Abstract
Background
Experimental findings have suggested that human cytomegalovirus (HCMV) infection of tumor cells may exert oncomodulatory effects that enhance tumor malignancy. However, controversial findings have been published on the presence of HCMV in malignant tumors. Here, we present the first study that systematically investigates HCMV infection in human nervous system tumors by highly sensitive immunohistochemistry in correlation with the HCMV serostatus of the patients.
Methods
Immunohistochemical and quantitative PCR-based methods to detect different HCMV antigens and genomic HCMV DNA were optimized prior to the investigation of pathological samples. Moreover, the pathological results were matched with the HCMV serostatus of the patients.
Results
HCMV immediate-early, late, and pp65 antigens could be detected in single cells from HCMV strain Hi91-infected UKF-NB-4 neuroblastoma cells after 1:1024 dilution with noninfected UKF-NB-4 cells. Genomic HCMV DNA could be detected in copy numbers as low as 430 copies/mL. However, we did not detect HCMV in tumors from a cohort of 123 glioblastoma, medulloblastoma, or neuroblastoma patients. Notably, we detected nonspecifically positive staining in tumor tissues of HCMV seropositive and seronegative glioblastoma patients. The HCMV serostatus of 67 glioblastoma patients matched the general epidemiological prevalence data for Western countries (72% of female and 57% of male glioblastoma patients were HCMV seropositive). Median survival was not significantly different in HCMV seropositive versus seronegative glioblastoma patients.
Conclusions
The prevalence of HCMV-infected tumor cells may be much lower than previously reported based on highly sensitive detection methods.
Keywords: cytomegalovirus, glioma, oncomodulation
Human cytomegalovirus (HCMV), a member of the herpes virus family, persists after primary (normally unrecognized) infection for the remainder of the patient's life. The seroprevalence ranges from ∼50% to 100% within a population, and higher prevalence levels are detected in females than in males. Lower socioeconomic status is associated with increased HCMV seroprevalence,1,2 and HCMV is also a major pathogen in immunocompromised individuals. Although HCMV reactivates with varying frequency and results in detectable virus levels in the blood and virus shedding, HCMV disease is an extremely rare event in immunocompetent individuals.3 During critical illness of immunocompetent patients, HCMV reactivation may be associated with prolonged hospitalization and death.4
For decades, it has been speculated that HCMV may play a role in the course of various cancer diseases.5,6 Experimental evidence has suggested that HCMV may cause oncomodulatory activity (ie, modulate the malignant properties) of established cancer (or other tumor-associated) cells and enhance cancer malignancy even though it is not a tumor virus with proven transformational potential.5,7 In 2002, the idea of HCMV-induced oncomodulation was strongly supported by a clinical study that reported on the detection of HCMV proteins and oligonucleotides in a very high percentage of gliomas.8 This was followed by further studies reporting the detection of HCMV in a high percentage of cancer tissues from different entities, including colorectal cancer, prostate cancer, medulloblastoma, and neuroblastoma.9–13 However, other groups have been skeptical and have not found HCMV in high fractions of tumors.14–19 Interestingly, a study correlating HCMV serostatus with the detection of HCMV in tumor tissues has been missing. To fill this gap, we present here a systematic study on the detection of HCMV by immunohistochemistry and quantitative (q)PCR in tissues from patients with nervous system tumors, including a subgroup of glioblastoma patients in which we performed a combined analysis of HCMV serostatus data with immunohistochemical HCMV detection in tumor tissues.
Material and Methods
Patient Data
In total, 200 paraffin-embedded tissue samples from 123 patients suffering from tumors of the nervous system, including neuroblastomas (n = 12), WHO grade IV medulloblastomas (n = 20), and WHO grade IV glioblastomas (n = 91), were investigated in our study. Additionally, normal brain tissue from autopsy cases and histologically normal-appearing brain tissue surrounding tumor samples were studied. Tissue samples from patients with confirmed HCMV encephalitis served as positive controls for HCMV infection. The samples were obtained from the tissue bank of the Edinger Institute (Neurological Institute), Frankfurt, Germany. Tissue samples were fixed in 4% buffered formalin (Roth) and embedded in paraffin. The histopathological diagnoses were performed according to the World Health Organization classification. The use of patient material was approved by the local ethics committee of the Goethe University Frankfurt/Main, Germany (GS-04/09 and GS-249/11).
