Abstract
Nanodiscs are self-assembled discoidal fragments of lipid bilayers 8 – 16 nm in diameter, stabilized in solution by amphipathic helical scaffold protein. As stable and highly soluble membrane mimetics with controlled lipid composition and ability to add affinity tags to the scaffold protein, Nanodiscs represent an attractive model system for solubilization, isolation, purification, and biophysical and biochemical studies of membrane proteins. We overview various approaches to the structural and functional studies of different classes of integral membrane proteins such as ion channels, transporters, GPCR and other receptors, membrane enzymes, blood coagulation cascade proteins, et cetera incorporated into Nanodiscs with the special focus on the advantages provided by homogeneity, ability to control oligomerization state of the target protein and lipid composition of the bilayer. Special attention is paid to the opportunities provided by Nanodisc system for the detailed studies of the role of different lipid properties and protein – lipid interactions in the functional behavior of membrane proteins.
Keywords: Nanodiscs, Lipoproteins, Lipid-protein interactions, GPCR, Cytochrome P450, Tissue Factor, Membrane protein solubilization
1. Introduction
The interactions between lipids and proteins are one of the most fundamental processes in living organisms, responsible for critical cellular events ranging from replication, cell division, signaling and movement. Forming the central coupling responsible for maintaining the functionality of the breadth of proteins, receptors and enzymes that find their natural home in biological membranes, the fundamental mechanisms of recognition of protein for lipid and vice versa has been a focal point of biochemical and biophysical investigations for many decades. Complexes of lipids and proteins, such as the various lipoprotein factions, play central roles in the trafficking of important proteins, small molecules and metabolites such as cholesterol, and are often implicated in disease states. Recently an engineered lipoprotein particle, termed the Nanodisc, a modified form of the human high density lipoprotein fraction, has served as a membrane miminic for the investigation of membrane proteins and studies of lipid-protein interactions. In this review, we summarize the current knowledge regarding this self-assembling lipid-protein complex and provide examples for its utility in the investigation of a large number of biological systems.
1.1 Development of the Nanodisc system
By the late 1990s, we had acquired one of the new atomic force microscopes (AFM) that could operate under water and had modified it to execute a form of force-volume imaging for soft biological samples. Looking for good candidates for AFM structural studies, we were struck by the pioneering work of Professor Ana Jonas at the University of Illinois in which she described the atherosclerotic process and the role of lipoproteins in reverse cholesterol transport. In this clinical process, the predominant circulating forms of high density lipoproteins (HDL) are balls of variable size containing cholesterol esters, lipids and proteins, although a transient form is roughly discoidal and stabilized by the amphipathic apolipoprotein A-I (Apo-AI). Because methods had been developed for reconstituting the larger HDL particles by the removal of detergents from a lipid-enriched Apo-AI fraction isolated from human blood (1, 2), we were able to image these HDL particles on atomically flat mica and show that they were extremely heterogeneous with peaks in the size distribution that corresponded to the lengths of the repeating Apo-AI protein (3). Having previously discovered that many eukaryotic proteins could be expressed in E. coli from synthetic genes (4), we undertook the synthesis of a recombinant Apo-AI coding sequence suitable for expression in this bacterial host and the modification of reconstitution procedures for production of uniformly sized nanoparticles. The result of this genetic engineering exercise was a set of “membrane scaffold proteins” (MSPs) that were capable of self-assembly into discoidal phospholipid bilayers (termed Nanodiscs) wrapped with an amphipathic helical belt surrounding the alkyl chains on the phospholipids.
The importance of these soluble Nanodiscs to studies of membrane proteins was recognized by others in our laboratory struggling with the challenge of making precise biophysical measurements on detergent-solubilized membrane proteins. The wild idea emerged that perhaps if these integral membrane proteins were included in the detergent-solubilized mixture of lipid and MSP, and the same process of self-assembly were initiated to form Nanodiscs, the membrane protein target would find their natural home in the bilayer. Thus, the discoidal particle would become a model system wherein the membrane protein would think it was in a native-like membrane environment, maintaining its naturalistic presentation in a monomeric state with full functionality and yet rendered soluble in aqueous solutions via its encircling MSP belt.
We tried this multi-component assembly idea with the simplest membrane proteins that we had on hand, mammalian cytochrome b5 and the cytochrome P450 reductase, which both have large soluble domains but simple helical anchors in the membrane. Working exceedingly well, subsequent fractionations allowed us to separate “free Nanodiscs” from “protein-containing Nanodiscs” and to quantitate the topology of surface protrusions relative to the Nanodisc surface (5, 6). Further experiments led to the development of a general method for assembling membrane proteins into Nanodiscs (7–9). While the public response to this bio-assembly system was initially somewhat muted due to concerns that deeply embedded and totally integral membrane proteins would not readily be incorporated, subsequent efforts over the next decade (including that by many independent laboratories) demonstrated the broad applicability of Nanodiscs for studying the breadth and complexity of membrane protein complexes. The individual sections of this review highlight current information on the structure and assembly process of Nanodiscs as well as the range of proteins and methods accessible with these technologies.
2. The Nanodisc self-assembly process
The Nanodisc is composed of one or more lipids and two copies of the amphipathic helical membrane scaffold protein (MSP). An initial goal of our research was to provide a structural determination of the lipid and protein factors that make up the Nanodisc. Various lengths of the membrane scaffold protein were generated by recombinant manipulation of the MSP sequence and its subsequent expression in E. coli. By using a combination of small-angle X-ray scattering, size-exclusion chromatography and direct analysis of the lipid-to-protein ratios, the Nanodisc structure was verified (7, 10). Together with the solid state NMR (11), these results proved that the earlier suggestions that the amphipathic helices of the MSP were directed parallel to the bilayer normal in a "picket fence" arrangement (12) were in error. Rather, the MSP encircles the discoidal membrane fragment in a "double belt" configuration as initially suggested by Wlodawer et al. (13). Data obtained for Nanodiscs of different sizes, generated by using varying length MSPs, provided us with overall structures similar to those proposed for nascent discoidal HDL particles. But, importantly, the different length MSP belts, and exact control of the assembly conditions, generated homogeneous and monodisperse populations with a size uniformity not evident in previous biological assemblies made with full-length Apo-AI. In the final derived Nanodisc structure, a circular fragment of the phospholipid bilayer is circumscribed by two copies of the MSP unit (7, 10), as illustrated in Fig. 1.
