Abstract
Noncoding microRNAs (miRNAs or miRs) have been revealed as critical epigenetic factors in the regulation of various cellular processes, including drug metabolism and disposition. However, research on miRNA functions is limited to the use of synthetic RNA and recombinant DNA agents. Herein, we show that novel pre-miRNA-27b (miR-27b) agents can be biosynthesized in Escherichia coli using recombinant RNA technology, and recombinant transfer RNA (tRNA)/mir-27b chimera was readily purified to a high degree of homogeneity (>95%) using anion-exchange fast protein liquid chromatography. The tRNA-fusion miR-27b was revealed to be processed to mature miRNA miR-27b in human carcinoma LS-180 cells in a dose- and time-dependent manner. Moreover, recombinant tRNA/miR-27b agents were biologically active in reducing the mRNA and protein expression levels of cytochrome P450 3A4 (CYP3A4), which consequently led to lower midazolam 1′-hydroxylase activity. These findings demonstrate that pre-miRNA agents can be produced by recombinant RNA technology for functional studies.
Introduction
MicroRNAs (miRNAs or miRs) comprise a large family of small noncoding RNAs (ncRNAs) that govern target gene expression in cells via translation inhibition or mRNA deadenylation/degradation mechanisms. Our recent study showed that miR-27b regulates the protein and mRNA expression of cytochrome P450 (P450) 3A4 (CYP3A4) (Pan et al., 2009a), the most abundant P450 enzyme expressed in human liver and intestine. Change in CYP3A4 expression, which is transcriptionally controlled by a number of nuclear receptors, such as vitamin D receptor (VDR/NR1I1) (Schmiedlin-Ren et al., 2001; Thummel et al., 2001), pregnane X receptor (PXR/NR1I2) (Lehmann et al., 1998; Xie et al., 2000), and retinoid X receptor-α (RXRα/NR2B1) (Wang et al., 2008), may lead to large variability in drug metabolism (Wang and LeCluyse, 2003; Thummel and Lin, 2014). MiR-148a was also found to influence CYP3A4 expression via the targeting of PXR (Takagi et al., 2008). Further studies suggest that other miRNAs, such as miR-34a and miR-577, could be involved in the modulation of CYP3A4 expression (Vuppalanchi et al., 2013; Lamba et al., 2014; Oda et al., 2014; Wei et al., 2014). In addition, other P450 enzymes (e.g., CYP1B1) and drug transporters (e.g., P-glycoprotein, breast cancer resistance protein, and multidrug resistance-associated proteins) may be regulated by miRNAs, such as miR-27b, miR-124, miR-519c, and miR-1291 (Tsuchiya et al., 2006; To et al., 2008; Zhu et al., 2008; Pan et al., 2009b, 2013; Haenisch et al., 2011; Li et al., 2011; Rieger et al., 2013; Shukla et al., 2013; Xie et al., 2013; Markova and Kroetz, 2014). These findings support the concept that miRNAs are critical epigenetic factors in the modulation of drug metabolism and disposition that might consequently alter drug efficacy (Yu, 2007; Yu and Pan, 2012; Ingelman-Sundberg et al., 2013; Yokoi and Nakajima, 2013).
Currently, viral or nonviral vector-based miRNA expression systems are heavily used for in vitro and in vivo studies on miRNA functions (Liu and Berkhout, 2011). These agents are literally recombinant DNA materials, and this approach is generally less efficient because it relies on the host cells or organisms to transcribe the DNA to miRNA precursors. RNA agents, such as the miRNA mimics and precursors as well as antisense antagomirs produced by chemical synthesis, are another major class of materials used in miRNA research (Ling et al., 2013). These synthetic RNA agents all consist of unnatural modifications, and it is unknown how chemical modifications may alter the biologic activity of miRNAs despite the mimics exhibiting a longer half-life. In vitro transcription (Beckert and Masquida, 2011) may produce RNA agents in variable lengths, whereas a large-scale production requires more but inexpensive RNA polymerases. Recently, tRNA (Ponchon and Dardel, 2007; Ponchon et al., 2009; Nelissen et al., 2012) and ribosomal RNA (Liu et al., 2010) have been used as scaffolds for successful production of recombinant RNAs in Escherichia coli (E. coli) for structural analyses. This recombinant RNA technology may also offer a novel means to biosynthesize RNA agents for functional studies.
In the present study, we show that novel pre-miRNA Homo sapiens pre-microRNA-27b (hsa-mir-27b) agents were successfully expressed in E. coli using recombinant RNA technology. Recombinant tRNA/mir-27b chimeras were isolated by anion-exchange fast protein liquid chromatography (FPLC). In addition, the purified tRNA/mir-27b agents were found to be processed to mature miR-27b in human carcinoma LS-180 cells, which consequently reduced CYP3A4 protein expression and resulted in a lower midazolam 1′-hydroxylase activity. These findings may offer novel clues for the development of natural pre-miRNA agents for functional studies in drug metabolism.
