Abstract
Background
Anaplastic oligodendroglial tumors are rare, and median survival varies widely. Analysis of 1p19q deletion is performed commonly and is an important prognostic factor. However, age and other clinical variables also carry prognostic value, and it is unclear how to incorporate them into clinical decision making or to combine them for prognostication.
Methods
We compiled a retrospective database of 1013 patients with newly diagnosed anaplastic oligodendrogliomas or oligoastrocytomas and performed a recursive partitioning analysis to generate independent prognostic classes among 587 patients with informative 1p19q status. Variables included for survival classification were age (continuous), history of prior low-grade glioma, 1p19q deletion status, histology (presence or absence of an astrocytic component), tumor lobe, tumor hemisphere, gender, extent of resection, postresection treatment, and performance status at diagnosis.
Results
Recursive partitioning analysis identified 5 prognostic groups based on hazard similarity: class I (age <60 y, 1p19q codeleted), class II (age <43 y, not codeleted), class III (age 43–59 y, not codeleted, frontal lobe tumor or age ≥60 y, codeleted), class IV (age 43–59 y, not codeleted, not frontal lobe tumor or age 60–69 y, not codeleted), and class V (age ≥70 y, not codeleted). Survival differences were highly significant (P < .0001), with medians ranging from 9.3 years (95% CI: 8.4–16.0) for class I to 0.6 years (95% CI: 0.5–0.9) for class V.
Conclusions
These 5 distinct classification groups were defined using prognostic factors typically obtained during routine management of patients with anaplastic oligodendroglial tumors. Validation in a prospective clinical trial may better differentiate patients with respect to treatment outcome.
Keywords: 1p19q deletion, anaplastic, anaplastic oligoastrocytoma, oligodendroglioma, prognosis, recursive partitioning analysis
Anaplastic oligodendroglial tumors, anaplastic oligodendrogliomas (AOs), and anaplastic oligoastrocytomas (AOAs) are rare World Health Organization grade III primary brain tumors.1 We previously compiled a retrospective dataset encompassing 1013 cases of AO/AOA diagnosed from 1981–2007 and published demographics, comparative outcomes, and changes in the initial postoperative treatment strategies over time.2,3 However, survival is highly variable, with some patients succumbing within months and others surviving for over 10 years from diagnosis.
Deletion of chromosomes 1p and 19q4 is a significant and favorable prognostic factor5–7 and is predictive of benefit from adding chemotherapy with procarbazine, lomustine, and vincristine (PCV) to radiotherapy.8,9 However, other clinical variables, such as age and performance status, also carry prognostic significance, as does extent of resection.7 It is unclear how to weigh molecular and clinical factors for relative importance and how to incorporate them into prognostication and treatment decisions.
Prognostic models have been generated for other brain tumors, such as glioblastoma,10–12 anaplastic astrocytoma,11 low-grade glioma,13 and primary central nervous system lymphoma.14 However, to our knowledge, no such model exists for anaplastic oligodendroglial tumors. In the current report, we performed recursive partitioning analysis (RPA) on our retrospective dataset2,3 to generate independent prognostic classes using routinely available clinical variables.
Materials and Methods
We generated a decision tree using RPA, also termed Classification and Regression Trees, to stratify patients into distinct risk groups to predict overall survival,15 using our dataset collected retrospectively following institutional review board approval.2 Classification and Regression Trees is a data-mining technique that searches among all the variables to partition the data into homogeneous subgroups. This algorithm first partitions (ie, divides) the data into 2 subgroups by searching through all possible prognostic variables to find the division that is most prognostic for survival. It is recursive in that it then repeats the search to identify additional partitions (subdivisions) within a previously identified subgroup. Each partition point is referred to as a node. The algorithm stops when the data have been divided into subgroups that are too small to divide further in a meaningful manner—for example, when <20 observations remain at any potential branch point or when <7 observations remain at a terminal node. The partitions and nodes form a “tree” that allows for a clinically intuitive visual display of the factors examined and any potential interactions.15 All analyses were conducted and completed at Memorial Sloan-Kettering Cancer Center by A.S.R. and K.S.P. using SAS version 9.2 and R version 2.3.1 software (the recursive PARTitioning package [RPART]).