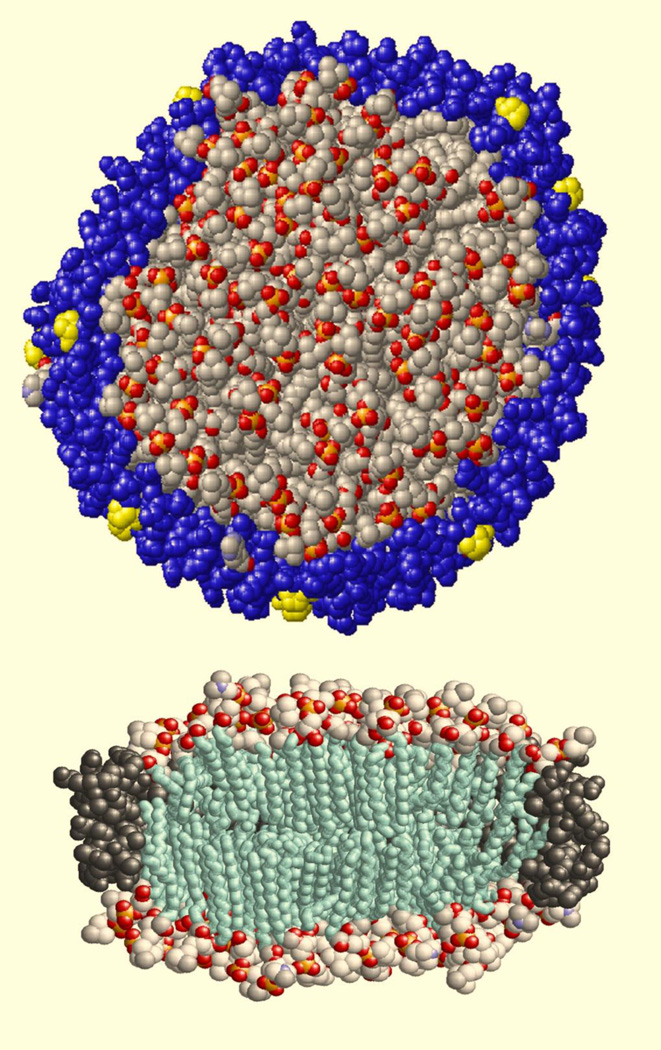
Figure 1.
Results of MD simulations of Nanodiscs assembled with DPPC molecules and scaffold protein MSP1D1 (14). Alignment of prolines shown in yellow can be seen at the left part of the top view (top), while the different conformations of the lipid acyl tails in the middle of Nanodisc and at the protein – lipid interface can be seen at the bottom.
Detailed reviews of these structural and biological investigations using native and altered MSP sequences wherein the number of amphipathic helices punctuated by prolines and glycines is varied, have been published (7, 10, 15). This set of MSPs that allow Nanodiscs to be assembled with of varying sizes, is summarized in Table 1. Analogous constructs incorporating hexahistidine or FLAG tags at the amino- or carboxy-terminus of MSP were created, allowing for convenient purification of the MSP protein from E. coli lysates or the assembled Nanodiscs following incorporation of membrane proteins. An engineered TEV protease or Factor X protease cleavage site was also incorporated, enabling facile removal of the affinity tag should this be desired, as is in cases where the membrane protein target carries the affinity label. This large collection of MSPs of varying composition, and the optimization of lipid-protein ratios for the self-assembly process, have enabled the generation of uniformly-sized Nanodiscs ranging in diameter from 9.8 to 17 nm (15).
Table 1.
Labels and amino acid sequences of membrane scaffold proteins used for self-assembly of Nanodiscs (adopted with permission from (15)).
| Abbreviation | Description | Amino Acid Sequence |
|---|---|---|
| H1 | Helix 1 | LKLLDNWDSVTSTFSKLREQLG |
| H1 (1-11) | Truncated Helix 1 | STFSKLREQLG |
| H1 (l-17) | Truncated Helix 1 | REQLG |
| H2 | Helix 2 | PVTQEFWDNLEKETEGLRQEMS |
| H3 | Helix 3 | KDLEEVKAKVQ |
| H4 | Helix 4 | PYLDDFQKKWQEEMELYRQKVE |
| H5 | Helix 5 | PLRAELQEGARQKLHELQEKLS |
| H6 | Helix 6 | PLGEEMRDRARAHVDALRTHLA |
| H7 | Helix 7 | PYSDELRQRLAARLEALKENGG |
| H8 | Helix 8 | ARLAEYHAKATEHLSTLSEKAK |
| H9 | Helix 9 | PALEDLRQGLL |
| H10 | Helix 10 | PVLESFKVSFLSALEEYTKKLNTQ |
| FX | Original N-terminus | MGHHHHHHIEGR |
| TEV | Modified N-terminus | MGHHHHHHH DYDIPTTENLYFQG |
| Scaffold variant |
protein | Abbreviated namea |
Composition |
|---|---|---|---|
| 1 | MSP1 | FX-H1-H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 2 | MSP2 | FX-H1- H2-H3-H4-H5-H6-H7-H8-H9-H10-GT- H1-H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 3 | MSP1E1 | FX-H1-H2-H3-H4-H4-H5-H6-H7-H8-H9-H10 | |
| 4 | MSP1E2 | FX-H1-H2-H3-H4-H5-H4-H5-H6-H7-H8-H9-H10 | |
| 5 | MSP1E3 | FX-H1-H2-H3-H4-H5-H6-H4-H5-H6-H7-H8-H9-H10 | |
| 6 | MSP1D1 | TEV- H1 (1-11)- H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 7 | MSP1D2 | TEV- H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 8 | MSP2N1 | TEV- H1 (l-11)- H2-H3-H4-H5-H6-H7-H8-H9-H10-GT- H1 (1-11)- H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 9 | MSP2N2 | TEV- H1 (1-11)- H2-H3-H4-H5-H6-H7-H8-H9-H10-GT- H2-H3-H4-H5-H6-H7-H8-H9-H10 | |
| 10 | MSP2N3 | TEV- H1 (1-11)- H2-H3-H4-H5-H6-H7-H8-H9-H10-GT- H1 (1-17)- H2-H3-H4-H5-H6-H7-H8-H9-H10 |
Design of the membrane scaffold proteins.
Abbreviated names: (1) MSP1, membrane scaffold protein 1 with N-terminal hexahistidine tag, followed by Factor X recognition site.
(2) MSP2, fusion of two MSP1 molecules with short Glycine-Threonine linker and N-terminal hexahistidine tag, followed by Factor X recognition site.
(3–5) MSP1E1, MSP1E2, MSP1E3, extended membrane scaffold proteins, obtained via insertion of one (residues 56-77 of original MSP1), two (residues 56-99), or three (residues 56-121) extra 22-mer amphipathic helices after the residue Q55 into original MSP1 (1) sequence, as described (10). All have N-terminal hexahistidine tag and Factor X recognition site.
(6) MSP1D1, MSP1 with the first 11 N-terminal amino acids removed which contains the N-terminal heptahistidine tag and TEV protease recognition site with linker.
(7) MSP1D2, MSP1 with the first 22 N-terminal amino acids removed contains the N-terminal heptahistidine tag and TEV protease recognition site with linker.
