1, respectively. (C) A Western blot of axonemes from the same strains was probed with an antibody to the IC140 component of the inner dynein arm I1. As expected, IC140 assembles onto axonemes in all of the strains and serves as a loading control for these Western blots.
Oda5p Assembly onto the Axoneme Is Defective in the oda10 Mutant
Because Oda5p, Oda8p, and Oda10p have been proposed to interact, we investigated whether localization of Oda5p is disrupted in the oda8 and oda10 mutant strains. Western blots revealed that Oda5p does assemble onto oda8 axonemes. In contrast, Oda5p fails to assemble onto axonemes of the oda10 mutant (Figure 6A), demonstrating that a functional Oda10 protein is required for proper localization of Oda5p. This provides the first biochemical evidence for an interaction between Oda5p and Oda10p.
The ODA-DC Can Assemble onto oda5, oda8, and oda10 Axonemes
To determine whether the ODA-DC can assemble onto the axoneme in the absence of Oda5p, as well as in the absence of ODA8 and ODA10 gene products, we probed Western blots of isolated axonemes by using an antibody to DC2, the 62-kDa component of the ODA-DC (Figure 6B). As expected, axonemes from oda1 and oda3, which do not assemble the ODA-DC, lack DC2, whereas axonemes from oda9, which are missing only the outer dynein arm, contain DC2 as do wild-type axonemes. Oda5, oda8, and oda10 axonemes also contain DC2. Therefore, DC2 can assemble onto the axoneme in these three mutants, suggesting that the entire ODA-DC can assemble onto axonemes independently of Oda5, Oda8, and Oda10 proteins.
Oda5p-associated Proteins
In an effort to identify proteins that interact with Oda5p, we used a quantitative mass spectrometry technique called ICAT (Han et al., 2001) to compare proteins present in wild-type versus oda5 flagellar fractions. ICAT allows one to determine the ratios of individual proteins in the two fractions being compared.
For the ICAT experiments, we wanted to analyze as comprehensive a set of flagellar proteins as possible. However, subassemblies of the outer dynein arm are known to remain in the cytoplasm of mutants unable to assemble the arm due to loss of an outer arm protein (Fowkes and Mitchell, 1998), and preassembled axonemal complexes are present in the flagellar matrix, even in nonregenerating flagella (Qin et al., 2004). Because the presence of any stable but unassembled Oda5-interacting proteins would compromise the ICAT analysis, we selectively removed the matrix proteins by treating the isolated flagella with the nonionic surfactant Tergitol Type NP-40, which, in contrast to Nonidet, disrupts but does not completely dissolve the flagellar membrane (Figure 7A). The axonemes and residual membrane were then collected by centrifugation, leaving the matrix in the supernatant. The resulting pellets were then washed, solubilized in SDS and urea, and labeled with the isotopically light (wild type) and isotopically heavy (oda5) forms of the ICAT reagent, which specifically labels cysteines. The labeled samples were combined, digested with trypsin, fractionated by cation exchange chromatography, and the labeled peptides purified by affinity chromatography. The purified, labeled peptides were analyzed by electrospray ionization MS/MS. The ratio of the isotopically heavy (oda5-1) to isotopically light (wild type) peptide in each peptide pair was determined from the signal intensities of the peaks as the pair eluted into the mass spectrometer. Peptide sequences were identified from the MS/MS spectra by searching the Chlamydomonas 20021010 expressed sequence tag (EST) database, or the BLAST nonredundant database with Chlamydomonas specified as the organism. Peptide and EST sequences were then used to search version 1.0 of the JGI Chlamydomonas genome database (http://genome.jgi-psf.org/chlre1/chlre1.home.html).
