Abstract
The A kinase anchoring protein 350 (AKAP350) is a multiply spliced type II protein kinase A anchoring protein that localizes to the centrosomes in most cells and to the Golgi apparatus in epithelial cells. In the present study, we sought to identify AKAP350 interacting proteins that could yield insights into AKAP350 function at the Golgi apparatus. Using yeast two-hybrid and pull-down assays, we found that AKAP350 interacts with a family of structurally related proteins, including FBP17, FBP17b, and cdc42 interacting protein 4 (CIP4). CIP4 interacts with the GTP-bound form of cdc42, with the Wiscott Aldrich Syndrome group of proteins, and with microtubules, and exerts regulatory effects on cytoskeleton and membrane trafficking. CIP4 is phosphorylated by protein kinase A in vitro, and elevation of intracellular cyclic AMP with forskolin stimulates in situ phosphorylation of CIP4. Our results indicate that CIP4 interacts with AKAP350 at the Golgi apparatus and that either disruption of this interaction by expressing the CIP4 binding domain in AKAP350, or reduction of AKAP350 expression by RNA interference leads to changes in Golgi structure. The results suggest that AKAP350 and CIP4 influence the maintenance of normal Golgi apparatus structure.
INTRODUCTION
Cyclic AMP-dependent protein kinase (A kinase) anchoring proteins (AKAPs) are a family of >50 proteins that are structurally diverse, but all contain an amphipathic helix that mediates their interaction with the regulatory subunits of protein kinase A (PKA) (reviewed in Alto et al., 2002). They also contain subcellular targeting domains that direct their localization, controlling PKA location within the cell. AKAPs also interact with other proteins involved in signaling pathways. Thus, they facilitate the specificity and kinetics of a particular pathway, by compartmentalizing its components. Scaffolding of multifunctional enzymes with their substrates accounts for the specific propagation of messenger pathways within cellular compartments.
AKAP350, AKAP450, and CG-NAP are members of a group of AKAPs derived from a multiply spliced gene on human chromosome 7 (Keryer et al., 1993; Dransfield et al., 1997; Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999). AKAP350 proteins localize to the centrosomes and the Golgi apparatus (Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999; Shanks et al., 2002a, b). The shortest 3′ splice variant of this gene is yotiao, an AKAP that interacts with the N-methyl-d-aspartate receptor ion channel in neurons, positioning this channel close to both its activator PKA and its inactivating regulator, protein phosphatase 1 (Lin et al., 1998; Westphal et al., 1999). We have described three further 3′ splice variants that differ in their carboxyl termini: AKAP350A, AKAP350B, and AKAP350C (Schmidt et al., 1999; Shanks et al., 2002b). CG-NAP and AKAP450 have the same carboxyl termini as AKAP350A, but differ in their amino terminal initiation sites. We will refer to the AKAP350A/CG-NAP/AKAP450 proteins throughout as AKAP350.
AKAP350 interacts with type II PKA through two separate RII binding motifs. The multiple pathways regulated by AKAP350 are apparent from the diverse proteins that it scaffolds at discrete locations. The nerve growth factor-activated protein kinase (Takahashi et al., 1999) interacts with AKAP350 both at the centrosomes and the Golgi apparatus. Casein kinase Iδ and ε (Sillibourne et al., 2002), the phospho-diesterase 4E (Tasken et al., 2001), and the small Ran GTPase (Keryer et al., 2003) associate with AKAP350 specifically at the centrosomes. Hypophosphorylated protein kinase Cε colocalizes with AKAP350 in the Golgi area (Takahashi et al., 2000). AKAP350 also interacts with the protein phosphatases PP1 and PP2A (Takahashi et al., 1999), calmodulin (Gillingham and Munro, 2000), and with the components of the γ-tubulin ring complex, GCP2, and GCP3 (Takahashi et al., 2002). We have recently described that AKAP350 interacts with different members of the chloride intracellular channel (CLIC) family at the Golgi apparatus, the centrosomes, and midbody of mammalian cells (Shanks et al., 2002a; Berryman and Goldenring, 2003) and with the members of the transforming acid coiled-coiled protein family (TACC3 and TACC4) at the centrosomes (Steadman et al., 2002).
The presence of AKAP350 at the Golgi apparatus seems to be cell type dependent. Primary cultured epithelial cells and epithelial derived cell lines have extensive Golgi staining for AKAP350, whereas the T lymphocyte-derived Jurkat cells only show centrosomal AKAP350 staining (Schmidt et al., 1999; Takahashi et al., 1999; Steadman et al., 2002). We have recently described a motif comprising amino acids 3259-3307 of AKAP350A that targets it to the Golgi apparatus; and using specific antibodies, we identified that AKAP350A was the main AKAP350 isoform in the Golgi apparatus of HCA-7 cells (Shanks et al., 2002b). AKAP350 and the brefeldin A-inhibited guanine nucleotide-exchange protein 2, BIG2 (Li et al., 2003), are presently the only AKAPs described in the Golgi. Several studies have implicated PKA in the regulation of Golgi function. PKA modulates transport from the cis- to the trans-Golgi, and from the Golgi to the cell surface (Muniz et al., 1996; Cobbold et al., 2002). PKA activity is necessary for the normal budding of vesicles at the Golgi (Muniz et al., 1997). Nevertheless, significant Golgi substrates for PKA have not been identified.
In this study, we report that AKAP350 interacts with the cdc42 interacting protein 4 (CIP4) at the Golgi apparatus. CIP4 itself has the characteristics of a scaffolding protein, interacting with the GTP-bound cdc42 (Aspenstrom, 1997), the Wiscott Aldrich Syndrome Protein (WASP), microtubules (Tian et al., 2000), and the cdc42 and Rac1 activating protein RICH. Recent studies suggest that CIP4 is involved in the regulation of the cytoskeleton and membrane trafficking (Aspenstrom, 1997; Tian et al., 2000; Dombrosky-Ferlan et al., 2003). We found that CIP4 is a PKA substrate in vitro and that stimulation of PKA in situ elicits CIP4 phosphorylation. Our results provide evidence that the presence of both CIP4 and AKAP350 at the Golgi is necessary for the maintenance of its normal structure.
MATERIALS AND METHODS
Materials
Oligonucleotides used were synthesized by Invitrogen (Carlsbad, CA). Advantage Taq was purchased from BD Biosciences Clontech (Palo Alto, CA). DNA sequencing was performed using dye terminator chemistry automated sequencing by either the Molecular Biology Core Facility at the Medical College of Georgia or the DNA Sequencing Core Facility at Vanderbilt University. Species-specific Cy5-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Prolong Antifade, 4′,6-diamidino-2-phenylindole (DAPI), and species-specific Alexa 488- and 568-conjugated secondary antibodies were purchased from Molecular Probes (Eugene, OR).
Antibodies
The monoclonal antibody 14G2 anti-AKAP350, rabbit anti-AKAP350A, and rabbit anti-CIP4 were prepared as we described previously (Schmidt et al., 1999; Tian et al., 2000; Shanks et al., 2002a). The monoclonal anti-p58 and anti-α-tubulin were purchased from Sigma-Aldrich (St. Louis, MO), monoclonal anti-Golgin97 was from Molecular Probes, rabbit anti-calreticulin was from Abcam (Cambridge, MA), monoclonal anti-cdc42 was from Santa Cruz Biotechnology (Santa Cruz, CA), and rabbit anti-RXXS/T(Pi) was purchased from Cell Signaling Technology (Beverly, MA). The rabbit anti-giantin was a gift from Dr. E.K.L. Chan (University of Florida, Gainesville, FL).
To generate the monoclonal anti-CIP4 antibody, mice were immunized with full-length glutathione S-transferase (GST)-CIP4 expressed in Escherichia coli, and hybridoma cell lines were produced following standard techniques at the National Institutes of Health Laboratory Animal Sciences Program. Lines producing antibody against the immunogen were cloned by limiting dilution, and the clones were screened for reactivity to FLAG-CIP4 (118-481). Two clones, AD191 and AD192, were chosen for further study due to their strong reactivity. Both clones were from the same line and have the isotype IgG1/k. In Western blots, both AD191 and AD192 detected endogenous CIP4 in Cos7 cells, as well as overexpressed myc-tagged CIP4 and CIP4 deletion mutants 1-417, 1-408, D (380-481), and D (280-481). These antibodies did not react with the CIP4 deletion mutant 1-118, the isolated SH3 domain of CIP4, or GST. We concluded that AD191 and AD192 recognize an epitope within amino acids 118-280 of CIP4 and are functionally equivalent. In the experiments we presented here, we used AD191.