Assessment of HCMV Serostatus
Anti-HCMV IgG, IgM, and IgA antibody screening was performed with the Architect 2000SR (Abbott Diagnostics) according to the manufacturers' instructions. While the detection of HCMV-IgG antibodies demonstrates HCMV seropositivity, the detection of IgM and/or IgG antibodies indicates a primary infection and/or HCMV reactivation.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections (3 µm) were subjected to immunohistochemistry using the following antibodies: a mouse antibody directed against the early nuclear, nonstructural DNA-binding p52 HCMV protein (43 kDa; IgG1; clone CCH2; dilution: 1:500; Dako); a mouse antibody directed against the HCMV immediate-early antigen (IAC) (76 kDa; IgG2a; clone DDG9; dilution 1:500; Dako); a mouse antibody directed against the HCMV pp65 antigen (IgG1; clone 26; dilution 1:200; Leica); and a mouse antibody directed against HCMV IEA (IgG1; clone MCA2147; dilution 1:50; Serotec). In addition, the anti-HCMV pp65 and anti HCMV IEA antibodies were used at dilutions of 1:20 and 1:5. Tissue labeling for all antigens was performed using the DiscoveryXT immunohistochemistry system (Ventana). After deparaffinization, the conditioning heat pretreatment with TE buffer was performed for either 36 minutes (clone CCH2 and DDG9 [Dako]) or 52 min (clone 26 [Leica] and clone MCA2147 [Serotec]) followed by a 4-minute blocking step using inhibitor D (Ventana). The primary antibodies were applied for 32 minutes at 37°C followed by one drop of Universal Secondary Antibody (Ventana) for another 32 minutes at 37°C. Next, an avidin-biotin blocker was applied to the samples for 4 minutes. For diaminobenzidine (DAB) visualization, the sections were incubated with one drop of I-View SA-HRP for 16 minutes, followed by incubation with DAB/H2O2 for an additional 8 minutes. All sections were finally incubated with a copper enhancer (Ventana) for 4 minutes and then washed, counterstained with hematoxylin, and mounted.
Quantitative PCR
DNA was extracted from 12 neuroblastomas and 10 glioblastomas using standard protocols. HCMV real-time PCR was performed using 10 µl of the extracted DNA as previously described.20 As positive controls, a series of 4 samples with diluted HCMV titers (430 000 HCMV copies per ml [cs/mL], 43 000 cs/mL, 4300 cs/mL, and 430 cs/mL) derived from HCMV strain Hi91-infected human foreskin fibroblasts and a sample of cerebellar tissue from a patient with HCMV encephalitis were included. Water was used as negative control, and glyceraldehyde 3-phosphate dehydrogenase was used as housekeeping gene.21
Cell Culture
UKF-NB-4 cells, derived from bone marrow metastases of a patient with MYCN-amplified stage IV neuroblastoma, were grown at 37°C in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 IU/mL penicillin, and 100 µg/mL streptomycin.22 The following established glioma cell lines were tested: LN18, U138, U87, LN428, D247, T98G, LN319, LNT229, A172, U251, and U373 (kindly provided by Dr. N. de Tribolet of University Hospital, Lausanne, Switzerland). The adherent human glioma cell lines were grown in Dulbecco's modified Eagle medium (Gibco Invitrogen) supplemented with 10% FCS (Provitro) and 1% penicillin/streptomycin (Sigma Aldrich). All cells were grown in an incubator atmosphere of 5% CO2 at 37°C (Heraeus, Thermo Scientific). Daoy medulloblastoma cells were a gift from T. Pietsch (Bonn, Germany). Cells were centrifuged, fixed in 4% buffered formalin (Roth), embedded in paraffin, and further processed in the same manner as patient material.
Virus Culture
Virus culture was performed as previously described.22 Stocks of HCMV strain Hi91 were prepared in human foreskin fibroblasts maintained in minimal essential medium with 4% FCS. Virus titers were determined by plaque titration. UKF-NB-4 cells were infected at a multiplicity of infection of 2. Seventy-two hours post infection, the HCMV-infected UKF-NB-4 cells were diluted with noninfected UKF-NB-4 cells in ratios of 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, and 1:128 and forwarded to paraffin-embedding and immunohistochemical analysis or qPCR analysis.
Statistical Aanalysis
The association of patient survival with HCMV serostatus was assessed by Kaplan-Meier analysis. Statistical significance levels were determined by log-rank and Wilcoxon tests. The association of serostatus and (i) sex as well as Karnofsky performance score (KPS) was tested using the likelihood-ratio test, (ii) age by Student' t test, and (iii) location of lesion by nonparametric Wilcoxon test. A significance level of alpha = 0.05 was selected for all tests. Statistical analysis was performed using JMP 8.0.1 software (SAS). Graphics were prepared using GraphPad Prism software (GraphPad Software).