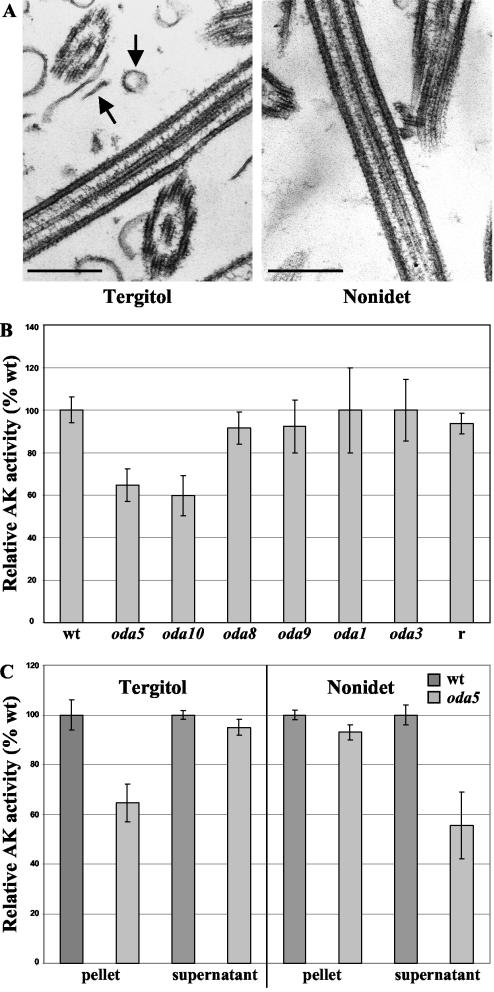
Figure 7.
Adenylate kinase activity is reduced in oda5 and oda10 mutant flagella. (A) Electron micrographs of flagella extracted with Tergitol and Nonidet P-40 detergents. Tergitol-extracted axonemes contain cosedimenting flagellar membranes (left, arrows), whereas Nonidet-extracted axonemes are virtually free of flagellar membranes (right). Bar, 300 nm. (B) Adenylate kinase activity was measured in Tergitol-treated axonemes from wild type, oda1, oda3, oda5, oda8, oda9, oda10, and an HA-tagged ODA5 rescued strain (r). The histogram shows AK activity as a percentage relative to that of wild type. Adenylate kinase activity is specifically reduced in oda5 and oda10 mutant axonemes, but it is restored to the wild-type level in the strain transformed with an HA-tagged ODA5 gene construct. (C) When flagella are treated with Tergitol, the Oda5p-associated AK activity remains in the pellet. However, when flagella are treated with Nonidet P-40, the Oda5p-associated AK activity is released into the supernatant. For each fraction, the AK activity of oda5 is shown as a percentage relative to that of wild type. Error bars in B and C represent the standard deviation calculated from three independent measurements.
Although the ICAT analysis identified only some of the known axonemal proteins, the results for these were as expected (Table 2). For example, peptides representing the α, β, and γ heavy chains of the outer dynein arm were present in oda5-1 flagella in amounts ranging from 0 to 0.24 times their wild-type levels. In contrast, peptides representing proteins of the ODA-DC (DC1), inner dynein arm (p28), and central-microtubule-pair-complex (PF6) were not reduced in oda5-1 compared with wild type. These results validate the ICAT approach for the quantitative comparison of flagellar fractions.
Table 2.
