Pretreatment mass spectrometry-based serum and plasma peptidomics have shown promising results for prediction of treatment outcome in patients with solid tumors. Limited sample sizes and absence of signature validation in many studies have prohibited clinical implementation thus far. This review indicates that this profiling approach enables treatment selection, but additional prospective studies are warranted.
Keywords: SELDI, MALDI, Cancer, Peptidomics, Personalized medicine, Blood
Abstract
Introduction.
Treatment selection tools are needed to enhance the efficacy of targeted treatment in patients with solid malignancies. Providing a readout of aberrant signaling pathways and proteolytic events, mass spectrometry-based (MS-based) peptidomics enables identification of predictive biomarkers, whereas the serum or plasma peptidome may provide easily accessible signatures associated with response to treatment. In this systematic review, we evaluate MS-based peptide profiling in blood for prompt clinical implementation.
Methods.
PubMed and Embase were searched for studies using a syntax based on the following hierarchy: (a) blood-based matrix-assisted or surface-enhanced laser desorption/ionization time-of-flight MS peptide profiling (b) in patients with solid malignancies (c) prior to initiation of any treatment modality, (d) with availability of outcome data.
Results.
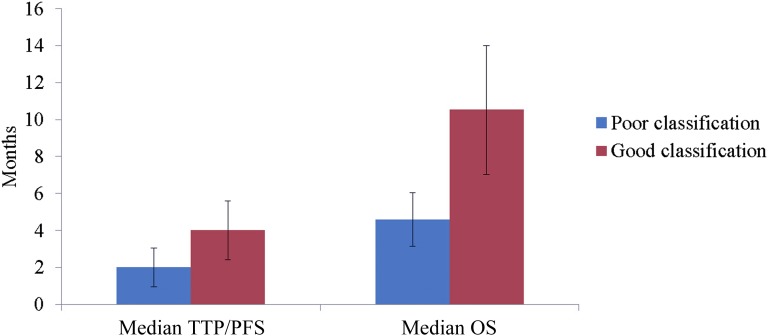
Thirty-eight studies were eligible for review; the majority were performed in patients with non-small cell lung cancer (NSCLC). Median classification prediction accuracy was 80% (range: 66%–93%) in 11 models from 14 studies reporting an MS-based classification model. A pooled analysis of 9 NSCLC studies revealed clinically significant median progression-free survival in patients classified as “poor outcome” and “good outcome” of 2.0 ± 1.06 months and 4.6 ± 1.60 months, respectively; median overall survival was also clinically significant at 4.01 ± 1.60 months and 10.52 ± 3.49 months, respectively.
Conclusion.
Pretreatment MS-based serum and plasma peptidomics have shown promising results for prediction of treatment outcome in patients with solid tumors. Limited sample sizes and absence of signature validation in many studies have prohibited clinical implementation thus far. Our pooled analysis and recent results from the PROSE study indicate that this profiling approach enables treatment selection, but additional prospective studies are warranted.
Implications for Practice:
Treatment selection tools are needed to enhance the efficacy of treatment in patients with solid tumors. Mass spectrometry-based peptidomics enables identification of predictive biomarkers, whereas the serum or plasma peptidome may provide easily accessible signatures associated with response to treatment. This review discusses 38 studies on blood-based peptidomics performed before initiation of systemic and/or local treatment and includes a pooled analysis based on pretreatment outcome classification in patients with non-small cell lung cancer. This analysis and recent results from the PROSE study indicate that this profiling technique enables treatment selection in patients with cancer, but additional prospective studies are warranted.
Introduction
Since 2003, more than 20 targeted therapies have reached clinical approval for treatment of patients with advanced solid tumors [1, 2], but their often temporary effectiveness in a limited selection of patients as well as their significant toxicities emphasize the need for a clinically applicable strategy to predict efficacy of these agents. Currently, mutation status guides treatment in some tumor types, but actual response to molecular-guided treatment is not guaranteed [3–5]. Because responses to targeted therapies depend on their effect on downstream signaling activities in tumor tissue, mass spectrometry-based (MS-based) proteomics provides a potential tool for prediction of response to these drugs by recognizing changes in protein abundance and activating post-translational modifications and protein-protein interactions [6, 7]. Blood has been suggested to provide the ideal biological sample for profiling of these effects due to the accessibility of the serum and plasma peptidome, consisting of proteins and peptides released by tissue (including tumor tissue) as a result of proteolytic cascades [8–10].
More than 20,000 research articles on matrix-assisted laser desorption/ionization time-of-flight MS (MALDI-TOF MS) and surface-enhanced laser desorption/ionization time-of-flight MS (SELDI-TOF MS) for profiling of the blood peptidome have been published since their development in the late 1980s and 1990s. Mass spectral peaks correspond to ions formed from peptides and proteins with a molecular weight of less than 20 kDa, and the peak amplitude indicates their abundance (Fig. 1) [11–13]. Several studies have reported on the potential for early detection by discriminating patients from healthy controls (HCs) based on differential serum proteome as a consequence of tumor biology [14–17], but clinical implementation has been hampered by the difficulties of other groups in reproducing the results [18, 19]. In the meantime, strict sample-handling procedures and unbiased study design have been shown to result in high intra- and interlaboratory reproducibility of mass spectra and algorithm-based classification concordance [20–22]. In this paper, we have systematically reviewed studies on MS-based serum and plasma peptidomics performed in patients with solid tumors in relation to outcome following systemic and local treatment and evaluated its appropriateness for prompt implementation in the clinic.
Figure 1.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry serum pattern. Representative example of a mass spectrum obtained from serum, showing several peptide peaks in the high- and low-intensity ranges.
Abbreviation: m/z, mass-to-charge ratio.
Materials and Methods
Literature Search
Search engines PubMed and Embase were used to identify studies published until September 2013 using serum or plasma MALDI- or SELDI-TOF MS peptidome profiling in patients with solid malignancies prior to initiation of any treatment modality, with availability of outcome data including response and progression-free survival (PFS) or overall survival (OS). For PubMed, the following syntax was applied: (“Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization”[MeSH] OR maldi[tiab] OR seldi[tiab] OR “surface enhanced laser desorption”[tiab] OR “matrix assisted laser desorption”[tiab] OR serum proteomic test) AND cancer[sb] AND (“Serum”[MeSH] OR serum[tiab] OR blood[subheading]). For Embase, the following syntax was used: “surface enhanced laser desorption ionization time of flight mass spectrometry”/exp OR “surface enhanced laser desorption ionization time of flight mass spectrometry” OR “matrix assisted laser desorption ionization time of flight mass spectrometry”/exp OR “matrix assisted laser desorption ionization time of flight mass spectrometry” OR “serum proteomic test”:ab,ti OR seldi:ab,ti OR maldi:ab,ti OR “surface enhanced laser desorption”:ab,ti OR “matrix assisted laser desorption”:ab,ti AND (“'serum”/exp OR “serum” OR serum:ab,ti OR “blood”/exp OR “blood”) AND (“neoplasm”/exp OR “neoplasm”). Subsequent limits were applied for studies in humans and studies published in English since 1995, with an available abstract. Based on results of the syntax-based search, we performed an additional PubMed search using the search term “VeriStrat,” a serum proteomic test.
Selection of Papers
Potentially relevant studies retrieved by the PubMed and Embase searches were independently reviewed for eligibility by three investigators (L.M.S., M.L., H.M.W.V.) and two investigators (M.L. and H.M.W.V.), respectively, according to aforementioned criteria. Unpublished studies were not considered eligible. Levels of evidence were not used to assess the value of each publication selected for inclusion.
