Abstract
CYP3A5 plays a prominent role in the metabolism of maraviroc, an approved drug for human immunodeficiency virus (HIV)-1 treatment and a candidate for HIV-1 prevention. We studied the effect of the CYP3A5 genotype on pharmacokinetics of maraviroc and a primary CYP3A5-dependent metabolite of maraviroc denoted as metabolite 1 (M1). Volunteers were screened for health status and CYP3A5 genotype (wild-type allele *1 and dysfunctional alleles *2, *3, *6, and *7) to obtain 24 evaluable subjects in three groups (n = 8 each): homozygous dysfunctional (two dysfunctional alleles), heterozygous (one *1 allele and one dysfunctional allele), and homozygous wild-type (two *1 alleles). Subjects received 300 mg maraviroc orally followed by blood collection for 32 hours. The homozygous wild-type group exhibited lower mean plasma maraviroc concentrations at almost all sampling times. The median (interquartile range) maraviroc area under the plasma concentration-time curves from time 0 to infinity (AUC0–inf) were 2099 (1422–2568) ng⋅h/ml, 1761 (931–2640) ng⋅h/ml, and 1238 (1065–1407) ng⋅h/ml for the homozygous dysfunctional, heterozygous, and homozygous wild-type groups, respectively. The homozygous wild-type group had 41% lower maraviroc AUC0–inf and 66% higher apparent clearance compared with the homozygous dysfunctional group (P = 0.02). The AUC0–inf ratios of maraviroc to M1 in heterozygous and homozygous wild-type subjects were lower by 51 and 64% relative to the homozygous dysfunctional group, respectively (P < 0.001). In conclusion, the lower maraviroc concentrations in the homozygous wild-type group indicate that maraviroc may be underdosed in people homozygous for the CYP3A5*1 allele, including almost one-half of African Americans.
Introduction
Maraviroc is a U.S. Food and Drug Administration (FDA)–approved drug used in combination with other anti-human immunodeficiency virus (HIV) drugs for treatment of HIV-1 infection. It inhibits HIV-1 entry by blocking the interaction between viral envelope glycoprotein (gp120) and the chemokine (C-C motif) receptor 5 (CCR5), a coreceptor that HIV-1 uses to enter host cells (Dorr et al., 2005). HIV-1 strains predominantly use CCR5 as a coreceptor during the transmission stage and in the early stages of HIV disease (Michael et al., 1997; Philpott, 2003). As the only available CCR5 antagonist on the market, maraviroc has been considered as a potential candidate for HIV-1 pre-exposure prophylaxis since maraviroc penetrates into the male and female reproductive tracts and colorectal tissues (Dumond et al., 2009; Brown et al., 2011), the primary sites of transmission through sexual exposure, and has favorable safety profiles (Gulick et al., 2014). Maraviroc is currently being investigated for reducing the acquisition of HIV-1 infection with once-daily oral administration alone or in combination with other anti-HIV drugs in people who are at risk (ClinicalTrials.gov identifier NCT01505114).
After oral administration in humans, the absorption of maraviroc has been predicted to be approximately 84% at 300 mg (Abel et al., 2008e, 2009a). Maraviroc undergoes extensive metabolism via N-dealkylation and oxygenation, which produces several oxidative metabolites (Walker et al., 2005). N-dealkylation is mediated by CYP3A4 (Hyland et al., 2008). According to a mass balance study using a 14C-labeled dose of maraviroc, the N-dealkylated product was the major circulating metabolite in blood, and the N-dealkylated metabolite recovered in excreta accounted for 7% of the administered dose (Abel et al., 2008e). Although the abundance of each individual oxidative metabolite in plasma was less than that of the N-dealkylated metabolite, collectively, the oxygenated products accounted for approximately 40% of the administered maraviroc dose (Walker et al., 2005; Abel et al., 2008e, 2009a), suggesting that oxidative metabolism plays an important role in maraviroc elimination. Our previous in vitro metabolism study demonstrated that both CYP3A4 and CYP3A5 exhibited activity toward maraviroc (Lu et al., 2012); however, CYP3A5 played a predominant role in the formation of an abundant oxidative metabolite denoted as metabolite 1 (M1), resulting from oxygen insertion on the difluorocyclohexane ring of maraviroc (Supplemental Fig. 1), with a 23-fold higher maximum formation rate and 4-fold higher intrinsic clearance compared with CYP3A4 (Lu et al., 2012).
The involvement of CYP3A5 in maraviroc oxidative metabolism indicates that maraviroc pharmacokinetics may be affected by CYP3A5 genetic polymorphisms that determine the expression and/or activity of CYP3A5. The CYP3A5*1 allele is referred to as the wild-type and it is associated with the highest CYP3A5 protein expression, whereas alternative alleles CYP3A5*3, CYP3A5*6, and CYP3A5*7 are considered to be nonfunctional or dysfunctional since the single-nucleotide alterations in these alleles lead to lower expression or no activity of CYP3A5 (Hustert et al., 2001; Kuehl et al., 2001; Lamba et al., 2002; Lin et al., 2002). In individuals who express CYP3A5, CYP3A5 is present in the intestines (Kolars et al., 1994) and liver with CYP3A5 content comprising up to greater than 50% of total hepatic CYP3A (Kuehl et al., 2001). Furthermore, CYP3A5 expression varies greatly with ethnicity due to differences in allele frequency distribution. CYP3A5*3 is abundantly present in European Americans, with 80–90% being CYP3A5 nonexpressors (van Schaik et al., 2002; Daly, 2006). By contrast, the majority of African Americans are CYP3A5 expressors due to carrying at least one wild-type CYP3A5*1 allele and approximately 45% are homozygous CYP3A5*1 carriers (Xie et al., 2004).
Maraviroc pharmacokinetic variations may lead to changes in antiviral efficacy since the currently recommended maraviroc dosing regimen delivers concentrations near the top but still within the steep portion of the exposure-response curve (FDA, 2007; Jacqmin et al., 2013). Maraviroc plasma trough concentrations (Cmin) and average concentrations (Cave) have been used for exposure-response analysis. Although maximal antiviral efficacy is achieved when Cmin and Cave are above 50 ng/ml and 100 ng/ml (Jacqmin et al., 2013), respectively, most studies had Cmin below 43 ng/ml with the recommended regimen (FDA, 2007). There is currently limited information on the pharmacokinetics of maraviroc in individuals with different CYP3A5 genotypes, which is not only necessary to understand pharmacokinetic variations among people who are on maraviroc for treatment of HIV-1 infection but is also extremely important to know if maraviroc is going to be used alone for HIV pre-exposure prophylaxis among people who are at high risk for HIV-1 infection. Therefore, we performed this clinical study to investigate the impact of CYP3A5 polymorphism on pharmacokinetics of maraviroc and the primary CYP3A5-dependent metabolite M1.
Materials and Methods
Study Design.
This was an open-label, single-dose study. Inclusion criteria included age 18–65 years, healthy with no acute medical illness, no evidence of hepatic or renal impairment (liver function tests values < 1.5 upper limit of normal, total bilirubin below the upper limit of normal, creatinine clearance > 60 ml/min), and negative HIV test results. Any concomitant medications or herbal supplements for which there is a known risk of pharmacokinetic or pharmacodynamics drug interactions, including those that inhibit CYP3A family enzymes, were prohibited for 4 weeks prior to and during the study. The study was reviewed and approved by the institutional review board of the Johns Hopkins Medical Institutions and was conducted in compliance with national and local institutional guidelines. All participants gave written informed consent.