Selected ICAT-labeled peptides identified by MS/MS
| Peptide | Ratio oda5/WT | Protein | ||
|---|---|---|---|---|
| Peptides from known outer dynein arm proteins absent or greatly reduced in oda5-1 | ||||
| CGYSVANGR | 0.050163 | α-DHC | ||
| ELEICK | 0 | α-DHC | ||
| SQAVDASEYEALCR | 0.136725 | α-DHC | ||
| TECYR | 0.239353 | α-DHC | ||
| AVDAWCAQVAATSDEK | 0.176057 | β-DHC | ||
| CPVYTTEAR | 0.0165195 | β-DHC | ||
| DFYGDCMK | 0.097077 | γ-DHC | ||
| Peptides from known axonemal proteins not reduced in oda5-1 | ||||
| CEAIEK | 1.951047 | Inner dynein arm p28 light chain | ||
| ETGICPIR | 1.002732 | Inner dynein arm p28 light chain | ||
| SVCIGAEQGLR | 1.329003 | DC1 | ||
| GGSCAFYESEQLR | 1.154318 | PF6 | ||
| Other peptides reduced in oda5-1 | ||||
| TVLFFDCPEEEMEK | 0.614294 | Adenylate kinase | ||
| ALDQAEQFESSIMPCK | 0.565586 | Adenylate kinase | ||
| CHVISAVAAPDDVYGK | 0.545381 | Adenylate kinase | ||
| CEALMK | 0.645473 | Adenylate kinase | ||
| VQALDFSCDER | 0.63913 | WD-repeat protein | ||
Four other peptides, all derived from the same predicted gene, were present in oda5-1 in amounts ranging from 0.54 to 0.64 their wild-type levels. This gene is predicted to encode an AK with a mass of ∼70 kDa and homology to AKs in other organisms. The cDNA sequence was derived from EST sequences, PCR-amplified cDNA clones, and predicted coding sequence in the JGI Chlamydomonas genome version 2.0 database (Figure 8B). Another peptide representing a previously unidentified WD-repeat protein was present in oda5-1 at about two-thirds its amount in wild type (Table 2); this protein (GreenGenie 492.9 in version 1.0 of the JGI Chlamydomonas genome) is represented in the EST database (20021010.5906.1).
Figure 8.
The flagellar AK gene structure and predicted cDNA and protein sequences. (A) Intron-exon structure of the AK gene. Black boxes indicate the exons; solid lines indicate the noncoding regions; arrow indicates the direction of transcription. (B) AK cDNA sequence and deduced protein sequence. Underlined and in bold are an upstream, in-frame stop codon, TAG; a consensus polyadenylation signal sequence, TGTAA, located 521 bp downstream of the stop codon (*); and the peptides identified by ICAT. The conserved AK domain is present in three nearly identical repeats; the shaded regions denote the AK P-loop motif and the AK signature motif in each of the three domains. Nucleotides 1–1075 and 1328–2619 have been confirmed by PCR amplification of cDNA; nucleotides 1076-1327 are derived from the JGI version 2.0 gene model for this gene; nucleotides 2620–2703 are derived from EST sequences. These sequences have been reported in GenBank/EMBL/DDBJ with accession no. AY452531. (C) Comparison of the C. reinhardtii flagellar AK and H. sapiens AK1 and AK5. The highly conserved AK domains, containing the P-loop motif and the AK signature motif, are indicated by black boxes, whereas unique sequences are represented by a line. The human AK5 enzyme occurs as a small isoform (AK5) and two longer isoforms called variant 1 and 2 (AK5–1 and AK5–2). The greatest percent identity/percent similarity between one of the three Chlamydomonas AK domains and one of the AK domains of each human AK isoform are indicated.
Flagellar AK Activity Is Reduced in oda5
AK activity has previously been observed within the Chlamydomonas flagellum (Watanabe and Flavin, 1976). To confirm the apparent reduction of this enzyme in oda5-1 flagella, we directly assayed AK enzymatic activity in Tergitol-extracted axonemes from wild-type, oda1, oda3, oda5-1, oda8, oda9, oda10-2, and oda5-2 rescued strains. Relative to its level in wild-type, AK activity was reduced by ∼30–50% in oda5-1 and oda10-2 but not in oda1, oda3, oda8, or oda9 (Figure 7B). Corresponding reductions in activity also were observed in oda5-2 and oda10-1 (our unpublished data). A strain that was rescued for the Oda- motility defect with an HA-tagged ODA5 gene construct had wild-type levels of AK activity, demonstrating that it also is rescued for the AK- defect. The ∼35–50% reduction in AK activity in oda5 axonemes correlates well with the 35–45% reduction in AK protein levels in oda5-1 as determined by ICAT. The similar reduction observed in oda10 provides additional evidence for a biochemical connection between Oda5p, Oda10p and AK. The loss of AK activity is not a general consequence of the failure to assemble the outer arm or ODA-DC, because AK activity is normal in oda1, oda3, and oda9 mutants lacking these structures.