Results
Immunocytochemical and qPCR-based Methods Detect HCMV in Human Neuroblastoma Cells With Similar Sensitivity
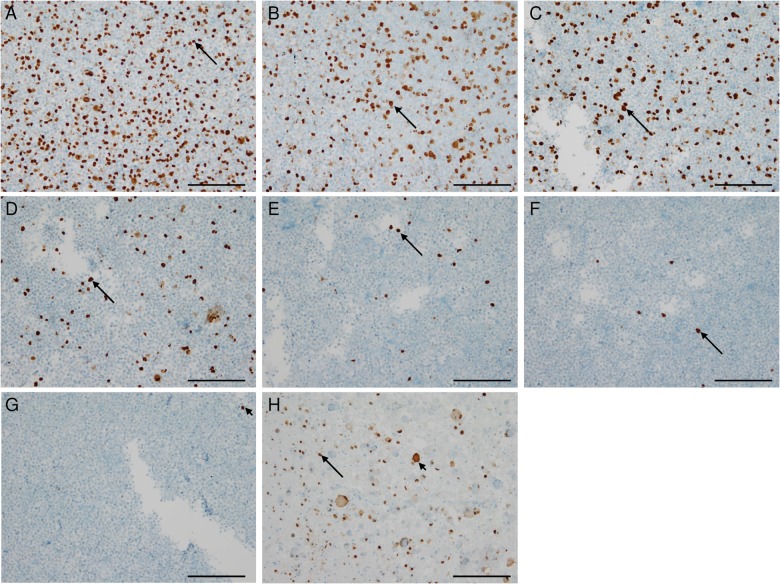
Human neuroblastoma cells (UKF-NB-4) were infected with HCMV Hi91 at a multiplicity of infection of 2, centrifuged, and paraffin-embedded for immunohistochemical analyses. Approximately 40% of HCMV-infected neuroblastoma cells stained positively for HCMV-IEA (Fig. 1A). Subsequently, infected cells were diluted at a ratio of 1:1 to 1:1024 with noninfected UKF-NB-4 cells finally showing only single HCMV-IEA-positive cells as assessed by immunocytochemistry (Fig. 1B–G). Cerebellar tissue from a patient with HCMV encephalitis served as positive control (Fig. 1H).
Fig. 1.
Immunocytochemistry (ICC) and immunohistochemistry (IHC) displaying detection limits in HCMV diagnostics. (A) ICC of paraffin-embedded HCMV strain Hi91-infected UKF-NB-4 neuroblastoma cells showing HCMV-IEA positive nuclei in 30%–40% of the cells (arrow). (B) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:1. (C) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:4. (D) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:16. (E) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:64. (F) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:256. (G) ICC of paraffin-embedded HCMV-infected UKF-NB-4 cells diluted with noninfected cells in a ratio of 1:1024. (H) Cerebellar tissue from a patient with HCMV encephalitis showing atypical HCMV-positive nuclei (arrow) of infected neurons and positive nuclei of surrounding glial cells (arrow-heads). (Scale 100 µm).
Lack of HCMV Antigen Detection by Immunohistochemical Analysis in Tumors of the Central Nervous System
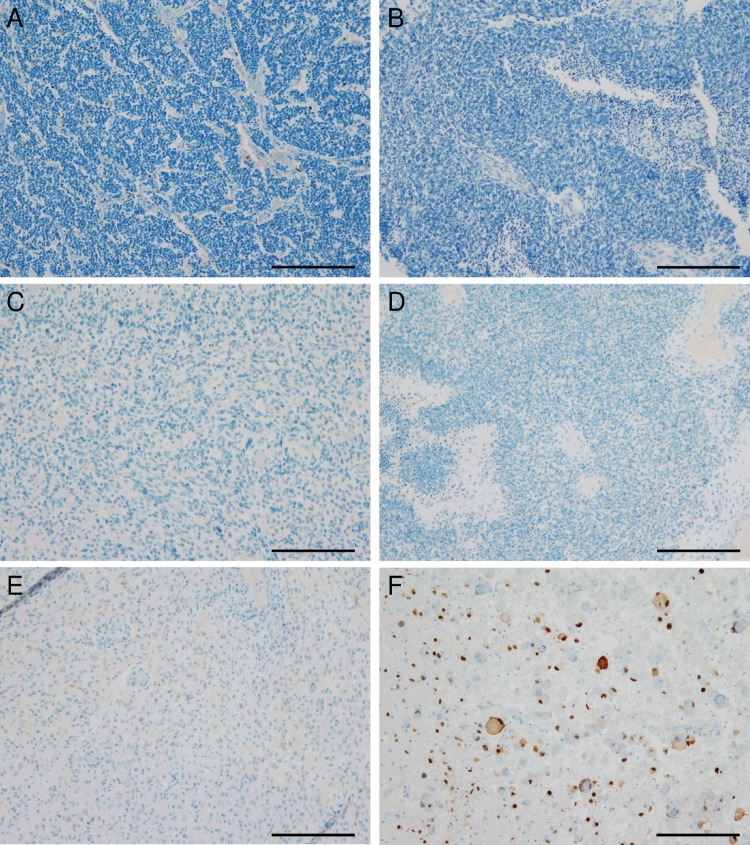
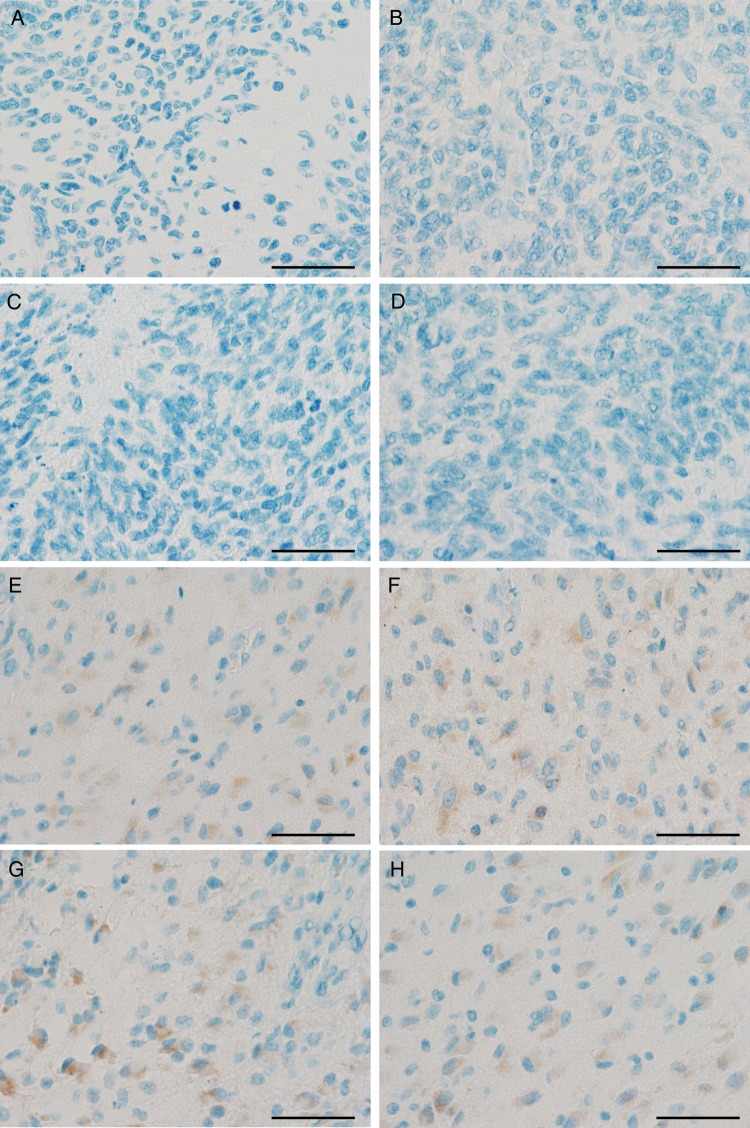
The immunohistochemical analyses revealed negative results for the HCMV-associated antigens pp65, early antigen, or IEA in 12 of 12 neuroblastomas (Fig. 2A), 20 of 20 medulloblastomas (Fig. 2B), and 89 of 91 glioblastoma samples (Fig. 2C and D). Only single glioblastoma cases showed weakly positive staining results (Fig. 2E), while cerebellar