Results
Number of Studies Meeting Selection Criteria
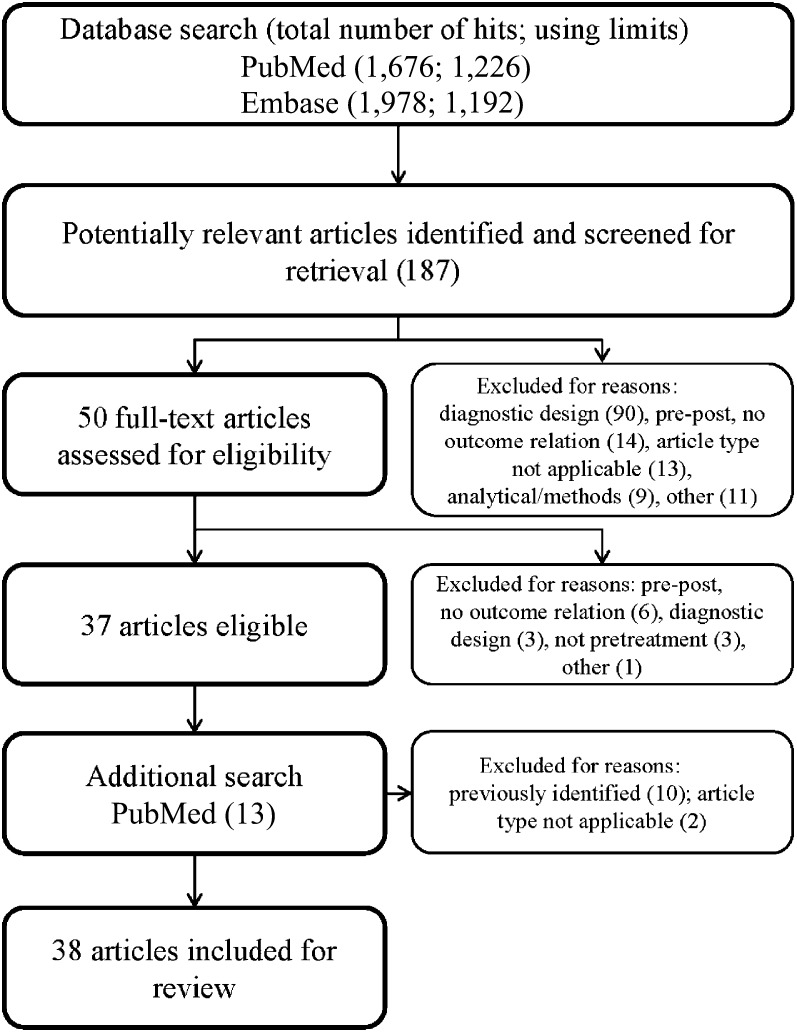
Using the PubMed and Embase syntaxes and aforementioned limits, 1,226 and 1,192 potentially relevant studies, respectively, were identified. Figure 2 depicts the subsequent stepwise selection of 37 eligible articles. With Embase, 74% of the PubMed-selected articles were found, but no additional eligible articles were found. One study that had not been identified by either syntax was found by an additional PubMed search for “VeriStrat,” resulting in a total of 38 included articles for discussion in this review. The keywords of the non-syntax-identified article did not include SELDI, MALDI, serum, blood, or cancer.
Figure 2.
Database search and article selection. Search syntaxes were constructed in consultation with an information specialist. Screening of titles and abstracts was performed independently by three investigators. Full-text articles were independently appraised for predefined selection criteria for inclusion in the review.
Investigated Tumor Types and Treatment Modalities in Eligible Studies
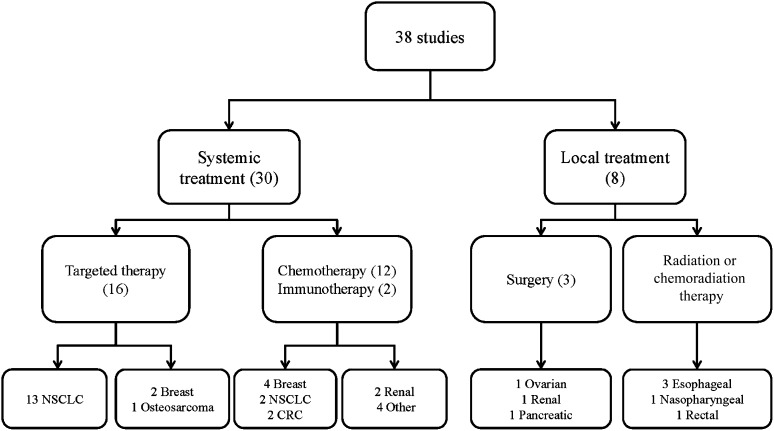
Thirty of 38 eligible studies investigated blood-based peptidomics before systemic treatment, mainly in non-small cell lung cancer (NSCLC) and in breast cancer. In four of eight studies investigating local treatment, profiling was applied prior to chemoradiation (Fig. 3). Study details are summarized in Table 1.
Figure 3.
Tumor types and treatments in eligible studies. Overview of tumor types and treatment modalities in 38 eligible articles reporting matrix-assisted or surface-enhanced laser desorption/ionization time-of-flight mass spectrometry peptide profiling in pretreatment serum or plasma.
Abbreviations: CRC, colorectal cancer; NSCLC, non-small cell lung cancer.
Table 1.
Eligible studies reporting MS-based peptidomics
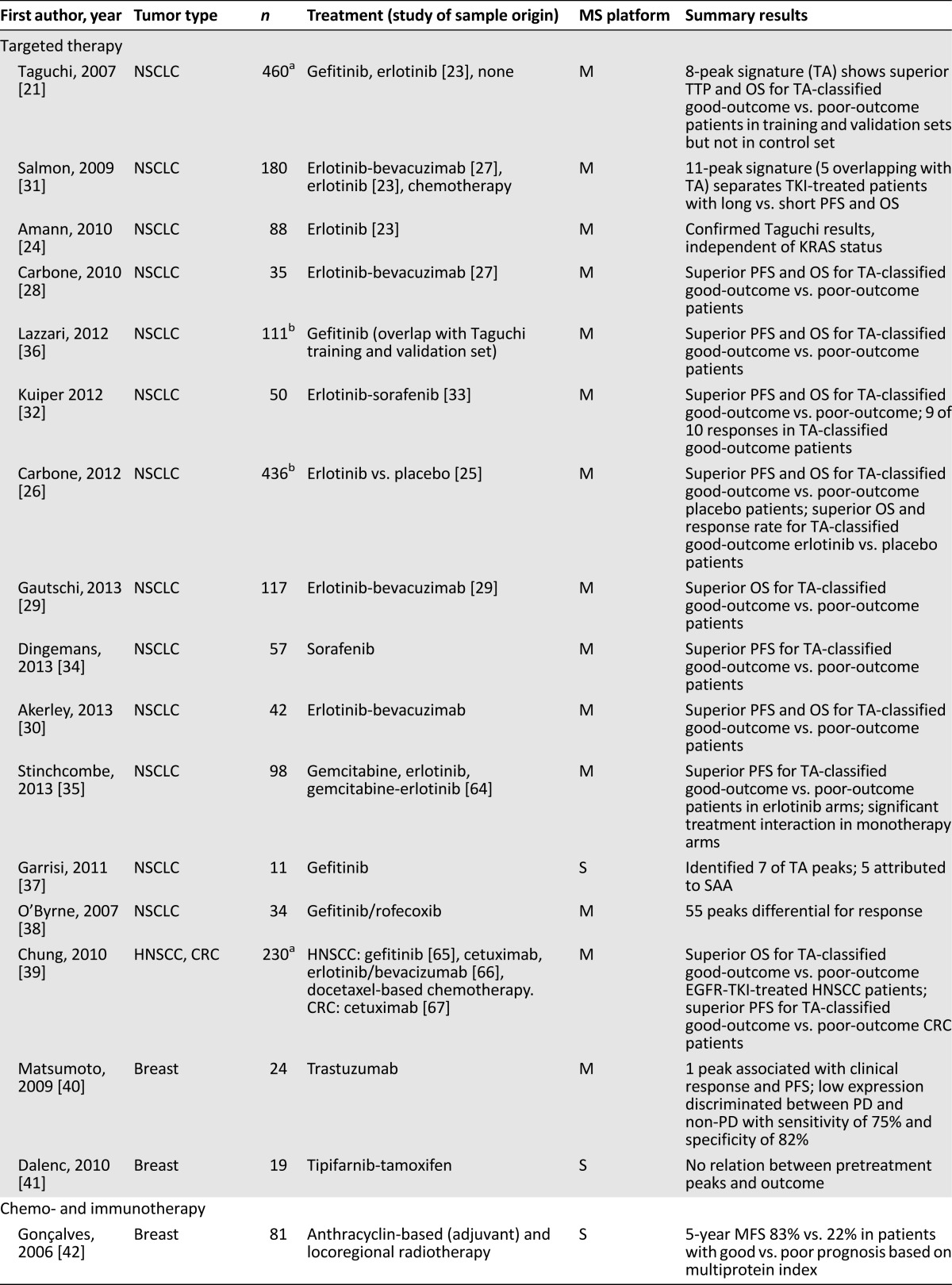
Studies Reporting Profiling Prior to Systemic Therapy
Targeted Therapy in NSCLC
Twelve of 16 included studies investigating profiling prior to targeted therapy are related to the study by Taguchi et al. in patients with advanced NSCLC treated with tyrosine kinase inhibitors (TKIs) directed against epidermal growth factor receptor (EGFR) [21]. In this study, a training set of pretreatment sera (n = 139, gefitinib) was supplemented with two validation cohorts (n = 67, gefitinib; n = 96, erlotinib; from ECOG-E3503) [23]. A control set of 158 patients who did not receive EGFR-targeting TKIs was included from three additional cohorts (2 advanced, 1 early stage). Eight differentially expressed mass-to-charge ratio (m/z) values or “peptide peaks” (5,843, 11,446, 11,530, 11,685, 11,759, 11,903, 12,452, and 12,580 Da) in the training set were used to construct an algorithm based on spectra from clinically most distinct patients in terms of time to progression (TTP) and OS. Patients with progressive disease (PD) within 1 month were classified as “poor outcome,” and patients with stable disease (SD) for more than 6 months were classified as “good outcome.” In the validation sets, TTP and OS were significantly longer in good-outcome patients compared with poor-outcome patients, with a median OS of 207 versus 92 days (hazard ratio [HR] of death: 0.50; p = .054) in the first validation cohort and 306 versus 107 days (HR: 0.41; p < .001) in the second validation cohort. No survival difference could be observed between both classifications in the control set. Interestingly, parallel application of the 8-peptide algorithm to plasma of 73 erlotinib-treated patients did not alter their serum-based outcome classification [21]. This algorithm has been commercialized as “VeriStrat” but will be further referred to in this article as the “Taguchi algorithm” (TA). Meanwhile, 12 additional studies have either applied or reported on the TA; 8 investigated nonoverlapping cohorts (Table 1).