Interestingly, we found that if wild-type flagella were extracted with Nonidet P-40 rather than Tergitol, the fraction of AK activity that is deficient in oda5-1 mutants is soluble. When wild-type and oda5 flagella were extracted with Tergitol, the AK specific activity in the resulting pellet was 35% less in oda5 than in wild type, whereas the AK specific activity in the soluble fraction was equivalent in wild type and oda5 (Figure 7C). However, when flagella were demembranated with Nonidet P-40, the amount of AK activity remaining in the axonemes was equivalent in wild type versus oda5, but now the relative amount of AK activity in the soluble fraction was reduced ∼40% in oda5 relative to wild type (Figure 7C). The most likely explanation for these results is that the AK is associated with the Oda5p complex via a Nonidet-sensitive bond. Alternatively, because Nonidet, but not Tergitol, completely removes the flagellar membrane (Figure 7A), the Oda5p-associated AK also may be connected to the flagellar membrane.
DISCUSSION
Loss of the ODA5 gene causes loss of the outer dynein arm, resulting in a slow, jerky swimming phenotype (Kamiya, 1988), together with a previously undescribed phenotype – reduction in the level of a newly identified flagellar AK. We have isolated the ODA5 gene and find that outer dynein arm assembly and adenylate kinase activity are restored when the gene is transformed back into the oda5-2 mutant, demonstrating that the defect in ODA5 is responsible for both the Oda- and AK- (adenylate kinase) phenotypes. Oda5p localizes to the flagellum and remains associated with the axoneme after Nonidet P-40 demembranation, indicating that it is an axonemal protein. Oda5p assembly onto the axoneme is independent of the outer arm and the ODA-DC, but dependent upon the Oda10 gene product. These results indicate that Oda5p is part of a complex with Oda10p and the newly identified flagellar AK.
Structure of Oda5p
The ODA5 cDNA predicts a novel 66-kDa protein containing extensive α-helical domains. There are five regions with a high probability of forming coiled-coils. Coiled coils commonly mediate protein–protein interactions, and it is likely that Oda5p interacts with itself or with another protein via these regions. The region between coiled-coils C and D in Figure 3B is largely hydrophobic, with 63 of 126 amino acids being hydrophobic. Just after predicted coiled-coil domain E is a stretch of 10 glycines (G434–G444), which would break the coiled-coil structure and could serve as a flexible hinge between the coiled-coil domains and the COOH terminus. A very acidic region (E619–E634) is located near the COOH terminus and may mediate interactions between Oda5p and the outer dynein arm or other axonemal components; Oda5p is released from the axoneme by 0.6 M KCl, indicating its association with the axoneme is ionic in nature. These features of Oda5p are reminiscent of the DC1 and DC2 subunits of the ODA-DC, which also have predicted coiled-coil structures and COOH-terminal charged regions.
Although the predicted mass of Oda5p is 66 kDa, our antibody to Oda5p detected a protein with a Mr of 76,000 on SDS-polyacrylamide gels. An anomalously high Mr is not uncommon for coiled-coil proteins and similarly was observed for DC1 and DC2 (see Koutoulis et al., 1997, for discussion).
Oda5p Is Not a Subunit of the Outer Arm or the ODA-DC
Although Oda5p, the outer dynein arms, and the ODA-DC are all released from the axoneme by 0.6 M KCl, Oda5p sediments separately from the outer arms and the ODA-DC in sucrose density gradients. Inasmuch as Oda5p is required for assembly of the outer dynein arm, it is likely that it associates directly or indirectly with the arm in vivo, but that this association is disrupted by the high-salt extraction. However, Oda5p also assembles onto the axoneme independently of the outer arm and the ODA-DC, as evidenced by its presence on the axonemes of several oda mutants lacking the latter structures. Conversely, the ODA-DC can assemble onto the axoneme in the absence of Oda5p. Therefore, Oda5p is not simply a previously unidentified subunit of the outer arm or the ODA-DC, but a separate and independently assembling component of the axoneme that is necessary for outer arm attachment to the doublet microtubules.