Yeast Two-Hybrid Assays
A rabbit parietal cell library in pAD-GAL4 (Lapierre et al., 1999) was screened with the fragment of AKAP350 located between the two RII interacting motif (nucleotides 3611-6813) [AKAP350(1076-2143)], cloned into pBD-GAL-Cam (Stratagene, La Jolla, CA) by using EcoRI and SalI sites. The Y190 yeast strain harboring the HIS3 and β-galactosidase reporter genes was used for screening of approximately 1 million clones as described previously (Shanks et al., 2002b). One positive colony was rescued into XL-1 Blue bacteria, and plasmid DNA was prepared using Miniprep kits (QIAGEN, Valencia, CA). The isolated clone and the bait both gave negative results when analyzed by themselves in yeast two-hybrid binary assays.
FBP17, FBP17b, CIP4, and its deletion constructs were cloned into pAD-GAL4 by using EcoRI and SalI restriction sites and tested for interaction with AKAP350(1076-2143)-pBD-GAL4-Cam, in the yeast-two hybrid assay. Positive results were recorded if β-galactosidase assay was positive within 3 h.
In Vitro Binding Assays
A full-length CIP4 cDNA cloned into pRK5 as described previously (Tian et al., 2000) was used as a template to clone CIP4 into the pET30c vector (Novagen, Madison, WI) by using EcoRI and SalI sites, to produce (His)6 fused to the carboxy terminus of CIP4. The protein was produced using BL21(DE3)pLysS bacteria and purified using Ni-beads (QIAGEN) as indicated by the manufacturer. To assess CIP4 interaction with AKAP350 in vitro, (His)6-CIP4 bound to Ni beads or Ni beads alone were incubated with a 100,000 × g gastric mucosal supernatant overnight at 4°C, in the presence of protease inhibitors (protease inhibitor cocktail for mammalian tissues; Sigma-Aldrich). After removing the nonbound material, the beads were washed three times for 10 min with 50 mM imidazole and protease inhibitors and eluted by heating 20 min at 65°C in sample buffer (100 mM Tris, pH 6.8, 2% SDS, 8% glycerol, 25 μM dithiothreitol). The starting material and the eluates were resolved by SDS-PAGE by using 3-10% gradient gels. The proteins were transferred for 2 h at 750 mA to nitrocellulose membranes for subsequent Western blotting with anti-AKAP350 antibody.
CIP4 in vitro phosphorylation was assessed by incubating (His)6-CIP4 with 50 mU of PKA catalytic subunit (Promega) in a reaction buffer (150 mM NaCl, 5 mM MgCl, 0.5 mM EGTA, 1 mM dithiothreitol, 20 μM ATP, 25 mM Tris, pH 7.4). The reactions were incubated at 30°C for 2 or 10 min, in the presence or absence of 1 or 10 μM H-89 (Calbiochem, San Diego, CA). The reactions were terminated by heating the samples in sample buffer, at 65°C for 20 min. The samples were resolved on 8% SDS-PAGE. Proteins were transferred to Immobilon membranes and analyzed by Western blot as described below.
Cell Treatments
Madin-Darby canine kidney (MDCK) cells were grown on glass coverslips to confluence in DMEM supplemented with 10% bovine calf serum. Cells treated with brefeldin A (BFA) (Calbiochem) were incubated in the same media containing 5 μg/ml BFA for 1, 10, or 60 min. Cells treated with nocodazole (Calbiochem) were incubated in media containing 1 or 15 μM nocodazole for 1 h at 37 C. In all cases, cells were washed three times with phosphate-buffered saline (PBS) and fixed and permeabilized in a fixative that preserves microtubules (4% paraformaldehyde, 0.1% Triton X-100, 80 mM K-PIPES, pH 7.2, 1 mM EGTA, 1 mM MgSO4, 30% glycerol) for 20 min at room temperature for analysis by immunofluorescence microscopy.
To assess modulation of CIP4 phosphorylation in situ, MDCK cells were grown to confluence in 15-cm petri dishes. To analyze the effect of PKA activation, cells were incubated for 10 min in DMEM without serum in the presence of different concentrations of forskolin (Sigma-Aldrich) (0.2, 1, or 10 μM). Control cells were incubated with the appropriate concentration of the vehicle (dimethyl sulfoxide). To verify whether the effect of cAMP increases was mediated by PKA, cells were preincubated with 10 μM myristoylated protein kinase A inhibitor (Calbiochem) for 20 min and then for 3 min in the presence of 5 μM forskolin (Sigma-Aldrich). After the incubations, the cells where scraped in cold PBS containing protease and phosphatase inhibitors (Sigma-Aldrich). Cell lysates and immunoprecipitates by using rabbit anti-CIP4 antibody were prepared as described below and analyzed by Western blot by using an antibody that recognizes the phosphorylated RXXS/T site [RXXS/T(Pi)] or monoclonal anti-CIP4.
Subcellular Fractionation and Immunoprecipitation
For analyzing CIP4 expression in MDCK, cells were grown to confluence and washed in PBS. The cells were scraped in PBS and pelleted at 200 × g. for 3 min at 4°C. The cells were then homogenized in ice-cold 250 mM sucrose, 10 mM HEPES, pH 7.4, containing protease inhibitors, by using a Potter Teflon on glass homogenizer. The homogenates were centrifuged for 5 min at 4°C at 1000 × g, and the postnuclear supernatant was centrifuged at 100,000 × g for 60 min. The pellet (P100) was resuspended in PBS, and the supernatant (SN) was also kept. For immunoprecipitating CIP4 by using the rabbit anti-CIP4 antibody, pelleted cells were resuspended in 150 mM NaCl, 1% CHAPS, 10 mM Tris, pH 7.4, with protease and phosphatase inhibitors and subjected to two freeze (-80 C)-thaw cycles. After the lysis, cells were spun down at 1000 × g for 5 min, and the pellet was discarded. Immunoprecipitations of the lysates by using the rabbit anti-CIP4 antibody were performed as described previously (Tian L et al., 2000). The samples were resolved using 8% SDS-PAGE and transferred overnight at 100 mA to Immobilon membranes for immunoblotting.
Western Blot Analysis
Blots were blocked with 5% nonfat milk in Tris-buffered saline, 0.05% Tween 20 (TBS-Tween) and then probed with the primary antibodies [14G2, 1:500 for AKAP350; rabbit anti-CIP4, 1:2000; mouse anti-CIP4, 1:50; rabbit anti-RXXS/T(Pi), 1:1000; rabbit anti-calreticulin, 1:2000] in 0.5% nonfat milk-TBS-Tween for 1 h at room temperature. After washing three times with TBS-Tween, blots were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (1:7500) in 1% milk-TBS-Tween for 30 min. Blots were finally washed three times with TBS-Tween, and three times with TBS, and immunoreactivity was detected with chemiluminescence (Supersignal; Pierce Chemical. Rockford, IL) and subsequent exposure to BioMax film (Eastman Kodak, Rochester, NY).
Immunofluorescence Microscopy
Cells were blocked with 17% donkey serum, 0.3% Triton X-100 in PBS, and then incubated simultaneously with rabbit anti-CIP4 (1:250), rabbit antigiantin (1:300), or rabbit anti-AKAP35A (1:80) and mouse anti-AKAP350 (14G2) (1:80), mouse anti-p58 (1:80), mouse anti-golgin-97 (1:100), mouse anti-α-tubulin (1:250), or mouse anti-cdc42 (1:100) for 2 h at room temperature. After washing with PBS, the cells were incubated with Alexa 568- and Alexa 488-conjugated anti-rabbit or anti-mouse IgG for 1 h. When cells were transfected with vectors containing green fluorescent protein (GFP) constructs, Alexa 568- and Cy5-conjugated anti-rabbit or anti-mouse antibodies were used. The cells were washed and finally incubated with 10 μM DAPI in 50 mM sodium phosphate for 10 min. Slides were mounted with Prolong Antifade and examined with an Axiophot microscope (Carl Zeiss, Jena, Germany) equipped with a SPOT digitizing camera.
GFP-AKAP350(1076-2143) Expression
The AKAP350(1076-2143) domain was cloned into pEGFP-C2 (BD Biosciences Clontech) by using EcoRI and SalI sites, generating a construct coding for GFP fused to the N terminus of AKAP350(1076-2143) [GFP-AKAP350(1076-2143)]. MDCK cells were grown overnight on glass coverslips and then transfected with this construct by using Effectene (QIAGEN). Twenty hours posttransfection, the cells were fixed and stained as described above.