As an internal validation, 10-fold cross-classification was performed within RPART by randomly dividing the full sample into 10 subsamples. In this way, the full model was “fitted” with 90% of the full sample, while the remaining 10% was used as a validation dataset. This procedure was repeated 10 times. The results from this 10-fold cross-validation were used to “prune” the full tree to a more parsimonious model.16
The tree in this analysis was built using the following collected variables of potential prognostic significance: age (as a continuous variable), gender, KPS at diagnosis (≥70 vs <70), histology (AO vs AOA), history of prior low-grade glioma (yes vs no), dominant lobe of tumor (frontal vs other), hemisphere of the tumor (right vs left vs bilateral), extent of resection (biopsy vs debulking), postoperative treatment (radiotherapy only vs chemotherapy only vs both), and 1p19q deletion status (codeleted vs not codeleted, ie, one or neither deleted). Among 1013 patients with newly diagnosed anaplastic oligodendroglial tumors treated with various strategies in the original dataset,2 there were 587 with informative deletion results at both 1p and 19q treated at diagnosis with the commonly used strategies of radiotherapy, chemotherapy, or both (Table 1). Recursive partitioning analysis was performed on these 587 cases because analysis of 1p19q deletion status is now widely available, routinely performed, and incorporated into clinical decision making.17 Deletion status of 1p19q is also clearly prognostic for survival5–7 and is predicative of benefit from alkylator chemotherapy.8,9 Therefore, cases with unknown deletion status were excluded. Survival time was measured as the interval from histologic diagnosis of AO/AOA to death or last follow-up. Independent prognostic classes were generated with final nodes grouped by similar hazards. Kaplan–Meier curves present the final classification of prognostic groups and were tested for significance by log-rank.
Table 1.
Patient characteristics
|
n = 587 |
||
|---|---|---|
| Age, y | ||
| Median | 41 | |
| Range | 18–83 | |
| Gender, n (%) | ||
| Men | 324 | (55) |
| Women | 263 | (45) |
| Histology | ||
| Anaplastic oligodendroglioma | 337 | (57) |
| Anaplastic oligoastrocytoma | 250 | (43) |
| Prior low-grade glioma | ||
| Yes | 81 | (14) |
| No | 498 | (85) |
| Unknown | 8 | (1) |
| Extent of resection | ||
| Resection | 509 | (87) |
| Biopsy | 60 | (10) |
| Unknown | 18 | (3) |
| KPS | ||
| ≥70% | 496 | (84) |
| <70% | 40 | (7) |
| Unknown | 51 | (9) |
| 1p19q deletion status | ||
| Codeleted | 280 | (48) |
| Not codeleted | 307 | (52) |
| Lobe | ||
| Frontal | 335 | (57) |
| Other | 252 | (43) |
| Hemisphere | ||
| Right | 259 | (44) |
| Left | 270 | (46) |
| Bilateral | 10 | (2) |
| Unknown | 48 | (8) |
| Initial postoperative treatment | ||
| Chemotherapy alone | 128 | (22) |
| Radiotherapy alone | 121 | (21) |
| Both | 338 | (58) |
Results
Clinical characteristics for the 587 cases are shown in Table 1. Only 3 variables contributed to the final tree (Fig. 1): age, deletion status, and tumor location. Among patients younger than 60 years, 1p19q codeletion was the strongest prognostic factor, and these patients survived longest. For patients younger than 60 but without codeleted tumors, the analysis identified another age band of 43–59 years. Tumor location was only prognostic among patients without codeleted tumors aged 43–59, in whom tumors with frontal lobe predominance were favorable. Among patients older than 60 years, codeletion was also favorable. For patients 60 and older without codeleted tumors, the analysis demonstrated another prognostic age band, with the very oldest (≥70) carrying the worst prognosis.
Fig. 1.
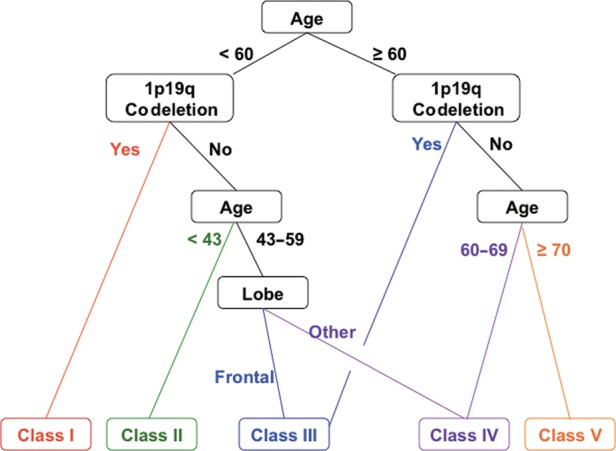
RPA results for all patients (n = 587) with informative 1p19q deletion status showing the final decision tree with terminal nodes and consolidation into 5 distinct prognostic classes using commonly available clinical variables. Age is in years; lobe refers to predominant tumor location; colors indicate route to various classes (Fig. 2); black lines and text are associated with >1 class.