Volunteers were screened for health status and genotyped for CYP3A5 (*1, *2, *3, *6, and *7). Healthy volunteers were enrolled into three groups (n = 8 each): homozygous dysfunctional (two dysfunctional alleles), heterozygous (one CYP3A5*1 allele and one dysfunctional allele), and homozygous wild-type (two CYP3A5*1 alleles). A single dose of a 300-mg maraviroc tablet was given to the volunteers orally along with 200 ml water under direct observation. Ten milliliters of venous blood was collected before (0 hours) and 1, 2, 3, 4, 6, 8, 10, 24, and 32 hours after the drug administration. Plasma was prepared by centrifugation of blood at 1500g for 10 minutes at 4°C and stored at −80°C until analysis.
CYP3A5 Genotyping Procedure.
Genomic DNA was extracted from blood samples using the QIAamp DNA Blood Mini Kit (Qiagen Inc, Valencia, CA). The following previously reported primers were used for amplification of polymorphic loci-containing fragments: forward primer 5′-CCTGCCTTCAATTTTTCACTG-3′ and reverse primer 5′-GCAATGTAGGAAGGAGGGCT-3′ for *3 (Kuehl et al., 2001), forward primer 5′-GCTGCATGTAT AGTGGAAGGAC-3′ and reverse primer 5′-GGAATTGTACCTTTTAAGTGGATG-3′ for *6 (Floyd et al., 2003), and forward primer 5′-AAATACTTCA CGAATACTATGATC-3′ and reverse primer 5′-CAGGGACATAATTGATTATCTTTG-3′ for *7 (Chou et al., 2001). iProof High-Fidelity DNA polymerase (Bio-Rad, Hercules, CA) was used for polymerase chain reactions (PCRs) with the following program: 1) 98°C for 30 seconds; 2) 98°C for 10 seconds; 3) 65.2°C for CYP3A5*3, 59.0°C for CYP3A5*6, and 57.8°C for CYP3A5*7; 4) 72°C for 45 seconds; 5) repeat steps 2–4 for 35 cycles; and 6) 72°C for 10 minutes. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Inc.) followed by Sanger sequencing for CYP3A5 genotype determination. Nonfunctional allele CYP3A5*2 with a nucleotide transition from C to A (van Schaik et al., 2002), located in the fragment of PCR product amplified for *7, was also examined from the sequenced PCR product.
Measurement of Plasma Concentrations of Maraviroc and M1.
The structures of maraviroc and M1 are shown in Supplemental Fig. 1. Plasma maraviroc concentrations were determined using a validated ultra-performance liquid chromatography-tandem mass spectrometry method as previously reported (Emory et al., 2014), with inter- and intra-assay precision of ≤5.4 and ≤6.0%, and inter- and intra-assay accuracy within ±10.2 and ±8.4%, respectively. The validation results exceeded standards recommended by the FDA (2013). M1 was quantitated using the calibration curve of 4-hydroxyphenyl maraviroc, which had the same precursor/daughter mass to charge (m/z) ratios with M1. Fifty microliters of plasma was mixed with 50 µl of 10 ng/ml internal standard [2H6]maraviroc followed by the addition of 500 µl acetonitrile and filtration by applying a vacuum. Filtrates were dried and reconstituted in 75 µl of mobile phases A (5% acetonitrile/0.1% formic acid in water) and B (5% water/0.1% formic acid in acetonitrile). Ten microliters was injected onto a Waters Acquity BEH C8 ultra-performance liquid chromatography column (1.7 µm, 50 × 2.1 mm) with a flow rate of 1 ml/min and analytes were detected over 8 minutes using a QTRAP5500 Quadrupole-Linear Ion Trap mass spectrometer (AB SCIEX, Framingham, MA). Mobile phases were programmed at 4% B from 0 to 0.5 minutes, 6% B at 4.0 minutes, 15% B from 4.1 to 5 minutes, 100% B from 5.1 to 6.5 minutes, and 4% B from 7 to 8 minutes. The parent/daughter ion transitions monitored were as follows: M1 and 4-hydroxyphenyl maraviroc, m/z 530.2 > 405.2; and [2H6]maraviroc, m/z 520.6 > 389.1. The calibration ranges were linear from 0.5 to 1000 ng/ml and 0.25 to 100 ng/ml for maraviroc and M1, respectively.
Pharmacokinetic and Statistical Analyses.
Pharmacokinetic parameters of maraviroc and M1 including the area under the concentration-time curve from time 0 to infinity (AUC0–inf), area under the curve from time 0 to 32 hours (AUC0–32), plasma peak concentration (Cmax), time to reach Cmax (Tmax), half-life (t1/2), and oral apparent clearance (CL/F) were estimated by WinNonlin software (version 6.1; Pharsight, Cary, NC) by performing noncompartmental analysis. Previous studies have shown that maraviroc pharmacokinetics is time independent with the AUC0-inf of maraviroc after single doses having similar values to the maraviroc area under the curve (AUC) during a dosing interval (12 hours) at steady state with the currently recommended dosing regimen (Abel et al., 2008f, 2009b). The predicted Cave values for multiple dosing maraviroc were estimated by AUC0–inf divided by 12 hours. The sample size of the study was estimated using power analysis based on our previous observation of 34% of coefficient of variation for the major oxidative metabolite formation activity in human liver microsomes to detect a 50% difference (with a 5% type I error for a two-sided test and 80% power) in the AUC of maraviroc between any two of the three CYP3A5 genotypic groups. Pharmacokinetic parameters among the three groups were compared using the Kruskal–Wallis test in R software (version 2.15.1; R Project for Statistical Computing, Vienna, Austria). Differences between the groups with different CYP3A5 genotypes were assessed using the Mann–Whitney–Wilcoxon test. P < 0.05 was considered statistically significant. Linear regression was performed in SigmaPlot (version 12.3; Systat Software, Inc., San Jose, CA).
Results
Genotyping Results and Subject Demographics.
A total of 34 subjects were evaluated for CYP3A5 genotype screening. Fourteen subjects were found to be homozygous dysfunctional (41.2%; 10 CYP3A5*3/*3, 2 CYP3A5*3/*6, 1 CYP3A5*6/*7, and 1 CYP3A5*7/*7), 10 were heterozygous (29.4%; 7 CYP3A5*1/*3, 2 CYP3A5*1/*6, and 1 CYP3A5*1/*7), and 10 were homozygous wild-type (29.4%; CYP3A5*1/*1). No CYP3A5*2 was found. We enrolled 25 healthy volunteers but 1 had no measurable maraviroc and M1 in all samples. For the 24 evaluable participants, the homozygous dysfunctional group had 5 CYP3A5*3/*3, 2 CYP3A5*3/*6, and 1 CYP3A5*6/*7; the heterozygous group had 7 CYP3A5*1/*3 and 1 CYP3A5*1/*6; and the homozygous wild-type group had 8 CYP3A5*1/*1.
The mean ± S.D. age and weight of the subjects were 41 ± 11 years and 78.2 ± 15.1 kg, respectively. Among the 24 volunteers, 17 were African Americans, 6 were European Americans, and 1 was Asian. Age, weight, body mass index, creatinine clearance, total protein, albumin, bilirubin, and aspartate transaminase activity were similar among different genotypic groups (Table 1). Alkaline phosphatase and alanine transaminase were lower in the homozygous wild-type group compared with the other two groups, but the differences were not statistically significant. There were six females, with two, one, and three of them distributing in homozygous dysfunctional, heterozygous, and homozygous wild-type groups, respectively.