Reduction of AKAP350 Expression by Short Interfering RNA (siRNA)
The 21 nucleotides RNA duplexes were synthesized using the Silencer siRNA kit (Ambion, Austin, TX). Four target sequences were chosen following the guidelines described by Elbashir et al. (2001). HeLa cells were transfected with the different oligos by using the siSPORT Lipid transfection agent (Ambion), following the manufacturer's instructions. AKAP350 expression was analyzed at 2, 3, and 4 d after the transfection. Two of the RNA duplexes induced a significant decrease in AKAP350 expression, as analyzed by immunofluo-rescence and Western blot: siRNA1 was specifically targeted to nucleotides 10917-10937 (AAATCCCTTGCCAGCACATGA), and siRNA2 to nucleotides 9421-9441 (AAACGAGAAAGTAGAAGAATT) of human AKAP350A. The control siRNA was designed scrambling the nucleotides of the siRNA1 target sequence (AACTATGCCACCGTGACCTAA).
Morphometric Analysis of the Golgi Apparatus
The Golgi apparatus was stained with antibodies to the Golgi proteins: p58 (cis-Golgi) (Saraste et al., 1987), Giantin (cis- and medial-Golgi) (Lindstedt et al., 1995), and golgin 97 (trans-Golgi) (Luke et al., 2003). The images were captured with a SPOT camera at 40× by using the same settings for each Golgi marker. The boundary of the Golgi complex defined by fluorescent staining by Golgi markers and that for the cell itself as assessed in a phase contrast image was outlined for each analyzed cell by using the drawing tool in the MetaMorph program (Universal Imaging, Dowingtown, PA). The average fluorescence intensity of each marker was determined both in the Golgi area and in the total cell area. With these measurements, we calculated both the percentage of area of the cell occupied by the Golgi membranes and the relative intensity of each fluorescent marker in the Golgi area compared with the average intensity over the entire cell area. The effects of the expression of the CIP4 interacting domain in AKAP350 [AKAP350(1076-2143)] and of the siRNA-induced down-regulation of AKAP350 expression on these parameters were analyzed using paired Student's t-test or analysis of variance with post hoc analysis of significant means by the Neuman-Keuls test, respectively.
RESULTS
FBP17, FBP17b, and CIP4 Interact with AKAP350
We screened a rabbit parietal cell library with a 3.2-kb segment of AKAP350, AKAP350(1076-2143). One positive clone was identified as a partial coding sequence with high homology with the human sequence of formin binding protein 17 (FBP17). The cloned rabbit sequence could represent either the rabbit homologue of FBP17 or a splice variant (Figure 1A). We designated this sequence FBP17b (GenBank accession no. AY345341). FBP17 was first identified in mouse as a binding protein for formins (Chan et al., 1996), and the human homologue was subsequently isolated as a fusion partner of the MLL gene at 11q23 (Fuchs et al., 2001). Using yeast two-hybrid binary assays, we confirmed the FBP17b interaction with AKAP350(1076-2143) and found that human FBP17 also interacts with the AKAP350(1076-2143) domain (Figure 1C). FBP17 and FBP17b contain an aminoterminal FER-CIP4 homology region, a central domain with high homology with the cdc42 interacting domain present in CIP4, and a carboxyl-terminal src homology (SH)3 domain, highly similar to the SH3 in CIP4 (Figure 1B). Because of the high homology between CIP4 and FBP17, we tested the capability of CIP4 to interact with AKAP350(1076-2143) (Figure 1C). The yeast two-hybrid binary assay was also positive for CIP4. We used deletion constructs of CIP4 in binary assays and found that the amino-terminal 118 amino acids (microtubule binding domain, MBD), but not the SH3 domain, were necessary for this interaction (Figure 1C). Interestingly the same domain is responsible for CIP4 interaction with microtubules (Tian et al., 2000). Using yeast two-hybrid binary assays, we tried to define further the domain in AKAP350 interacting with CIP4, but AKAP350(1076-2143) was the smallest construct that was specifically positive with CIP4.
Figure 1.
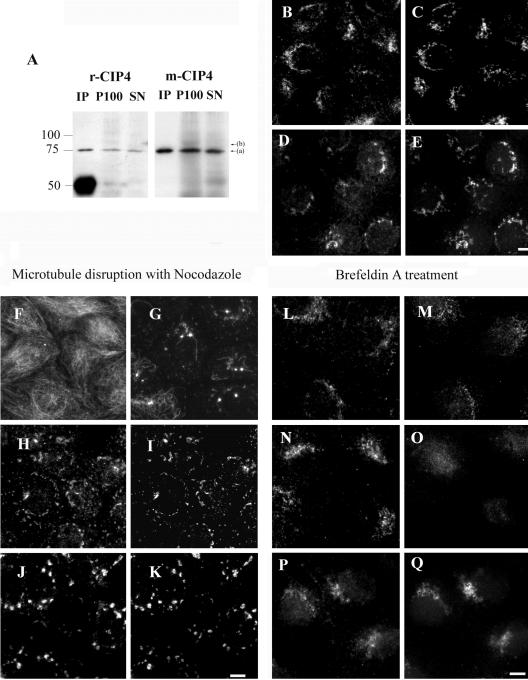
FBP17b, FBP17, and CIP4 interact with AKAP350. (A) Genomic organization of human FBP17. The 15 exons are denoted, with the dashed boxes indicating the exons that are spliced out in the FBP17B transcript. The splicing does not interrupt the exons comprising the MBD, the cdc42 interacting domain (CID), or the SH3 domain. (B) Diagram showing identity (top numbers) and similarity (bottom numbers) between FBP17 exons 1-15, and CIP4 exons 1-14. (C) FBP17, FBP17b, CIP4, and the CIP4 deletion constructs of the MBD or the C-terminal SH3 domain were cloned in pBD-Gal4 vector. The interaction of these proteins with AKAP350 was assessed in yeast two-hybrid binaries assays with pAD-AKAP350(1076-2143). The results confirm FBP17b interaction and indicate that FBP17 and CIP4 also interact with AKAP350(1076-2143) and that the MBD, but not the SH3 domain, in CIP4 is necessary for the interaction. (D) In vitro confirmation of AKAP350/CIP4 interaction. (His)6-tagged CIP4 was attached to Ni beads. A 100,000 × g gastric mucosal supernatant (SM) was incubated with His-CIP4 Ni beads (CIP4) or Ni beads alone (blank). After incubation, samples were washed and eluted with sample buffer. Samples were subjected to electrophoresis in 3-10% SDS-PAGE followed by Western blotting with anti-AKAP350 monoclonal antibody (14G2). Bar, 5 μm.
To confirm the interactions observed in the yeast two-hybrid assays, we performed in vitro binding assays by using recombinant GST-FBP17b or (His)6-CIP4 incubated with a 100,000 × g gastric mucosal supernatant. Both CIP4 (Figure 1D) and FBP17b (our unpublished data) pulled down AKAP350. Sequential immunoblotting demonstrated that calreticulin in the 100,000 × g supernatant did not associate with either CIP4-bound or control beads (our unpublished data).
CIP4 Colocalizes with AKAP350 at the Golgi in MDCK Cells
We prepared constructs coding for the fusion proteins FPB17-GFP and CIP4-GFP. When transfected in HeLa and MDCK cells, the FBP17-GFP showed a distribution indicative of plasma membrane association, whereas the CIP4-GFP showed perinuclear distribution, compatible with Golgi localization (our unpublished data). Because previous investigations had demonstrated a prominent association of AKAP350 with the Golgi apparatus (Shanks et al., 2002a, b), we sought to investigate the possible interaction of CIP4 and AKAP350. We analyzed the endogenous expression of CIP4 by using two anti-CIP4 antibodies (see MATERIALS AND METHODS). The Western blot of MDCK cellular extracts showed a main immunoreactive band present in both the particulate and soluble fractions, with an apparent molecular mass of 75 kDa, higher than the predicted molecular mass for CIP4 (63 kDa) (Figure 2A). The Western blot of HeLa and HCA-7 cell extracts, as well as the myc-tagged CIP4 overexpressed in COS-7, also showed a major immunoreactive band of 75 kDa (our unpublished data). This difference is probably due to the presence of secondary structures, because the protein produced in bacteria also showed a higher molecular mass (our unpublished data). The 100,000 × g microsomes (P100) of MDCK cells also showed a weaker band recognized by both antibodies, with a molecular mass of 83 kDa. Treatment of the samples with alkaline phosphatase did not modify this band, suggesting that it did not correspond to phosphorylated CIP4. There are at least five splice variants for human CIP4, including CIP4a (CIP4), CIP4b (FELIC), CIP4c, CIPh (CIP4/2), and the human salt tolerant protein (HSTP) (Tsuji and Tsuji, 2000; Chang et al., 2002; Wang et al., 2002; Dombrosky-Ferlan et al., 2003). Only CIP4 shows a ubiquitous distribution, so it is likely that the 75-kDa band present in the three different cell lines analyzed corresponds to CIP4, and the 83-kDa band might correspond to a different splice variant present in MDCK cells.