Oda5p Interacts with Oda10p
The inability of oda5, oda8, and oda10 mutants to complement in temporary dikaryons suggested that the Oda5, Oda8, and Oda10 proteins interact in a complex (Kamiya, 1988). Our finding that Oda5p is missing from axonemes of the oda10 mutant demonstrates that a functional Oda10p is required for the localization of Oda5p and provides the first biochemical evidence for an interaction between these two proteins. In contrast, Oda5p was not missing from axonemes of the oda8 mutant. It is possible that Oda8p is associated with Oda5p and Oda10p, but not required for binding of Oda5p to the axoneme (Figure 9).
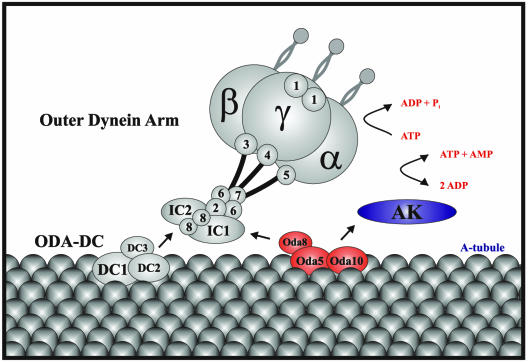
Figure 9.
Model for assembly of the Oda5p/Oda8p/Oda10p complex in the wild-type axoneme. The outer dynein arm attaches to the A-tubule via the ODA-DC and the Oda5p/Oda8p/Oda10p complex; the flagellar AK is held near the outer arm by its association with the latter. α, β, and γ indicate the α, β, and γ dynein heavy chains; numbers indicate dynein LC subunits.
A Flagellar AK Associates with the Oda5p/Oda10p Complex
ICAT analysis revealed that a flagellar AK was reduced 35–45% in oda5-1 compared with wild type. AK catalyzes the reversible reaction: 2 ADP ↔ ATP + AMP. Direct enzymatic assays confirmed that flagellar AK activity was reduced in oda5 and indicated that the AK activity was similarly reduced in oda10 but not in the other oda mutants. Therefore, a portion of the newly identified AK is dependent upon Oda5p and Oda10p for its incorporation into the flagellar structure. Because the flagellar AK is not reduced in other mutants that lack the outer arms or the outer arms together with the ODA-DC, it is not simply a subunit of either of these structures. Rather, it seems to be specifically associated with the Oda5p/Oda10p complex. AK activity is not reduced in oda8, consistent with our finding that Oda5p is assembled onto oda8 axonemes.
A model illustrating the assembly of these complexes onto the axoneme is shown in Figure 9. Outer dynein arm assembly requires both the ODA-DC and the Oda5p/Oda8p/Oda10p complex, whereas assembly of the outer arm-associated AK requires only the Oda5p/Oda8p/Oda10p complex. Our biochemical results and the lack of cytoplasmic complementation between oda5, oda8, and oda10 can be accounted for as follows: If either the ODA5 or ODA10 gene is disrupted, then the remaining subunits of the Oda5p/Oda8p/Oda10p complex are unstable, and no partial complex is formed. In this case, neither outer arm assembly nor adenylate kinase assembly occurs. If the ODA8 gene is disrupted, then Oda5p (and possibly Oda10p) are stable, forming a partial complex, which is targeted to the axoneme. Assembly of the partial complex is sufficient for binding of adenylate kinase but not the outer arm.
The Flagellar AK
The flagellar AK identified by ICAT analysis is encoded by a gene having five exons (Figure 8A). The NH2 terminus, predicted by exons 1 and 2, is unique, whereas the third, fourth, and fifth exons are each predicted to encode a nearly identical domain that closely resembles the conserved domain of other AKs. The conserved domain of AKs has two conserved motifs, the AK signature motif and the ATP-binding motif or P-loop. The AK signature motif constitutes part of the catalytic cleft and contains conserved arginine and aspartic acid residues. The P-loops of AKs deviate from the usual P-loop consensus sequence ([AG]-X4-G-K-[ST]) in that the last position is occupied by a glycine instead of a serine or threonine (Prosite PDOC00017). The flagellar AK identified here contains the conserved aspartic acid and arginine in the signature motif and the conserved glycine substitution within the P-loop motif in each of its three AK domains (Figure 8B). In a phylogenetic comparison with the known human AKs, the Chlamydomonas flagellar AK was most closely related to the cytosolic isoforms AK1 and AK5 (our unpublished results). Interestingly, although it is unusual for AKs to have repeats of the conserved AK domain, two isoforms of human AK5 (AK5 variant 1 and 2) both contain two AK domains (Figure 8C).