Multivariate analysis by Amann et al. in 41 patients from the aforementioned erlotinib validation cohort [23] indicated that the algorithm predicted survival in patients with wild-type EGFR and independent of KRAS mutation status. However, the sample size of mutant tumors was small (n = 12) [24]. In 2012, Carbone et al. applied the TA to plasma profiles available for 441 of 731 patients from the randomized placebo-controlled BR.21 study [25], which established the role of erlotinib in patients with advanced NSCLC. Prognostic properties of the TA were confirmed in the placebo arm, showing superior median PFS and OS for good-outcome patients compared with patients classified as poor outcome (OS: 6.6 vs. 3.1 months; HR: 0.44; p < .0001). No significant correlation was found with EGFR or KRAS mutation status. Patients classified as good outcome seemed to benefit more from erlotinib than from placebo (OS: 10.5 vs. 6.6 months; HR: 0.63; p = .002), whereas OS for poor-outcome patients was not significantly different between arms (4.0 vs. 3.1 months; HR: 0.77; p = .11); the survival curves separated at times longer than 4 months. Multivariate adjusted analyses showed similar relative benefit from erlotinib for both classifications by a nonsignificant interacting p value, indicating a prognostic value of the algorithm. Nevertheless, erlotinib-treated patients classified as good outcome had a significantly higher response rate (RR) than poor-outcome patients (11.5% vs. 1.1%; p = .002) [26].
The TA has been applied to serum profiles of patients treated with erlotinib plus bevacizumab (erl/bev) in three studies. Analyzing 35 patient sera from a phase I/II study [27], Carbone et al. found median OS to be significantly longer for good-outcome patients (61 weeks) than for poor-outcome patients (24 weeks) (HR: 0.14, p = .007); median and PFS was also significantly longer at 24 weeks versus 8 weeks, respectively (HR: 0.045, p = .003). The TA classified all 8 patients with partial response (PR) and 6 of 7 patients with SD at more than 16 weeks as having good outcome, whereas 2 of 6 patients with initial PD were classified as poor outcome [28]. Gautschi et al. confirmed the superior median OS for good-outcome patients versus poor-outcome patients (13.4 vs. 6.2 months; HR: 0.48; p = .003) in 117 patients receiving first-line treatment with erl/bev. In contrast to the previous study, median PFS was not significantly different between good versus poor outcome classification (4.0 vs. 3.2 months; HR: 0.77, p = .263), suggesting, according to the investigators, that TA performance may be dependent on prior treatment [29]. This is supported by a phase II study by Akerley et al. with erl/bev in 42 previously untreated patients. Median PFS and OS were superior patients classified as good outcome compared with those classified as poor outcome (PFS: 18.9 vs. 6.3 weeks; p = .0035; OS: 71.4 vs. 19.9 weeks; HR: 0.27; p = .0015). PFS for study treatment plus subsequent chemotherapy was also significantly longer in good-outcome patients, underscoring the prognostic properties of the algorithm. Nonetheless, 9 of 10 observed responses, as assessed by the Response Evaluation Criteria in Solid Tumors (RECIST), occurred in patients with good-outcome classification [30]. Salmon et al. [31] developed an alternative 11-peak signature based on 37 erl/bev-treated patients [27]. Five peaks overlapped with the TA. In a validation set of 82 erlotinib-treated patients [23], compound scores from the MS features and their peak intensities could distinguish between patients with long and short PFS and OS, discriminating between patients with compound scores lower than the median and equal to or greater than the median, whereas no significant difference was found in a control set of 61 chemotherapy-treated patients [31].
TA-based sorting of patient groups has been shown in the context of several other TKI-based treatment regimens. Analysis of TA performance by Kuiper et al. [32] in serum from 50 chemotherapy-naïve patients treated with erlotinib plus sorafenib [33] again confirmed superior outcomes for patients classified as good outcome versus poor outcome (OS: 13.7 vs. 5.6 months; HR: 0.30; p = .009; PFS: 5.5 vs. 2.7 months; HR: 0.40; p = .035), whereas the objective RR was not significantly different [32]. For sorafenib monotherapy, as reported by Dingemans et al. in 55 pretreated patients, PFS was significantly longer in TA-discerned good-outcome patients versus poor-outcome patients (2.6 vs. 1.5 months; HR: 1.4; p = .029), whereas OS was not significantly different (6.0 vs. 2.5 months; HR: 1.3; p = .166), probably due to a relatively large variation in OS duration [34]. Stinchcombe et al. applied the TA to sera of 98 elderly patients treated in a randomized phase II trial of first-line therapy with gemcitabine, erlotinib, or combination [64]. Superior median OS was observed for good-outcome patients versus poor-outcome patients in the erlotinib monotherapy arm only (n = 32; 255 vs. 51 days; HR: 0.40; p = .014), whereas PFS was significantly longer in good-outcome patients for both erlotinib-containing arms. The absence of prognostic power in the gemcitabine arm (n = 28) may have been due to crossing over to erlotinib after progression. Interestingly, a significant treatment interaction was demonstrated for patients in the monotherapy arms, indicating that good-outcome patients had more benefit from erlotinib, whereas poor-outcome patients benefited more from gemcitabine. In line with this observation, the authors point to the median PFS and OS of 22 and 51 days, respectively, in erlotinib-treated patients classified as poor outcome, suggesting that this drug is not an acceptable first-line treatment option for these patients [35].
Three additional studies have reported results of profiling prior to gefitinib-based treatment. Lazzari et al. [36] confirmed superior prognosis for TA-discerned good-outcome classification in 111 previously described patients [21]. During treatment, 88% of patients maintained their baseline classification, whereas at treatment withdrawal, largely due to PD, only 26% of good-outcome patients shifted to poor outcome classification [36]. Garrisi et al. identified 7 of 8 TA peaks from spectra of 11 NSCLC patients prior to gefitinib and 10 HCs. Of these, five could be attributed to isoforms of the acute-phase protein serum amyloid A (SAA), but SAA serum concentrations determined by enzyme-linked immunosorbent assay were not related to TTP [37]. From profiles of 34 patients treated with gefitinib plus rofecoxib, O’Byrne et al. found 55 differential peaks between responders (n = 3) and patients with SD/PD (n = 31) and 90 between patients with disease control (n = 14) and nonresponders [38].
Targeted Therapy in Tumors Other Than NSCLC
Hypothesizing that the TA would reflect EGFR dependency regardless of the primary tumor site or the EGFR-targeting agent used, Chung et al. profiled patients with advanced head and neck squamous cell carcinoma (HNSCC; 55 treated with gefitinib [65], 32 treated with erl/bev [66], 21 treated with cetuximab, and 34 treated with docetaxel-based chemotherapy) and 88 cetuximab-treated patients with colorectal cancer (CRC) [67]. For patients with HNSCC, median OS was superior for those classified as good outcome versus poor outcome treated with gefitinib (36.7 vs. 18.0 weeks; HR: 0.41; p = .007) and erl/bev (39.5 vs. 29.1 weeks; HR: 0.20; p = .02), whereas no significant difference was found in cetuximab-treated patients (38.3 vs. 11.9 weeks; HR: 0.26; p = .06) and chemotherapy-treated patients (39.4 vs. 15.5 weeks; HR: 0.88; p = .76). OS data were not available for the CRC patients, but PFS was significantly longer for good-outcome patients versus poor-outcome patients (8.9 vs. 8.4 weeks; HR: 0.51; p = .007) due to the observed separation beyond the 9-week response assessment point [39].
Two eligible studies were performed in patients with advanced breast cancer. Aiming to identify glycosylation biomarkers for trastuzumab, Matsumoto et al. analyzed plasma N-glycan profiles of 24 patients. One peak was significantly lower in patients with PD [40]. From profiles of 19 patients treated with tamoxifen plus tipifarnib, a farnesyltransferase inhibitor, Dalenc et al. identified 1 peptide that was significantly associated with TTP in serum obtained after 8 weeks of treatment, whereas pretreatment sera did not provide such association [41].