Figure 2.
CIP4 colocalizes with AKAP350 at the Golgi. (A) Cellular extracts from MDCK cells were analyzed for immunoreactivity with the rabbit anti-CIP4 (r-CIP4) antibody and compared with the monoclonal anti-CIP4 antibody. The Western blot of r-CIP4 immunoprecipitate (IP), and the 100,000 × g microsomal pellet (P100) and supernatant (SN) detect a main band with an apparent molecular mass of approximately 75 kDa (a). We also observed a higher band with weaker immunoreactivity at 83 kDa (b). Both antibodies recognized the same bands. The lower mass band in the immunoprecipitated (IP) lane blotted with r-CIP4 corresponds to the heavy chain of the antibody used in the immunoprecipitation. (B-E) MDCK cells were fixed and dual stained with r-CIP4 (B and D) and anti-AKAP350 (14G2) (C) or antibody against the Golgi apparatus marker p58 (E). The immunofluorescence images show extensive areas of colocalization of CIP4 and AKAP350 (upper row) in Golgi elements. The arrow indicates centrosomal staining of AKAP350 that did not show any CIP4 immunoreactivity. (F-K) MDCK cells were treated with 15 μM nocodazole for 1 h, fixed, and dual-stained with r-CIP4 (H and J) and anti-AKAP350 (I) or anti-p58 (K) antibodies. F and G show α-tubulin staining in control or nocodazole-treated cells respectively. (L-Q) MDCK cells were treated with 5 μM BFA for different periods. The figures show the dual staining with r-CIP4 (L and N) and anti-cdc42 (M and O) antibodies of control cells (L and M) or cells treated with BFA for 1 min (N and O) and cells treated for 1 h with BFA stained with r-CIP4 (P) and 14G2 anti-AKAP350 (Q) antibodies. Bar, 5 μm.
The subcellular localization of CIP4 in MDCK cells was investigated using dual staining with the rabbit anti-CIP4 and either anti-AKAP350 (14G2) or anti-p58 (cis-Golgi marker) antibodies. CIP4 was observed in the perinuclear area (Figure 2, B and D) where it costained with both AKAP350 (Figure 2C) and p58 (Figure 2E). In addition, we observed some cytosolic staining for CIP4. In most of the cells there was no costaining of CIP4 with AKAP350 at the centrosomes. Microtubules are required to determine the localization and organization of the Golgi apparatus (reviewed in Rios and Bornens, 2003). Nocodazole (15 μM) induced a great disorganization of microtubules (Figure 2G) and Golgi vesiculation (Figure 2K). After the microtubule disruption, CIP4 remained colocalized with AKAP350 and p58 (Figure 2, H-K).
CIP4 interacts with the GTP bound form of cdc42. A significant fraction of cdc42 is localized to the Golgi and is rapidly removed by BFA (Erickson et al., 1996). MDCK cells dual stained for CIP4 and cdc42 showed perinuclear localization for both proteins (Figure 2, L and M). Treatment with BFA for 1 min shifted cdc42 to the cytosol, but only slightly modified CIP4 distribution (Figure 2, O and N). Even after 60 min of BFA treatment, CIP4 still had a Golgi-like localization (Figure 2P). AKAP350 staining after BFA treatment had the same pattern as CIP4 (Figure 2Q). These studies indicate that, in contrast to cdc42, CIP4 association with the Golgi membranes is not directly dependent on the BFA-sensitive ARF GTPases.
CIP4 Is Phosphorylated by PKA
To assess phosphorylation of CIP4, we used an antibody that recognizes phosphorylated serine or threonine with an arginine at position-3 [RXXS/T(Pi)], a good phosphorylation site for PKA as well as for cGMP-dependent protein kinase, protein kinase C, Akt, and RSK. The Western blot analysis of CIP4 immunoprecipitates from MDCK lysates by using this antibody showed an immunoreactive band that disappeared if the samples were treated with alkaline phosphatase (our unpublished data), indicating that CIP4 was phosphorylated, and the antibody was specifically recognizing the phosphorylated CIP4.
We incubated recombinant (His)6-CIP4 with the catalytic subunit of PKA in the presence or absence of the PKA inhibitor H-89 (Figure 3A). The anti-RXXS/T(Pi) antibody recognized a main band migrating at ∼83 kDa, the apparent molecular mass for the recombinant CIP4, when (His)6-CIP4 was incubated with the catalytic subunit of PKA. CIP4 was not immunoreactive with the anti-RXXS/T(Pi) antibody when incubated without the catalytic subunit of PKA, or when 10 μM H-89 was added to the reaction. Beads without recombinant CIP4 incubated with the catalytic subunit of PKA did not show the 83-kDa band.
Figure 3.
CIP4 is phosphorylated by PKA. (A) In vitro phosphorylation. (His)6-tagged CIP4 attached to Ni beads was incubated with the catalytic subunit of PKA in the appropriate buffer at 30°C for 10 min. The experiment was also performed in the presence of the PKA inhibitor H-89 (1 and 10 μM). As controls, we incubated Ni beads without bound protein with PKA (first lane) and (His)6-CIP4-Ni beads without PKA (second lane). The reactions were terminated by heating the samples in sample buffer at 65°C for 20 min. The samples were resolved by SDS-PAGE and transferred to Immobilon membranes, and CIP4 phosphorylation was analyzed using an antibody that recognizes the RXXS/T(Pi) site. (B and C) In situ analysis of CIP4 phosphorylation. MDCK cells were grown to confluence and stimulated with different doses of forskolin (F) for 10 min (B), or alternatively were preincubated for 20 min in the presence or absence of 10 μM myristoylated PKI and then stimulated with 5 μM forskolin for 3 min (C). After the incubations, cells were scraped and lysed in the presence of protease and phosphatase inhibitors, and CIP4 immunoprecipitates were prepared as described and analyzed for phosphorylation as in A. (A-C) Representative of three separate experiments. For quantification of phosphorylated CIP4, we performed the densitometry of the bands using the NIH Image J program and divided the density of each RXXT/S(Pi) immunoreactive band by that of the same band with the monoclonal anti-CIP4 antibody. The analysis of the in situ phosphorylation indicated that forskolin induced an increase on CIP4 phosphorylation in the RXXS/T site of 88%, and this increase was prevented by preincubation with the PKA inhibitor.
For assessing the modulation of the in situ phosphorylation by PKA, we incubated MDCK cells with 0.2, 1, or 10 μM forskolin for 10 min. We found that 1 μM forskolin increased the immunoreactivity of CIP4 with the RXXS/T(Pi) antibody, and the increase was higher at 10 μM forskolin (Figure 3B). Incubating cells with 5 μM forskolin, we found that at 3 min there was already an increase in CIP4 phosphorylation level, which was prevented by the presence of the cell-permeant PKA inhibitor myristoylated PKI (Figure 3C). These results were consistently observed in three different experiments with an 88% average increase in CIP4 phosphorylation in cells stimulated with forskolin. As Figure 3C shows, myristoylated PKI induced a small increase in phosphorylated CIP4, which may indicate that other pathways are also involved in the regulation of CIP4 phosphorylation.
