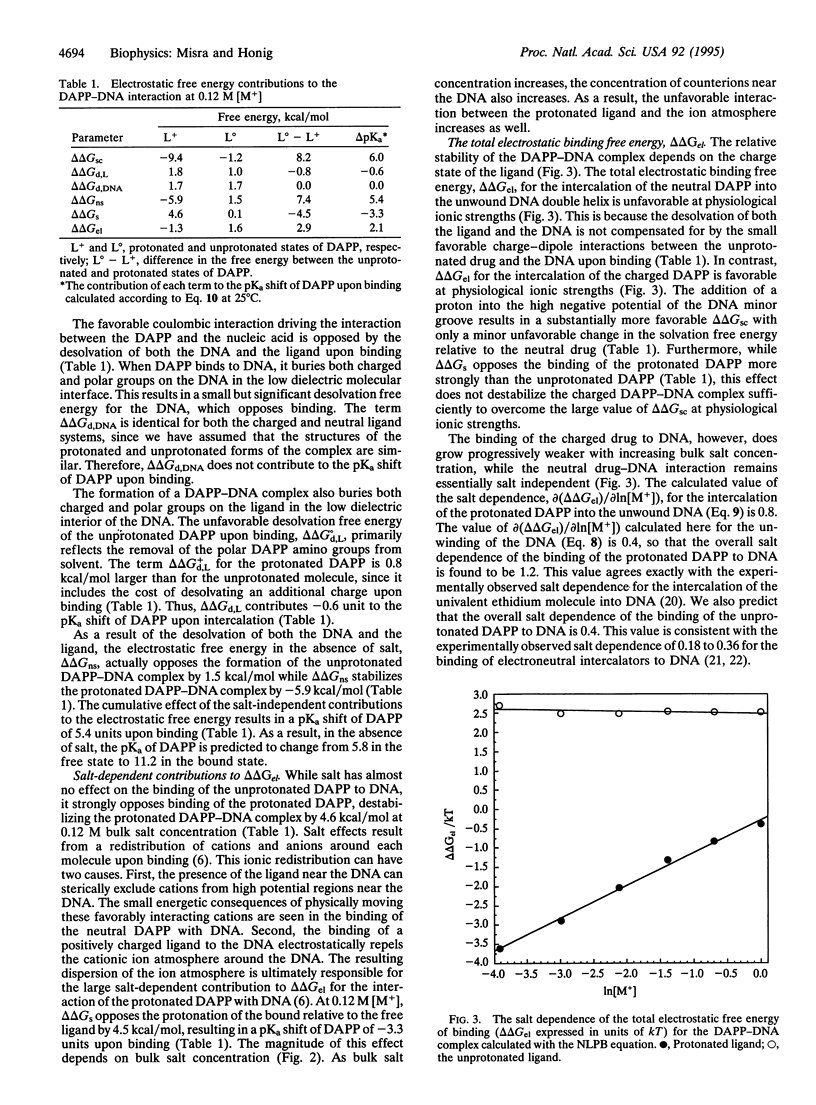
Abstract
A model based on the nonlinear Poisson-Boltzmann equation is used to study the electrostatic contribution to the binding free energy of a simple intercalating ligand, 3,8-diamino-6-phenylphenanthridine, to DNA. We find that the nonlinear Poisson-Boltzmann model accurately describes both the absolute magnitude of the pKa shift of 3,8-diamino-6-phenylphenanthridine observed upon intercalation and its variation with bulk salt concentration. Since the pKa shift is directly related to the total electrostatic binding free energy of the charged and neutral forms of the ligand, the accuracy of the calculations implies that the electrostatic contributions to binding are accurately predicted as well. Based on our results, we have developed a general physical description of the electrostatic contribution to ligand-DNA binding in which the electrostatic binding free energy is described as a balance between the coulombic attraction of a ligand to DNA and the disruption of solvent upon binding. Long-range coulombic forces associated with highly charged nucleic acids provide a strong driving force for the interaction of cationic ligands with DNA. These favorable electrostatic interactions are, however, largely compensated for by unfavorable changes in the solvation of both the ligand and the DNA upon binding. The formation of a ligand-DNA complex removes both charged and polar groups at the binding interface from pure solvent while it displaces salt from around the nucleic acid. As a result, the total electrostatic binding free energy is quite small. Consequently, nonpolar interactions, such as tight packing and hydrophobic forces, must play a significant role in ligand-DNA stability.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Boger D. L., Invergo B. J., Coleman R. S., Zarrinmayeh H., Kitos P. A., Thompson S. C., Leong T., McLaughlin L. W. A demonstration of the intrinsic importance of stabilizing hydrophobic binding and non-covalent van der Waals contacts dominant in the non-covalent CC-1065/B-DNA binding. Chem Biol Interact. 1990;73(1):29–52. doi: 10.1016/0009-2797(90)90107-x. [DOI] [PubMed] [Google Scholar]
- Burke T. G., Mishra A. K., Wani M. C., Wall M. E. Lipid bilayer partitioning and stability of camptothecin drugs. Biochemistry. 1993 May 25;32(20):5352–5364. doi: 10.1021/bi00071a010. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Janin J. The price of lost freedom: entropy of bimolecular complex formation. Protein Eng. 1989 Oct;3(1):1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Rashin A., Fine R., Honig B. On the calculation of electrostatic interactions in proteins. J Mol Biol. 1985 Aug 5;184(3):503–516. doi: 10.1016/0022-2836(85)90297-9. [DOI] [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Harvey S. C. Treatment of electrostatic effects in macromolecular modeling. Proteins. 1989;5(1):78–92. doi: 10.1002/prot.340050109. [DOI] [PubMed] [Google Scholar]