Studies Reporting Profiling Prior to Chemo- or Immunotherapy
Four eligible studies performed postoperative profiling prior to adjuvant chemotherapy in three in patients with breast cancer and one patient with ovarian cancer [42–45]. Although the candidate biomarkers identified in these studies most likely have prognostic relevance, a relationship to chemosensitivity cannot be excluded.
Mazouni et al. evaluated profiles from 39 patients with HER2-positive breast cancer receiving neoadjuvant paclitaxel-anthracyclin-based chemotherapy [70]. Two peptides were differentially expressed in patients with pathological complete response compared with patients with residual disease [46]. Plasma profiles of 27 osteosarcoma patients treated with preoperative cisplatin-doxorubicin and high-dose methotrexate have been investigated by Li et al., who considered patients with tumor necrosis of 90% or higher in the resection specimen as responders. The resulting 56-peak model predicted response with overall accuracy of 85% [47].
Two studies reported profiling prior to palliative chemotherapy in patients with NSCLC. From patterns of 27 previously untreated patients treated with cisplatin-gemcitabine plus the proteasome inhibitor bortezomib [71], Voortman et al. identified 6 differential peptides based on relatively short versus long PFS in the most distinct 22 patients. When applied to all samples, the signature showed a significantly shorter median PFS in patients classified as having short versus predicted long PFS (120 vs. 191 days; p = .036). Median OS of these patients was also significantly different at 144 versus 436 days (p = .036). Five differential peaks were identified between patients responding with PR (n = 9) and non-PR, of which two overlapped with the previous signature. This signature revealed a significantly shorter PFS and a nonsignificantly shorter OS for patients with non-PR [48]. Han et al. investigated profiles of 93 patients treated with first-line cisplatin-docetaxel chemotherapy. Considering only patients with PR as chemotherapy-sensitive, 76% of patients showed initial resistance. From a training set of 62 patients, a 5-peptide model was constructed that accurately separated chemotherapy-resistant patients from chemotherapy-sensitive patients in 84% of the blinded test set (n = 31) [49]. A similar approach by the same investigators in 56 SCLC patients treated with cisplatin-etoposide resulted in a 2-peak model performing with 80% accuracy [50].
Two eligible studies performed profiling prior to first-line palliative chemotherapy in patients with advanced CRC. Yuan et al. analyzed sera of 70 patients treated with 5-fluorouracil (5-FU) and oxaliplatin (FOLFOX, n = 44) and 5-FU and irinotecan (FOLFIRI, n = 26). Regarding patients with PR or SD as responders, a signature of the six most discriminating peaks for FOLFOX and the seven (nonoverlapping) peaks for FOLFIRI was constructed. Their performance in a randomly chosen test set is summarized in Table 2 [51]. Helgason et al. identified 1 differential peptide in profiles of “polar opposite” responders (20 PR, 10 PD) out of 40 patients treated with capecitabine-oxaliplatin [52]. A similar approach in 68 patients with advanced gastric cancer eligible for response evaluation upon treatment with first-line epirubicin, cisplatin, and capecitabine did not result in differential peaks, but low intensity of 1 peptide (11,600 Da, SAA) was significantly correlated to longer OS [53].
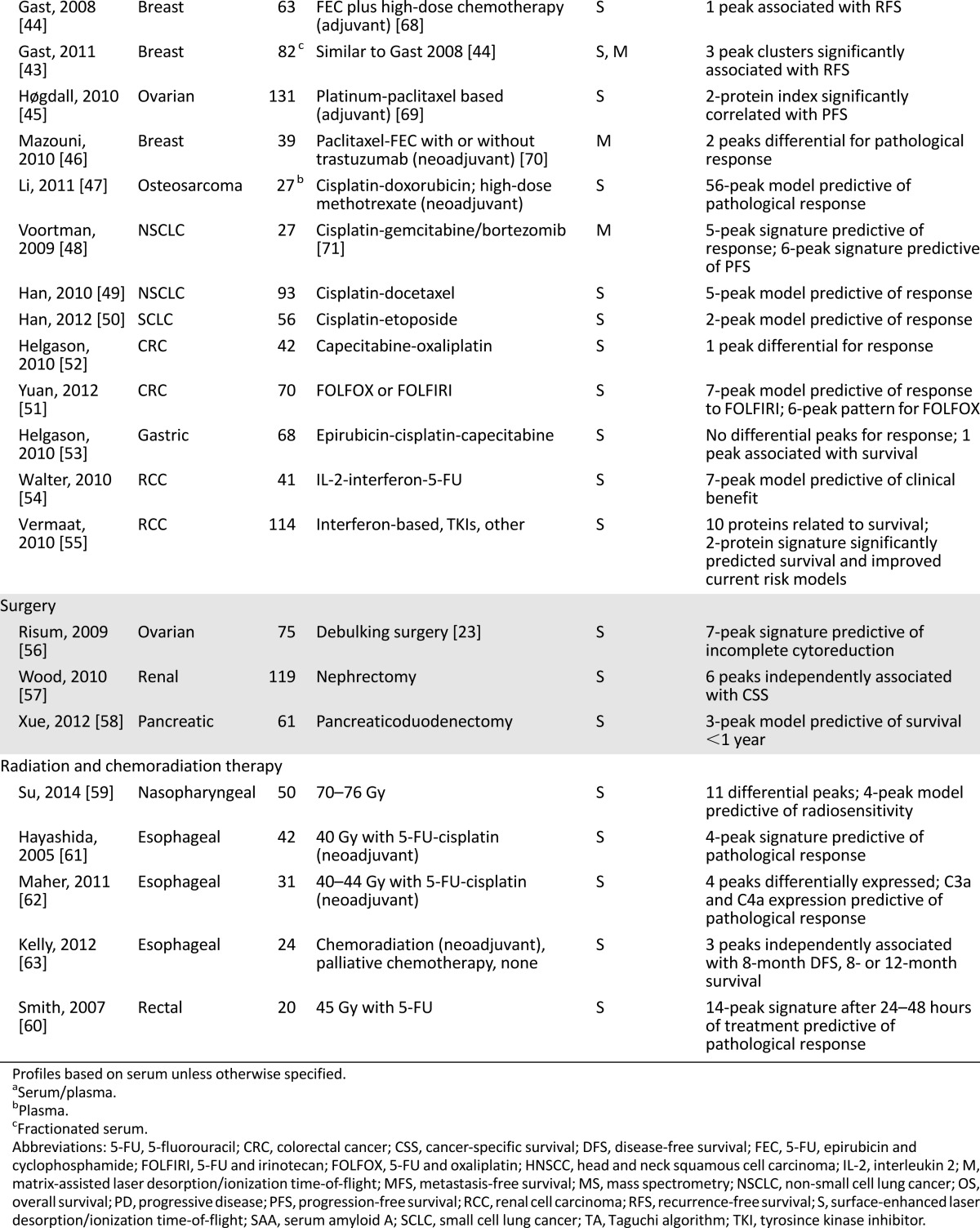
Table 2.
MS-based classifier characteristics
Since the introduction of targeted agents, immunotherapy has been decreasingly applied in metastatic renal cell carcinoma (mRCC), despite its ability to induce durable responses. Walter et al. investigated serum profiles of 41 patients treated with interleukin-2, interferon, and 5-FU. Constructed from a pattern of 25 proteins associated with response, including SAA isoforms and transthyretin (TTR), a rule base could classify patients as having predicted clinical benefit versus PD with accuracy of 66% [54]. Vermaat et al. identified 10 proteins significantly associated with OS in serum from 114 patients with mRCC, with 73% obtained prior to first-line interferon-based treatment. Serum concentrations of identified proteins apolipoprotein-A2 (Apo-A2), SAA, and TTR were predictive of survival; SAA and TTR levels were able to improve the commonly used prognostic mRCC model [55].
Studies Reporting Profiling Prior to Local and Combination Treatment
Surgery
Ultimately, predicted nonresponders to surgery might receive additional adjuvant or neoadjuvant systemic treatment to improve their outcome or palliative treatment only. Risum et al. analyzed preoperative serum profiles and CA-125 levels to predict incomplete primary debulking surgery in 75 patients with stage III–IV ovarian cancer. A panel of 7 of 10 prespecified proteins were combined into a single-valued ovarian cancer risk index (OvaRI). Overall accuracy was 72% for the OvaRI and 67% for CA-125, but combined analysis did not improve the predictive power of either analysis [56]. Wood et al. identified 6 peaks independently associated with cancer-specific survival but not with DFS after nephrectomy in serum of 119 patients with RCC; one peak (1,525 Da) was identified as an SAA fragment [57]. From 40 patients with pancreatic cancer who planned to undergo pancreaticoduodenectomy, eventually followed by adjuvant therapy in 87.5% of patients, Xue et al. developed a 3-peptide panel that, when combined with CA 19-9, predicted survival of less than 1 year with an area under the receiver operating characteristic curve of 0.96. In the verification set of 21 patients, predictive accuracy was 76%. One peptide was identified as ApoC-II, for which serum levels significantly correlated to short survival in an independent validation set [58].