TABLE 1.
Demographic information of the healthy volunteers
Data are presented as the mean ± S.D.
| Group | No CYP3A5*1 (n = 8) | One CYP3A5*1 (n = 8) | Two CYP3A5*1 (n = 8) |
|---|---|---|---|
| Age (yr) | 45.1 ± 12.7 | 39.4 ± 11.7 | 38.0 ± 8.8 |
| Weight (kg) | 84.0 ± 15.6 | 76.7 ± 14.9 | 73.8 ± 14.5 |
| Body mass index (kg/m2)a | 26.4 ± 4.5 | 24.9 ± 3.6 | 23.8 ± 3.6 |
| Creatine clearance (ml/min)b | 114.3 ± 28.8 | 103.2 ± 15.2 | 100.1 ± 30.2 |
| Total protein (g/dl) | 7.0 ± 0.4 | 7.5 ± 0.4 | 7.1 ± 0.3 |
| Albumin (g/dl) | 4.6 ± 0.3 | 4.6 ± 0.1 | 4.5 ± 0.3 |
| Bilirubin (mg/dl) | 0.5 ± 0.2 | 0.4 ± 0.3 | 0.5 ± 0.2 |
| Alkaline phosphatase (IU/L) | 73.3 ± 16.7 | 74.9 ± 17.0 | 54.9 ± 14.3 |
| Alanine transaminase (IU/L)b | 17.5 ± 4.5 | 18.5 ± 6.8 | 13.9 ± 3.4 |
| Aspartate transaminase (IU/L) | 19.6 ± 2.9 | 22.4 ± 4.9 | 20.0 ± 2.9 |
| Female/male volunteers (n) | 2/6 | 1/7 | 3/5 |
Calculated as weight/(height × height).
Calculated as [(140 − age [yr]) × weight (kg)]/[72 × serum creatinine concentration (mg/dl)] (multiply by 0.85 for female sex).
Effect of CYP3A5 Polymorphism on the Pharmacokinetics of Maraviroc and M1.
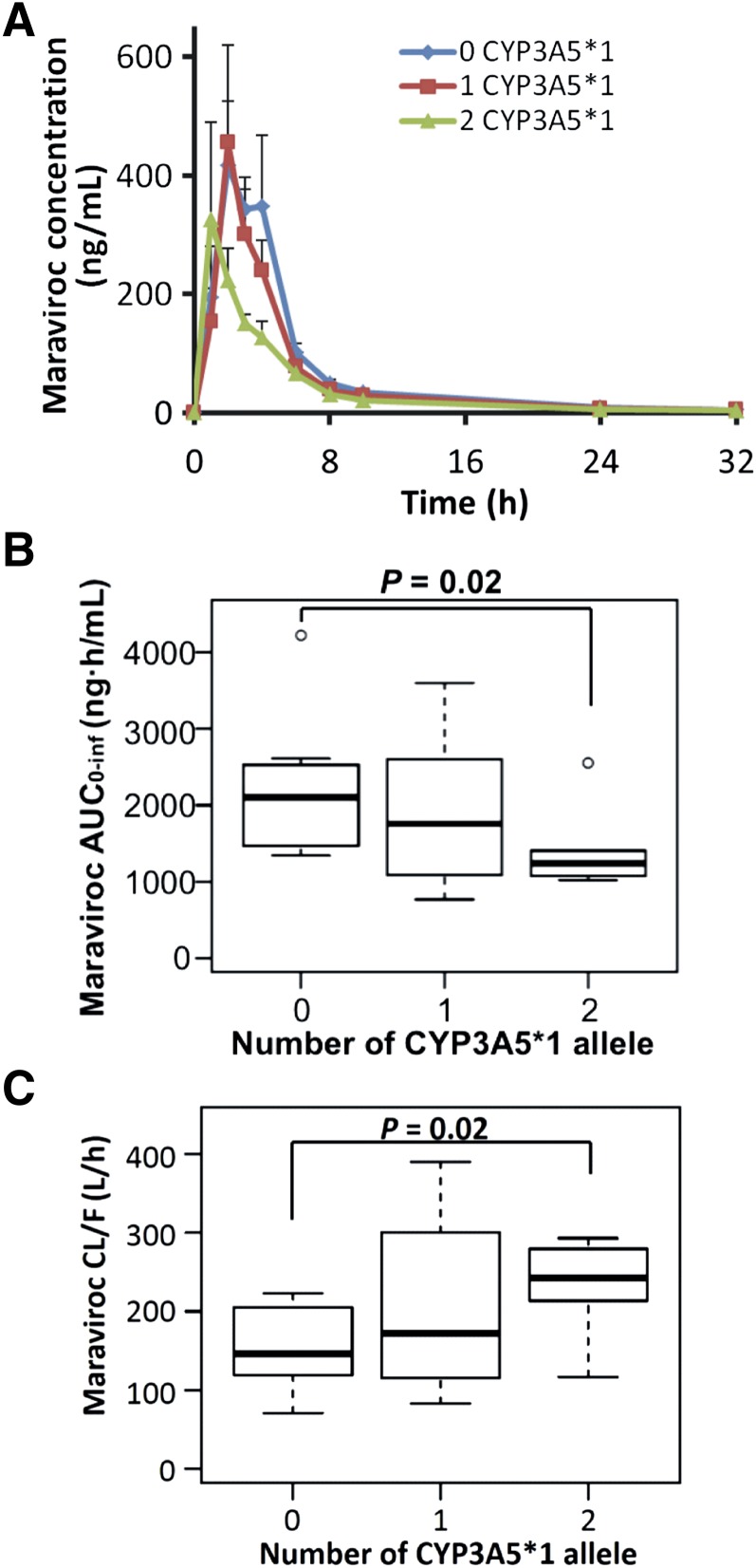
The maraviroc concentration-time profiles of the homozygous dysfunctional group were similar to those of the heterozygous group (Fig. 1A). By contrast, the homozygous wild-type group exhibited lower mean plasma maraviroc concentrations at almost all blood collection times. Compared with the homozygous dysfunctional group, the homozygous wild-type group had 41% lower maraviroc AUC0–inf (P = 0.02; Fig. 1B), 66% higher CL/F (P = 0.02; Fig. 1C), and 33% lower but not significantly different Cmax (P = 0.33). The heterozygous group exhibited large interindividual variability in AUC0–inf and CL/F, which was not statistically different from either of the other two groups. The medians and interquartile ranges (IQRs) of pharmacokinetic parameters of maraviroc and M1 are shown in Table 2. Tmax and t1/2 were not statistically different among the three groups. The percentage of the extrapolated AUC from the time of the last measurable concentration to infinity is small, as reflected by the values of AUC0–32 being very close to values of AUC0–inf. Using values of AUC0–inf from this single-dose maraviroc study, we estimated values of maraviroc Cave at steady state if maraviroc was administered for multiple doses. The medians of Cave in the homozygous dysfunctional and heterozygous groups were well above 100 ng/ml, the threshold value below which there is an increased risk of virological failure. In comparison, the median of predicted Cave in the CYP3A5 homozygous wild-type group dropped to the threshold at a value of 103 ng/ml, and a one-half of people in this group (4 of 8) had Cave falling below 100 ng/ml (Table 2).
Fig. 1.