tissue derived from a patient with HCMV encephalitis (positive control) (Fig. 2F) showed the expected positive staining results. Additionally, established human medulloblastoma and high-grade glioma cells lines were negative for HCMV antigens (data not shown). To rule out that HCMV infection in tumors of the nervous system might be undetected due to the antibody concentration being too low, we increased the antibody concentrations for pp65 and IEA up to extraordinarily high concentrations of 1:20 and 1:5, which are usually not used in routine diagnostic procedures. Higher concentrated antibodies directed against HCMV-IEA (Serotec) and pp65 (Leica) neither led to positivity in former negative cases (Fig. 3A–D) nor to increased staining intensity or frequency in positive glioblastoma cases (Fig. 3E and F).
Fig. 2.
Tumors of the nervous system are virtually negative for HCMV antigens. (A) Neuroblastoma stained with the DAKO-antibody against HCMV early antiegen (EA) and HCMV immediate-early antigen (IEA) showing only HCMV-negative nuclei. (B) Human medulloblastoma stained with the DAKO antibody against HCMV EA/IEA also being completely negative. (C and D) Two different glioblastoma specimens also being negative for HCMV EA/IEA. (E) Glioblastoma being slightly positive for HCMV in some areas. (F) Specimen of a patient suffering from cerebellar HCMV encephalitis showing HCMV EA/IEA-positive nuclei of neurons and surrounding glial cells. (Scale 100 µm).
Fig. 3.
Increased anti-HCMV antibody concentrations do not lead to stronger staining intensities or frequencies in tumors of the nervous system in either positive or negative cases. Negative glioblastomas were stained for pp65 antigen (A: antibody dilution 1:200; B: antibody dilution 1:5) and immediate early antigen (IEA); (C: antibody dilution 1:50; D: antibody dilution 1:5) using different antibody concentrations. Slightly HCMV-positive glioblastomas were stained for pp65 antigen (E: antibody dilution 1:200; F: antibody dilution 1:5), and IEA (G: antibody dilution 1:50; H: antibody dilution 1:5) using different antibody concentrations (Scale 50 µm).