Materials and Methods
Chemicals and Materials.
Midazolam (MDZ) and its metabolite 1’-hydroxymidazolam (1′-HO-MDZ) were purchased from Cambridge Isotope Laboratories (Tewksbury, MA) and Cayman Chemical Company (Ann Arbor, MI), respectively. 1-α-hydroxycholecalciferol (1α-VD3) was bought from EMD Millipore (Billerica, MA). Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Restriction enzymes including SalI and AatII as well as the T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). All other chemicals and organic solvents were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific Inc. (Waltham, MA).
Prediction of RNA Secondary Structure.
The secondary structures of pre-miRNAs, tRNA and chimeric RNAs (Fig. 1A) were predicted using the CentroidFold (http://www.ncrna.org/centroidfold) (Sato et al., 2009), Centroidhomfold (http://www.ncrna.org/centroidhomfold) (Hamada et al., 2011), and RNAstructure (http://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html) (Reuter and Mathews, 2010).
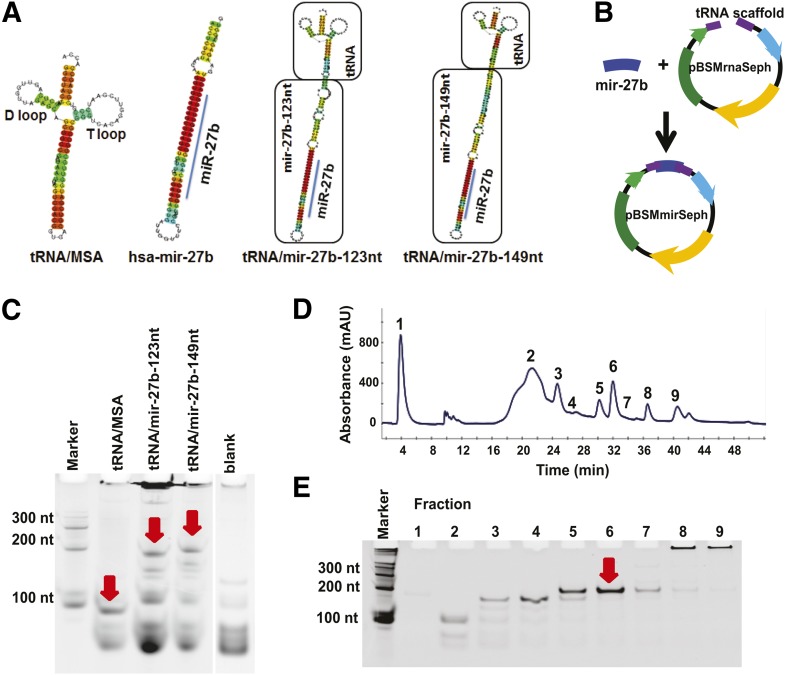
Fig. 1.
Design and production of recombinant tRNA-fusion pre-miR-27b agents. (A) Secondary structures of tRNA/MSA, hsa-mir-27b, and chimeric tRNA/mir-27b-123nt and tRNA/mir-149nt were predicted by CentroidFold. Consistent results were obtained using Centroidhomfold and RNAstructure. (B) The target hsa-mir-27b inserts encoding mir-27b-123nt and mir-27b-149nt were ligated into the pBSMrnaSeph vector linearized by endonucleases SalI and AatII to produce the recombinant pre-miRNA expression plasmids (pBSMmirSeph). (C) The tRNA/mir-27b-123nt and -149nt chimeras were successfully expressed in E. coli. The arrows indicate the recombinant ncRNA bands at expected sizes. Total RNAs (1 μg per lane) were analyzed by denaturing polyacrylamide (8%) gel electrophoresis. Untreated bacteria (blank) served as control. (D) FPLC traces during the purification of chimeric tRNA/mir-27b-149nt. Total RNAs were separated using anion-exchange FPLC and monitored at 260 nm. (E) Denaturing polyacrylamide (8%) gel electrophoresis indicated that fraction #6 consisted of high-purity (>95%) recombinant tRNA/mir-27b-149nt. Similar results were obtained for tRNA/mir-27b-123nt (data not shown).
Construction of Plasmids.