Therefore, RPA identified 5 distinct risk groups based on hazard similarity (Table 2): class I (age <60 y, 1p19q codeleted, tumor in any location); class II (age <43 y, not codeleted, tumor in any location); class III (age 43–59 y, not codeleted, frontal lobe tumor; or age ≥60 y and codeleted, tumor in any location); class IV (not codeleted and either age 43–59 y with tumor not in the frontal lobe or age 60–69 y with tumor in any location); and class V (age ≥70 y, not codeleted, tumor in any location). Kaplan–Meier survival analysis confirmed the existence of 5 distinct risk classification groups identified by RPA, with the best outcome for class I (median, 9.3 y; 95% CI: 8.4, 16 y), the worst for class V (median, 0.6 y; 95% CI: 0.5, 0.9 y), and with classes II, III, and IV intermediate (Table 2 and Fig. 2). The log-rank P-value for overall differences among the 5 curves was <.0001. Although the confidence intervals around the median for classes I and II overlapped (Table 2), the hazards remained significantly different (P = .02; Fig. 2).
Table 2.
Independent prognostic classes
| Class | n | Age, y | 1p19q Codeletion | Dominant Lobe of Tumor | Median Overall Survival (95% CI) |
|---|---|---|---|---|---|
| I | 256 | <60 | Yes | Any | 9.3 (8.4–16.0) |
| II | 174 | <43 | No | Any | 8.9 (5.5–10.8) |
| III | 40 | 43–59 | No | Frontal | 4.3 (2.9–6.0) |
| 24 | ≥60 | Yes | Any | ||
| IV | 48 | 43–59 | No | Not frontal | 2.0 (1.6–2.3) |
| 27 | 60–69 | No | Any | ||
| V | 18 | ≥70 | No | Any | 0.6 (0.5–0.9) |
Fig. 2.
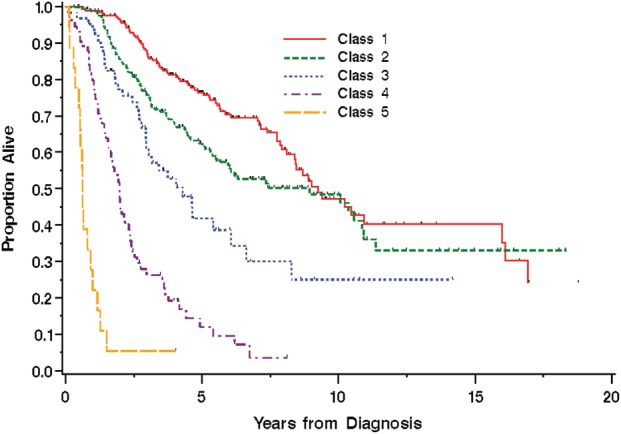
Kaplan–Meier survival curves by prognostic class formed by groups of similar hazard (hazards for classes I–V: 0.59, 0.85, 1.2–1.7, 3.4–3.5, 8.2, respectively).
Discussion
We identified 5 independent prognostic classes using readily available clinical variables. Only 3 variables were identified in this classification schema: age, 1p19q codeletion status, and tumor location. The most powerful variable was age <60 or ≥60 years. Most prospective trials recognize the prognostic importance of age, and some use it as a stratification factor in randomization. However, seminal phase III studies used age cutoffs of 406,9 or 505,8 in randomization procedures or multivariate analyses.7 Our results (using age as a continuous variable) suggest instead that 60 years may be the most important cut point, and future trials may need to consider this in the statistical design. Additional age bands, <43, 43–59, and ≥70 years, had prognostic value but were less significant than 1p19q status. Although the confidence intervals for classes I and II overlap, they are statistically and clinically distinct; separating them retains clinical value.