Expression of the CIP4 Binding Domain of AKAP350 Altered Golgi Structure
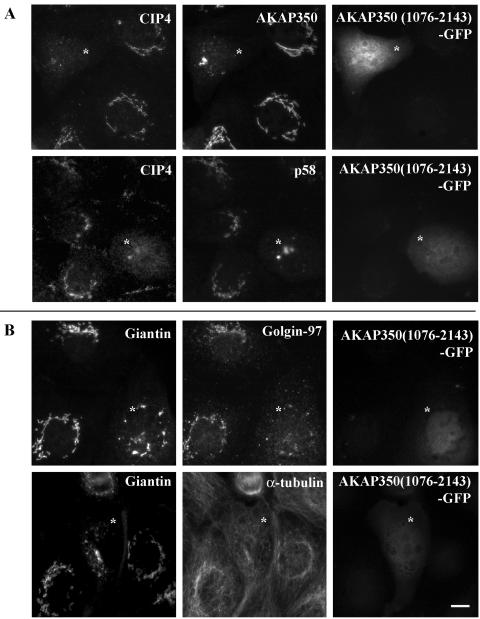
To examine further the interaction of CIP4 with AKAP350 in situ, we transfected MDCK cells with AKAP350(1076-2143) fused to GFP. This construct contained the CIP4 binding domain, but it did not localize to the Golgi apparatus or centrosomes, because it did not contain the distinct targeting domains (Shanks et al., 2002b). As was expected if CIP4 interacts in situ with this construct, in 90% of the transfected cells CIP4 increased its cytosolic distribution with a decrease in colocalization with both AKAP350 and p58 on Golgi membranes (Figure 4A). These changes in CIP4 localization were accompanied with changes in the pattern of p58 (Figure 4A), giantin, and golgin-97 (Figure 4B) distribution. The staining patterns for the cis- and cis-medial-Golgi proteins p58 and giantin (Saraste et al., 1987; Lindstedt et al., 1995) suggested a modification in the Golgi stacking, with some cells showing a very compact pattern (Figure 4A), whereas other cells demonstrated vesiculated Golgi membranes (Figure 4B). The staining for AKAP350 in the transfected cells showed the same patterns. In contrast, the trans-Golgi protein golgin-97 (Luke et al., 2003) showed a very dispersed pattern in >80% of the transfected cells. Staining for microtubules with α-tubulin antibodies did not show major changes in the cells expressing the CIP4 binding domain of AKAP350. Even in cells showing modifications in the Golgi morphology, greater intensity microtubule still was observed in association with regions where the Golgi membrane elements were present (Figure 4B).
Figure 4.
AKAP350(1076-2143) expression alters the structure of the Golgi apparatus. MDCK cells were transiently transfected with GFP-AKAP350(1076-2143) and grown for 20 h. Fixed cells were dual stained with r-CIP4 and anti-AKAP350 or anti-p58 antibodies (A), or with anti-giantin and anti-golgin-97 or anti-α-tubulin antibodies (B). The viability of the cells was confirmed by analysis of the DAPI staining and the phase contrast image of the cells (our unpublished data). The stars indicate the positions of cells expressing AKAP350(1076-2143)-GFP. Bar, 5 μm.
We obtained quantitative descriptions of these observations in fluorescent images of Golgi marker staining in transfected and nontransfected cells (Table 1). We compared two parameters as reflections of the compactness or dispersal of Golgi membranes stained with the fluorescent markers: We calculated the percentage of the total cell area occupied by Golgi staining. Table 1 demonstrates the quantitation of the two patterns of staining for giantin in transfected cells. In the first set of cells, we observed a significant decrease in the area of the cell occupied by the giantin-staining membranes and an increase in the intensity of staining within these membranes, all consistent with the condensation of Golgi membranes (Figure 4A). In the second morphology, Golgi areas and intensity of staining within the Golgi were unchanged, consistent with the observation of regional vesiculation (Figure 4B). In contrast with patterns for the cis- and medial-Golgi marker, quantitation of the trans-Golgi marker golgin-97 staining demonstrated a significant increase in the percentage of area of the cell stained and a decrease in the average staining intensity in the cells expressing AKAP350(1076-2143), all consistent with a general dispersal of golgin-97 staining in transfected cells (Figure 4B).
Table 1.
Effect of expression of AKAP350A(1076-2143) on the Golgi apparatus
| Control | AKAP350A(1076-2143) | |
|---|---|---|
| Giantin (morphology 1) | ||
| Golgi (% area per cell) | 18 ± 3 | 5 ± 1* |
| Golgi I/total I | 3.3 ± 0.6 | 6.5 ± 1.1* |
| Giantin (morphology 2) | ||
| Golgi (% area per cell) | 15 ± 1 | 12 ± 1 |
| Golgi I/total I | 3.4 ± 0.5 | 3.0 ± 0.6 |
| Golgin-97 | ||
| Golgi (% area per cell) | 41 ± 6 | 63 ± 7** |
| Golgi I/total I | 1.44 ± 0.14 | 1.25 ± 0.07* |
The Golgi and total cell areas and the average fluorescence intensity (I) for giantin and golgin-97 were measured in cells expressing AKAP350A(1076-2143)-GFP and in nontransfected cells from the same field (control). The giantin staining in the cells expressing AKAP350A(1076-2143) presented in two major patterns, both of which indicated a modification of the Golgi morphology, and each pattern comprising ∼40% of the transfected cells. The “morphology 1” was similar to p58 staining in Figure 4A, and the quantification indicated there was a reduction of the Golgi area to ∼30% the area in the control cells. The “morphology 2” showed modification of the stacking pattern and some vesiculation (Figure 4B), without changes in the morphometric parameters analyzed. Golgin-97 staining in the transfected cells presented a dispersed pattern in ∼80% of the transfected cells (Figure 4B). There was no significant change in the total fluorescence intensity for either giantin or golgin-97 (our unpublished data), indicating no significant changes in the expression levels of these Golgi proteins. The results of the analysis of 10 cells from each group are presented as the means ± S.E.M.
p < 0.05
p < 0.01
AKAP350 RNA Interference Alters Golgi Apparatus Morphology
Using an antibody specific for AKAP350A, we previously described the localization of this splice variant in the Golgi apparatus in the epithelial cell line HCA-7 (Shanks et al., 2002b). We induced a reduction in AKAP350 expression in HeLa cells by RNA interference, by using in vitro synthesized 21-base RNA duplexes targeted to two specific sequences in AKAP350A (siRNA1 and siRNA2). The control was designed using a scrambled sequence of the siRNA1. The immunofluorescence analysis of the transfected cells showed that siRNA1 and siRNA2 induced a reduction of AKAP350A expression 48 h after transfection (our unpublished data). The Western blot of these cells with the pan-anti-AKAP350 14G2 monoclonal antibody demonstrated a reduction to ∼20% of the control levels for the cells transfected with the siRNA1 and to ∼40% in the cells transfected with the siRNA2 (Figure 5). As Figure 5 shows, CIP4 total expression was not altered by AKAP350 RNA interference.
Figure 5.
siRNA1 and siRNA2 induce a decrease in AKAP350 expression. HeLa cells were transfected with RNA duplexes targeted to two different AKAP350 specific sequences (siRNA1 and siRNA2) or with the RNA duplexes synthesized using a scrambled sequence of the siRNA1 target sequence (siRNA control) and allowed to grow for 48 h. The cells were scraped and lysed in the presence of protease inhibitors. The samples were resolved by SDS-PAGE and transferred to nitrocellulose or Immobilon membranes for the analysis of AKAP350 or CIP4 expression. The densitometry of the bands indicated a reduction of total AKAP350 protein to 20% of control with the siRNA1 and to 40% of control with the siRNA2 transfected cells. The RNA interference for AKAP350 expression did not modify the CIP4 protein levels.
The reduction of AKAP350 expression induced a redistribution of CIP4, with loss of the perinuclear (Golgi) localization (Figure 6). The analysis of the staining for Golgi proteins p58 and golgin-97 (Figure 6) indicated that AKAP350 depletion induced a modification of the structure of the Golgi apparatus. We also induced AKAP350A RNA interference by cloning the target sequence used to design the siRNA1 upstream of the H1 promoter in the pSUPER vector (Brummelkamp et al., 2002), and transiently transfecting HeLa cells with this vector. Using this system, we observed both the redistribution of CIP4 and changes in the Golgi morphology (our unpublished data). The immunostaining with anti-α-tubulin (Figure 6) and phalloidin (our unpublished data) did not show any remarkable difference between the control and the AKAP350-depleted cells, but we cannot rule out modifications in the microtubule or actin cytoskeleton dynamics that would not be evident using this morphological methodology.
Figure 6.
Reduction of AKAP350A expression by RNA interference alters CIP4 distribution and induces vesiculation of the Golgi apparatus. HeLa cells were transfected with the AKAP350A-targeted RNA duplexes siRNA1 and siRNA2 or with the siRNA control, and allowed to grow for 72 h. After fixation, cells were dual stained with the 14G2 anti-AKAP350 and r-CIP4 antibodies (first column), anti-AKAP350A and anti-p58 (second column), anti-AKAP350A and anti-golgin-97 (third column), and anti-AKAP350A and anti-α-tubulin (fourth column). Approximately 90% of the cells transfected with siRNA1 and siRNA2 showed a decrease in AKAP350A staining that was more prominent in the cells expressing the siRNA1. Approximately 40% of the siRNA1 and 20% of the siRNA2 cells showed no AKAP350A staining. The reduction in AKAP350 expression was confirmed by Western blot (see Figure 5). The viability of the cells was confirmed by evaluation of the DAPI staining and the phase contrast images of the cells (our unpublished data). Bar, 2.5 μm.