To express the pre-miR-27b agents using tRNA scaffold (Fig. 1A), the DNA fragments encoding 123-nt and 149-nt hsa-mir-27b were first amplified from human genomic DNA by PCR using the primers 5′- ACG CGT CGA CCC AGC GAT GAC CTC TCT AAC -3′ (forward) and 5′- CAT CGA CGT CCT TAA CTG TCC CCA TCT CAC C -3′ (reverse), and 5′- ACG CGT CGA CCG TCC CTT TAT TTA TGC CCA GC -3′ (forward) and 5′- CAT CGA CGT CCG GCT CCA ACT TAA CTG TCC -3′ (reverse), respectively. The amplicon was cloned into the vector pBSMrnaSeph (kindly provided by Dr. Luc Ponchon, Université Paris Descartes, France) (Ponchon and Dardel, 2007; Ponchon et al., 2009) (Fig. 1B) after being linearized by restriction endonucleases SalI and AatII. All hsa-mir-27b inserts were confirmed by Sanger sequencing analysis.
Expression of Recombinant ncRNAs.
Recombinant ncRNA chimera and control tRNAs were expressed using HST08 E. coli strain (Clontech, Mountain View, CA), as described (Ponchon and Dardel, 2007; Ponchon et al., 2009). Total RNAs were isolated from E. coli using the Tris-HCl-saturated phenol extraction method, quantitated using NanoDrop (Thermo Scientific, Waltham, MA), and analyzed by denaturing urea (6 M) polyacrylamide (8%) gel electrophoresis to assess the expression of recombinant ncRNAs.
Purification of Recombinant ncRNAs.
Recombinant ncRNAs were purified using a NGC QUEST 10PLUS CHROM FPLC System (Bio-Rad, Hercules, CA) consisting of two dual-piston pumps, a MULTI UV/visible detector, and a BioFrac fraction collector. For the isolation of tRNA/mir-27b, a UNO Q1 anion-exchange column (Bio-Rad) was equilibrated with five column volumes (at a flow rate 1.0 ml/min for 5 minutes) of buffer A (10-mM sodium phosphate, pH = 7.0). Total RNAs (∼1.0 mg) were then loaded onto the column and separated using a gradient elution method at the same flow rate 1.0 ml/min, 10 ml of 100% buffer A, 5 ml of 0–50% buffer B (buffer A consisting of 1 M sodium chloride), 5 ml of 50% buffer B, and 30 ml of 50–80% buffer B, followed by 1 ml of 80–100% buffer B, 2 ml of 100% buffer B, 2 ml of 0–100% buffer A, and 5 ml of 100% buffer A. For the purification of control methionyl-tRNA with a Sephadex apatamer (tRNA/MSA), a UNO Q6 anion-exchange column (Bio-Rad) was equilibrated with five column volumes (at a flow rate 6.0 ml/min for 5 minutes) of buffer A. After total RNAs (∼5.0 mg) were loaded onto the column and tRNA/MSA was separated using a gradient elution method at the same flow rate of 6.0 ml/min, 6 ml of 100% buffer A, 6 ml of 0–50% buffer B, 48 ml of 50% buffer B, and 60 ml of 50–60% buffer B, followed by 6 ml of 60–100% buffer B, 12 ml of 100% buffer B, 6 ml of 0–100% buffer A, and 12 ml of 100% buffer A. FPLC traces were monitored at 260 nm using the UV/visible detector. After being analyzed on a denaturing PAGE gel, the fractions containing pure chimeric RNAs were pooled. Recombinant ncRNAs were precipitated with ethanol, reconstituted with nuclease-free water, and then desalted and concentrated with Amicon ultra–0.5-ml centrifugal filters (30 KD; EMD Millipore). The purity of ncRNAs was further validated using PAGE and quantified using NanoDrop before other experiments.
Cell Culture and Transfection.
Human colon carcinoma LS-180 cells were purchased from American Tissue Culture Collection (Manassas, VA) and cultured in Eagle’s minimum essential medium (American Tissue Culture Collection) supplemented with 10% fetal bovine serum (Life Technologies, Carlsbad, CA) at 37°C in 5% carbon dioxide. Cells were transfected with purified recombinant ncRNAs using Lipofectamine 2000 (Life Technologies).
Reverse Transcription-Quantitative Real-Time Polymerase Chain Reaction
Regular reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR) analyses of mRNA levels and stem-loop RT-qPCR analysis of miRNA levels were conducted using the methods described previously (Pan et al., 2009a; Li et al., 2011), except that total RNAs were isolated from LS-180 cells using ZR RNA MiniPrep kit (Zymo Research, Irvine, CA), cDNA was synthesized from total RNA with iScript Reverse Transcription Supermix (Bio-Rad), and qPCR was conducted using SsoAdvanced SYBR Green Supermix (Bio-Rad) on a CFX96 Touch real-time PCR system (Bio-Rad). The same primers (Pan et al., 2009a; Li et al., 2011) were used for the analysis of mature miR-27b, U74, CYP3A4, VDR, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Primers 5′- ACC TCT CTA ACA AGG TGC AG GCT TAG -3′ (forward) and 5′- CAC CTT CTC TTC AGG TGC AGA AC -3′ (forward) were used for the analysis of pre-miRNA mir-27b. U74 was used as internal control for miR-27b, and GAPDH was used for other analytes. The relative expression was calculated using the equation, 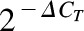 where ∆CT was the difference in CT value between the analyte and internal standard, and then normalized to the control treatment.
where ∆CT was the difference in CT value between the analyte and internal standard, and then normalized to the control treatment.