In addition, 1p19q deletion status, while important, was surprisingly not the most prognostic variable. Tumor location was important, but performance status, extent of resection, treatment, and histology did not contribute to the final model; however, these factors are interrelated. For example, extent of resection may have been partially addressed in the dataset because frontal tumors are more likely amenable to greater resection than tumors in other locations. Similarly, physician bias may have led to more aggressive treatment, including debulking resection rather than biopsy, in patients with higher KPS. Approximately 85% of patients had KPS ≥70, and patients were not further subdivided into those with the very best performance groups (KPS 90–100). Others have suggested that tumors of mixed histology behave between those of pure oligodendroglial and pure astrocytic varieties. By contrast, our results indicate that 1p19q status exerts a more powerful influence on survival than traditional histology and are consistent with similar conclusions from a phase III study (NOA-04) of anaplastic gliomas conducted by the Neuro-oncology Working Group of the German Cancer Society.7
Our analysis was limited by the retrospective nature of the original dataset and the lack of central review of either pathology or imaging, as detailed elsewhere.2 Accordingly, we did not attempt to differentiate gross total from subtotal resection, instead characterizing the extent of surgical excision as either biopsy or debulking. In addition, diagnostic criteria for gliomas have changed over time, especially for oligodendrogliomas.18 For example, the most recent World Health Organization criteria would now diagnose AOAs with necrosis as grade IV glioblastoma with an oligodendroglial component,1,19 and it is possible that such cases contributed to the poor prognosis in classes IV and V. However, histology (AO vs AOA) did not contribute to our final model, suggesting that the potential confounding effect was limited. As an alternative explanation, 1p19q codeletion is more common among AOs than AOAs, and codeletion may have been a surrogate for histology, especially in the absence of central pathology review. We also did not capture tumor size, which is prognostic in other brain tumors13 and may be important in AO/AOA. Finally, we did not perform testing for IDH mutation or MGMT promoter methylation, which are emerging as independent molecular prognostic factors.7,19,20 Recent results suggest that 1p19q codeleted tumors are a subset of those with IDH mutation; accordingly, lack of IDH mutation testing was likely most relevant for patients whose tumors did not have codeletion.21 However, IDH mutation may also have been partially addressed by age, as older patients almost never have IDH mutant tumors.22 Unfortunately, archival tissue was not available for either central pathology review or molecular interrogation, and the cost and logistical difficulties of locating, transporting, and analyzing tissue were insurmountable obstacles. However, IDH mutation and MGMT promoter methylation studies are not always widely available to community oncologists, and differences in laboratory method for both IDH23,24 and MGMT25 testing can lead to discordant results. Moreover, other molecular abnormalities of potential prognostic significance have been recently discovered, and which of these can or should be routinely analyzed is an emerging area of investigation in neuro-oncology.26 For example, we also did not test for mutations in CIC,27,28 ATRX29,30 (which appears mutually exclusive with 1p19q codeletion),31 FUBP1,28,30 or NHE-132; for Glioma-CpG island methylator phenotype (G-CIMP)33,34 (which appears to result from IDH mutation35–37 and may lead to and be more important than MGMT methylation34); or for “intrinsic glioma subtype.”38 Rather, our purpose was to generate a model using data routinely available in the clinic. At this time, we viewed 1p19q status as the most widely used molecular marker.17
As described in Materials and Methods, the current classification tree was subjected to robust internal validation. Ideally, an independent dataset for external validation is required. However, after carefully considering the other available studies, we concluded that the heterogeneity among existing datasets would likely preclude the validity of such analysis. For example, Radiation Therapy Oncology Group (RTOG) 9402 and European Organisation for Research and Treatment of Cancer (EORTC) 26951 were prospective randomized phase III studies, whereas ours was not. In addition, treatment in those studies always included radiotherapy (with or without PCV), whereas ours also included patients who did not receive radiotherapy as part of the initial treatment regimen, and also captured patients who received initial chemotherapy with temozolomide rather than PCV. RTOG 9402 excluded patients with KPS <60 and then grouped them by KPS 60–70 or 80–100, and EORTC 26951 excluded patients with an Eastern Cooperative Oncology Group performance status >2 and then grouped them by 0–1 or 2. By contrast, we had no limit on performance status and categorized KPS as <70 or ≥70. Finally, EORTC 26951 also excluded patients over age 70, which would have eliminated patients in our RPA class V.
In conclusion, the RPA prognostic model presented here is simple yet powerful. Although the model appears complex, we found that combining 3 routinely available variables (age, 1p19q codeletion status, and tumor location) powerfully predicted survival. Recursive partitioning analyses for other primary brain tumors are important. Our RPA may be a useful prognostic tool for patients with anaplastic oligodendroglial tumors.
Funding
Funding for development of the initial dataset was described previously. In addition, K.S.P. and A.S.R. were supported in part by P30 CA008748 and A.B.L. in part by P30 CA013696-40. Funding sources had no access to patient data and no role in conceiving or designing the study or in data collection, analysis, or interpretation of results.
Acknowledgments
We thank the Oligodendroglioma Study Group, from which this work originated. This study was presented as an abstract at the 2011 Society for Neuro-Oncology annual meeting.