Reduced maraviroc exposure and increased clearance in individuals homozygous for the wild-type CYP3A5*1 allele. (A) Mean (± S.E.) plasma concentrations of maraviroc after administration of a single oral dose of 300 mg maraviroc. (B and C) Box plots of maraviroc AUC0–inf (B) and oral apparent clearance (C) (n = 8 in each group).
TABLE 2.
Pharmacokinetic parameters of maraviroc and metabolite M1
Data are presented as the median (IQR).
| Group | No CYP3A5*1 (n = 8) | One CYP3A5*1 (n = 8) | Two CYP3A5*1 (n = 8) |
|---|---|---|---|
| Maraviroc | |||
| Cmax (ng/ml) | 505 (348–959) | 435 (210–889) | 339 (237–552) |
| Tmax (h) | 2.5 (2.0–3.8) | 2.0 (1.2–3.0) | 2.0 (1.0–3.5) |
| AUC0–32 (ng⋅h/ml) | 2056 (1454–2416) | 1718 (1200–2513) | 1180 (1038–1350)* |
| AUC0-inf (ng⋅h/ml) | 2099 (1422–2568) | 1761 (931–2640) | 1238 (1065–1407)* |
| t1/2 (h) | 7.6 (7.1–8.7) | 7.4 (6.5–10.8) | 8.7 (8.0–9.7) |
| CL/F (l/h) | 146 (117–212) | 173 (114–343) | 243 (213–282)* |
| Cave (ng/ml)a | 175 (126–207) | 147 (103–214) | 103 (90–117)* |
| Cave <100 ng/mlb | 0% (0/8) | 25% (2/8) | 50% (4/8) |
| M1 | |||
| Cmax (ng/ml) | 10.1 (6.5–18.5) | 20.7 (7.6–30.1) | 15.9 (15.2–18.9) |
| Tmax (h) | 3.0 (2.3–3.8) | 2.0 (1.3–3.0) | 2.0 (1.0–2.0) |
| AUC0–32 (ng⋅h/ml) | 40.4 (36.0–48.2) | 90.1 (49.2–117.9) | 61.0 (56.2–70.6)* |
| AUC0–inf (ng⋅h/ml) | 41.1 (32.2–53.1) | 91.9 (38.7–133.8) | 64.5 (57.4–82.9)* |
| AUC0–inf ratio (marviroc/M1) | 49.5 (35.3–64.4) | 24.0 (19.8–25.7)*** | 17.8 (15.7–22.5)*** |
Plasma Cave values for multiple dosing maraviroc at steady state were estimated as AUC0–inf of maraviroc after a single dose of 300 mg maraviroc divided by the dosing interval (12 hours) of the current recommended maraviroc dosing regimen for HIV-1 treatment.
Percentage of individuals who had Cave below 100 ng/ml.
P < 0.05; ***P < 0.001, compared with homozygous dysfunctional group of individuals who did not carry a wild-type allele CYP3A5*1.
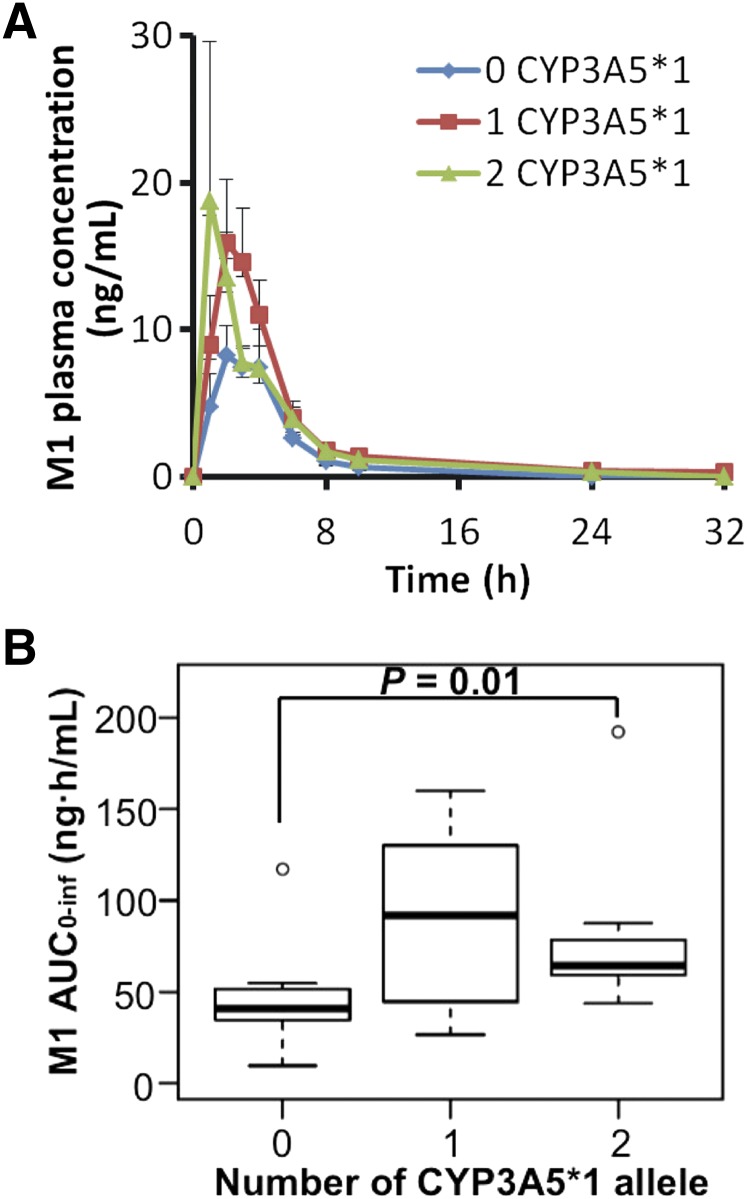
For metabolite M1, the plasma concentrations are lower in the homozygous dysfunctional group compared with the other two groups (Fig. 2A). The homozygous wild-type group exhibited 57% higher M1 AUC0–inf compared with the homozygous dysfunctional group (P = 0.01), whereas the heterozygous group was not statistically different from the homozygous dysfunctional group (P = 0.10) (Fig. 2B). The M1 Cmax was not different among the three genotypic groups. Median Tmax in the homozygous wild-type group was 1 hour earlier than in the homozygous dysfunctional group, although it was not statistically significant (P = 0.06).
Fig. 2.
The CYP3A5 homozygous dysfunctional group has reduced plasma M1 concentrations. (A) Mean (± S.E.) plasma concentrations of M1 after maraviroc administration. (B) AUC0–inf of metabolite M1 (n = 8 for each of the three groups).