Potential Pitfalls in the Morphological Assessment of HCMV Immunohistochemistry in Human Samples
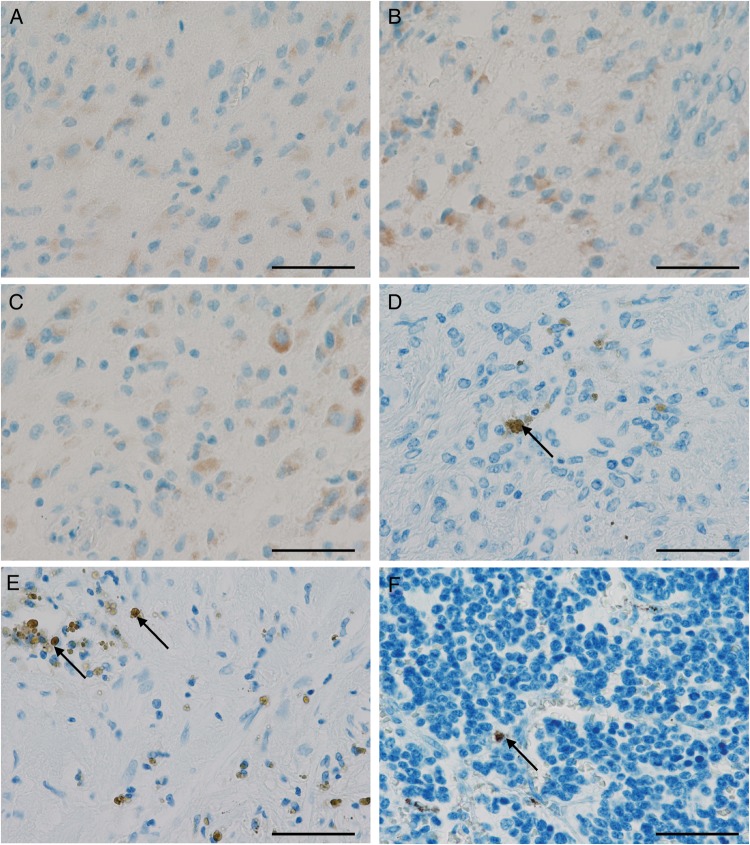
At first glance, single cases seemed to reveal positive staining signals for all antibodies used in our study (Fig. 4A and B). However, the staining positivity was largely restricted to gemistocytes, a subtype of reactive or neoplastic astrocytes that often cross-react in immunohistochemical analyses with various antibodies (Fig. 4B and C). Other positive signals were derived from hematoidin and hemosiderin pigments related to old bleeding (Fig. 4D and E) as well as formalin pigment (Fig. 4F) as fixation artefacts. The negative (Fig. 4G) and isotype (Fig. 4H) controls of the respective samples exhibited similar staining results, thereby indicating unspecificity of the staining in these specimens. Notably, high concentrations of the antibodies directed against HCMV antigens resulted in positive staining, which was interpreted as nonspecific background staining due to its presence in tumors from both seropositive and seronegative patients.
Fig. 4.
Old intratumoral bleeding and fixation artefacts are potential pitfalls in immunohistochemistry-based HCMV diagnostics. (A) Positive glioblastoma case with gemistocytic cells being positive for HCMV-pp65 antigen and also for (B) HCMV immediate early antigen (IEA). (C) Same glioblastoma case stained without primary antibody also showing a similar positive signal indicating an unspecific reaction. (D and E) Neuroblastoma with hematoidin artefacts appearing like positive tumor cells (arrows). (F) Neuroblastoma with formalin pigments (arrow). (Scale 50 µm).
HCMV is Undetectable in Human Neuroblastoma and Glioblastoma Specimens Using qPCR
The number of PCR cycles necessary to detect HCMV in tissue culture-derived control samples was inversely correlated with the virus concentrations (26.23 cycles for 430 000 cs/mL, 28.97 for 43 000 cs/mL, 30.95 for 4300 cs/mL, and 35.51 for 430 cs/mL). In addition, we detected HCMV (562 cs/mL) in a cerebellar tissue sample from a patient with HCMV encephalitis that served as a positive control. However, the analysis of DNA extracted from patient tumor samples (12 neuroblastomas, 10 glioblastomas) did not result in detectable virus levels.