Radiation and Chemoradiation Therapy
Su et al. analyzed serum profiles of 50 patients with nasopharyngeal cancer in relation to response to 7–8 weeks of radiation therapy, as assessed by nasopharyngoscopy and computed tomography scanning. Only patients exhibiting rapid regression were considered radiation sensitive. A 4-peptide pattern derived from 11 differential peaks predicted radiosensitivity in 78% of patients [59].
Three eligible studies evaluated profiling in relation to pathological response to neoadjuvant 5-FU-based chemoradiation [60–62]. Smith et al. analyzed sera taken before and during course of treatment from 20 patients with rectal cancer in relation to response according to the Mandard tumor regression grade (TRG). Although the “optimal” pretreatment model performed poorly, a 14-peptide classifier in serum taken 24–48-hours after treatment resulted in sensitivity of 88% and specificity of 80% [60]. Applying an alternative 3-grade scale for response, Hayashida et al. constructed a 4-peptide signature from profiles of 27 patients with squamous cell esophageal cancer. This performed with 93% accuracy in an independent validation set (n = 15) at sensitivity of 100% and specificity of 80% [61]. In 31 patients with mixed histological-type esophageal cancer, Maher et al. identified 4 differential peptides between TRG-based responders and nonresponders; 2 were identified as complement C3a and C4a. Their baseline serum levels predicted response witrun -1th sensitivity of 79% and specificity of 83% in 87% of patients [62]. From profiles of 24 patients with esophageal cancer, for whom treatment included neoadjuvant chemoradiation (n = 11) and palliative chemotherapy (n = 8), Kelly et al. identified TTR (14,029 Da), Apo-A1 (27,665 Da) and SAA (11,670 Da) as independently associated with 8-month DFS and survival at 8 and 12 months, respectively [63].
MS-Based Classification Models
Fourteen studies developed an MS-based classification model. Differential peptide peaks from these models and their performance in terms of overall accuracy, sensitivity, and specificity are summarized in Table 2. Median accuracy was 81% (range: 66%–93%) in 11 of 14 studies reporting performance of the classifier. Additional mass peaks associated with outcome that were not validated or applied into an MS-based prediction model are listed in supplemental online Table 1.
Pooled Analysis of Survival Times in NSCLC Studies
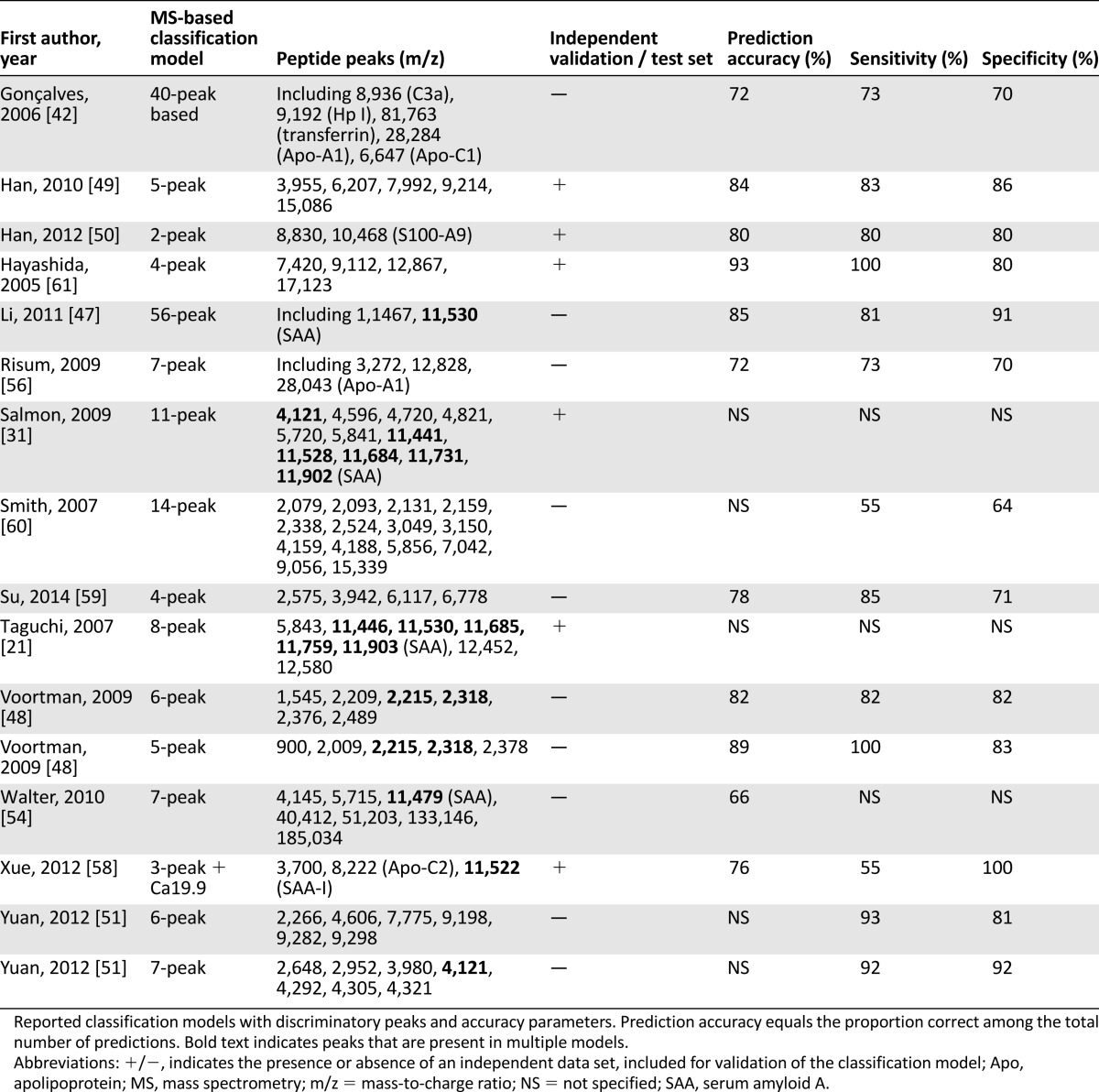
As an example of the potential power of serum and plasma peptidomics for prediction of treatment outcome in NSCLC, we pooled TTP/PFS and OS times available for the treated patients (933 received targeted therapy, 87 received chemotherapy-based treatment) from 9 discussed NSCLC studies (Table 3). This analysis revealed median PFS and OS in poor-outcome patients of 2.00 ± 1.06 and 4.58 ± 1.45 months, respectively, whereas in good-outcome patients, median PFS was 4.01 ± 1.60 months and median OS was 10.52 ± 3.49 months (Fig. 4). Aware of the small number of chemotherapy-treated patients, we also specified outcome for treatment type by algorithm status. PFS for poor-outcome patients was significantly shorter for those treated with targeted therapy than for chemotherapy-treated patients (1.87 vs. 3.94 months, p = .002), whereas OS was not different (4.28 vs. 4.73 months, p = .459). For patients classified as good outcome treated with targeted therapy and chemotherapy, median PFS and OS did not differ (PFS: 3.85 vs. 4.37 months; p = .579; OS: 9.92 vs. 11.98 months; p = .651).
Table 3.
Outcome of non-small cell lung cancer patients in pooled analysis
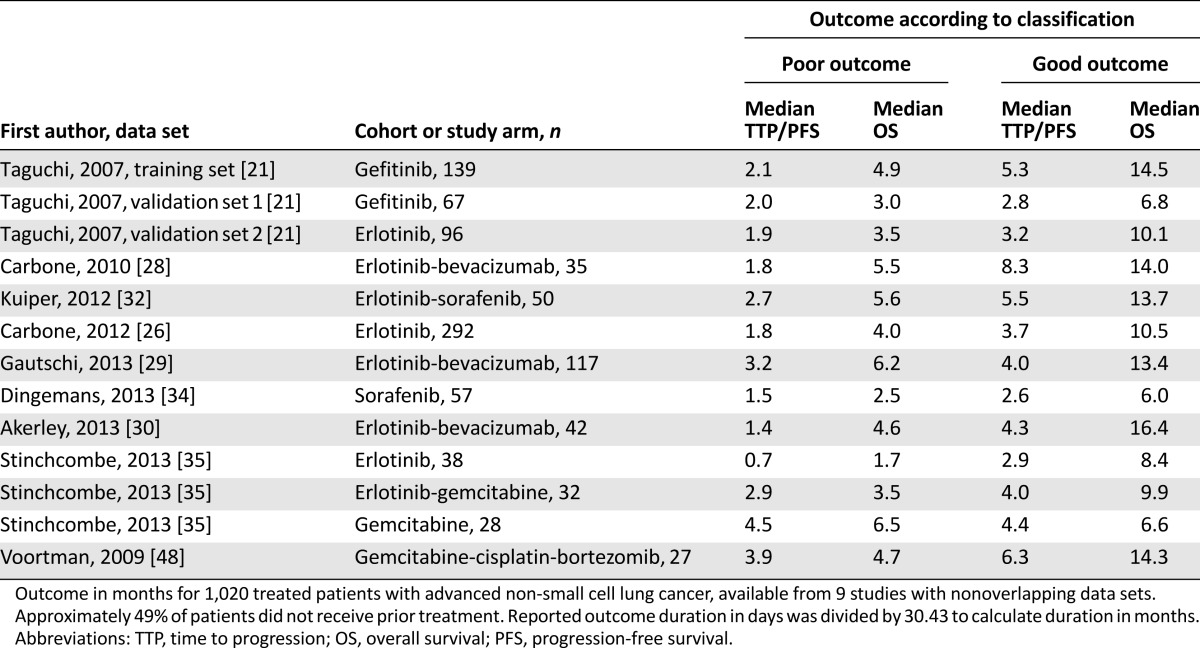
Figure 4.