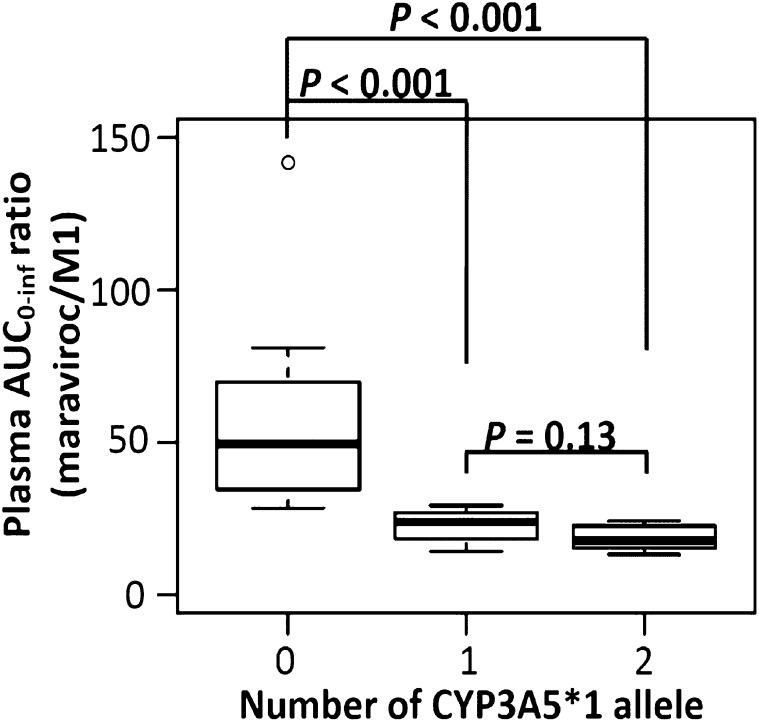
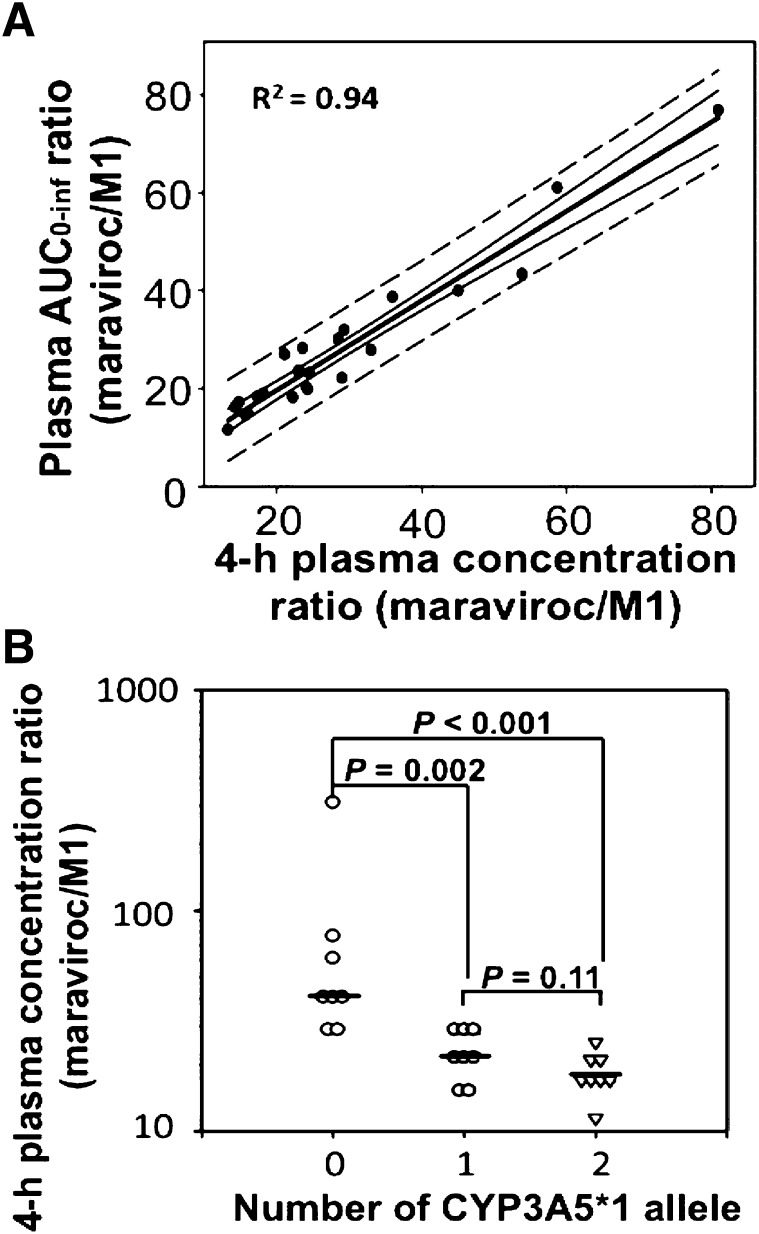
The influence of the CYP3A5 genotype was more evident in the AUC0–inf ratio of maraviroc to metabolite M1 (AUCmaraviroc/AUCM1). The values of AUCmaraviroc/AUCM1 were statistically significant among the three groups (Kruskal–Wallis test, P < 0.001). Compared with the homozygous dysfunctional group that had a median AUCmaraviroc/AUCM1 at 49.5 (IQR, 35.3–64.4), the homozygous wild-type group and the heterozygous group were 64 and 52% lower with a median at 17.8 (IQR, 15.7–22.5) and 24.0 (IQR, 19.8–25.7), respectively, and the differences were both statistically significant (P < 0.001; Fig. 3; Table 2). To identify a simple method to assess the maraviroc/M1 ratio using a single blood sample, the correlation between AUC0–inf ratios and plasma concentration ratios of maraviroc to M1 was examined at all sample collection times and the strongest correlation was observed at 4 hours (R2 = 0.94; P < 0.001; Fig. 4A). Similar to the AUC0–inf ratio, the 4-hour plasma concentration ratios in the homozygous wild-type and heterozygous groups were 46% (P = 0.001) and 56% (P < 0.001) lower compared with the homozygous dysfunctional group (Fig. 4B).
Fig. 3.
Carrying the CYP3A5*1 allele caused a reduction of the AUCmaraviroc/AUCM1 ratio. Statistical testing was performed using the Wilcoxon rank sum test in R software. Bonferroni post hoc test significance level of P < 0.017.
Fig. 4.
Four-hour plasma concentration ratios were strongly correlated with plasma AUCmaraviroc/AUCM1 ratios. (A) Scatter plot of AUC0–inf ratios against 4-hour plasma concentration ratios with fitted linear regression line (middle thick black line), 95% confidence interval (next outer curved lines), and the prediction band (broken lines). An extreme value (x = 310; y = 142) from an individual with genotype of CYP3A5*3/*3 was excluded (R2 = 0.87 when included). (B) CYP3A5 genotype effect on the 4-hour plasma concentration ratio of maraviroc to M1. The horizontal lines represent medians.
Discussion
Since we previously showed that CYP3A5 played a substantial role in the oxidative metabolism of maraviroc (Lu et al., 2012), we conducted this study to evaluate the impact of the CYP3A5 genotype on the pharmacokinetics of maraviroc and the primary CYP3A5 metabolite M1 after an oral dose of maraviroc in healthy volunteers. Maraviroc has time-independent pharmacokinetics with minimal accumulation in AUC at steady state compared with that after a single dose (Abel et al., 2008f, 2009b) and HIV-1 infection status does not change maraviroc pharmacokinetics (FDA, 2007). Therefore, data from this study are relevant to the clinical situations in which maraviroc is administrated twice daily in HIV-1–infected patients for treatment and for the potential use in HIV-1 prevention. The values of maraviroc Cmax and AUC in the CYP3A5 homozygous dysfunctional group from this study are comparable to those reported in previous maraviroc pharmacokinetic studies in which most participants were of European ancestry (Abel et al., 2008a,f; Pozniak et al., 2008).