(8–10) New MSP2 proteins based on MSP1 deletion mutants rather than original MSP1. All three have the same first MSP1D1 molecule, but vary in extent of truncation in the second MSP.
Further examination of the effect of varying MSP unit lengths, by determinations of the amounts of radiolabeled lipid and protein in the Nanodisc fractions on size exclusion columns, illustrated an interesting trend first described by Denisov et al. (10) was later expanded (15). It was found that insertion of extra helices in the central portion of the MSP unit (MSP1E1, MSP1E2, MSP1E3) resulted in Nanodiscs of increasing size but deletions of the N-terminal hexa-histidine tag and the subsequent 22 amino acids of the MSP protein did not significantly decrease the size of the Nanodisc formed. This suggested that the first 22 amino acids are marginally, if at all, involved in the self-assembly process and the resultant stabilization of the discoidal nanoparticle. Additional truncations from the N-terminal end beyond the first 22 amino acids led to a gradual decrease in lipid-protein ratio accompanied by an increase in the heterogeneity of the assembled Nanodiscs and a corresponding appearance of aggregated fractions. Further systematic studies of the lipid: protein ratio in Nanodiscs assembled from different MSP constructs has shown that the number of lipids per Nanodisc (N) and the number of amino acids in the scaffold protein (M) can be described a simple relationship (10, 15):
NL S = (0.423 M − 9.75)2
where S represents the mean surface area (measured in Å2) per lipid used to form the Nanodisc. The quadratic relationship between the number of lipid molecules per Nanodisc and the length of the MSP unit provides a confirmation of the flat two-dimensional morphology of Nanodisc particles. The similar sizes of Nanodiscs formed using the same MSP unit and varying lipids provides a clear indication that the length of the protein’s amphipathic helix is the sole determinant of the diameter of Nanodiscs with the different optimized lipid-protein stoichiometry reflecting the different surface areas occupied by individual lipids.
2.1 Molecular Dynamics simulations of Nanodisc structure
One structural question relates to the azimuthal alignment of the two MSP belts. The involvement of the N-terminal segment of the MSP protein in binding the lipid bilayer and the alignment of the two MSP units were examined by Molecular Dynamics (MD) simulations of Nanodiscs generated from various MSP units listed in Table 1 (14). Simulations of Nanodisc formed with MSP1Δ(1–11) and MSP1 Δ(1–22) scaffold proteins, which lacking the first 11 or 22 N-terminal residues respectively, resulted in discoidal structures with little deformation of either the lipid bilayer or the scaffold proteins. Notably, the two MSP units and their interspersed proline residues retain alignment with respect to each other with virtually no misalignment (14). Inspection of the hydrophobic tail groups of the DPPC lipids showed them effectively covered by the encircling MSP1 Δ(1–11) or MSP1Δ (1–22) and as well as in all other modified membrane scaffold proteins analyzed. Within each amphipathic MSP unit, the majority of the hydrophobic residues are on the interior side of the helices, making contact with the lipid hydrophobic tail and with the hydrophilic residues are oriented outwardly, effectively shielding water from the hydrophobic interior of the Nanodisc. These simulations also indicated that the presence of a hydrophilic solvent, such as water, is needed to maintain the discoidal Nanodisc structure. It was also noted that the diffusion of sodium ions was unhindered, with no obvious adhesion to the scaffold protein or lipid head groups. Additional MD simulations of Nanodiscs with bacteriorhodopsin (bR), an integral membrane protein that contains seven trans-membrane helices, indicated that Nanodisc structure does not assume any out-of-plane deformation with the addition of bR (14). However, the bR-containing Nanodisc adopts a slightly rectangular shape that is likely due to perturbations of the lipid bilayer caused by protein-lipid interactions. These initial models and subsequent improved models have been used to design MSP units optimal for individual membrane proteins and to plan experiments with membrane-embedded or membrane-associated proteins.
The results of these MD simulations also allow determining the average number of gauche isomers per lipid molecule in Nanodiscs for both the bulk, at the center, as well as the boundary lipids interacting with the membrane scaffold protein (14). Because the initial configuration of the lipid bilayer corresponds to the liquid crystalline state, it includes a high number of gauche isomers, the same for the bulk and boundary lipids. As the temperature of simulation is 27°C (i.e. below the phase transition for DPPC), the bilayer is equilibrated in the gel state as seen by a significant decrease in the gauche isomers fraction. But this decrease is much less pronounced for the boundary lipids (i.e., those within 15 Å from the nearest heavy atom of MSP). In this new equilibrium, the number of gauche isomers per lipid molecule is 2 for the bulk lipids and almost 5 for the boundary lipids with both numbers being in general agreement with those estimated from spectroscopic measurements for gel and liquid crystal phases (16), and from FTIR spectroscopy in reconstituted lipoproteins (17). This statistical analysis of MD simulations does not depend on the assumed thickness of boundary lipid layer, between 13–18 Å (where 15 Å is the average thickness of two lipid layers), if the mean area per lipid is 56 Å2. Yet, as can be seen in Figure 1, the thickness of the bilayer is higher in the center of the Nanodisc. While the strict quantitative comparison of all structural parameters derived from MD simulation with experimental results is a challenge, this analysis suggests a specific mode of structural perturbation of the lipid bilayer at the protein-lipid interface in Nanodiscs. The main reason for such perturbation is the hydrophobic mismatch between the average thickness of the protein helix of 10–11 Å, and the hydrophobic thickness of the lipid bilayer, 14–19 Å for DMPC and DPPC, depending on the phase state (18, 19). An important aspect of such perturbation is also the difference in hydrophobic mismatch below and above the main transition temperature, and the thermodynamic perturbation of lipid bilayer phase state by the scaffold protein.
2.2 Nanodisc bilayer phase transition and MSP interactions
The properties of the lipid bilayer nanoscale fragment containing 120–650 lipids have been studied further by observing the main thermotropic phase transition in Nanodiscs formed with DPPC and DMPC (15, 20, 21). For this purpose, Nanodiscs provide a unique system for the evaluation of fundamental physical properties of nanoscale lipid bilayers surrounded by protein. This is important as the protein weight percentage in biological membranes varies from 20–80% suggesting that a significant fraction of the lipid is in direct contact with protein (22). Such interfacial, or boundary, lipids are often perturbed by interactions with proteins, and the character and magnitude of this perturbation depend on the protein and lipid structures as well as the bilayer phase state (23, 24). Additionally, biological membranes often contain segregated domains in the submicron size range, which are usually formed with different lipid and protein contents. The properties of these discrete submicron domains are the subject of intensive experimental and theoretical studies (25, 26) that are difficult to interpret because of heterogeneities in natural biological membranes.