Immunoblot Analysis.
LS-180 cell were treated with 2.5 µM 1α-VD3 or vehicle for 96 hours and then transfected with 50 nM tRNA/mir-27b or tRNA/MSA for 72 hours. Cell lysates were prepared for immunoblot analysis using RIPA lysis buffer (Rockland Immunochemicals, Gilbertsville, PA) consisting of complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany); total RNAs were isolated for qPCR analysis as described already here. Protein concentrations were determined using the BCA Protein Assay Kit (Pierce, Rockford, IL). Whole-cell proteins (50 µg/lane) were separated on 10% SDS-PAGE and electrophoretically transferred onto polyvinylidene fluoride membranes (Bio-Rad). After being incubated with selective antibody against CYP3A4 (BD Biosciences, San Jose, CA), VDR, or GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) and subsequently with a horseradish peroxidase rabbit anti-mouse IgG (BD Bioscience) or a peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA), proteins were visualized with an enhanced chemiluminescence detection system (Bio-Rad), and images were acquired by ChemiDoc MP Imaging System (Bio-Rad).
Midazolam 1′-Hyroxylation Activity Assay.
Cellular MDZ metabolism was conducted as reported (Schmiedlin-Ren et al., 2001). Briefly, LS-180 cells were first treated with 2.5-µM 1α-VD3 for 96 hours and then transfected with 50 nM tRNA/mir-27b or control tRNA/MSA. At 72 hours post-transfection, 5 µM MDZ was administered to the media and cells were maintained at 37°C. Then 200 μl of media was collected at different time points. After adding 200 μl of media containing 100-nM harmine (internal standard), analytes were extracted with 3 ml of ethyl acetate. The extracts were evaporated and reconstituted for liquid chromatography–tandem mass spectrometry quantitation of MDZ and 1′-HO-MDZ using a AB SCIEX 4000 QTRAP System (AB SCIEX LLC, Framingham, MA) coupled with a Prominence LC-20AD HPLC system (Shimadzu, Columbia, MD), as we described previously (Granvil et al., 2003; Felmlee et al., 2008). All data were collected and processed using the Analyst software (AB SCIEX).
Statistical Analysis.
All values were mean ± S.D. Depending on the number of groups and variances, data were compared with unpaired Student’s t test or two-way analysis of variance (GraphPad Prism, San Diego, CA). Difference was considered statistically significant if the probability was less than 0.05 (P < 0.05).
Results and Discussion
To better maintain the hairpin structure of hsa-mir-27b (97 nucleotides (nt) in length) and evaluate the impact of length on the expression of recombinant ncRNAs, we extended the 5′ and 3′ flanking sequences of hsa-mir-27b to 123 nt and 149 nt. The secondary structures of resultant tRNA/mir-27b-123nt (227 nt in total) and tRNA/mir-27b-149nt (253 nt) chimeras as well as the tRNA scaffold and hsa-mir-27b were thus predicted using different algorithms (CentroidFold, Centroidhomfold and RNAstructure). All predictions consistently showed conservation of the stem-loop structure of hsa-mir-27b within chimeric ncRNAs (Fig. 1A), suggesting that chimeric tRNA/mir-27b would be accessible by cellular endoribonucleases such as Drosha and Dicer for the production of mature miR-27b. Therefore, their corresponding DNA segments were inserted into the pBSMmirSeph vector (Fig. 1B) linearized by endonucleases SalI and AatII, and all clones were confirmed by Sanger sequencing analyses before the expression of recombinant ncRNAs.
To assess whether recombinant tRNA/mir-27b was expressed, total RNAs were isolated from bacteria at 16 hours post-transformation with hsa-mir-27b expression plasmid and subjected to RNA electrophoretic mobility assay. A successful expression of chimeric tRNA/mir-27b and tRNA/MSA was indicated by the appearance of new RNA bands ∼200 nt and ∼100 nt long, respectively, in E. coli transformed with ncRNA expression plasmids, compared with untreated bacteria (Fig. 1C). It is noteworthy that the electrophoretic mobility of chimeric tRNA/pre-miRNA and tRNA/MSA is greater than that indicated by the single-stranded RNA markers. This result is likely due to the presence of “double-stranded” stem structure in these ncRNAs (Fig. 1A). Whereas tRNA/MSA was consistently expressed at a high level (∼10 mg/liter of culture) (Ponchon and Dardel, 2007; Ponchon et al., 2009), the expression levels of tRNA/mir-27b were relatively low (<500 μg/liter culture), and thus the size of hsa-mir-27b showed minimal effect on tRNA/mir-27b expression (Fig. 1C). This result is also different from that of the tRNA/mir-1291, which was usually expressed at greater than 10 mg/liter in culture, whereas the levels were sharply decreased with the increase of mir-1291 length (unpublished data). Indeed, the lower expression of recombinant tRNA/mir-27b was associated with the occurrence of other RNA fragments (e.g., ∼150 nt, Fig. 2A), suggesting that tRNA/mir-27b might be susceptible to bacterial RNases. Therefore, further study is warranted to improve the expression level of recombinant tRNA/mir-27b chimera.