- Jain S. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. VIII. Structures of two ethidium/dinucleoside monophosphate crystalline complexes containing ethidium: cytidylyl(3'-5') guanosine. J Biomol Struct Dyn. 1984 Mar;1(5):1179–1194. doi: 10.1080/07391102.1984.10507511. [DOI] [PubMed] [Google Scholar]
- Jayaram B., Sharp K. A., Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989 May;28(5):975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- Jones R. L., Wilson W. D. Effect of ionic strength on the pKa of ligands bound to DNA. Biopolymers. 1981 Jan;20(1):141–154. doi: 10.1002/bip.1981.360200110. [DOI] [PubMed] [Google Scholar]
- Klapper I., Hagstrom R., Fine R., Sharp K., Honig B. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase: effects of ionic strength and amino-acid modification. Proteins. 1986 Sep;1(1):47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- Lundbäck T., Cairns C., Gustafsson J. A., Carlstedt-Duke J., Härd T. Thermodynamics of the glucocorticoid receptor-DNA interaction: binding of wild-type GR DBD to different response elements. Biochemistry. 1993 May 18;32(19):5074–5082. doi: 10.1021/bi00070a015. [DOI] [PubMed] [Google Scholar]
- Misra V. K., Hecht J. L., Sharp K. A., Friedman R. A., Honig B. Salt effects on protein-DNA interactions. The lambda cI repressor and EcoRI endonuclease. J Mol Biol. 1994 Apr 29;238(2):264–280. doi: 10.1006/jmbi.1994.1286. [DOI] [PubMed] [Google Scholar]
- Misra V. K., Sharp K. A., Friedman R. A., Honig B. Salt effects on ligand-DNA binding. Minor groove binding antibiotics. J Mol Biol. 1994 Apr 29;238(2):245–263. doi: 10.1006/jmbi.1994.1285. [DOI] [PubMed] [Google Scholar]
- Phillips K., Phillips S. E. Electrostatic activation of Escherichia coli methionine repressor. Structure. 1994 Apr 15;2(4):309–316. doi: 10.1016/s0969-2126(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Phillips S. E., Manfield I., Parsons I., Davidson B. E., Rafferty J. B., Somers W. S., Margarita D., Cohen G. N., Saint-Girons I., Stockley P. G. Cooperative tandem binding of met repressor of Escherichia coli. Nature. 1989 Oct 26;341(6244):711–715. doi: 10.1038/341711a0. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994 Feb 11;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Krishnamoorthy C. R., Wang Y. H., Smith J. C. Mechanism of intercalation: ion effects on the equilibrium and kinetic constants for the interaction of propidium and ethidium with DNA. Biopolymers. 1985 Oct;24(10):1941–1961. doi: 10.1002/bip.360241008. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Luty B. A., Davis M. E., McCammon J. A. Poisson-Boltzmann analysis of the lambda repressor-operator interaction. Biophys J. 1992 Nov;63(5):1280–1285. doi: 10.1016/S0006-3495(92)81723-X. [DOI] [PMC free article] [PubMed] [Google Scholar]