Here, we found that compared with the CYP3A5 homozygous dysfunctional group, maraviroc AUC0–inf in the CYP3A5 homozygous wild-type group decreased by 41%. The reduction is similar in magnitude to the decrease (45%) in the maraviroc AUC caused by the concomitant use of CYP3A4 inducer, efavirenz. As a result, the maraviroc package inserts recommend doubling the maraviroc dose when coadministered with efavirenz (FDA, 2007; Abel et al., 2008a, 2009a). Furthermore, the median maraviroc Cave in the CYP3A5 homozygous wild-type (103 ng/ml) was very near the targeted Cave value associated with virological success (100 ng/ml). This indicates that nearly one-half of those individuals with wild-type homozygous CYP3A5 alleles fall below the target concentration for treatment.
CYP3A, including both CYP3A4 and CYP3A5, played a predominant role in maraviroc metabolism (Lu et al., 2012; Tseng et al., 2014). Hepatic metabolism has been thought to be responsible for nonrenal clearance, which accounts for 77% of maraviroc total clearance (Abel et al., 2008e, 2009a). Previous clinical studies have shown that CYP3A inhibitors such as ketoconazole and HIV-1 protease inhibitors significantly increased maraviroc exposure (Abel et al., 2008b). Among the HIV-1 protease inhibitors, ritonavir, saquinavir, and nelfinavir exhibit nonselective inhibition toward CYP3A4 and CYP3A5, whereas amprenavir and indinavir preferentially inhibit CYP3A4 with a weaker inhibitory effect on CYP3A5 (Granfors et al., 2006). If maraviroc is used in combination with drugs that are not CYP3A inhibitors for HIV-1 treatment or is used alone for HIV-1 pre-exposure prophylaxis, a difference in maraviroc exposure between the CYP3A5 homozygous dysfunctional and homozygous wild-type groups is expected based on our study results. If maraviroc is used concurrently with drugs that selectively inhibit CYP3A4, the difference in maraviroc exposure between the CYP3A5 homozygous dysfunctional and homozygous wild-type groups could be even greater than what we observed in this study. Lower maraviroc exposure in the CYP3A5 homozygous wild-type group may indicate the potential for failure of HIV-1 treatment or prevention and the development of drug resistance.
The reduced maraviroc exposure in homozygous carriers for CYP3A5*1 is an impactful finding since CYP3A5 is more frequently expressed in people with African ancestry, especially when combined with the fact that African Americans are disproportionately infected with HIV-1 and are at greater risk of acquiring HIV-1 infection compared with European Americans (Moore, 2011). Maraviroc was approved based on the MOTIVATE trials comprised of 85% individuals with European ancestry (Gulick et al., 2008) and most previous maraviroc pharmacokinetic studies were conducted in participants with European ancestry, with the median percentage of participants with European ancestry at 78.5% (Abel et al., 2008a,b,c,d,f; Chan et al., 2008; Pozniak et al., 2008; Dumond et al., 2009; Andrews et al., 2010; Ramanathan et al., 2010; Brown et al., 2011; Kakuda et al., 2011; Gruber et al., 2013; Mora-Peris et al., 2013; Taiwo et al., 2013; Vourvahis et al., 2013). A few studies compared maraviroc concentrations in individuals with African ancestry with individuals with European ancestry and concluded that there were no differences or higher concentrations in individuals with African ancestry (FDA, 2007; Okoli et al., 2012), which may be due to the use of CYP3A inhibitors such as ritonavir in the optimized background therapy in which CYP3A5 activity may have been inhibited in individuals who are carrying CYP3A5*1 allele. Nevertheless, a recent maraviroc exposure-response relationship assessment suggested that ethnicity is a prognostic factor of virologic success in the final model, with an odds ratio of 0.35 for people with African ancestry versus European ancestry when controlling for other prognostic factors (Jacqmin et al., 2013). Our finding of lower maraviroc exposure in homozygous CYP3A5*1 allele carriers indicates that people with this genotype may be at risk of HIV-1 treatment failure. Maraviroc concentrations associated with HIV-1 prevention have not been established, but this pharmacokinetic difference causing lower maraviroc concentrations in many African Americans warrants caution in future development of maraviroc as HIV-1 pre-exposure prophylaxis.
The absolute bioavailability of a 100-mg oral dose of maraviroc was 23% and has been predicted to be 33% at 300 mg (Abel et al., 2009a). The low bioavailability may be due to extensive presystemic first-pass intestine and hepatic metabolism. In this study, we found that oral apparent clearance increased but t1/2 did not change in the CYP3A5 homozygous wild-type group compared with the CYP3A5 homozygous dysfunctional group. Since CYP3A5 is present in the gut (Kivistö et al., 1996) and represents up to more than 50% of total CYP3A content in liver (Kuehl et al., 2001) in individuals who express CYP3A5, we suspect that the reduced maraviroc AUC in the CYP3A5 homozygous wild-type group may be a result of decreased bioavailability due to enhanced CYP3A5-mediated maraviroc oxidative metabolism in the gut and liver of those with this genotype, which was suggested by the lower AUCmaraviroc/AUCM1 ratio compared with the homozygous dysfunctional group. The higher AUCmaraviroc/AUCM1 ratio in the homozygous dysfunctional group is concordant with our previous in vitro observation that CYP3A5 was the major enzyme responsible for M1 formation. Besides M1, CYP3A5 was involved in the formation of several other oxidative metabolites (M2–M6) (Lu et al., 2012), which we did not measure in this study. Therefore, the CYP3A5 activity toward maraviroc may, at least partially, contribute to the reduced maraviroc AUC0–inf in the CYP3A5 homozygous wild-type group. Investigations of the pharmacokinetics of other CYP3A5 substrate drugs such as tacrolimus have also suggested that CYP3A5 polymorphism could affect drug bioavailability (Asberg et al., 2013). In addition, we observed a strong correlation between AUC0–inf ratios and 4-hour plasma concentration ratios of maraviroc to M1 in this study, which indicates that measuring concentration ratios of maraviroc/M1 at 4 hours after an oral maraviroc dose may be used as a simple method to predict the AUCmaraviroc/AUCM1 ratios to evaluate the CYP3A5 genotypic effect on maraviroc pharmacokinetics if intensive samplings are impractical under certain circumstances.
Previous maraviroc clinical studies demonstrated large interpatient pharmacokinetic variability (50% coefficient of variation) for maraviroc (FDA, 2007); however, the reasons were not clear. Our results suggest that the CYP3A5 genotype contributes to the variability of maraviroc exposure. We observed large variability in maraviroc AUC0–inf in the heterozygous group. The assignment of a wild-type allele was based on the absence of the common dysfunctional alleles CYP3A5*2, CYP3A5*3, CYP3A5*6, and CYP3A5*7. Besides these variants, other CYP3A5 nonfunctional alleles have been found with much lower frequency (van Schaik et al., 2002; Lee and Goldstein, 2005); however, we did not screen for these alleles in this study. Maraviroc pharmacokinetic variations could also be introduced by CYP3A4*22, an allele with 5.3% frequency in European Americans correlating with decreased CYP3A4 activity (Wang et al., 2011; Elens et al., 2013), or induction/inhibition of CYP3A activity by food, juice, unknown herbal supplements, or previous medications. In addition, maraviroc is a substrate for P-glycoprotein (also known as multidrug-resistance protein 1) (Walker et al., 2005), a drug efflux pump located on intestinal epithelial cells that limits drug absorption, and organic anion transporting polypeptide 1B1 (Siccardi et al., 2010), an influx transporter located on the basolateral membrane of hepatocytes mediating hepatic uptake of drugs. The polymorphism-dependent expression of these transporters has been reported to affect maraviroc plasma concentrations (Hoffmeyer et al., 2000; Schaeffeler et al., 2001; Walker et al., 2005; Fung and Gottesman, 2009; Siccardi et al., 2010). The polymorphisms in these genes, which we did not evaluate in this study, may also contribute to the maraviroc pharmacokinetic variations.