We quantitated the morphological changes we observed at the Golgi apparatus on cells transfected with control, siRNA1, and siRNA2 RNA duplexes in the same manner as described in the previous section. The morphometric analysis indicated that the decrease of AKAP350 expression by using either of the constructs induced a significant increase in both the cis- and trans-Golgi area, with a decrease in the relative average intensity of its markers p58 and golgin-97 (Table 2). These results are consistent with the observations in Figure 6 showing dispersal of Golgi elements in response to a reduction in AKAP350. This analysis also showed that the siRNA1 induced a greater effect on golgin-97 distribution, compared with siRNA2, likely due to the different levels of AKAP350 expressed in each group of cells (Figure 5). The p58 and golgin-97 total fluorescence intensity was significantly lower in the cells with decreased AKAP350 expression (our unpublished data).
Table 2.
Effect of the decrease of AKAP350 expression on the Golgi apparatus
| siRNA control | siRNA1 | siRNA2 | |
|---|---|---|---|
| p58 | |||
| n | 34 | 31 | 29 |
| Golgi (% area per cell) | 9.7 ± 0.5 | 15.2 ± 1.3* | 16.9 ± 3.0* |
| Golgi I/total I | 2.7 ± 0.2 | 1.6 ± 0.1** | 1.6 ± 0.1** |
| golgin-97 | |||
| n | 48 | 52 | 25 |
| Golgi (% area per cell) | 7.9 ± 0.8 | 17.9 ± 1.4** | 12.1 ± 0.9*# |
| Golgi I/total I | 3.8 ± 0.2 | 2.2 ± 0.1** | 2.9 ± 0.1**# |
The Golgi and total cell areas as well as the average fluorescence intensity (I) for p58 and golgin-97 were measured in HeLa cells transfected for 72 h with the RNA duplexes targeted to specific sequences of AKAP350 (siRNA1 and siRNA2) or with the siRNA control. The decrease in AKAP350 levels induced dispersal of the Golgi apparatus, as stained with p58 and golgin-97 (Figure 6). The results are presented as the means ± SEM of the indicated number of cells analyzed (n).
p < 0.05
p < 0.01 compared with the controls
p < 0.05 compared with siRNA1
DISCUSSION
AKAP350 is a large anchoring protein that in epithelial cells localizes to the Golgi apparatus and the centrosomes. Many of the proteins that interact with AKAP350 localize to the Golgi apparatus. Among them, PKA, PP2A, and casein kinase I have regulatory effects on the function and dynamics of this organelle (Muniz et al., 1997; Lowe et al., 2000; Yu and Roth, 2002; Ghosh and Kornfeld, 2003). We have recently described that the family of CLICs can interact with AKAP350, and specifically CLIC5B interacts with AKAP350 at the Golgi apparatus (Shanks et al., 2002a). However, previous studies only have speculated about AKAP350 function at the Golgi, and none of the interacting proteins so far described were known substrates for the enzymes scaffolded by AKAP350. In the present study, we describe that AKAP350 interacts with CIP4. CIP4 is present at the Golgi apparatus in epithelial derived cells and is a substrate for PKA. Our studies suggest that the presence of CIP4 and AKAP350 at the Golgi apparatus is necessary for the maintenance of normal Golgi structure.
Using the AKAP350(1076-2143) domain as a bait in yeast two-hybrid assays, we found that the structurally related proteins FBP17 and CIP4 interact with AKAP350, and these interactions were confirmed by the pull-down assays. FBP17 interacts both with formins, which are involved in actin nucleation and microtubule regulation, with sorting nexin 2, and with the CD95 ligand (Chan et al., 1996; Fuchs et al., 2001; Ghadimi et al., 2002). CIP4 and FBP17 seems to localize to different regions of the cell. Using GFP fusion constructs to express FBP17 and CIP4 in MDCK or HeLa cells, we found that GFP-FBP17 distributed mainly to the plasma membrane, whereas GFP-CIP4 showed perinuclear localization (Larocca and Goldenring, unpublished data). In this study, we focused on AKAP350 interaction with CIP4. At least five CIP4 splice variants have been described. Among them, CIP4 is the most ubiquitously expressed. FELIC also interacts with cdc42 but lacks the SH3 domain that allows CIP4 interaction with WASP and RICH. Even though their localization when expressed in macrophages is different, both CIP4 and FELIC have an inhibitory effect on cell migration (Dombrosky-Ferlan et al., 2003). The CIP4/2 splice variant interacts with the TC10 GTPase, which phylogenetically is very close to cdc42 and is necessary for the insulin-stimulated exocytotic insertion of the GluT4-containing vesicles in adipocytes (Chang et al., 2002). Thus, at least three of the CIP4 splice variants have a role in regulating membrane trafficking. The amino-terminal domain that is responsible for CIP4 binding to microtubules is required for binding AKAP350(1076-2143). This domain is also present in FELIC and CIP4/2, so they are also potential AKAP350 interactors. Nevertheless, no Golgi or centrosomal distribution has been described for these splice variants.
We have demonstrated that CIP4 interacts with AKAP350. CIP4 and AKAP350 colocalize at the Golgi apparatus in the epithelial derived HCA-7 (Shanks and Goldenring, unpublished data), MDCK, and HeLa cell lines. In cells treated with nocodazole, the Golgi apparatus was vesiculated, and the Golgi-derived elements lost their perinuclear localization. Both CIP4 and AKAP350 staining acquired the same pattern as the Golgi marker p58, similar to the pattern described for other Golgi proteins (Walenta et al., 2001). CIP4 was first described as an interactor with the constitutively active mutant of cdc42 (Aspenstrom, 1997). BFA prevents ARF activation, inducing the rapid redistribution to the cytosol of the COPI coat and other peripheral membrane proteins, including cdc42 (Erickson et al., 1996). MDCK cells are relatively resistant to BFA. At the dose we used, BFA induces tubulation and functional alterations of the trans-Golgi, but it does not alter the morphology of the Golgi stacks (Hunziker et al., 1991; Wagner et al., 1994). After 1 min of BFA treatment, cdc42 acquires a cytosolic distribution, whereas CIP4 remains localized at the Golgi. Even after 1 h of BFA treatment, CIP4 still showed Golgi localization. Thus, CIP4 interaction with the Golgi membranes does not depend directly either on ARF1 or the presence of cdc42 at the Golgi.
The results presented here indicate that the presence of AKAP350A and CIP4 at the Golgi is necessary for the maintenance of the normal structure of the Golgi apparatus. First, displacement of CIP4 from the Golgi by expression of AKAP350(1076-2143), which contains the CIP4-binding domain of AKAP350 but not the Golgi targeting domain, induced morphological alterations in the Golgi stacking and dispersion of the trans-Golgi. Second, decreasing the expression of AKAP350 by RNA interference elicited a dispersion of cis- and trans-Golgi elements. All of these results indicate that AKAP350 and CIP4 are critical regulators of Golgi structure. AKAP350 and CIP4 at the Golgi might influence the normal positioning, architecture, and dynamics of the Golgi apparatus through the scaffolding of critical regulators. PKA regulates multiple aspects of Golgi function. In vitro studies showed that PKA stimulates the association of activated ARF1 with the Golgi membranes (Martin et al., 2000). In accordance with this finding, PKA is necessary for the normal budding of vesicles at the Golgi (Muniz et al., 1997). PKA is also required for normal endosome-to-Golgi trafficking (Birkeli et al., 2003) and the retrograde transport of endoplasmic reticulum proteins bearing a carboxyl-terminal KDEL sequence from the Golgi back to the endoplasmic reticulum (Cabrera et al., 2003). AKAP350 also interacts with PP2A (Takahashi et al., 1999), and we have noted colocalization of PP2A with AKAP350 at the Golgi in parietal cells (Schmidt and Goldenring, unpublished data). PP2A regulates the postmitotic reassembly of the Golgi by dephosphorylation of GM130, which can then interact with p115 and form the GM130-p115-golgin complex (Lowe et al., 2000). Scaffolding of PKA and PP2A by AKAP350 at the Golgi may facilitate the maintenance of normal vesicle trafficking between the Golgi, endosomal compartments, and the endoplasmic reticulum. The reduction of AKAP350 expression by RNA interference induced a decrease in the levels of the trans-Golgi protein golgin-97 and the endoplasmic reticulum chaperone calreticulin (our unpublished data), results consistent with alterations in Golgi trafficking.