To understand the organization of subdomains in monodisperse Nanodiscs, we characterized the thermotropic properties of lipid bilayers assembled with the MSP1D1 and MSP1E3 scaffold proteins which correspond to ~ 140–340 lipids per disc (20, 21). Measurements of small-angle X-ray scattering (SAXS) at different temperatures allowed monitoring the size changes of Nanodiscs due to the linear thermal expansion of DMPC and DPPC bilayers in Nanodiscs. The coefficient of lateral thermal expansion coefficients, CT = (1/A)(dA/dT) measured for DPPC and DMPC below phase transition, are 0.002–0.003 K−1 , almost twice lower than those reported for giant unilamellar vesicles (27). Such differences are attributed partly to the confinement provided by the scaffold protein, and partly to the inability of the boundary lipids to undergo the same phase changes as the unrestricted lipids in the center of Nanodisc. Consistent with this confinement, the temperatures of the phase transition in Nanodiscs is higher than those observed for liposomes by 2–4 K (20). In addition, differential scanning calorimetry (DSC) shows significantly lower values of melting enthalpy and broader peaks for nanoscale lipid bilayers in Nanodiscs than those observed for the pure DMPC and DPPC. Comparison of DSC data and also results of Laurdan fluorescence measurements in Nanodiscs of different sizes allowed to suggest that the layer of 1 or 2 lipids directly interacting with the scaffold protein at the periphery of Nanodisc is virtually excluded from the main thermotropic transition, in agreement with other observations cited in the original study (20), as well as with the MD results described earlier in this review.
In larger Nanodiscs, such as the ~ 17 nm version assembled with a MSP fusion containing sixteen amphipathic helices in the scaffold protein and ~ 650 lipids per disc, the main phase transition from gel to liquid crystalline state of DPPC is indicated as a sharp increase of Nanodisc size at 39°C (15), which is close to the phase transition documented in multilamellar vesicles (28). The lateral thermal expansion coefficient CT for DPPC in large Nanodiscs is 0.005 K−1 for the liquid crystalline phase and 0.007 K−1 above phase transition, both values slightly higher than reported for smaller Nanodiscs (20), but similar to those measured for DMPC liposomes (27). This suggests that the high elasticity and flexibility of longer scaffold proteins in Nanodiscs allows for almost unrestricted expansion of the lipid bilayer with increasing temperature. It was also noted that the thermally induced phase transition is reversible in Nanodiscs if the temperature of the sample did not exceed 55°C for more than an hour. In this case the SAXS curves measured for the control (sample kept at 22°C) and the experimental sample were virtually identical to those measured before heating. This confirms the overall stability of these Nanodiscs in solution and provides additional support to the reported method of generation of large lipoprotein particles suitable for solubilization of functional complexes of membrane proteins.
3. Nanodiscs for analysis of lipid-protein interactions
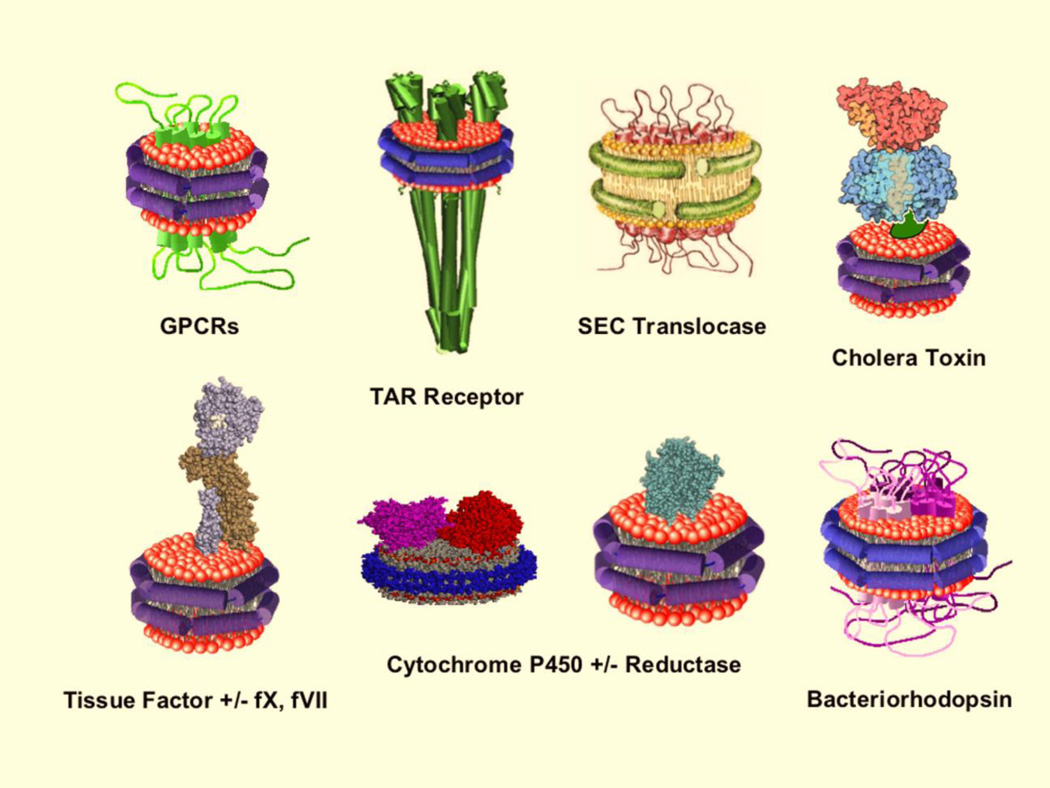
As reviewed recently by Bayburt and Sligar (29), Nanodiscs have provided unprecedented opportunities for studying a wide spectrum of membrane proteins that have been self-assembled into Nanodiscs, including many investigations that probe specific lipid requirements for activity or the lipid-protein interface. Included in this collection are integral membrane proteins, such as G-protein coupled receptors (GPCRs) (30–35), ion channels (36–39), transporters (40–45), pores and toxins (46, 47), receptors (48–51), cytochrome c oxidase (52); each incorporated into Nanodiscs with precisely controlled compositions. A pictorial "Rogues Gallery" of these embedded proteins is represented in Figure 2.
Figure 2.
Representative examples of integral membrane proteins successfully self-assembled into Nanodiscs. This include G-protein coupled receptors (29, 30, 33, 34, 53, 54), multi-subunit chemotactic receptors (48, 49, 55), complex assemblies such as the SEC translocase (40, 41, 56), many different cytochromes P450 both with and without co-incorporated reductase (9, 57–63), human tissue factor and the enzymes of the blood coagulation cascade (64–67), active trimeric bacteriorhodopsin (68) and cholera toxin binding to gangliosides (69, 70).