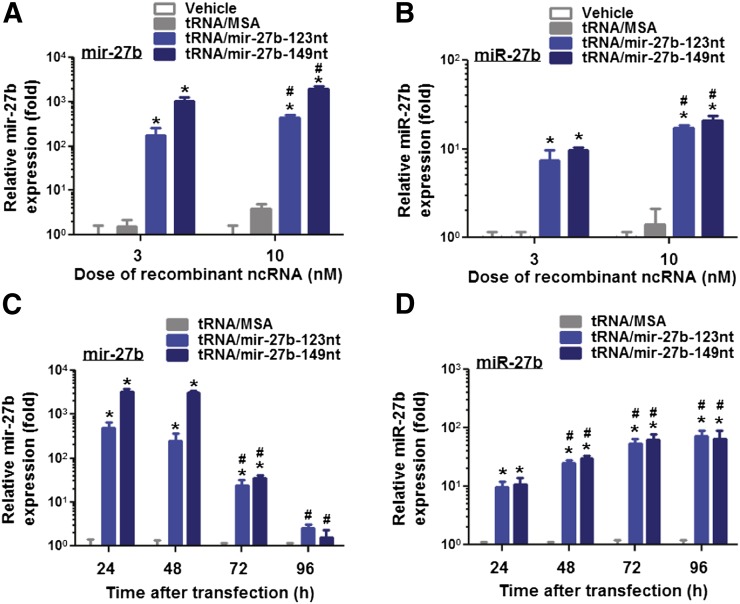
Fig. 2.
Recombinant pre-miRNA mir-27b chimera is processed to mature miRNA miR-27b in human LS-180 cells. The levels of mir-27b (A) and miR-27b (B) were increased in a dose-dependent manner in LS-180 cells at 24 hours post-transfection with the purified tRNA/mir-27b (3 and 10 nM), as determined by selective qPCR analysis. In addition, mir-27b (C) and miR-27b (D) levels varied in LS-180 cells at 24, 48, 72, and 96 hours post-transfection with 10 nM chimeric tRNA/mir-27b. Cells treated with the same doses of tRNA/MSA or vehicles were used as controls. GAPDH and U74 were used as internal control for the quantification of mir-27b and miR-27b, respectively. Values are mean ± S.D. of triplicate treatments with separate cultures and analyzed using two-way analysis of variance. *P < 0.01, compared with the corresponding control treatment (vehicle or tRNA/MSA) at the same dose or time point. #P < 0.01 compared with data obtained from the same tRNA/mir-27b agent at a lower dose (3 nM) or early time point (24 hours).
An anion-exchange FPLC method was thus developed for the purification of recombinant ncRNAs. Elution with low to high concentrations of sodium chloride solution successfully separated the recombinant tRNA/mir-27b (eluted at 31.2 minutes; Fig. 1D) from other small RNAs, tRNAs, and 5S ribosomal RNA using the Q1 column. Likewise, tRNA/MSA (18.5 minutes; data not shown) was well separated from other RNAs using the Q6 column. The purified tRNA/mir-27b (Fig. 1E) and tRNA/MSA (data not shown) indeed showed a high degree of homogeneity (>95%). A higher yield was also achieved for tRNA/MSA using the anion-exchange FPLC method (i.e., ∼0.5 mg of tRNA/MSA from 5 mg of total RNAs obtained from 200-ml culture; 10% recombinant RNA/total RNAs) than that using affinity chromatography (unpublished data). In contrast, around 100 µg of tRNA/mir-27b was generally purified from 5 mg of total RNAs obtained from 200 ml of culture, equivalent to a 2% yield of recombinant RNA/total RNAs, which is due mainly to the low expression level of tRNA/mir-27b (Fig. 1C). Indeed, the FPLC purification yields of other high-level expressing pre-miRNAs, including mir-1291, were all comparable (unpublished data), suggesting that the anion-exchange FPLC described in this study is a reliable method for the purification of recombinant ncRNAs.