CIP4 itself might constitute a family of scaffolding proteins. Besides interacting with cdc42, CIP4 also interacts with WASP, microtubules (Tian et al., 2000), the Src kinase Lyn (which phosphorylates WASP in a process regulated by cdc42 (Guinamard et al., 1998; Dombrosky-Ferlan et al., 2003), huntingtin (Holbert et al., 2002), and with the potential GTPase activating protein RICH (Richnau and Aspenstrom, 2001). Different studies suggest that, upon activation of ARF1, cdc42 is recruited to the Golgi together with the coatomer proteins, and leads to the assembly of a specific pool of actin necessary for vesicle release and/or targeting, in a process requiring WASP/Arp2/3 (reviewed in Stammer, 2002). CIP4 may be modulating the Golgi actin cytoskeleton, by interacting with its regulators. Interestingly, treatment of MDCK cells with latrunculin B, an inhibitor of actin nucleation, induced similar morphological changes on the Golgi as observed with the displacement of CIP4 by expression of the AKAP350(1076-2143) domain (Valderrama et al., 2001; our unpublished data). Staining with phalloidin of the AKAP350(1076-2143)-expressing cells or cells with reduction in AKAP350 expression did not show changes in the perinuclear actin. However, the loss of CIP4 interaction with AKAP350 could affect the formation of transient dynamic microfilaments at the Golgi, which would not be evident with the phalloidin staining. We found that CIP4 was phosphorylated by PKA in vitro, and stimulation of PKA increased CIP4 phosphorylation in situ. Future investigations will address whether modulation of CIP4 phosphorylation by PKA may regulate the interaction of CIP4 with cytoskeletal elements.
AKAP350 interacts with CIP4 in its amino terminus, which is also implicated in CIP4 interaction with microtubules (Tian et al., 2000). Thus, association with AKAP350 could modulate CIP4 interaction with microtubules. There is a tight relationship between microtubules and the Golgi apparatus. Microtubules interact with the Golgi stacks essentially at the cis face, a process involving GMAP210 (Infante et al., 1999) and the Hook3 protein (Walenta et al., 2001). Microtubules are necessary for Golgi positioning and morphology and for trafficking between Golgi and endoplasmic reticulum. There is strong evidence as well that the Golgi might function as a microtubule nucleating organelle (reviewed in Rios et al., 2003). AKAP350 is implicated in the microtubule nucleation at the centrosomes through its interaction with components of the γ-tubulin ring complex and the Ran small GTPase (Takahashi et al., 2002, Keryer et al., 2003). Similarly, AKAP350 and CIP4 may modulate microtubule interactions with the Golgi apparatus. We observed that both AKAP350(1076-2143) expression and the reduction in AKAP350 expression induced a decrease in the density of perinuclear microtubules. However, this alteration may be secondary to the changes in the Golgi apparatus, because there was still more intense staining for α-tubulin in the area where the Golgi membrane elements remained.
The Golgi matrix was proposed as a structure with self-organizing properties, which may ensure the architecture and polarity of the Golgi despite the magnitude of the membrane flux through the organelle (reviewed in Shorter and Warren, 2002). “Golgins” denominate the Golgi localized proteins with predicted coiled-coil domains (Barr and Short, 2003). Different golgins can interact in complexes that are involved in tethering COPI vesicles and maintaining the normal endoplasmic reticulum to Golgi and intra-Golgi membrane flow. Disruption of golgin interactions with Golgi membranes during mitosis is thought to mediate the Golgi disassembly (reviewed in Shorter et al., 2002). Consistent with the function of the golgins in the maintenance of the Golgi structure, a reduction in the expression of the Golgin-45 by RNA interference leads to the collapse of the Golgi structure (Short et al., 2001). Interestingly, AKAP350 structural and behavioral properties are consistent with its classification as a golgin. AKAP350 contains multiple coiledcoil domains throughout the protein, especially from amino acids 739-2270, and the loss of AKAP350A expression causes disorganization of the Golgi apparatus
In summary, we have delineated the interaction of AKAP350 with CIP4 at the Golgi apparatus. Displacement of CIP4 from the Golgi by expression of the CIP4 binding domain of AKAP350 caused disruption of the normal Golgi structure. Reduction of AKAP350 expression by RNA interference induced dispersal of Golgi elements. These results indicate that both AKAP350 and CIP4 are important in the maintenance of Golgi apparatus integrity.
Acknowledgments
We thank Dr. Ed Chan for the gift of giantin antibody. We thank Drs. Brent Steadman, Lynne Lapierre, and Jeff Franklin for support of this work. This study was supported by grants to J.R.G. from National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (DK-48370 and DK-43405) and a Veterans Administration Merit Award.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0757. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0757.
References
- Alto, N., Carlisle Michel, J.J., Dodge, K.L., Langeberg, L.K., and Scott, J.D. (2002). Intracellular targeting of protein kinases and phosphatases. Diabetes 51, S385-S388. [DOI] [PubMed] [Google Scholar]
- Aspenstrom, P. (1997). A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7, 479-487. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., and Short, B. (2003). Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15, 405-413. [DOI] [PubMed] [Google Scholar]
- Birkeli, K.A., Llorente, A., Torgersen, M.L., Keryer, G., Tasken, K., and Sandvig, K. (2003). Endosome-to-Golgi transport is regulated by protein kinase A type II alpha. J. Biol. Chem. 278, 1991-1997. [DOI] [PubMed] [Google Scholar]
- Berryman, M.A., and Goldenring, J.R. (2003). CLIC4 is enriched at cell-cell junctions and colocalizes with AKAP350 at the centrosome and midbody of cultured mammalian cells. Cell Motil. Cytoskeleton 56, 159-172. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- Cabrera, M., Muniz, M., Hidalgo, J., Vega, L., Martin, M.E., and Velasco, A. (2003). The retrieval function of the KDEL receptor requires PKA phosphorylation of its C-terminus. Mol. Biol. Cell 14, 4114-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D.C., Bedford, M.T., and Leder, P. (1996). Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 15, 1045-1054. [PMC free article] [PubMed] [Google Scholar]
- Chang, L., Adams, R.D., and Saltiel, A.R. (2002). The TC10-interacting protein CIP4/2 is required for insulin-stimulated Glut4 translocation in 3T3L1 adipocytes. Proc. Natl. Acad. Sci. USA 99, 12835-12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold, C., Ponnambalam, S., Francis, M.J., and Monaco, A.P. (2002). Novel membrane traffic steps regulate the exocytosis of the Menkes disease ATPase. Hum. Mol. Gen. 11, 2855-2866. [DOI] [PubMed] [Google Scholar]
- Dransfield, D.T., Yeh, J.L., Bradford, A.J., and Goldenring, J.R. (1997). Identification and characterization of a novel A-kinase-anchoring protein (AKAP120) from rabbit gastric parietal cells. Biochem. J. 322, 801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrosky-Ferlan, P., Grishin, A., Botelho, R.J., Sampson, M., Wang, L., Rudert, W.A., Grinstein, S., and Corey, S.J. (2003). Felic (CIP4b), a novel binding partner with the Src kinase Lyn and Cdc42, localizes to the phagocytic cup. Blood 101, 2804-2809. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Erickson, J.W., Zhan, Ch., Kahn, R.A., Evans, T., and Cerione, R.A. (1996). Mammalian cdc42 is a Brefeldin A-sensitive component of the Golgi apparatus. J. Biol. Chem. 271, 26850-26854. [DOI] [PubMed] [Google Scholar]
- Fuchs, U., et al. (2001). The human formin-binding protein 17 (FBP17) interacts with sorting nexin, SNX2, and is an MLL-fusion partner in acute myelogeneous leukemia. Proc. Natl. Acad. Sci. USA 98, 8756-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadimi, M.P., Sanzenbacher, R., Thiede, B., Wenzel, J., Jing, Q., Plomann, M., Borkhardt, A., Kabelitz, D., and Janssen, O. (2002). Identification of interaction partners of the cytosolic polyproline region of CD95 ligand (CD178). FEBS Lett. 519, 50-58. [DOI] [PubMed] [Google Scholar]