The large size range of those successfully self-assembled into Nanodiscs includes some with only one trans-membrane helix and others with as many as 24 trans-membrane helices. These assemblies have allowed for the detailed investigation of the effect of bilayer properties (structure, charge, etc.) on the physical, chemical and biochemical properties in well-defined oligomeric states associated with the functioning of these proteins in biological systems. They have also have provided multiple opportunities for studying interactions of proteins with their surrounding lipids (33, 43, 54, 71–73) as well as interactions of membrane lipids and embedded proteins with soluble interacting proteins (i.e., blood coagulation factors) (64, 66, 74). Nanodiscs without any incorporated proteins have been also used as nanoscale mimics of the biomembrane surface and its interactions with soluble proteins and peptides (75).
3.1 Use of Nanodiscs as a controlled membrane surface
Many biochemical processes occur on the surface of a membrane and require specific lipid compositions for full functionality. One example is the blood coagulation cascade that operates to inhibit blood loss following traumatic injury and depends on the recruitment of various serine proteases in the circulatory system to membrane surfaces. The mechanism of molecular recognition for these proteases involves both protein–protein interactions, in which the integral membrane protein tissue factor (TF) provides a surface for association, and protein-lipid interactions, in which the gamma-carboxyglutamate domain (Gla) interacts through bridging calcium ions with negatively charged lipids (45). In the past, difficulties in precisely quantitating the stoichiometry and affinity for these associations arose because typical liposomal preparations loaded with TF and anionic lipids, such as phosphatidyl serine (PS), displayed significant mobility in their lipids and proteins that complicated analysis. In more recent analyses, Nanodiscs have provided the controlled membrane surface with precise lipid compositions needed to mechanistically appreciate these interactions. The ability to derivatize Nanodiscs with various anchors, either biotin, nucleic acid or hexahistidine, presented the means for affixing Nanodiscs to sensor surfaces as needed to monitor associations with blood coagulation factors. In one particular study (66), Nanodiscs loaded with varying levels of PS in a pure phosphatidyl choline (PC) background were attached to sensor chips suitable for surface plasmon resonance readout. The binding affinity and stoichiometry for human factor VII and factor X as a function of the PS/(PC+PS) mole percent were readily determined. In another study (67) more complex lipid mixtures were accommodated in the Nanodisc platform to provide for the molecular analysis of the synergies of particular lipids in the coagulation factor activation cascade. This work led to the ABC "anything but choline" hypothesis describing the ability of smaller head group lipids, such as PG, to dramatically accelerate the initiation of the clotting process. Nanodiscs provides a stable supported lipid bilayer platform and hence have provided and excellent vehicle for structural investigations. For example, they have been used extensively in both solid state and solution NMR spectroscopy. This has provided a detailed picture of how the Gla domains, calcium and the anionic lipid head groups of phosphoserine come together to form a high affinity complex (65, 76, 77).
3.2 Membrane protein libraries using Nanodiscs
The most direct method for incorporating integral membrane proteins into the Nanodisc bilayer is to self-assemble it from a detergent-solubilized mixture of MSP and lipid, maintaining the correct overall stoichiometry of the component parts. Problematically, this necessitates the isolation and purification of the target protein, often a process that inactivates membrane proteins as they are separated from their native environment. To circumvent this problem, we demonstrated some years ago that one could detergent-solubilize a native membrane preparation in the presence of excess MSP and lipid whereupon detergent removal would distribute the starting membrane protein population into individual Nanodiscs, thus forming a soluble library of the components of the starting preparation (9, 59). In collaboration with Professor W. Kline and Dr. K. Wilcox at Northwestern University, Mr. Marty in our laboratory has advanced this concept to develop a protocol for generating Soluble Membrane Protein Libraries (SMPL) and pioneered their use in experiments seeking a potential intervention targets for Alzheimer's disease. These experiments have focused on identifying the synthetic receptor proteins, which interact with the soluble amyloid beta oligomers (known as amyloid beta derived diffusible ligands, ADDLs) that are potent neurotoxins involved in Alzheimer’s disease (78) In previous studies, the identification of synaptic receptor protein(s) has proven difficult and very controversial (79) since this integral membrane protein is restricted to synaptic junctions. Extending our earlier studies inserting either purified membrane proteins or mixtures of membrane proteins from over-expression systems (9, 59) into Nanodiscs, it is now possible to array the population of membrane proteins from synaptic vesicles into Nanodiscs. We are using this concept to isolate and identify ADDL-binding proteins using co-immunoprecipitation capture and Difference Imaging Gel Electrophoresis (DIGE) high-throughput proteomics. This same approach can also be used to isolate the protein-protein binding partners present in cell-cell and cell-viral recognition events and to further define the critical players in cellular communication and viral entry.
A first example using Nanodiscs to define the associations between membrane components and soluble interacting proteins (termed the interactome) have been published recently for the membrane-embedded bacterial SecYEG channel, maltose transporter MalFGK2 and membrane integrase YidC (56). Modifications of the lipid composition in the Nanodisc during the reconstitution process have proved to be a critical parameter for the successful isolation of SecA as a bona fide SecYEG interactor. This suggests that the Nanodisc system may be an excellent tool to test whether specific lipidic environments are critical for protein binding.
3.3 Protein-protein interactions using Nanodiscs
Interactions of human hepatic P450s and their electron transfer partners operate in the complex environment of the endoplasmic reticulum membrane. One important question with these various P450s is the potential role of specific phospholipids in controlling their metabolic activities. Some early investigations with membrane reconstitution systems suggested that trace amounts of phospholipids were required for optimal activity (80), while others suggested that the presence of specific phospholipids might stabilize some kinetically distinguishable conformations of P450s, such as the major drug-metabolizing isoform CYP3A4, and directly modulate its affinity for particular substrates (81, 82). Subsequent recent investigations (71) attempted to define the role of specific phospholipids in maintaining high catalytic turnover.
With the Nanodisc system representing a controllable membrane environment, we have an ideal platform for mechanistically assessing the role of PL in and other anionic lipids in hepatic drug metabolism. Many of our previous Nanodisc assemblies have routinely used mixed lipids ranging in composition from 100% POPC to 100% POPS (66) and others have used mixtures of PA, PS, PE head groups in PC backgrounds to evaluate the role of these in controlling X, Y and Z (64, 66, 74). Extending these previous experiments to analysis of P450 systems, we co-assembled human CYP3A4 together with its P450 reductase and varying lipids in Nanodiscs and quantitated NADPH-driven testosterone hydroxylation. Our initial analysis found that the steady-state rate of testosterone hydroxylation increases by 40% in Nanodiscs containing 30% POPS/70% POPC compared to Nanodiscs containing pure POPC, and that the overall coupling (defined as the fraction of oxygen used for product formation) increases by 60%. Even larger effects of the anionic lipid POPS were observed for the CYP3A4 hydroxylation of bromocriptine with the product formation rate and overall coupling increasing by 80% and 100%, respectively (Grinkova et al., unpublished results). The molecular origin of this effect is in the process of being evaluated via the careful measurement of the functional properties (spin state, rate of autoxidation, uncoupling and product formation) of various CYP3A4 states as a function of substrate loading and various anionic lipid compositions. It is possible there exists a synergy between phospholipids manifested through the increased accessibility of the proteins to the negatively charged head group, as was previously described for the case of blood coagulation factors binding to Nanodisc surfaces (66, 67).