To delineate whether chimeric tRNA/mir-27b can be processed into mature miR-27b, human colon carcinoma LS-180 cells were transfected with the purified recombinant tRNA/mir-27b and control tRNA/MSA, which is an excellent cell model system for studying CYP3A4 regulation (Schmiedlin-Ren et al., 2001; Thummel et al., 2001) and comprised the necessary machinery for miR-27b biogenesis (Pan et al., 2009a). Selective stem-loop RT and regular qPCR assays were used to quantify mature miRNA miR-27b and pre-miRNA mir-27b, respectively (Pan et al., 2009a; Li et al., 2011) (Fig. 2). The selectivity of stem-loop RT-qPCR for the analysis of mature miR-27b was supported by a low CT value (<25) for the cDNA prepared from total RNAs of tRNA/mir-27b–transfected cells versus much higher CT values for the same amount of cDNAs prepared from the purified tRNA/mir-27b (>30) and total RNAs of untreated or tRNA/MSA-transfected cells (>28), as well as the decrease in mir-27b levels versus increase of miR-27b over time in the cells after transfection with tRNA/mir-27b (Fig. 2, C and D). The data showed a sharp increase in hsa-mir-27b levels in the cells transfected with tRNA/mir-27b (Fig. 2, A and C), indicating successful delivery of recombinant tRNA/mir-27b into the cells. Such an increase of greater than 100-fold of hsa-mir-27b levels persisted 72 hours post-transfection (Fig. 2C), highlighting the stability of tRNA/mir-27b within the cells. Consequently, the levels of mature miR-27b were increased in a dose- (Fig. 2B) and time- (Fig. 2D) dependent manner in LS-180 cells transfected with tRNA/mir-27b. In addition, the levels of mature miR-27b formed from the same dose of tRNA/mir-27b-123nt and tRNA/mir-27b-149nt (Fig. 2B) at the same time points (Fig. 2D) were not significantly different. Together, these results support the production of mature miR-27b from recombinant tRNA/mir-27b, despite not knowing exactly how chimeric tRNA/mir-27b is processed in the cells and what ribonucleases are involved in the processes.
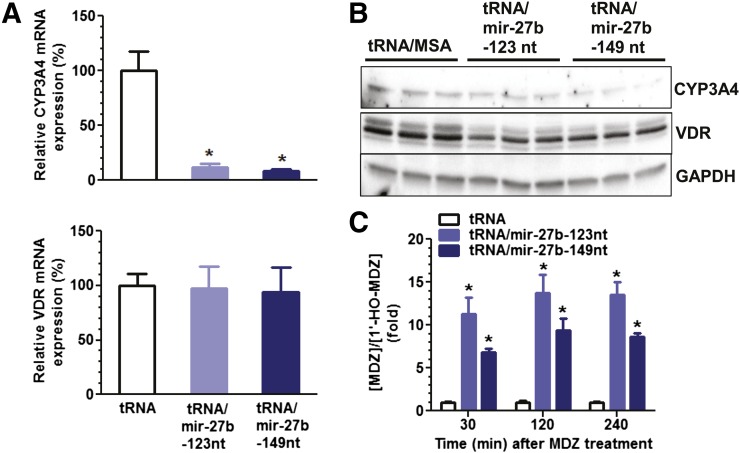
Our previous studies have demonstrated that the well-conserved miR-27b can regulate CYP3A4 expression in human carcinoma cells through targeting of the 3-untranslated regions of CYP3A4 and VDR/NR1I1 (Pan et al., 2009a). Therefore, we evaluated whether recombinant tRNA/mir-27b is effective in the modulation of CYP3A4 expression in LS-180 cells. RT-qPCR analyses (Fig. 3A) using gene-specific primers showed that CYP3A4 mRNA levels were reduced around 10-fold by both tRNA/mir-27b-123nt and tRNA/mir-27b-149nt in LS-180 cells, whereas VDR mRNA levels were not altered. Immunoblot analyses (Fig. 3B) using selective antibodies revealed that CYP3A4 and VDR protein levels were suppressed 30–50% and ∼20%, respectively, in LS-180 cells after transfection with chimeric tRNA/mir-27b. These results indicate that recombinant tRNA/mir-27b is biologically active in the inhibition of CYP3A4 expression in LS-180 cells after being processed to mature miR-27b (Fig. 2), which may involve post-transcriptional and transcriptional regulatory mechanisms (Pan et al., 2009a; Yu, 2009). Furthermore, we investigated the consequent effect of altered CYP3A4 expression on cellular drug metabolism capacity. MDZ, a widely used CYP3A4 probe drug that is not transported by P-glycoprotein or other transporters, was used to assess cellular CYP3A4 enzymatic activity. MDZ and 1′-HO-MDZ, the major metabolite produced by CYP3A4, were quantified by specific and sensitive liquid chromatography–tandem mass spectrometry method. Our data (Fig. 3C) revealed that the [MDZ]/[1′-HO-MDZ] metabolic ratios were increased more than 5-fold in LS-180 cells at various time points after transfection with either tRNA/mir-27b-123nt or tRNA/mir-149nt, which indicated a reduction of CYP3A4 enzymatic activity. Together, these results show that recombinant mir-27b chimeras are functional in the modulation of CYP3A4 protein outcome and consequently influence cellular drug metabolism capacity.