This investigation involved a small number of subjects in each of the three CYP3A5 genotypic groups (n = 8). The results merit confirmation via studies with a larger sample size.
In conclusion, we observed a reduction of maraviroc AUC0–inf almost by one-half and 66% higher maraviroc oral clearance in the CYP3A5 homozygous wild-type group. These findings raise concern that maraviroc may be underdosed in people who are homozygous for the CYP3A5*1 allele, which includes nearly one-half of all African Americans, a population with a disproportionately large incidence of HIV-1 infection.
Supplementary Material
Acknowledgments
The authors thank all of the study participants for their time and dedication. The authors also thank the Drug Development Unit and the Clinical Pharmacology Analytical Laboratory in the Division of Clinical Pharmacology at Johns Hopkins University, especially Jennifer Breakey, Mark Marzinke, and Joshua Emory, for their effort and support.
Abbreviations
- AUC
area under the curve
- AUC0–inf
area under the concentration-time curve from time 0 to infinity
- AUC0–32
area under the curve from time 0 to 32 hours
- AUCmaraviroc/AUCM1
AUC0–inf ratio of maraviroc to metabolite M1
- Cave
plasma average concentration
- CCR5
chemokine (C-C motif) receptor 5
- CL/F
apparent oral clearance (clearance divided by bioavailability F)
- Cmin
plasma trough concentration
- FDA
U.S. Food and Drug Administration
- HIV
human immunodeficiency virus
- IQR
interquartile range
- m/z
mass to charge
- M1
metabolite 1
- PCR
polymerase chain reaction
- Tmax
time to reach Cmax
- t1/2
half-life
Authorship Contributions
Participated in research design: Lu, Fuchs, Hendrix, Bumpus.
Conducted experiments: Lu, Fuchs, Hendrix, Bumpus.
Performed data analysis: Lu, Hendrix, Bumpus.
Wrote or contributed to the writing of the manuscript: Lu, Fuchs, Hendrix, Bumpus.
Footnotes
This research was supported by the Johns Hopkins Institute for Clinical and Translational Research (ICTR), which is funded in part by the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (NCATS) [Grant UL1TR001079], a component of the NIH, and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Johns Hopkins ICTR, NCATS, or NIH. The AB SCIEX QTRAP5500 Quadrupole-Linear Ion Trap console was purchased with proceeds from the NIH National Center for Research Resources [Grant 1S10RR27733 (to W.C.H.)]. The Waters Acquity ultra-performance liquid chromatograph interfaced with mass spectrometry was purchased with funds from Pendleton Enterprises [(to C.W.H.)].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abel S, Back DJ, Vourvahis M. (2009a) Maraviroc: pharmacokinetics and drug interactions. Antivir Ther 14:607–618 [PubMed] [Google Scholar]
- Abel S, Davis JD, Ridgway CE, Hamlin JC, Vourvahis M. (2009b) Pharmacokinetics, safety and tolerability of a single oral dose of maraviroc in HIV-negative subjects with mild and moderate hepatic impairment. Antivir Ther 14:831–837 [DOI] [PubMed] [Google Scholar]
- Abel S, Jenkins TM, Whitlock LA, Ridgway CE, Muirhead GJ. (2008a) Effects of CYP3A4 inducers with and without CYP3A4 inhibitors on the pharmacokinetics of maraviroc in healthy volunteers. Br J Clin Pharmacol 65 (Suppl 1):38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Russell D, Taylor-Worth RJ, Ridgway CE, Muirhead GJ. (2008b) Effects of CYP3A4 inhibitors on the pharmacokinetics of maraviroc in healthy volunteers. Br J Clin Pharmacol 65 (Suppl 1):27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Russell D, Whitlock LA, Ridgway CE, Muirhead GJ. (2008c) Effect of maraviroc on the pharmacokinetics of midazolam, lamivudine/zidovudine, and ethinyloestradiol/levonorgestrel in healthy volunteers. Br J Clin Pharmacol 65 (Suppl 1):19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Russell D, Whitlock LA, Ridgway CE, Muirhead GJ. (2008d) The effects of cotrimoxazole or tenofovir co-administration on the pharmacokinetics of maraviroc in healthy volunteers. Br J Clin Pharmacol 65 (Suppl 1):47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Russell D, Whitlock LA, Ridgway CE, Nedderman AN, Walker DK. (2008e) Assessment of the absorption, metabolism and absolute bioavailability of maraviroc in healthy male subjects. Br J Clin Pharmacol 65 (Suppl 1):60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, van der Ryst E, Rosario MC, Ridgway CE, Medhurst CG, Taylor-Worth RJ, Muirhead GJ. (2008f) Assessment of the pharmacokinetics, safety and tolerability of maraviroc, a novel CCR5 antagonist, in healthy volunteers. Br J Clin Pharmacol 65 (Suppl 1):5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews E, Glue P, Fang J, Crownover P, Tressler R, Damle B. (2010) Assessment of the pharmacokinetics of co-administered maraviroc and raltegravir. Br J Clin Pharmacol 69:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asberg A, Midtvedt K, van Guilder M, Storset E, Bremer S, Bergan S, Jelliffe R, Hartmann A, Neely MN. (2013) Inclusion of CYP3A5 genotyping in a nonparametric population model improves dosing of tacrolimus early after transplantation. Transpl Int 26:1198–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KC, Patterson KB, Malone SA, Shaheen NJ, Prince HM, Dumond JB, Spacek MB, Heidt PE, Cohen MS, Kashuba AD. (2011) Single and multiple dose pharmacokinetics of maraviroc in saliva, semen, and rectal tissue of healthy HIV-negative men. J Infect Dis 203:1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PL, Weatherley B, McFadyen L. (2008) A population pharmacokinetic meta-analysis of maraviroc in healthy volunteers and asymptomatic HIV-infected subjects. Br J Clin Pharmacol 65 (Suppl 1):76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou FC, Tzeng SJ, Huang JD. (2001) Genetic polymorphism of cytochrome P450 3A5 in Chinese. Drug Metab Dispos 29:1205–1209 [PubMed] [Google Scholar]
- Daly AK. (2006) Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet 45:13–31 [DOI] [PubMed] [Google Scholar]
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, et al. (2005) Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49:4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumond JB, Patterson KB, Pecha AL, Werner RE, Andrews E, Damle B, Tressler R, Worsley J, Kashuba AD. (2009) Maraviroc concentrates in the cervicovaginal fluid and vaginal tissue of HIV-negative women. J Acquir Immune Defic Syndr 51:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elens L, Nieuweboer A, Clarke SJ, Charles KA, de Graan AJ, Haufroid V, Mathijssen RH, van Schaik RH. (2013) CYP3A4 intron 6 C>T SNP (CYP3A4*22) encodes lower CYP3A4 activity in cancer patients, as measured with probes midazolam and erythromycin. Pharmacogenomics 14:137–149 [DOI] [PubMed] [Google Scholar]
- Emory JF, Seserko LA, Marzinke MA. (2014) Development and bioanalytical validation of a liquid chromatographic-tandem mass spectrometric (LC-MS/MS) method for the quantification of the CCR5 antagonist maraviroc in human plasma. Clin Chim Acta 431:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2007) Selzentry (Maraviroc): Clinical Pharmacology and Biopharmaceutics Review(s), US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research, Rockville, MD [Google Scholar]