3.4 Protein-lipid interaction studies using Nanodiscs
In addition to these previously mentioned studies, interactions of phosphatidylinositol (PI) binding proteins with membrane bilayers have been assessed in Nanodiscs generated predominantly with DOPC and minor amounts (1.25%) of PI (75). Using pull-down assays, NMR and fluorescence polarization methods, dissociation constants for the PI-binding proteins from the Nanodisc surface have been measured to be Kd ~ 30 nM, a tighter binding to PI in bilayers than previously defined in earlier studies with DPC micelles (Kd ~100 nM). Control experiments performed with Nanodiscs containing no PI show negligible binding, confirming the stoichiometric 1:1 interaction of PI-binding proteins with single molecules of PI incorporated into the lipid bilayer.
Modulations of proteorhodopsin (PR) by different lipid composition have also been assessed in Nanodiscs constructed with DMPC or POPC (54). Controlled assembly of PR monomers have provided the opportunity to compare its photophysical properties with the known properties of oligomeric PR in native membranes. Kinetic measurements have shown that the M and N states, which are key intermediates in the photocycle, have longer lifetimes in DMPC bilayers and shorter lifetimes in POPC bilayers. These results have been attributed to the better modeling of physiological conditions in E. coli membrane by the unsaturated longer hydrophobic tails of POPC. The authors also suggest that the difference in hydrophobic mismatch between the bilayer thickness and the length of the PR trans-membrane helices also plays an important role in the observed variations of the photophysical properties of this protein. It thus appears that the composition of the lipid bilayer is a vital component of the mechanism that controls the photophysical properties of PR and that Nanodiscs will be a very useful tool for the further analysis in this field.
Possible effect of anionic lipids on the yield of photoinitiated phosphorylation of monomeric rhodopsin incorporated in POPC/POPS Nanodiscs have been studied by Bayburt et al. (33). The results of this work show that the presence of POPS is not necessary for the efficient phosphorylation of rhodopsin by the rhodopsin kinase GRK1. However, arrestin binding was significantly stronger in the presence of 20% - 50% POPS and could be substantially inhibited by increasing ionic strength. These results suggest the dominant role of electrostatic repulsion between positively charged arrestin and POPC head groups which could be efficiently decreased by the presence of negatively charged POPS lipids in the Nanodisc bilayer, or by high ionic strength. Thus, incorporation of monomeric rhodopsin into Nanodiscs with different lipid composition allowed to directly prove that the rhodopsin monomers are as functional as oligomers with respect to arrestin binding and phosphorylation and to evaluate the role of anionic lipids in the functional properties of this prototype GPCR.
Further extending the analysis of bR and membrane interactions, the effect of lipid composition on bR unfolding has recently been studied by single-molecule atomic force spectroscopy (73). Unfolding of single bR molecules as a function of applied strength from the AFM tip attached to the terminal amino acid was compared for the native purple membrane fragments and for single bR molecules incorporated into Nanodiscs with DMPC. Somewhat unexpectedly, there was no difference between unfolding intermediates, consistently with the similar optical properties observed for bR in Nanodiscs and in purple membranes. This pioneering experiment opens a new venue for the direct straightforward measurements of the strength and stability of protein–lipid interactions for the integral membrane proteins.
Using epitope-tagged MSP proteins as a means of immunoprecipitating Nanodiscs containing embedded glycosphingolipid ganglioside (GM1) and immobilizing them on sensorchips, tightly bound proteins associated with GM1 have been evaluated by a number of physical means (69,70). Initial studies with surface plasmon resonance identified several tightly binding proteins in E. coli extracts. Subsequent mass-spectroscopic analysis identified one of these as the heat-labile enterotoxin LTB (69). These results demonstrate the effectiveness of approaches combining Nanodisc immunoprecipitations with mass-spectroscopy for clarification of proteins interacting with components in membrane surface. Extensions of this can be widely used for studies of protein–protein, protein–small molecule and protein–lipid interactions (83).
Reconstitution of an ABC transporter MsbA into Nanodiscs of different size and composition revealed the details of the functional interactions of the enzyme with the membrane bilayer (43). Enzymatic activity of MsbA varied up to 4-fold in Nanodiscs assembled with different lipids and also depended on the size of the Nanodisc particles. The highest rate was observed in DMPC discs of smaller size, while incorporation into the negatively charged DMPG discs resulted in the lowest ATPAse activity.
Effect of the membrane bilayer on the midpoint potential of human cytochrome P450 CYP3A4 have been measured by potentiometric titration of this heme enzyme incorporated into Nanodiscs assembled with zwitterionic POPC lipids (84). For both substrate free and substrate bound proteins, the measured absolute values of midpoint potentials turned out to be higher by 50 – 100 mV than for soluble isozymes CYP101 and CYP102. The same effect of the lipid bilayer has been observed later for the flavoprotein cytochrome P450 reductase (85). In addition, the presence of the negatively charged lipid POPS shifted midpoint potentials of both flavins to the more negative values, thus favoring the more efficient electron transfer to the cytochrome P450.
Application of Nanodiscs for the studies of the membrane-active peptides by solution NMR have been recently described for the peptide antibiotic Aam-I from fungi (72). This hydrophobic and poorly soluble in water 15N-labeled hexadecapeptide have been incorporated into Nanodiscs formed with various lipids by stepwise titration using stock solution in methanol up to 1:30 peptide-to-lipid molar stoichiometry. Comparison of 2D 1H-15N correlation spectra of Aam-I in Nanodiscs assembled with different lipids reveal broadened signals with the estimated rotational correlation time ~ 40 ns, lower than expected for Nanodiscs at the same conditions, i.e. 55 – 60 ns, indicating the presence of additional mobility of the peptide incorporated into bilayer. At the same time, many signals undergo the selective broadening and disappear from the 2D –spectra in the samples made with DMPC and POPC lipids. The detailed comparison of these signals allows for the site-selective assignment of the mobile fragments of Aam-I in Nanodiscs with different composition and suggests the presence of several conformational states at equilibrium.