Fig. 3.
Recombinant pre-mir-27b is effective in the modulation of CYP3A4 expression. (A) qPCR analyses revealed that recombinant tRNA/mir-27b significantly (*P < 0.001, one-way analysis of variance) reduced CYP3A4 mRNA expression levels in LS-180 cells compared with the tRNA scaffold. GAPDH was used as an internal control. (B) Western blot analyses showed that LS-180 cells transfected with tRNA/mir-27b had lower CYP3A4 protein expression levels than the cells transfected with control tRNA. GAPDH was used as a loading control. (C) CYP3A4 enzymatic activity, as measured by [MDZ]/[1′-HO-MDZ] metabolic ratio at various time points after exposure to MDZ (at 72 hours post-transfection), was significantly (*P < 0.001, two-way analysis of variance) lower in LS-180 cells treated with tRNA/mir-27b than the tRNA/MSA control. MDZ and metabolite concentrations were determined by the liquid chromatography–tandem mass spectrometry method. Values are mean ± S.D. of triplicate treatments with separate cultures. *P < 0.05 compared with the control tRNA/MSA treatment.
In summary, this study demonstrates a successful production of novel, biologically active mir-27b agents using the tRNA scaffold. Recombinant tRNA/mir-27b chimeras are readily purified to a high degree of homogeneity using an anion-exchange FPLC method developed in the present study, despite their expression levels being relatively lower than those of control tRNA/MSA and other pre-miRNAs (unpublished data). Our data show that the mir-27b chimeras are processed to mature miR-27b in human carcinoma LS-180 cells in a dose- and time-dependent manner. In addition, our results indicate that recombinant mir-27b suppresses CYP3A4 and VDR protein expression and results in a lower cellular drug metabolism capacity. These findings suggest that pre-miRNAs can be produced using tRNA based recombinant RNA technology, which represents novel ncRNA agents for studying miRNA functions in the control of drug metabolism and disposition.
Abbreviations
- 1α-VD3
1-α-hydroxycholecalciferol
- FPLC
fast protein liquid chromatography
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase hsa-mir-27b, Homo sapiens pre-microRNA-27b
- MDZ
midazolam
- miR or miRNA
microRNA
- ncRNA
noncoding RNA
- nt
nucleotides
- P450
cytochrome P450
- RT-qPCR
reverse transcription–quantitative real-time polymerase chain reaction
- tRNA
transfer RNA
- tRNA/MSA
methionyl-tRNA with a Sephadex apatamer
- 1′-HO-MDZ
1′-hydroxymidazolam
Authorship Contributions
Participated in research design: Yu, Li, Wang, Wu, Huang.
Conducted experiments: Li, Wang, Wu.
Contributed new reagents or analytical tools: Yu, Li, Wang, Wu.
Performed data analysis: Li, Wang, Wu, Huang, Yu.
Wrote or contributed to the writing of the manuscript: Yu, Li, Wang, Wu, Huang.
Footnotes
This project was supported in part by the National Institutes of Health National Cancer Institute [Grant 1U01-CA17531 to A.M.Y.] and the Natural Science Foundation of China [81320108027 to M.H.].