The flagellar AK is solubilized from the wild-type flagellum by treatment with Nonidet P-40 but not Tergitol. Because the AK is not predicted to contain transmembrane domains, and because its presence in the flagellum is dependent upon Oda5p, which is an axonemal protein, it is probable that the AK also is an axonemal component and that it is associated with the axoneme via interactions that survive the Tergitol treatment but are disrupted by Nonidet P-40.
AK activity previously has been reported in cilia and flagella from several organisms (Watanabe and Flavin, 1976; Schoff et al., 1989; Nakamura et al., 1999; Noguchi et al., 2001), although neither the specific AK nor its location in the flagellum were determined. The apparent association of the AK with Oda5p and Oda10p in the current study, and the fact that the latter are required for outer dynein arm assembly, suggest that the AK is held in proximity to the outer arm by the Oda5p/Oda10p complex (Figure 9). We propose that the outer dynein arm— one of the major ATP-hydrolyzing structures of the axoneme—is intimately associated with an ATP-regenerating system to achieve efficient conversion of ADP to ATP and AMP, thus ensuring that both high-energy phosphate bonds of ATP are readily accessible to this important force-generating machine.
That the flagellar AK is reduced but not absent in the oda5 and oda10 mutants indicates that the AK is located at additional sites within the axoneme. One likely site for the remaining flagellar AK is the inner dynein arm system. Such a localization would place the AK at two major sites of ATP utilization in the axoneme.
A WD-Repeat Protein Also Is Reduced in oda5 Flagella
ICAT analysis also identified one peptide, from a predicted WD-repeat protein, that was reduced in oda5-1 flagella by about the same amount as the AK. It is tempting to speculate that this protein may be associated with the AK. The protein has homologues in other flagellated organisms, including H. sapiens, Mus musculus, Macaca fascicularis, Anopheles gambiae, and Drosophila melanogaster. Further characterization of the protein will be required to clarify its relationship to the flagellar AK.
Utility of ICAT for Comparing Proteins in Flagella of Different Mutants
Our ICAT analysis correctly reported the relative levels, in wild-type versus oda5 flagella, of all peptides that were identified as being derived from known axonemal proteins, as well as those from the previously undescribed flagellar AK. The main shortcoming of the ICAT approach was its inability to identify a more comprehensive set of flagellar proteins, as evidenced by its failure to identify peptides from a greater percentage of known axonemal proteins. This was most likely due to the long duty cycle (∼14 s) of the mass spectrometer used, which prevented many peptides from being selected for fragmentation for MS/MS analysis. This problem should be greatly alleviated by newly available instruments, which have much faster data acquisition rates and thus can analyze a much larger number of peptides from a sample of the same complexity. With the use of such instruments, ICAT and similar quantitative proteomic approaches (Aebersold and Mann, 2003) are likely to become very valuable for identifying specific proteins whose levels are altered in mutant versus wild-type flagella.
Supplementary Material
Acknowledgments
We are grateful to Carolyn Silflow for providing the p3 × HA cassette, to Carol Dieckmann for arg- strains, to William Snell for the gametic cDNA library, to Susan Dutcher for help with the GreenGenie program, and to Toshiki Yagi for assistance with motion analysis. These studies were supported by a National Institutes of Health grant GM-30626 (to G.W.), by the Robert W. Booth fund at The Greater Worcester Community Foundation (to G.W.), by the Japanese Ministry of Education, Culture, Sports, Science and Technology (to R.K.), and by CREST of Japan Science and Technology Corporation (to R.K.). We also acknowledge grant 5 P30 DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases for use of University of Massachusetts Diabetes and Endocrinology Research Center supported core facilities.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0820. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0820.