Pooled analysis of non-small cell lung cancer studies. Pooled PFS and OS analysis according to pretreatment mass spectrometry-based classification of patients reported in Table 3.
Abbreviations: OS, overall survival; PFS, progression-free survival; TTP, time to progression.
Discussion
We have systematically reviewed available literature on pretreatment MS-based serum and plasma peptidomics in adult patients with solid malignancies in relation to treatment outcome to evaluate classification accuracy and readiness of this approach for clinical implementation.
Together, these results support the proof-of-concept for MS-based pretreatment profiling to influence clinical decision making in NSCLC, for example, by preferential chemotherapy for TA-classified poor-outcome patients, as has been suggested previously.
Thirteen of 38 included studies investigated profiling prior to targeted therapy in patients with advanced NSCLC. These were mostly related to a study published in 2007 by Taguchi et al. [21] in which an eight-peptide signature indicative of PFS and OS in patients treated with EGFR TKIs was identified. Five of these peaks have been identified as isoforms of the acute-phase protein SAA [32, 37, 72]. Several studies confirmed the prognostic properties of this algorithm in patients treated with targeted therapy-containing regimens [26, 30, 32], and some also indicated predictive potential [21, 35, 39]. We performed a pooled analysis of TTP/PFS and OS times available from 9 NSCLC studies. This revealed clinically significant median PFS and OS in poor-outcome patients, indicative of the potential of blood-based peptidomics, for consideration of withholding treatment in these patients (Fig. 4). In addition, the longer PFS for chemotherapy versus targeted therapy in patients classified as poor outcome (3.94 vs. 1.87 months; p = .002) in this exploratory analysis may point in the same predictive direction as the results of the recently presented PROSE study [73]. In this TA-stratified study, 285 patients with advanced NSCLC were randomized between second-line treatment with erlotinib or chemotherapy. In patients classified as poor outcome, OS was significantly shorter for those treated with erlotinib than for chemotherapy-treated patients (3.0 vs. 6.4 months; HR: 1.72; p = .022), whereas no difference was found in patients classified as good outcome (11.0 vs. 10.9 months; HR: 1.06; p = .714). Together, these results support the proof-of-concept for MS-based pretreatment profiling to influence clinical decision making in NSCLC, for example, by preferential chemotherapy for TA-classified poor-outcome patients, as has been suggested previously [73, 74].
Promising results of pretreatment MS-based peptidomics have also been obtained in other tumor types and treatment modalities, including neoadjuvant treatment for osteosarcoma and esophageal cancer, with prediction accuracies of the reported classifiers of 85% and 93%, respectively [47, 61].
Limitations of the reviewed studies included small sample sizes (median n = 59), variable design and outcome parameters, and absence of independent validation sets in most studies. In fact, validation of the signature was attempted in an independent cohort in only 6 of 14 studies reporting construction of an MS-based prediction model based on differential peaks (Table 2). One of our concerns that we were not able to address in detail in this review regards the quality of the applied data analysis methods and algorithm construction. Variability in sample collection and preparation may be responsible for the lack of peak reproducibility in studies investigating similar tumor types (Table 2). In recent years it has become clear that the use of a serum pool as a control and strict handling rules are crucial because several preanalytical factors, including clotting time and temperature, may influence reproducibility [75, 76]. The majority of studies did not provide details on sample handling and analysis and did not report the use of a prespecified collection protocol. In addition, they did not provide information on the exact interval between serum or plasma collection and treatment initiation.
In the discussed studies, SAA and apolipoprotein were the most frequently identified proteins in patients with a variety of tumors and treatments. It is known that patients with more advanced disease or poorer prognosis have increased blood levels of these markers.
Ideally, the identity or “source protein” of peptides constituting an MS-based blood signature would be known. This may enhance understanding of the underlying tumor biology and would facilitate validation and clinical implementation of these biomarkers. It is important to realize that a large part of the serum peptides are degradation products from circulating nontumor-derived proteins, resulting from differential protease activity in the tumor microenvironment [17], which may even involve or reflect an acute-phase host response [32]. Some of the peptides may also be generated from ex vivo protease activity during the clotting process required for serum preparation in the tube on venipuncture [17]. In the discussed studies, SAA and apolipoprotein were the most frequently identified proteins in patients with a variety of tumors and treatments. It is known that patients with more advanced disease or poorer prognosis have increased blood levels of these markers [55, 77, 78]. Consequently, corresponding mass peaks of these proteins are most likely prognostic markers. Alternative blood-based methods are in development, including genomics-, transcriptomics- and phosphoproteomics-based profiling [79–81], but investigation is required to determine their advantages over serum and plasma peptidomics in terms of predictive value and accuracy.
Conclusion
Pretreatment MS-based serum and plasma peptidomics have shown promising results for prediction of treatment outcome in patients with solid tumors. Limited sample size and lack of signature validation in most studies have prohibited clinical implementation thus far, despite although the technology within the field has matured. Our pooled analysis and results from the PROSE study indicate that this blood-based profiling approach enables treatment selection in patients with cancer, but additional prospective studies are warranted. Future studies should be designed to facilitate clinical decision making. They should have sufficient sample sizes and encompass uniform and strict sample collection and handling protocols and include validation efforts for the identified putative biomarkers using patient cohorts that are independent, from multiple centers, and preferably from multiple countries. Moreover, consensus on criteria to evaluate clinical implementation of proposed treatment selection tools is needed.
See http://www.TheOncologist.com for supplemental material available online.
This article is available for continuing medical education credit at CME.TheOncologist.com.
Supplementary Material
Acknowledgments
We thank VU University Medical Center Amsterdam Medical Information Specialist René Otten for his contribution to the syntax of the search terms. Mariette Labots and Lisette M. Schütte contributed equally to this paper.
Author Contributions
Conception/Design: Mariette Labots, Lisette M. Schütte, Johannes C. van der Mijn, Connie R. Jiménez, Henk M.W. Verheul
Provision of study material or patients: Mariette Labots
Collection and/or assembly of data: Mariette Labots, Lisette M. Schütte, Johannes C. van der Mijn, Connie R. Jiménez, Henk M.W. Verheul
Data analysis and interpretation: Mariette Labots, Lisette M. Schütte, Johannes C. van der Mijn, Thang V. Pham, Henk M.W. Verheul
Manuscript writing: Mariette Labots, Lisette M. Schütte
Final approval of manuscript: Mariette Labots, Lisette M. Schütte, Johannes C. van der Mijn, Thang V. Pham, Connie R. Jiménez, Henk M.W. Verheul
Disclosures
The authors indicated no financial relationships.
References
- 1.Keefe DM, Bateman EH. Tumor control versus adverse events with targeted anticancer therapies. Nat Rev Clin Oncol. 2012;9:98–109. doi: 10.1038/nrclinonc.2011.192. [DOI] [PubMed] [Google Scholar]
- 2.U.S. Food and Drug Administration Hematology/oncology (cancer) approvals & safety notifications: 2014. http://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm279174.htmAccessed March 3, 2014.