- FDA (2013) Guidance for Industry: Bioanalytical Method Validation, US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research, Center for Veterinary Medicine, Rockville, MD [Google Scholar]
- Floyd MD, Gervasini G, Masica AL, Mayo G, George AL, Jr, Bhat K, Kim RB, Wilkinson GR. (2003) Genotype-phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics 13:595–606 [DOI] [PubMed] [Google Scholar]
- Fung KL, Gottesman MM. (2009) A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta 1794:860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granfors MT, Wang JS, Kajosaari LI, Laitila J, Neuvonen PJ, Backman JT. (2006) Differential inhibition of cytochrome P450 3A4, 3A5 and 3A7 by five human immunodeficiency virus (HIV) protease inhibitors in vitro. Basic Clin Pharmacol Toxicol 98:79–85 [DOI] [PubMed] [Google Scholar]
- Gruber VA, Rainey PM, Lum PJ, Beatty GW, Aweeka FT, McCance-Katz EF. (2013) Interactions between alcohol and the HIV entry inhibitor Maraviroc. J Int Assoc Provid AIDS Care 12:375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick RM, Fatkenheuer G, Burnside R, Hardy WD, Nelson MR, Goodrich J, Mukwaya G, Portsmouth S, Heera JR. (2014) Five-year safety evaluation of maraviroc in HIV-1-infected treatment-experienced patients. J Acquir Immune Defic Syndr 65:78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, et al. MOTIVATE Study Teams (2008) Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, et al. (2000) Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA 97:3473–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, et al. (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779 [DOI] [PubMed] [Google Scholar]
- Hyland R, Dickins M, Collins C, Jones H, Jones B. (2008) Maraviroc: in vitro assessment of drug-drug interaction potential. Br J Clin Pharmacol 66:498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacqmin P, Wade JR, Weatherley B, Snoeck E, Marshall S, McFadyen L. (2013) Assessment of maraviroc exposure-response relationship at 48 weeks in treatment-experienced HIV-1-infected patients in the MOTIVATE studies. CPT Pharmacometrics Syst Pharmacol 2:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuda TN, Abel S, Davis J, Hamlin J, Schöller-Gyüre M, Mack R, Ndongo N, Petit W, Ridgway C, Sekar V, et al. (2011) Pharmacokinetic interactions of maraviroc with darunavir-ritonavir, etravirine, and etravirine-darunavir-ritonavir in healthy volunteers: results of two drug interaction trials. Antimicrob Agents Chemother 55:2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivistö KT, Bookjans G, Fromm MF, Griese EU, Münzel P, Kroemer HK. (1996) Expression of CYP3A4, CYP3A5 and CYP3A7 in human duodenal tissue. Br J Clin Pharmacol 42:387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolars JC, Lown KS, Schmiedlin-Ren P, Ghosh M, Fang C, Wrighton SA, Merion RM, Watkins PB. (1994) CYP3A gene expression in human gut epithelium. Pharmacogenetics 4:247–259 [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Goldstein JA. (2005) Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics 6:357–371 [DOI] [PubMed] [Google Scholar]
- Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE. (2002) Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol 62:162–172 [DOI] [PubMed] [Google Scholar]
- Lu Y, Hendrix CW, Bumpus NN. (2012) Cytochrome P450 3A5 plays a prominent role in the oxidative metabolism of the anti-human immunodeficiency virus drug maraviroc. Drug Metab Dispos 40:2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael NL, Chang G, Louie LG, Mascola JR, Dondero D, Birx DL, Sheppard HW. (1997) The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med 3:338–340 [DOI] [PubMed] [Google Scholar]
- Moore RD. (2011) Epidemiology of HIV infection in the United States: implications for linkage to care. Clin Infect Dis 52 (Suppl 2):S208–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Peris B, Croucher A, Else LJ, Vera JH, Khoo S, Scullard G, Back D, Winston A. (2013) Pharmacokinetic profile and safety of 150 mg of maraviroc dosed with 800/100 mg of darunavir/ritonavir all once daily, with and without nucleoside analogues, in HIV-infected subjects. J Antimicrob Chemother 68:1348–1353 [DOI] [PubMed] [Google Scholar]
- Okoli C, Siccardi M, Thomas-William S, Dufty N, Khonyongwa K, Ainsworth J, Watson J, Cook R, Gandhi K, Hickinbottom G, et al. (2012) Once daily maraviroc 300 mg or 150 mg in combination with ritonavir-boosted darunavir 800/100 mg. J Antimicrob Chemother 67:671–674 [DOI] [PubMed] [Google Scholar]
- Philpott SM. (2003) HIV-1 coreceptor usage, transmission, and disease progression. Curr HIV Res 1:217–227 [DOI] [PubMed] [Google Scholar]
- Pozniak AL, Boffito M, Russell D, Ridgway CE, Muirhead GJ. (2008) A novel probe drug interaction study to investigate the effect of selected antiretroviral combinations on the pharmacokinetics of a single oral dose of maraviroc in HIV-positive subjects. Br J Clin Pharmacol 65 (Suppl 1):54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan S, Abel S, Tweedy S, West S, Hui J, Kearney BP. (2010) Pharmacokinetic interaction of ritonavir-boosted elvitegravir and maraviroc. J Acquir Immune Defic Syndr 53:209–214 [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Eichelbaum M, Brinkmann U, Penger A, Asante-Poku S, Zanger UM, Schwab M. (2001) Frequency of C3435T polymorphism of MDR1 gene in African people. Lancet 358:383–384 [DOI] [PubMed] [Google Scholar]
- Siccardi M, D’Avolio A, Nozza S, Simiele M, Baietto L, Stefani FR, Moss D, Kwan WS, Castagna A, Lazzarin A, et al. (2010) Maraviroc is a substrate for OATP1B1 in vitro and maraviroc plasma concentrations are influenced by SLCO1B1 521 T>C polymorphism. Pharmacogenet Genomics 20:759–765 [DOI] [PubMed] [Google Scholar]
- Taiwo B, Acosta EP, Ryscavage P, Berzins B, Lu D, Lalezari J, Castro J, Adeyemi O, Kuritzkes DR, Eron JJ, et al. (2013) Virologic response, early HIV-1 decay, and maraviroc pharmacokinetics with the nucleos(t)ide-free regimen of maraviroc plus darunavir/ritonavir in a pilot study. J Acquir Immune Defic Syndr 64:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng E, Walsky RL, Luzietti RA, Jr, Harris JJ, Kosa RE, Goosen TC, Zientek MA, Obach RS. (2014) Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos 42:1163–1173 [DOI] [PubMed] [Google Scholar]
- van Schaik RH, van der Heiden IP, van den Anker JN, Lindemans J. (2002) CYP3A5 variant allele frequencies in Dutch Caucasians. Clin Chem 48:1668–1671 [PubMed] [Google Scholar]
- Vourvahis M, Plotka A, Mendes da Costa L, Fang A, Heera J. (2013) Pharmacokinetic interaction between maraviroc and fosamprenavir-ritonavir: an open-label, fixed-sequence study in healthy subjects. Antimicrob Agents Chemother 57:6158–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DK, Abel S, Comby P, Muirhead GJ, Nedderman AN, Smith DA. (2005) Species differences in the disposition of the CCR5 antagonist, UK-427,857, a new potential treatment for HIV. Drug Metab Dispos 33:587–595 [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11:274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR. (2004) Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics 5:243–272 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.