Abbreviations used: DHC, dynein heavy chain; IC, intermediate chain; LC, light chain; ODA-DC, outer dynein arm-docking complex; RFLP, restriction fragment length polymorphism; AK, adenylate kinase; ICAT, isotope-coded affinity tagging; JGI, Joint Genome Institute.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Aebersold, R., and Mann, M. (2003). Mass spectrometry-based proteomics. Nature 422, 198-207. [DOI] [PubMed] [Google Scholar]
- Afzelius, B.A., and Mossberg, B. (1995). Immotile-cilia syndrome (primary ciliary dyskinesia), including Kartagener Syndrome. In: The Metabolic and Molecular Bases of Inherited Disease, vol. III, ed. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, New York: McGraw-Hill Book Company, 3943-3954. [Google Scholar]
- Ahmed, N.A., and Mitchell, D.R. (2003). oda16-1 contains a gene that rescues slow-swimming Chlamydomonas (Abstract). Mol. Biol. Cell 14, 436a. [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- Berger, B., Wilson, D.B., Wolf, E., Tonchev, T., Milla, M., and Kim, P.S. (1995). Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92, 8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, A.B., Patel-King, R.S., Benashski, S.E., McCaffery, J.M., Goldstein, L.S., and King, S.M. (1999). Drosophila roadblock and Chlamydomonas LC 7, a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146, 165-180. [PMC free article] [PubMed] [Google Scholar]
- Brokaw, C.J. (1994). Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil. Cytoskeleton 28, 199-204. [DOI] [PubMed] [Google Scholar]
- Casey, D.M., Inaba, K., Pazour, G.J., Takada, S., Wakabayashi, K., Wilkerson, C.G., Kamiya, R., and Witman, G.B. (2003). DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol. Biol. Cell 14, 3650-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy, R., Purton, S., and Rochaix, J.D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8, 2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella, L.M., and King, S.M. (2001). Dynein motors of the Chlamydomonas flagellum. Int. Rev. Cytol. 210, 227-268. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. (1995). Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47, 531-540. [DOI] [PubMed] [Google Scholar]
- Fernandez, E., Schnell, R., Ranum, L.P., Hussey, S.C., Silflow, C.D., and Lefebvre, P.A. (1989). Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86, 6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes, M.E., and Mitchell, D.R. (1998). The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell 9, 2337-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, D.K., Eng, J., Zhou, H., and Aebersold, R. (2001). Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19, 946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use, San Diego: Academic Press. [DOI] [PubMed]
- Hoops, H.J., and Witman, G.B. (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97, 902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R. (1988). Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J. Cell Biol. 107, 2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R. (2000). Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods 22, 383-387. [DOI] [PubMed] [Google Scholar]
- Kindle, K.L., Schnell, R.A., Fernandez, E., and Lefebvre, P.A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109, 2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., Otter, T., and Witman, G.B. (1985). Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc. Natl. Acad. Sci. USA 82, 4717-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., and Patel-King, R.S. (1995). Identification of a Ca(2+)-binding light chain within Chlamydomonas outer arm dynein. J. Cell Sci. 108, 3757-3764. [DOI] [PubMed] [Google Scholar]
- Koutoulis, A., Pazour, G.J., Wilkerson, C.G., Inaba, K., Sheng, H., Takada, S., and Witman, G.B. (1997). The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137, 1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.B., Lin, S., Jia, H., Wu, H., Roe, B.A., Kulp, D., Stormo, G.D., and Dutcher, S.K. (2003). Analysis of the Chlamydomonas reinhardtii genome structure using large-scale sequencing of regions on linkage groups I and III. J. Eukaryot. Microbiol. 50, 145-155. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and Brown, K.S. (1994). Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes. J. Cell Sci. 107, 635-644. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and Kang, Y. (1991). Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 113, 835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.R., and Rosenbaum, J.L. (1985). A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J. Cell Biol. 100, 1228-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Iitsuka, K., and Fujii, T. (1999). Adenylate kinase is tightly bound to axonemes of Tetrahymena cilia. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 124, 195-199. [DOI] [PubMed] [Google Scholar]