References
- Beckert B, Masquida B. (2011) Synthesis of RNA by in vitro transcription. Methods Mol Biol 703:29–41 [DOI] [PubMed] [Google Scholar]
- Felmlee MA, Lon HK, Gonzalez FJ, Yu AM. (2008) Cytochrome P450 expression and regulation in CYP3A4/CYP2D6 double transgenic humanized mice. Drug Metab Dispos 36:435–441 [DOI] [PubMed] [Google Scholar]
- Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, Krausz KW, Gonzalez FJ. (2003) Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug Metab Dispos 31:548–558 [DOI] [PubMed] [Google Scholar]
- Haenisch S, Laechelt S, Bruckmueller H, Werk A, Noack A, Bruhn O, Remmler C, Cascorbi I. (2011) Down-regulation of ATP-binding cassette C2 protein expression in HepG2 cells after rifampicin treatment is mediated by microRNA-379. Mol Pharmacol 80:314–320 [DOI] [PubMed] [Google Scholar]
- Hamada M, Yamada K, Sato K, Frith MC, Asai K. (2011) CentroidHomfold-LAST: accurate prediction of RNA secondary structure using automatically collected homologous sequences. Nucleic Acids Res 39:W100–W106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Zhong XB, Hankinson O, Beedanagari S, Yu AM, Peng L, Osawa Y. (2013) Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos 41:1725–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba V, Ghodke Y, Guan W, Tracy TS. (2014) microRNA-34a is associated with expression of key hepatic transcription factors and cytochromes P450. Biochem Biophys Res Commun 445:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan YZ, Seigel GM, Hu ZH, Huang M, Yu AM. (2011) Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, -519c and -520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem Pharmacol 81:783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12:847–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Stepanov VG, Strych U, Willson RC, Jackson GW, Fox GE. (2010) DNAzyme-mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol 10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Berkhout B. (2011) miRNA cassettes in viral vectors: problems and solutions. Biochim Biophys Acta 1809:732–745 [DOI] [PubMed] [Google Scholar]
- Markova SM, Kroetz DL. (2014) ABCC4 is regulated by microRNA-124a and microRNA-506. Biochem Pharmacol 87:515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen FH, Leunissen EH, van de Laar L, Tessari M, Heus HA, Wijmenga SS. (2012) Fast production of homogeneous recombinant RNA: towards large-scale production of RNA. Nucleic Acids Res 40:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Nakajima M, Tsuneyama K, Takamiya M, Aoki Y, Fukami T, Yokoi T. (2014) Retinoid X receptor α in human liver is regulated by miR-34a. Biochem Pharmacol 90:179–187 [DOI] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. (2009a) MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos 37:2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YZ, Morris ME, Yu AM. (2009b) MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 75:1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YZ, Zhou A, Hu Z, Yu AM. (2013) Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab Dispos 41:1744–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc 4:947–959 [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F. (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4:571–576 [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger JK, Klein K, Winter S, Zanger UM. (2013) Expression variability of absorption, distribution, metabolism, excretion-related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos 41:1752–1762 [DOI] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T. (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res 37:W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. (2001) Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos 29:1446–1453 [PubMed] [Google Scholar]
- Shukla U, Tumma N, Gratsch T, Dombkowski A, Novak RF. (2013) Insights into insulin-mediated regulation of CYP2E1: miR-132/-212 targeting of CYP2E1 and role of phosphatidylinositol 3-kinase, Akt (protein kinase B), mammalian target of rapamycin signaling in regulating miR-132/-212 and miR-122/-181a expression in primary cultured rat hepatocytes. Drug Metab Dispos 41:1769–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. (2008) Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283:9674–9680 [DOI] [PubMed] [Google Scholar]
- Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. (2001) Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol Pharmacol 60:1399–1406 [DOI] [PubMed] [Google Scholar]
- Thummel KE, Lin YS. (2014) Sources of interindividual variability. Methods Mol Biol 1113:363–415 [DOI] [PubMed] [Google Scholar]
- To KK, Zhan Z, Litman T, Bates SE. (2008) Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 28:5147–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66:9090–9098 [DOI] [PubMed] [Google Scholar]
- Vuppalanchi R, Liang T, Goswami CP, Nalamasu R, Li L, Jones D, Wei R, Liu W, Sarasani V, Janga SC, et al. (2013) Relationship between differential hepatic microRNA expression and decreased hepatic cytochrome P450 3A activity in cirrhosis. PLoS ONE 8:e74471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, LeCluyse EL. (2003) Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin Pharmacokinet 42:1331–1357 [DOI] [PubMed] [Google Scholar]
- Wang K, Chen S, Xie W, Wan YJ. (2008) Retinoids induce cytochrome P450 3A4 through RXR/VDR-mediated pathway. Biochem Pharmacol 75:2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Jiang S, Zhang Y, Wang X, Peng X, Meng C, Liu Y, Wang H, Guo L, Qin S, et al. (2014) The effect of microRNAs in the regulation of human CYP3A4: a systematic study using a mathematical model. Sci Rep 4:4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. (2000) Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 406:435–439 [DOI] [PubMed] [Google Scholar]
- Xie X, Miao L, Yao J, Feng C, Li C, Gao M, Liu M, Gong L, Wang Y, Qi X, et al. (2013) Role of multiple microRNAs in the sexually dimorphic expression of Cyp2b9 in mouse liver. Drug Metab Dispos 41:1732–1737 [DOI] [PubMed] [Google Scholar]
- Yokoi T, Nakajima M. (2013) microRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol 53:377–400 [DOI] [PubMed] [Google Scholar]
- Yu AM. (2007) Small interfering RNA in drug metabolism and transport. Curr Drug Metab 8:700–708 [DOI] [PubMed] [Google Scholar]
- Yu AM. (2009) Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol 5:1513–1528 [DOI] [PubMed] [Google Scholar]
- Yu AM, Pan YZ. (2012) Noncoding microRNAs: small RNAs play a big role in regulation of ADME? Acta Pharmaceutica Sinica B 2:93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. (2008) Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol 76:582–588 [DOI] [PMC free article] [PubMed] [Google Scholar]