- 3.Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 4.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: Standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 5.Russo A, Rizzo S. Biomarkers and efficacy: Are we nearly there yet? Ann Oncol. 2011;22:1469–1470. doi: 10.1093/annonc/mdr258. [DOI] [PubMed] [Google Scholar]
- 6.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 8.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: Spinning biologic trash into diagnostic gold. J Clin Invest. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpova MA, Moshkovskii SA, Toropygin IY, et al. Cancer-specific MALDI-TOF profiles of blood serum and plasma: Biological meaning and perspectives. J Proteomics. 2010;73:537–551. doi: 10.1016/j.jprot.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Petricoin EF, Belluco C, Araujo RP, et al. The blood peptidome: A higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6:961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 11.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez CR, Piersma S, Pham TV. High-throughput and targeted in-depth mass spectrometry-based approaches for biofluid profiling and biomarker discovery. Biomarkers Med. 2007;1:541–565. doi: 10.2217/17520363.1.4.541. [DOI] [PubMed] [Google Scholar]
- 13.Albrethsen J. The first decade of MALDI protein profiling: A lesson in translational biomarker research. J Proteomics. 2011;74:765–773. doi: 10.1016/j.jprot.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 15.Sidransky D, Irizarry R, Califano JA, et al. Serum protein MALDI profiling to distinguish upper aerodigestive tract cancer patients from control subjects. J Natl Cancer Inst. 2003;95:1711–1717. doi: 10.1093/jnci/djg099. [DOI] [PubMed] [Google Scholar]
- 16.Honda K, Hayashida Y, Umaki T, et al. Possible detection of pancreatic cancer by plasma protein profiling. Cancer Res. 2005;65:10613–10622. doi: 10.1158/0008-5472.CAN-05-1851. [DOI] [PubMed] [Google Scholar]
- 17.Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: Comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 19.Ransohoff DF. Lessons from controversy: Ovarian cancer screening and serum proteomics. J Natl Cancer Inst. 2005;97:315–319. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 20.Semmes OJ, Feng Z, Adam BL, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 21.Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: A multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 22.Knol JC, Jimenez CR. MALDI-TOF serum profiling using semiautomated serum peptide capture with magnetic reversed phase (C18) beads. Methods Mol Biol. 2011;790:3–16. doi: 10.1007/978-1-61779-319-6_1. [DOI] [PubMed] [Google Scholar]
- 23.Brahmer JR, Lee JW, Traynor AM, et al. Dosing to rash: A phase II trial of the first-line erlotinib for patients with advanced non-small-cell lung cancer an Eastern Cooperative Oncology Group Study (E3503) Eur J Cancer. 2014;50:302–308. doi: 10.1016/j.ejca.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amann JM, Lee JW, Roder H, et al. Genetic and proteomic features associated with survival after treatment with erlotinib in first-line therapy of non-small cell lung cancer in Eastern Cooperative Oncology Group 3503. J Thorac Oncol. 2010;5:169–178. doi: 10.1097/JTO.0b013e3181c8cbd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 26.Carbone DP, Ding K, Roder H, et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J Thorac Oncol. 2012;7:1653–1660. doi: 10.1097/JTO.0b013e31826c1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbst RS, Johnson DH, Mininberg E, et al. Phase I/II trial evaluating the anti-vascular endothelial growth factor monoclonal antibody bevacizumab in combination with the HER-1/epidermal growth factor receptor tyrosine kinase inhibitor erlotinib for patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:2544–2555. doi: 10.1200/JCO.2005.02.477. [DOI] [PubMed] [Google Scholar]
- 28.Carbone DP, Salmon JS, Billheimer D, et al. VeriStrat classifier for survival and time to progression in non-small cell lung cancer (NSCLC) patients treated with erlotinib and bevacizumab. Lung Cancer. 2010;69:337–340. doi: 10.1016/j.lungcan.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautschi O, Dingemans AM, Crowe S, et al. VeriStrat® has a prognostic value for patients with advanced non-small cell lung cancer treated with erlotinib and bevacizumab in the first line: Pooled analysis of SAKK19/05 and NTR528. Lung Cancer. 2013;79:59–64. doi: 10.1016/j.lungcan.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Akerley W, Boucher K, Rich N, et al. A phase II study of bevacizumab and erlotinib as initial treatment for metastatic non-squamous, non-small cell lung cancer with serum proteomic evaluation. Lung Cancer. 2013;79:307–311. doi: 10.1016/j.lungcan.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Salmon S, Chen H, Chen S, et al. Classification by mass spectrometry can accurately and reliably predict outcome in patients with non-small cell lung cancer treated with erlotinib-containing regimen. J Thorac Oncol. 2009;4:689–696. doi: 10.1097/JTO.0b013e3181a526b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuiper JL, Lind JS, Groen HJ, et al. VeriStrat(®) has prognostic value in advanced stage NSCLC patients treated with erlotinib and sorafenib. Br J Cancer. 2012;107:1820–1825. doi: 10.1038/bjc.2012.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lind JS, Dingemans AM, Groen HJ, et al. A multicenter phase II study of erlotinib and sorafenib in chemotherapy-naive patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:3078–3087. doi: 10.1158/1078-0432.CCR-09-3033. [DOI] [PubMed] [Google Scholar]
- 34.Dingemans AM, Mellema WW, Groen HJ, et al. A phase II study of sorafenib in patients with platinum-pretreated, advanced (stage IIIb or IV) non-small cell lung cancer with a KRAS mutation. Clin Cancer Res. 2013;19:743–751. doi: 10.1158/1078-0432.CCR-12-1779. [DOI] [PubMed] [Google Scholar]
- 35.Stinchcombe TE, Roder J, Peterman AH, et al. A retrospective analysis of VeriStrat status on outcome of a randomized phase II trial of first-line therapy with gemcitabine, erlotinib, or the combination in elderly patients (age 70 years or older) with stage IIIB/IV non-small-cell lung cancer. J Thorac Oncol. 2013;8:443–451. doi: 10.1097/JTO.0b013e3182835577. [DOI] [PubMed] [Google Scholar]
- 36.Lazzari C, Spreafico A, Bachi A, et al. Changes in plasma mass-spectral profile in course of treatment of non-small cell lung cancer patients with epidermal growth factor receptor tyrosine kinase inhibitors. J Thorac Oncol. 2012;7:40–48. doi: 10.1097/JTO.0b013e3182307f17. [DOI] [PubMed] [Google Scholar]
- 37.Garrisi VM, Bongarzone I, Mangia A, et al. Characterization of a serum protein pattern from NSCLC patients treated with gefitinib. Clin Biochem. 2011;44:936–940. doi: 10.1016/j.clinbiochem.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 38.O’Byrne KJ, Danson S, Dunlop D, et al. Combination therapy with gefitinib and rofecoxib in patients with platinum-pretreated relapsed non small-cell lung cancer. J Clin Oncol. 2007;25:3266–3273. doi: 10.1200/JCO.2006.09.2791. [DOI] [PubMed] [Google Scholar]
- 39.Chung CH, Seeley EH, Roder H, et al. Detection of tumor epidermal growth factor receptor pathway dependence by serum mass spectrometry in cancer patients. Cancer Epidemiol Biomarkers Prev. 2010;19:358–365. doi: 10.1158/1055-9965.EPI-09-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto K, Shimizu C, Arao T, et al. Identification of predictive biomarkers for response to trastuzumab using plasma FUCA activity and N-glycan identified by MALDI-TOF-MS. J Proteome Res. 2009;8:457–462. doi: 10.1021/pr800655p. [DOI] [PubMed] [Google Scholar]
- 41.Dalenc F, Doisneau-Sixou SF, Allal BC, et al. Tipifarnib plus tamoxifen in tamoxifen-resistant metastatic breast cancer: A negative phase II and screening of potential therapeutic markers by proteomic analysis. Clin Cancer Res. 2010;16:1264–1271. doi: 10.1158/1078-0432.CCR-09-1192. [DOI] [PubMed] [Google Scholar]
- 42.Gonçalves A, Esterni B, Bertucci F, et al. Postoperative serum proteomic profiles may predict metastatic relapse in high-risk primary breast cancer patients receiving adjuvant chemotherapy. Oncogene. 2006;25:981–989. doi: 10.1038/sj.onc.1209131. [DOI] [PubMed] [Google Scholar]
- 43.Gast MC, Zapatka M, van Tinteren H, et al. Postoperative serum proteomic profiles may predict recurrence-free survival in high-risk primary breast cancer. J Cancer Res Clin Oncol. 2011;137:1773–1783. doi: 10.1007/s00432-011-1055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gast MC, van Tinteren H, Bontenbal M, et al. Haptoglobin phenotype is not a predictor of recurrence free survival in high-risk primary breast cancer patients. BMC Cancer. 2008;8:389. doi: 10.1186/1471-2407-8-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Høgdall E, Fung ET, Christensen IJ, et al. Proteomic biomarkers for overall and progression-free survival in ovarian cancer patients. Proteomics Clin Appl. 2010;4:940–952. doi: 10.1002/prca.200900171. [DOI] [PubMed] [Google Scholar]