- Noguchi, M., Sawada, T., and Akazawa, T. (2001). ATP-regenerating system in the cilia of Paramecium caudatum. J. Exp. Biol. 204, 1063-1071. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., and Witman, G.B. (1999a). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Koutoulis, A., Benashski, S.E., Dickert, B.L., Sheng, H., Patel-King, R.S., King, S.M., and Witman, G.B. (1999b). LC2, the Chlamydomonas homologue of the t complex-encoded protein Tctex2, is essential for outer dynein arm assembly. Mol. Biol. Cell 10, 3507-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., and Witman, G.B. (2000). Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22, 285-298. [DOI] [PubMed] [Google Scholar]
- Pfister, K.K., Fay, R.B., and Witman, G.B. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell. Motil. 2, 525-547. [DOI] [PubMed] [Google Scholar]
- Porter, M.E., Bower, R., Knott, J.A., Byrd, P., and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., and Sale, W.S. (2000). The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151, F37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Diener, D.R., Geimer, S., Cole, D.G., and Rosenbaum, J. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- Sager, R., and Granick, S. (1953). Nutritional studies with Chlamydomonas reinhardtii. Ann. N.Y. Acad. Sci. 466, 18-30. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Mitchell, D.R., and Kamiya, R. (1991). A Chlamydomonas outer arm dynein mutant missing the alpha heavy chain. J. Cell Biol. 113, 615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, H., Takada, S., King, S.M., Witman, G.B., and Kamiya, R. (1993). A Chlamydomonas outer arm dynein mutant with a truncated beta heavy chain. J. Cell Biol. 122, 653-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoff, P.K., Cheetham, J., and Lardy, H.A. (1989). Adenylate kinase activity in ejaculated bovine sperm flagella. J. Biol. Chem. 264, 6086-6091. [PubMed] [Google Scholar]
- Silflow, C.D., LaVoie, M., Tam, L.W., Tousey, S., Sanders, M., Wu, W., Borodovsky, M., and Lefebvre, P.A. (2001). The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153, 63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow, C.D., Lefebvre, P.A., McKeithan, T.W., Schloss, J.A., Keller, L.R., and Rosenbaum, J.L. (1982). Expression of flagellar protein genes during flagellar regeneration in Chlamydomonas. Cold Spring Harb. Symp. Quant. Biol. 46, 157-169. [DOI] [PubMed] [Google Scholar]
- Takada, S., Sakakibara, H., and Kamiya, R. (1992). Three-headed outer arm dynein from Chlamydomonas that can functionally combine with outer-arm-missing axonemes. J. Biochem. 111, 758-762. [DOI] [PubMed] [Google Scholar]
- Takada, S., Wilkerson, C.G., Wakabayashi, K., Kamiya, R., and Witman, G.B. (2002). The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell 13, 1015-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi, K., Takada, S., Witman, G.B., and Kamiya, R. (2001). Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil. Cytoskeleton 48, 277-286. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., and Flavin, M. (1976). Nucleotide-metabolizing enzymes in Chlamydomonas flagella. J. Biol. Chem. 251, 182-192. [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., Koutoulis, A., Pazour, G.J., and Witman, G.B. (1995). The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129, 169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., and Witman, G.B. (1994). Molecular analysis of the gamma heavy chain of Chlamydomonas flagellar outer-arm dynein. J. Cell Sci. 107, 497-506. [DOI] [PubMed] [Google Scholar]
- Witman, G.B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. [DOI] [PubMed] [Google Scholar]
- Witman, G.B., Carlson, K., Berliner, J., and Rosenbaum, J.L. (1972). Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54, 507-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., Wilkerson, C.G., and King, S.M. (1994). The biochemistry, genetics and molecular biology of flagellar dyneins. In: Microtubules, ed. J.S. Hyams and C.W. Lloyd, New York: Wiley-Liss, 229-249.
- Yang, P., and Sale, W.S. (1998). The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9, 3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.