4. Summary and Future Directions
In this brief review we have attempted to provide examples that speak to the utility of the Nanodisc system for the investigations of membrane proteins and their interaction with lipids. While many membrane mimetic models can provide critical biochemical, biophysical and structural input (86–88), the Nanodisc system has been shown to be a robust means for stabilizing and investigating protein and lipid interactions. Detailed experiments have allowed one to study the interactions of the membrane proteins with a membrane, the mechanism of action of membrane proteins, and the three-dimensional structure in a native lipid membrane environment. No other experimental system currently allows for such precise control over the environment of membrane proteins. Since Nanodiscs contain a small patch of membrane of known composition, they are also ideally suited for studying the properties of bilayers such as lipid phase transitions and the binding and transport of small molecules such as cholesterol. As the critical membrane scaffold protein components are now readily available, many laboratories are now independently using this technology. The future promises to bring numerous additional advances in the use of these lipoprotein particles in structure determination, the formulation and delivery of therapeutics and elucidation of mechanism.
Acknowledgements
Our work is supported by grants from the National Institutes of Health GM33775 and GM31756
REFERENCES
- 1.Jonas A, Kezdy KE, Wald JH. Defined apolipoprotein A-I conformations in reconstituted high density lipoprotein discs. J Biol Chem. 1989;264:4818–4824. [PubMed] [Google Scholar]
- 2.Wald JH, Krul ES, Jonas A. Structure of apolipoprotein A-I in three homogeneous, reconstituted high density lipoprotein particles. J Biol Chem. 1990;265:20037–20043. [PubMed] [Google Scholar]
- 3.Carlson JW, Jonas A, Sligar SG. Imaging and manipulation of high-density lipoproteins. Biophys J. 1997;73:1184–1189. doi: 10.1016/S0006-3495(97)78150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck von Bodman S, et al. Synthesis, bacterial expression, and mutagenesis of the gene coding for mammalian cytochrome b5. Proc Natl Acad Sci U S A. 1986;83:9443–9447. doi: 10.1073/pnas.83.24.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayburt TH, Carlson JW, Sligar SG. Single molecule height measurements on a membrane protein in nanometer-scale phospholipid bilayer disks. Langmuir. 2000;16:5993–5997. [Google Scholar]
- 6.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc Natl Acad Sci U S A. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters. 2002;2:853–856. [Google Scholar]
- 8.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Civjan NR, et al. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–558. doi: 10.2144/03353rr02. 560,562-563. [DOI] [PubMed] [Google Scholar]
- 10.Denisov IG, et al. Directed Self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. Structural analysis of nanoscale self-assembled discoidal lipid bilayers by solid-state NMR spectroscopy. Biophys J. 2006;91:3819–3828. doi: 10.1529/biophysj.106.087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips JC, et al. Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys J. 1997;73:2337–2346. doi: 10.1016/S0006-3495(97)78264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wlodawer A, et al. High-density lipoprotein recombinants: evidence for a bicycle tire micelle structure obtained by neutron scattering and electron microscopy. FEBS Lett. 1979;104:231–235. doi: 10.1016/0014-5793(79)80821-2. [DOI] [PubMed] [Google Scholar]
- 14.Shih AY, et al. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinkova YV, Denisov IG, Sligar SG. Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Design & Selection. 2010;23:843–848. doi: 10.1093/protein/gzq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendelsohn R, et al. Quantitative determination of conformational disorder in the acyl chains of phospholipid bilayers by infrared spectroscopy. Biochemistry. 1989;28:8934–8939. doi: 10.1021/bi00448a037. [DOI] [PubMed] [Google Scholar]
- 17.Lins L, et al. Structure of the apolipoprotein A-IV/lipid discoidal complexes: an attenuated total reflection polarized Fourier transform infrared spectroscopy study. Biochim Biophys Acta. 1993;1149:267–277. doi: 10.1016/0005-2736(93)90210-q. [DOI] [PubMed] [Google Scholar]
- 18.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tristram-Nagle S, Nagle JF. Lipid bilayers: thermodynamics, structure, fluctuations, and interactions. Chem Phys Lipids. 2004;127:3–14. doi: 10.1016/j.chemphyslip.2003.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denisov IG, et al. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 22.Gennis RB. Biomembranes. Springer-Verlag, New York: Molecular structure and function; 1989. [Google Scholar]
- 23.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Marsh D. Electron spin resonance in membrane research: protein-lipid interactions from challenging beginnings to state of the art. Eur Biophys J. 2010;39:513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 26.He HT, Marguet D. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu Rev Phys Chem. 2011;62:417–436. doi: 10.1146/annurev-physchem-032210-103402. [DOI] [PubMed] [Google Scholar]
- 27.Needham D, Evans E. Structure and mechanical properties of giant lipid (DMPC) vesicle bilayers from 20 degrees C below to 10 degrees C above the liquid crystal-crystalline phase transition at 24 degrees C. Biochemistry. 1988;27:8261–8269. doi: 10.1021/bi00421a041. [DOI] [PubMed] [Google Scholar]
- 28.Cevc G, editor. Phospholipids Handbook. New York: Marcel Dekker, Inc; 1993. [Google Scholar]
- 29.Bayburt TH, Sligar SG. Membrane protein assembly into nanodisks. FEBS Letters. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitz AJ, et al. Functional reconstitution of β2-adrenergic receptors utilizing self-assembling nanodisc technology. Biotechniques. 2006;40:601–602. doi: 10.2144/000112169. 604,606,608,610,612. [DOI] [PubMed] [Google Scholar]
- 31.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Nat Acad Sci USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whorton MR, et al. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayburt TH, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knepp AM, et al. Direct measurement of thermal stability of expressed CCR5 and stabilization by small molecule ligands. Biochemistry. 2011;50:502–511. doi: 10.1021/bi101059w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldsmith BR, et al. Biomimetic chemical sensors using nanoelectronic readout of olfactory receptor proteins. ACS Nano. 2011;5:5408–5416. doi: 10.1021/nn200489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raschle T, et al. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer Nanodiscs. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu T-Y, et al. Solution NMR spectroscopic characterization of human VDAC-2 in detergent micelles and lipid bilayer nanodiscs. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbamem.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shenkarev ZO, et al. Lipid-protein Nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J Am Chem Soc. 2010;132:5628–5629. doi: 10.1021/ja9097498. [DOI] [PubMed] [Google Scholar]
- 39.Shenkarev ZO, et al. NMR structural and dynamical investigation of the isolated voltage-sensing domain of the potassium channel KvAP: Implications for voltage gating. J Am Chem Soc. 2010;132:5630–5637. doi: 10.1021/ja909752r. [DOI] [PubMed] [Google Scholar]
- 40.Alami M, et al. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO Journal. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalal K, Duong F. Reconstitution of the SecY translocon in nanodiscs. Methods in Molecular Biology. 2010;619:145–156. doi: 10.1007/978-1-60327-412-8_9. [DOI] [PubMed] [Google Scholar]