- 46.Mazouni C, Baggerly K, Hawke D, et al. Evaluation of changes in serum protein profiles during neoadjuvant chemotherapy in HER2-positive breast cancer using an LC-MALDI-TOF/MS procedure. Proteomics. 2010;10:3525–3532. doi: 10.1002/pmic.201000057. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Dang TA, Shen J, et al. Plasma proteome predicts chemotherapy response in osteosarcoma patients. Oncol Rep. 2011;25:303–314. doi: 10.3892/or.2010.1111. [DOI] [PubMed] [Google Scholar]
- 48.Voortman J, Pham TV, Knol JC, et al. Prediction of outcome of non-small cell lung cancer patients treated with chemotherapy and bortezomib by time-course MALDI-TOF-MS serum peptide profiling. Proteome Sci. 2009;7:34. doi: 10.1186/1477-5956-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han M, Liu Q, Yu J, et al. Identification of candidate molecular markers predicting chemotherapy resistance in non-small cell lung cancer. Clin Chem Lab Med. 2010;48:863–867. doi: 10.1515/CCLM.2010.169. [DOI] [PubMed] [Google Scholar]
- 50.Han M, Dai J, Zhang Y, et al. Support vector machines coupled with proteomics approaches for detecting biomarkers predicting chemotherapy resistance in small cell lung cancer. Oncol Rep. 2012;28:2233–2238. doi: 10.3892/or.2012.2037. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Tan CW, Shen H, et al. Identification of the biomarkers for the prediction of efficacy in first-line chemotherapy of metastatic colorectal cancer patients using SELDI-TOF-MS and artificial neural networks. Hepatogastroenterology. 2012;59:2461–2465. doi: 10.5754/hge12127. [DOI] [PubMed] [Google Scholar]
- 52.Helgason HH, Engwegen JY, Zapatka M, et al. Identification of serum proteins as prognostic and predictive markers of colorectal cancer using surface enhanced laser desorption ionization-time of flight mass spectrometry. Oncol Rep. 2010;24:57–64. doi: 10.3892/or_00000828. [DOI] [PubMed] [Google Scholar]
- 53.Helgason HH, Engwegen JY, Zapatka M, et al. Serum proteomics and disease-specific biomarkers of patients with advanced gastric cancer. Oncol Lett. 2010;1:327–333. doi: 10.3892/ol_00000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter M, Heinze C, Steiner T, et al. Immunochemotherapy-associated protein patterns in tumour tissue and serum of patients with metastatic renal cell carcinoma. Arch Physiol Biochem. 2010;116:197–207. doi: 10.3109/13813455.2010.513392. [DOI] [PubMed] [Google Scholar]
- 55.Vermaat JS, van der Tweel I, Mehra N, et al. Two-protein signature of novel serological markers apolipoprotein-A2 and serum amyloid alpha predicts prognosis in patients with metastatic renal cell cancer and improves the currently used prognostic survival models. Ann Oncol. 2010;21:1472–1481. doi: 10.1093/annonc/mdp559. [DOI] [PubMed] [Google Scholar]
- 56.Risum S, Høgdall E, Engelholm SA, et al. A proteomics panel for predicting optimal primary cytoreduction in stage III/IV ovarian cancer. Int J Gynecol Cancer. 2009;19:1535–1538. doi: 10.1111/IGC.0b013e3181a840f5. [DOI] [PubMed] [Google Scholar]
- 57.Wood SL, Rogers M, Cairns DA, et al. Association of serum amyloid A protein and peptide fragments with prognosis in renal cancer. Br J Cancer. 2010;103:101–111. doi: 10.1038/sj.bjc.6605720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue A, Chang JW, Chung L, et al. Serum apolipoprotein C-II is prognostic for survival after pancreatic resection for adenocarcinoma. Br J Cancer. 2012;107:1883–1891. doi: 10.1038/bjc.2012.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su F, Zhu X, Liang Z, et al. Analysis of serum proteome profiles in nasopharyngeal carcinoma with different radiosensitivity. Clin Transl Oncol. 2014;16:147–152. doi: 10.1007/s12094-013-1052-y. [DOI] [PubMed] [Google Scholar]
- 60.Smith FM, Gallagher WM, Fox E, et al. Combination of SELDI-TOF-MS and data mining provides early-stage response prediction for rectal tumors undergoing multimodal neoadjuvant therapy. Ann Surg. 2007;245:259–266. doi: 10.1097/01.sla.0000245577.68151.bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashida Y, Honda K, Osaka Y, et al. Possible prediction of chemoradiosensitivity of esophageal cancer by serum protein profiling. Clin Cancer Res. 2005;11:8042–8047. doi: 10.1158/1078-0432.CCR-05-0656. [DOI] [PubMed] [Google Scholar]
- 62.Maher SG, McDowell DT, Collins BC, et al. Serum proteomic profiling reveals that pretreatment complement protein levels are predictive of esophageal cancer patient response to neoadjuvant chemoradiation. Ann Surg. 2011;254:809–816; discussion 816–817. doi: 10.1097/SLA.0b013e31823699f2. [DOI] [PubMed] [Google Scholar]
- 63.Kelly P, Paulin F, Lamont D, et al. Pre-treatment plasma proteomic markers associated with survival in oesophageal cancer. Br J Cancer. 2012;106:955–961. doi: 10.1038/bjc.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stinchcombe TE, Peterman AH, Lee CB, et al. A randomized phase II trial of first-line treatment with gemcitabine, erlotinib, or gemcitabine and erlotinib in elderly patients (age ≥70 years) with stage IIIB/IV non-small cell lung cancer. J Thorac Oncol. 2011;6:1569–1577. doi: 10.1097/JTO.0b013e3182210430. [DOI] [PubMed] [Google Scholar]
- 65.Cohen EE, Kane MA, List MA, et al. Phase II trial of gefitinib 250 mg daily in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:8418–8424. doi: 10.1158/1078-0432.CCR-05-1247. [DOI] [PubMed] [Google Scholar]
- 66.Seiwert T, Davis D, Yan D, et al. pKDR/KDR ratio predicts response in a phase I/II pharmacodynamics study of erlotinib and bevacizumab for recurrent or metastatic head and neck cancer (HNC) Proc Am Soc Clin Oncol. 2007;25:6021. [Google Scholar]
- 67.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 68.Rodenhuis S, Bontenbal M, Beex LV, et al. High-dose chemotherapy with hematopoietic stem-cell rescue for high-risk breast cancer. N Engl J Med. 2003;349:7–16. doi: 10.1056/NEJMoa022794. [DOI] [PubMed] [Google Scholar]
- 69.Karlsen MA, Sandhu N, Hogdall C, et al. Evaluation of HE4, CA125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) as diagnostic tools of epithelial ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2012;127:379–383. doi: 10.1016/j.ygyno.2012.07.106. [DOI] [PubMed] [Google Scholar]
- 70.Buzdar AU, Valero V, Ibrahim NK, et al. Neoadjuvant therapy with paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide chemotherapy and concurrent trastuzumab in human epidermal growth factor receptor 2-positive operable breast cancer: an update of the initial randomized study population and data of additional patients treated with the same regimen. Clin Cancer Res. 2007;13:228–233. doi: 10.1158/1078-0432.CCR-06-1345. [DOI] [PubMed] [Google Scholar]
- 71.Voortman J, Smit EF, Honeywell R, et al. A parallel dose-escalation study of weekly and twice-weekly bortezomib in combination with gemcitabine and cisplatin in the first-line treatment of patients with advanced solid tumors. Clin Cancer Res. 2007;13:3642–3651. doi: 10.1158/1078-0432.CCR-07-0061. [DOI] [PubMed] [Google Scholar]
- 72.Milan E, Lazzari C, Anand S, et al. SAA1 is over-expressed in plasma of non small cell lung cancer patients with poor outcome after treatment with epidermal growth factor receptor tyrosine-kinase inhibitors. J Proteomics. 2012;76:91–101. doi: 10.1016/j.jprot.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 73.Gregorc V, Novello S, Lazzari C, et al. Predictive value of a proteomic signature in patients with non-small-cell lung cancer treated with second-line erlotinib or chemotherapy (PROSE): A biomarker-stratified, randomised phase 3 trial. Lancet Oncol. 2014;15:713–721. doi: 10.1016/S1470-2045(14)70162-7. [DOI] [PubMed] [Google Scholar]
- 74.Molina-Pinelo S, Pastor MD, Paz-Ares L. VeriStrat: A prognostic and/or predictive biomarker for advanced lung cancer patients? Expert Rev Respir Med. 2014;8:1–4. doi: 10.1586/17476348.2014.861744. [DOI] [PubMed] [Google Scholar]
- 75.Timms JF, Arslan-Low E, Gentry-Maharaj A, et al. Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem. 2007;53:645–656. doi: 10.1373/clinchem.2006.080101. [DOI] [PubMed] [Google Scholar]
- 76.West-Nørager M, Kelstrup CD, Schou C, et al. Unravelling in vitro variables of major importance for the outcome of mass spectrometry-based serum proteomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:30–37. doi: 10.1016/j.jchromb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 77.Cho WC, Yip TT, Yip C, et al. Identification of serum amyloid a protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res. 2004;10:43–52. doi: 10.1158/1078-0432.ccr-0413-3. [DOI] [PubMed] [Google Scholar]
- 78.Cho WC, Yip TT, Cheng WW, et al. Serum amyloid A is elevated in the serum of lung cancer patients with poor prognosis. Br J Cancer. 2010;102:1731–1735. doi: 10.1038/sj.bjc.6605700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van der Mijn JC, Jiménez CR, Piersma SR, et al. Kinase activity of tumor-derived exosomes as a potential biomarker for response to treatment. Cancer Res. 2012;72(suppl):176a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.