Conflict of interest statement. Relevant to this publication, A.B.L., T.F.C., N.A.P., J.G.C., H.I.R., P.Y.W., F.G.K., M.C.C., and W.P.M. consulted for and/or received honoraria from Schering Corp and/or Merck & Co; A.B.L. consulted for Bristol-Myers Squibb; A.B.L., J.G.C., D.S., P.Y.W., M.C.C., L.E.A., and N.A.P. consulted for Sigma-Tau Pharmaceuticals; N.A.P., M.C.C., and S.A.W. served on a Speaker's Bureau for Merck & Co (formerly Schering Corp). L.E.A. is now a full-time employee of and stockholder in F. Hoffmann–La Roche Ltd.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- 2.Lassman AB, Iwamoto FM, Cloughesy TF, et al. International retrospective study of over 1000 adults with anaplastic oligodendroglial tumors. Neuro Oncol. 2011;13(6):649–659. doi: 10.1093/neuonc/nor040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panageas KS, Iwamoto FM, Cloughesy TF, et al. Initial treatment patterns over time for anaplastic oligodendroglial tumors. Neuro Oncol. 2012;14(6):761–767. doi: 10.1093/neuonc/nos065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 5.Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24(18):2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 6.van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24(18):2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 7.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 8.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 10.Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. doi: 10.1215/S1152851703000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran WJ, Jr., Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85(9):704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 12.Scott JG, Bauchet L, Fraum TJ, et al. Recursive partitioning analysis of prognostic factors for glioblastoma patients aged 70 years or older. Cancer. 2012;118(22):5595–5600. doi: 10.1002/cncr.27570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pignatti F, van den Bent M, Curran D, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20(8):2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 14.Abrey LE, Ben-Porat L, Panageas KS, et al. Primary central nervous system lymphoma: the Memorial Sloan-Kettering Cancer Center prognostic model. J Clin Oncol. 2006;24(36):5711–5715. doi: 10.1200/JCO.2006.08.2941. [DOI] [PubMed] [Google Scholar]
- 15.Therneau TM, Atkinson EJ. Technical report series No. 61, An introduction to recursive partitioning using the RPART routines. Department of Health Science Research, Mayo Clinic, Rochester, MN;: 1997. [Google Scholar]
- 16.Breiman L, Friedman JH, Olshen RA, et al. Classification and Regression Trees. Belmont, CA: Wadsworth International Group; 1984. [Google Scholar]
- 17.Abrey LE, Louis DN, Paleologos N, et al. Survey of treatment recommendations for anaplastic oligodendroglioma. Neuro Oncol. 2007;9(3):314–318. doi: 10.1215/15228517-2007-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burger PC. What is an oligodendroglioma? Brain Pathol. 2002;12(2):257–259. doi: 10.1111/j.1750-3639.2002.tb00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16(5):1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 20.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27(35):5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 23.Preusser M, Capper D, Hartmann C. IDH testing in diagnostic neuropathology: review and practical guideline article invited by the Euro-CNS research committee. Clin Neuropathol. 2011;30(5):217–230. doi: 10.5414/np300422. [DOI] [PubMed] [Google Scholar]
- 24.van den Bent MJ, Hartmann C, Preusser M, et al. Interlaboratory comparison of IDH mutation detection. J Neurooncol. 2013;112(2):173–178. doi: 10.1007/s11060-013-1056-z. [DOI] [PubMed] [Google Scholar]
- 25.van den Bent MJ, Erdem-Eraslan L, Idbaih A, et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19(19):5513–5522. doi: 10.1158/1078-0432.CCR-13-1157. [DOI] [PubMed] [Google Scholar]
- 26.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59(8):1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 27.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226(1):7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weller M, Pfister SM, Wick W, et al. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol. 2013;14(9):e370–e379. doi: 10.1016/S1470-2045(13)70168-2. [DOI] [PubMed] [Google Scholar]
- 32.Blough MD, Al-Najjar M, Chesnelong C, et al. DNA hypermethylation and 1p loss silence NHE-1 in oligodendroglioma. Ann Neurol. 2012;71(6):845–849. doi: 10.1002/ana.23610. [DOI] [PubMed] [Google Scholar]
- 33.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bent MJ, Gravendeel LA, Gorlia T, et al. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. Clin Cancer Res. 2011;17(22):7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 35.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulholland S, Pearson DM, Hamoudi RA, et al. MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer. 2012;131(5):1104–1113. doi: 10.1002/ijc.26499. [DOI] [PubMed] [Google Scholar]
- 38.Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]