- 42.Dalal K, et al. Structure, binding, and activity of Syd, a SecY-interacting protein. J Biol Chem. 2009;284:7897–7902. doi: 10.1074/jbc.M808305200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai T, et al. Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 2011;585:3533–3537. doi: 10.1016/j.febslet.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie TK, Kwon H, Atkins WM. Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid Nanodiscs and inhibition by the antibodies MRK16 and UIC2. J Biol Chem. 2011;286:39489–39496. doi: 10.1074/jbc.M111.284554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez FJ, Orelle C, Davidson AL. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2010;132:9513–9515. doi: 10.1021/ja104047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalal K, Duong F. The SecY complex: conducting the orchestra of protein translocation. Trends Cell Biol. 2011;21:506–514. doi: 10.1016/j.tcb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Katayama H, et al. Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc Natl Acad Sci USA. 2010;107:3453–3457. doi: 10.1073/pnas.1000100107. S3453/3451-S3453/3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boldog T, et al. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glueck JM, Koenig BW, Willbold D. Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Analyt Biochem. 2011;408:46–52. doi: 10.1016/j.ab.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 51.Mi L-Z, et al. Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid Nanodiscs. Biochemistry. 2008;47:10314–10323. doi: 10.1021/bi801006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Näsvik Öjemyr L, et al. Reconstitution of respiratory oxidases in membrane nanodiscs for investigation of proton-coupled electron transfer. FEBS Letters. 2012 doi: 10.1016/j.febslet.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayburt TH, et al. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 54.Ranaghan MJ, et al. Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers. J Am Chem Soc. 2011;133:18318–18327. doi: 10.1021/ja2070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang XX, et al. Nanodiscs and SILAC-based mass spectrometry to identify a membrane protein interactome. J Proteome Res. 2012;11:1454–1459. doi: 10.1021/pr200846y. [DOI] [PubMed] [Google Scholar]
- 57.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Denisov IG, Sligar SG. Cytochromes P450 in Nanodiscs. Biochim Biophys Acta, Proteins and Proteomics. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan H, et al. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch Biochem Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 60.Duan H, Schuler MA. Heterologous expression and strategies for encapsulation of membrane-localized plant P450s. Phytochemistry Rev. 2006;5:507–523. [Google Scholar]
- 61.Frank DJ, Denisov IG, Sligar SG. Analysis of heterotropic cooperativity in cytochrome P450 3A4 using α-naphthoflavone and testosterone. J Biol Chem. 2011;286:5540–5545. doi: 10.1074/jbc.M110.182055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grinkova YV, Denisov IG, Sligar SG. Functional reconstitution of monomeric CYP3A4 with multiple cytochrome P450 reductase molecules in Nanodiscs. Biochem Biophys Research Communications. 2010;398:194–198. doi: 10.1016/j.bbrc.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grinkova YV, et al. The ferrous-oxy complex of human aromatase. Biochem Biophys Res Comm. 2008;372:379–382. doi: 10.1016/j.bbrc.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrissey JH, et al. Blood clotting reactions on nanoscale phospholipid bilayers. Thrombosis Res. 2008;122:S23–S26. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonas A, Krajnovich DJ. Effect of cholesterol on the formation of micellar complexes between bovine A-I apolipoprotein and L-alpha-dimyristoyl-phosphatidylcholine. J Biol Chem. 1978;253:5758–5763. [PubMed] [Google Scholar]
- 66.Shaw AW, et al. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 67.Jonas A, Maine GT. Kinetics and mechanism of phosphatidylcholine and cholesterol exchange between single bilayer vesicles and bovine serum high-density lipoprotein. Biochemistry. 1979;18:1722–1728. doi: 10.1021/bi00576a014. [DOI] [PubMed] [Google Scholar]
- 68.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 69.Borch J, Roepstorff P, Moeller-Jensen J. Nanodisc-based co-immunoprecipitation for mass spectrometric identification of membrane-interacting proteins. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O110.006775. O110 006775, 006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borch J, et al. Nanodiscs for immobilization of lipid bilayers and membrane receptors: Kinetic analysis of cholera toxin binding to a glycolipid receptor. Anal Chem. 2008;80:6245–6252. doi: 10.1021/ac8000644. [DOI] [PubMed] [Google Scholar]
- 71.Jonas A. Microviscosity of lipid domains in human serum lipoproteins. Biochim Biophys Acta. 1976;486:10–22. doi: 10.1016/0005-2760(77)90065-0. [DOI] [PubMed] [Google Scholar]
- 72.Shenkarev ZO, et al. Lipid-protein nanodiscs: Possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Moscow) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- 73.Zocher M, et al. Single-molecule force spectroscopy from Nanodiscs: An assay to quantify folding, stability, and interactions of native membrane proteins. ACS Nano. 2012;6:961–971. doi: 10.1021/nn204624p. [DOI] [PubMed] [Google Scholar]
- 74.Morrissey JH, et al. Protein-membrane interactions: blood clotting on nanoscale bilayers. J Thrombos Haemostas. 2009;7:169–172. doi: 10.1111/j.1538-7836.2009.03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kobashigawa Y, et al. Phosphoinositide-incorporated lipid-protein nanodiscs: A tool for studying protein-lipid interactions. Analyt Biochem. 2011;410:77–83. doi: 10.1016/j.ab.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 76.Boettcher JM, et al. Atomic view of calcium-induced clustering of phosphatidylserine in mixed lipid bilayers. Biochemistry. 2011;50:2264–2273. doi: 10.1021/bi1013694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morrissey JH, et al. Protein-phospholipid interactions in blood clotting. Thrombosis Res. 2010;125:S23–S25. doi: 10.1016/j.thromres.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Abeta42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilcox K, et al. Abeta oligomer-induced synapse degeneration in Alzheimer's disease. Cell Mol Neurobiology. 2011;31:939–948. doi: 10.1007/s10571-011-9691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 81.Jonas A, Jung RW. Fluidity of the lipid phase of bovine serum high density lipoprotein from fluorescence polarization measurements. Biochem Biophys Res Commun. 1975;66:651–657. doi: 10.1016/0006-291x(75)90559-8. [DOI] [PubMed] [Google Scholar]
- 82.Matz CE, Jonas A. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J Biol Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 83.Marty MT, Das A, Sligar SG. Ultra-thin layer MALDI mass spectrometry of membrane proteins in nanodiscs. Analyt Bioanalyt Chemistry. 2012;402:721–729. doi: 10.1007/s00216-011-5512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P 450 3A4. J Am Chem Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Das A, Sligar SG. Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer. Biochemistry. 2009;48:12104–12112. doi: 10.1021/bi9011435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jonas A, Hesterberg LK, Drengler SM. Incorporation of excess cholesterol by high density serum lipoproteins. Biochim Biophys Acta. 1978;528:47–57. doi: 10.1016/0005-2760(78)90051-6. [DOI] [PubMed] [Google Scholar]
- 87.Raschle T, et al. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serebryany E, Zhu GA, Yan ECY. Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim Biophys Acta. 2012;1818:225–233. doi: 10.1016/j.bbamem.2011.07.047. [DOI] [PubMed] [Google Scholar]