HCMV Serostatus of Glioblastoma Patients Is Not Associated With Patient Survival
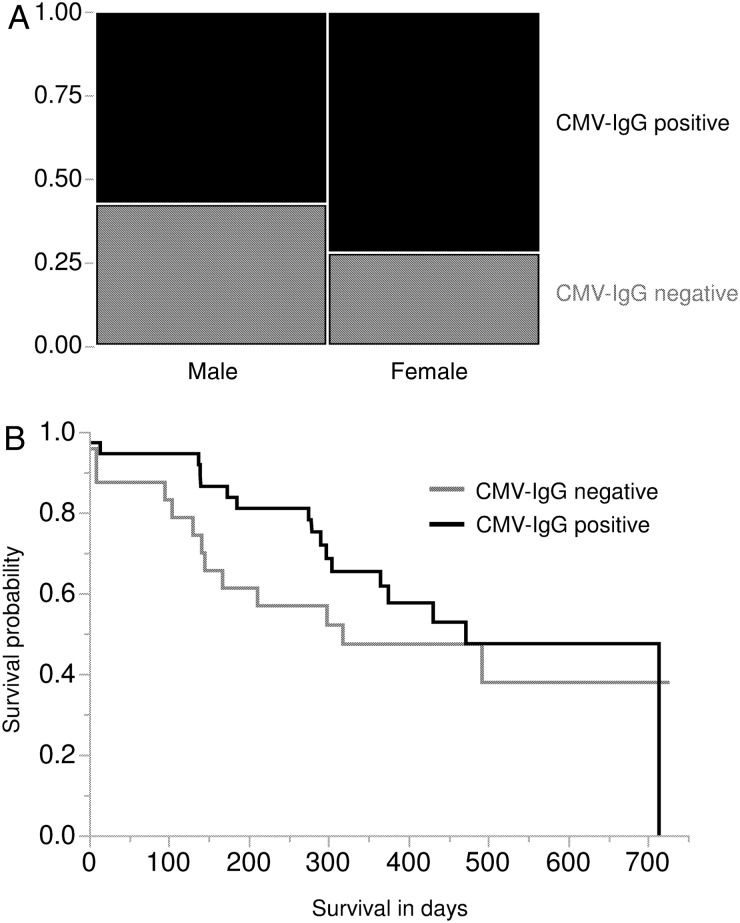
The HCMV serostatus of 67 patients in the glioblastoma cohort was determined, and clinical outcome data were available from 61 of them. Twenty-four of 67 (36%) glioblastoma patients were HCMV seronegative, and 43 of 67 (64%) patients were seropositive, as indicated by IgM and IgG status. Seventy-two percent (23/32) of female patients and 57% (20/35) of male patients were HCMV seropositive (Fig. 5A). Both overall and sex-related epidemiological values were in accordance with previously published HCMV prevalence data for Western countries.23 Furthermore, we determined whether the HCMV serostatus of patients was related to their clinical course of glioblastoma disease. Median survival times were increased in HCMV-seropositive glioblastoma patients, although the differences did not reach statistical significance (Fig. 5B). No significant differences in patient age, sex, KPS, and location of lesion as well as progression-free and overall survival were detected between HCMV-seropositive and HCMV-seronegative individuals (Table 1). We also analyzed if the anti-HCMV IgG titers correlated with glioblastoma patient survival, but no significant association of anti-HCMV IgG levels with patient survival was detected.
Fig. 5.
Patient survival is not associated with the HCMV serostatus of glioblastoma patients. (A) Contingency analysis of HCMV serostatus by sex. (B) Kaplan Meyer survival analysis investigating dependence of HCMV serostatus and patient survival. Log-rank P = .362; Wilcoxon P = .125.
Table 1.
Patient data
| Seropositive GBM Patients | Seronegative GBM Patients | P value | |
|---|---|---|---|
| Sex (male/female) | 20/23 | 15/9 | .207* |
| Median age (range) y | 60 (34–80) | 57.5 (36–74) | .518** |
| Karnofsky performance score median (range) | 80% (30%–100%) | 90% (80%–100%) | .179* |
| Location of lesion (supra-/infratentorial) | 43/0 | 24/0 | >.999*** |
| Median overall survival (range) | 304 (0–714) days | 284 (0–725) days | .362**** |
| Median progression-free survival (range) d | 242 (0–1433) days | 152 (9–1178) days | .397**** |
*Likelihood-ratio test.
**Student' t test.
***Nonparametric Wilcoxon test.
****Kaplan-Meier analysis/log-rank test.
Discussion
There is a long-standing controversy about how many tumors of nervous system HCMV can be detected and whether the presence of HCMV in tumor tissues may be associated with enhanced malignancy.8,18 While a couple of groups have failed to detect HCMV in a high percentage of glioblastomas by using immunohistochemical methods,15,16 others have reported the expression of various HCMV proteins in glioblastomas by using optimized immunohistochemistry techniques.10,24 Similar discrepancies were reported in molecular pathological studies showing a very high percentage (94%, 16/17) of HCMV-positive brain tumors as assessed by classical Sanger sequencing,25 whereas highly sensitive RNA deep sequencing methods failed to detect relevant amounts of HCMV (P < .05 parts per million) in a large cohort of 167 brain tumors.18 We combined nucleic acid and protein-based analyses but did not detect HCMV in tumors of the nervous system (Fig. 2), although we were able to detect low HCMV copy numbers or single HCMV-positive tumor cells in experimental systems by our approach (Figs. 1 and 3).