- Ghosh, P., and Kornfeld, S. (2003). AP-1 binding to sorting signals and release from clathrin-coated vesicles is regulated by phosphorylation. J. Cell Biol. 160, 699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro, S. (2000). The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinamard, R., Aspenstrom, P., Fougereau, M., Chavrier, P., and Guillemot, J.C. (1998). Tyrosine phosphorylation of the Wiskott-Aldrich syndrome protein by Lyn and Btk is regulated by CDC42. FEBS Lett. 434, 431-436. [DOI] [PubMed] [Google Scholar]
- Holbert, S., Dedeoglu, A., Humbert, S., Saudou, F., Ferrante, R.J., and Neri, C. (2002). Cdc-42-interacting protein 4 binds to huntingtin: neuropathololgic and biological evidence for a role in Huntignton's disease. Proc. Natl. Acad. Sci. USA 100, 2712-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker, W., Whitney, J.A., and Mellman, I. (1991). Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell 67, 617-627. [DOI] [PubMed] [Google Scholar]
- Infante, C., Ramos-Morales, F., Fedriani, C., Bornens, M., and Rios, R.M. (1999). GMAP-210, A cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J. Cell Biol. 145, 83-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer, G., Di Fiore, B., Celati, C., Lechtreck, K.F., Mogensen, M., Delouvee, A., Lavia, P., Bornens, M., and Tassin, A.M. (2003). Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell 14, 4260-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer, G., Rios, R.M., Landmark, B.F., Skalhegg, B., Lohmann, S.M., and Bornens, M. (1993). A high-affinity binding protein for the regulatory subunit of cAMP-dependent protein kinase II in the centrosome of human cells. Exp. Cell Res. 204, 230-40. [DOI] [PubMed] [Google Scholar]
- Lapierre, L.A., Tuma, P.L., Navarre, J., Goldenring, J.R., and Anderson, J.M. (1999). VAP-33 localizes to both an intracellular vesicle population and with occludin at the tight junction. J. Cell Sci. 112, 3723-3732. [DOI] [PubMed] [Google Scholar]
- Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J., and Vaughan, M. (2003). Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2). Proc. Natl. Acad. Sci. USA 100, 1627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.J., Wyszynski, M., Madhaven, R., Sealock, R., Kim, J.U., and Sheng, M. (1998). Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 18, 2017-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt, A.D., Foquet, M., Renz, M., Seelig, H.P., Glick, B.S., and Hauri, H.P. (1995). A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golgi apparatus. Proc. Natl. Acad. Sci. USA 92, 5102-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Gonatas, N.K., and Warren, G. (2000). The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149, 341-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke, M.R., Kjer-Nielsen, L., Brown, D.L., Stow, J.L., and Gleeson, P.A. (2003). GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 278, 4216-4226. [DOI] [PubMed] [Google Scholar]
- Martin, M.E., Hidalgo, J., Rosa, J.L., Crottet, P., and Velasco, A. (2000). Effect of protein kinase A activity on the association of ADP-ribosylation factor 1 to Golgi membranes. J. Biol. Chem. 275, 19050-19059. [DOI] [PubMed] [Google Scholar]
- Muniz, M., Martin, M.E., Hidalgo, J., and Velasco, A. (1996). A regulatory role for cAMP-dependent protein kinase in protein traffic along the exocytic route. J. Biol. Chem. 271, 30935-30941. [DOI] [PubMed] [Google Scholar]
- Muniz, M., Martin, M.E., Hidalgo, J., and Velasco, A. (1997). Protein kinase A activity is required for the budding of constitutive transport vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. USA 94, 14461-14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richnau, N., and Aspenstrom, P. (2001). Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J. Biol. Chem. 276, 35060-35070. [DOI] [PubMed] [Google Scholar]
- Rios, R.M., and Bornens, M. (2003). The Golgi apparatus at the cell centre. Curr. Opin. Cell Biol. 15, 60-66. [DOI] [PubMed] [Google Scholar]
- Saraste, J., Palade, G.E., and Farquhar, M.G. (1987). Antibodies to rat pancreas Golgi subfractions: identification of a 58-kD cis-Golgi protein. J. Cell Biol. 105, 2021-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, P.H., Dransfield, D.T., Claudio, J.O., Hawley, R.G., Trotter, K.W., Milgram, S.L., and Goldenring, J.R. (1999). AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 274, 3055-3066. [DOI] [PubMed] [Google Scholar]
- Shanks, R.A., Larocca, M.C., Berryman, M., Edwards, J.C., Urushidani, T., Navarre, J., and Goldenring, J.R. (2002b). AKAP350 at the Golgi apparatus. II. Association of AKAP350 with a novel chloride intracellular channel (CLIC) family member. J. Biol. Chem. 277, 40973-40980. [DOI] [PubMed] [Google Scholar]
- Shanks, R.A., Steadman, B.T., Schmidt, P.H., and Goldenring, J.R. (2002a). AKAP350 at the Golgi apparatus. I. Identification of a distinct Golgi apparatus targeting motif in AKAP350. J. Biol. Chem. 277, 40967-40972. [DOI] [PubMed] [Google Scholar]
- Short, B., Preisinger, C., Korner, R., Kopajtich, R., Byron, O., and Barr, F.A. (2001). A GRASP55-Rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (2002). Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18, 379-420. [DOI] [PubMed] [Google Scholar]
- Sillibourne, J.E., Milne, D.M., Takahashi, M., Ono, Y., and Meek, D.W. (2002). Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol. 322, 785-797. [DOI] [PubMed] [Google Scholar]
- Stammer, M. (2002). Regulating the actin cytoskeleton during vesicular transport. Curr. Opin. Cell Biol. 14, 428-433. [DOI] [PubMed] [Google Scholar]
- Steadman, B.T., Schmidt, P.H., Shanks, R.A., Lapierre, L.A., and Goldenring, J.R. (2002). Transforming acidic coiled-coil-containing protein 4 interacts with centrosomal AKAP350 and the mitotic spindle apparatus. J. Biol. Chem. 277, 30165-30176. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Mukai, H., Oishi, K., Isagawa, T., and Ono, Y. (2000). Association of immature hypophosphorylated protein kinase C epsilon with an anchoring protein CG-NAP. J. Biol. Chem. 275, 34592-34596. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Shibata, H., Shimakawa, M., Miyamoto, M., Mukai, H., and Ono, Y. (1999). Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J. Biol. Chem. 274, 17267-17274. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Yamagiwa, A., Nishimura, T., Mukai, H., and Ono, Y. (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 13, 3235-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasken, K.A., Collas, P., Kemmner, W.A., Witczak, O., Conti, M., and Tasken, K. (2001). Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 276, 21999-22002. [DOI] [PubMed] [Google Scholar]
- Tian, L., Nelson, D.L., and Stewart, D.M. (2000). Cdc42-interacting protein 4 mediates binding of the Wiskott-Aldrich syndrome protein to microtubules. J. Biol. Chem. 275, 7854-7861. [DOI] [PubMed] [Google Scholar]
- Tsuji, E., and Tsuji, Y. (2000). Molecular cloning and chromosomal localization of human salt-tolerant protein. Genetica 108, 259-262. [DOI] [PubMed] [Google Scholar]
- Valderrama, F., Duran, J.M., Babia, T., Barth, H., Renau-Piqueras, J., and Egea, G. (2001). Actin microfilaments facilitate the retrograde transport from the Golgi complex to the endoplasmic reticulum in mammalian cells. Traffic 2, 717-726. [DOI] [PubMed] [Google Scholar]
- Wagner, M., Rajasekaran, A.K., Hanzel, D.K., Mayor, S., and Rodriguez-Boulan, E. (1994). Brefeldin A causes structural and functional alterations of the trans-Golgi network of MDCK cells. J. Cell Sci. 107, 933-943. [DOI] [PubMed] [Google Scholar]
- Walenta, J.H., Didier, A.J., Liu, X., and Kramer, H. (2001). The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152, 923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Rudert, W.A., Grishin, A., Dombrosky-Ferlan, P., Sullivan, K., Deng, X., Whitcomb, D., and Corey, S. (2002). Identification and genetic analysis of human and mouse activated Cdc42 interacting protein-4 isoforms. Biochem. Biophys. Res. Commun. 293, 1426-1430. [DOI] [PubMed] [Google Scholar]
- Westphal, R.S., Tavalin, S.J., Lin, J.W., Alto, N.M., Fraser, I.D., Langeberg, L.K., Sheng, M., and Scott, J.D. (1999). Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 285, 93-96. [DOI] [PubMed] [Google Scholar]
- Witczak, O., Skalhegg, B.S., Keryer, G., Bornens, M., Tasken, K., Jahnsen, T., and Orstavik, S. (1999). Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 18, 1858-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S., and Roth, M.G. (2002). Casein Kinase I regulates membrane binding by ARF GAP1. Mol. Biol. Cell 13, 2559-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]