The appropriate interpretation of immunohistochemical stainings may be complicated by frequently encountered pigments such as hematoidin, hemosiderin, and formalin precipitates (Fig. 4) as well as the unspecific staining predominantly seen in gemistocytic cells (Fig. 3). Notably, nonspecific background staining was observed in glioblastoma tissues from HCMV seropositive and seronegative patients. It is known that glial cells with gemistocytic morphology frequently react in an unspecific manner with many antibodies. After careful review of many articles claiming high HCMV infection rates in human tumors of the nervous system, it became evident that only single cells with gemistocytic morphology displayed positive results for HCMV and the vast majority of surrounding cells remained largely unstained.26 Since it is not clear if these gemistocytic cells are of neoplastic or reactive origin, only tissue-based genetic analyses, such as in-situ hybridization techniques for amplifications or deletions and immunohistochemical methods for detecting tumor-specific mutated proteins, may confirm the neoplastic origin of gemistocytic cells.
Based on the very high fraction of HCMV-positive glioblastoma tissues detected by studies using optimized techniques, glioblastomas might be expected to be overrepresented among HCMV-seropositive individuals.8,10,24,27 However, previous studies did not detect a relationship between HCMV seroprevalence and glioblastoma incidence.28–30 These findings are consistent with our own findings that HCMV infections do not appear to be overrepresented among glioma patients. Currently, clinical studies are investigating HCMV as a target for the treatment of tumors of the nervous system. In one of the most recent data collections, glioblastoma patients who were reported to display HCMV-positive tumors and were treated with antiviral drugs showed unexpectedly long survival times.31 However, this study has severe shortcomings in both conception and realization; most importantly, however, it contradicted the controlled study published by the same group.32–34 A more recent study suggested that a combination of autologous HCMV-specific T cells and chemotherapy may be beneficial for glioblastoma patients.35 However, the tumors were not investigated for HCMV in that study. Thus, it remains unclear whether the therapeutic effects may be a direct consequence of targeting HCMV-positive cancer cells or whether they may have resulted from other (off-target) effects. Our results suggest that the latter may be more likely case. Nevertheless, the beneficial clinical effects of anti-HCMV therapy for cancer patients, which have been reported by different groups,31,35 also warrant attention if they do not exert their effects through the targeting of HCMV-infected cancer cells.
Taken together, our results indicate that HCMV cannot be detected in large fractions of human tumors of the nervous system, which corroborates long-standing pathological findings and current deep sequencing data.15,18,16 Notably, we have detected HCMV IEA by PCR in neuroblastoma tissues from 4 of 8 patients in previous experiments.36 In light of our current findings, it is likely that this was due to the invasion of HCMV-infected immune cells, such as monocytes/macrophages. In addition, correlation of clinicopathological parameters with HCMV serostatus indicates no association of HCMV infection with development or progression of glioblastomas. Both diagnostics of HCMV in tumors of the nervous system and earlier clinical trials focusing on HCMV as a target for antitumor treatment have to be (re-)evaluated with caution. It still remains unclear, however, whether HCMV may enhance malignancy and serve as a therapeutic target in rare cases of HCMV-infected cancers.
Funding
The work was supported by the Hilfe für krebskranke Kinder Frankfurt e.V., the Frankfurter Stiftung für krebskranke Kinder, and the Kent Cancer Trust.
Acknowledgments
We thank Maika Dunst and Cornelia Zachskorn for technical assistance.
Conflict of interest statement. The authors declare no conflict of interest.
References
- 1.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20(4):202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 2.Sinclair J. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J Clin Virol. 2008;41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- 4.Limaye AP, Kirby KA, Rubenfeld GD, et al. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA. 2008;300:413–422. doi: 10.1001/jama.300.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinatl J, Jr, Cinatl J, Vogel JU, et al. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39(4):259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 6.Michaelis M, Doerr HW, Cinatl J., Jr. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cinatl J, Jr, Vogel JU, Cinatl J, et al. Long-term productive human cytomegalovirus infection of a human neuroblastoma cell line. Int J Cancer. 1996;65(1):90–96. doi: 10.1002/(SICI)1097-0215(19960103)65:1<90::AID-IJC16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Cobbs CS, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62(12):3347–3350. [PubMed] [Google Scholar]
- 9.Baryawno N, Rahbar A, Wolmer-Solberg N, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2012;121(10):4043–4055. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dziurzynski K, Chang SM, Heimberger AB, et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012;14(3):246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnsen JI, Baryawno N, Söderberg-Nauclér C. Is human cytomegalovirus a target in cancer therapy? Oncotarget. 2011;2(12):1329–1338. doi: 10.18632/oncotarget.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157(2):193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolmer-Solberg N, Baryawno N, Rahbar A, et al. Frequent detection of human cytomegalovirus in neuroblastoma: a novel therapeutic target? Int J Cancer. 2013;133(10):2351–2361. doi: 10.1002/ijc.28265. [DOI] [PubMed] [Google Scholar]
- 14.Cosset E, Petty TJ, Dutoit V, et al. Comprehensive metagenomic analysis of glioblastoma reveals absence of known virus despite antiviral-like type I interferon gene response. Int J Cancer. 2014;135(6):1381–1389. doi: 10.1002/ijc.28670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau SK, Chen YY, Chen WG, et al. Lack of association of cytomegalovirus with human brain tumors. Mod Pathol. 2005;18(6):838–843. doi: 10.1038/modpathol.3800352. [DOI] [PubMed] [Google Scholar]
- 16.Sabatier J, Uro-Coste E, Pommepuy I, et al. Detection of human cytomegalovirus genome and gene products in central nervous system tumors. Br J Cancer. 2005;92(4):747–750. doi: 10.1038/sj.bjc.6602339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sehic D, Forslund O, Sandén E, et al. Absence of Epstein-Barr and cytomegalovirus infection in neuroblastoma cells by standard detection methodologies. Pediatr Blood Cancer. 2013;60(9):E91–E93. doi: 10.1002/pbc.24535. [DOI] [PubMed] [Google Scholar]
- 18.Tang KW, Alaei-Mahabadi B, Samuelsson T, et al. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita Y, Ito Y, Isomura H, et al. Lack of presence of the human cytomegalovirus in human glioblastoma. Mod Pathol. 2014;27(7):922–929. doi: 10.1038/modpathol.2013.219. [DOI] [PubMed] [Google Scholar]
- 20.Preiser W, Buxbaum S, Fleckenstein C, et al. Measurement of CVM DNA levels and CMV-specific CD4+ lymphocyte counts in patient specimens: Requirements for test quality and practicality in a routine diagnostic service. In: Prösch S, Cinatl J, Scholz M, editors. New Aspects of CMV-Related Immunopathology. Vol. 24. Karger: Basel; 2003. pp. 133–148. [Google Scholar]
- 21.Griscelli F, Barrois M, Chauvin S, et al. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J Clin Microbiol. 2001;39(12):4362–4369. doi: 10.1128/JCM.39.12.4362-4369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaelis M, Barth S, Breitling R, et al. Selection of a highly invasive neuroblastoma cell population through long-term human cytomegalovirus infection. Oncogenesis. 2012;1:e10. doi: 10.1038/oncsis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro SC, Hall B, Whybin LR, et al. Diagnosis of and screening for cytomegalovirus infection in pregnant women. J Clin Microbiol. 2005;43(9):4713–4718. doi: 10.1128/JCM.43.9.4713-4718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheurer ME, Bondy ML, Aldape KD, et al. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116(1):79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharjee B, Renzette N, Kowalik TF. Genetic analysis of cytomegalovirus in malignant gliomas. J Virol. 2012;86(12):6815–6824. doi: 10.1128/JVI.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell DA, Xie W, Schmittling R, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 2008;10(1):10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Söderberg-Nauclér C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–223. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Amirian ES, Marquez-Do D, Bondy ML, et al. Anti-human-cytomegalovirus immunoglobulin G levels in glioma risk and prognosis. Cancer Med. 2013;2(1):57–62. doi: 10.1002/cam4.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehrer S, Green S, Ramanathan L, et al. No consistent relationship of glioblastoma incidence and cytomegalovirus seropositivity in whites, blacks, and Hispanics. Anticancer Res. 2012;32(3):1113–1115. [PubMed] [Google Scholar]
- 30.Sjöström S, Hjalmars U, Juto P, et al. Human immunoglobulin G levels of viruses and associated glioma risk. Cancer Causes Control. 2011;22(9):1259–1266. doi: 10.1007/s10552-011-9799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Söderberg-Nauclér C, Rahbar A, Stragliotto G. Survival in patients with glioblastoma receiving valganciclovir. N Engl J Med. 2013;369(10):985–986. doi: 10.1056/NEJMc1302145. [DOI] [PubMed] [Google Scholar]
- 32.Stragliotto G, Rahbar A, Solberg NW, et al. Effects of valganciclovir as an add-on therapy in patients with cytomegalovirus-positive glioblastoma: a randomized, double-blind, hypothesis-generating study. Int J Cancer. 2013;133(5):1204–1213. doi: 10.1002/ijc.28111. [DOI] [PubMed] [Google Scholar]
- 33.Weller M, Soffietti R, Brada M. The legend of cytomegalovirus and glioblastoma lives on. Neuro Oncol. 2013;16(1):166. doi: 10.1093/neuonc/not204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick W, Wick A, Platten M. Good maths is needed to understand CMV data in glioblastoma. Neuro Oncol. 2014;16(1):165. doi: 10.1093/neuonc/not212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuessler A, Smith C, Beagley L, et al. Autologous T cell therapy for cytomegalovirus as a consolidative treatment for recurrent glioblastoma. Cancer Res. 2014;74(13):3466–3476. doi: 10.1158/0008-5472.CAN-14-0296. [DOI] [PubMed] [Google Scholar]
- 36.Michaelis M, Baumgarten P, Mittelbronn M, et al. Oncomodulation by human cytomegalovirus: novel clinical findings open new roads. Med Microbiol Immunol. 2011;200(1):1–5. doi: 10.1007/s00430-010-0177-7. [DOI] [PubMed] [Google Scholar]