Abstract
Cytochrome P450 (P450) protein-protein interactions resulting in modulation of enzyme activities have been well documented using recombinant isoforms. This interaction has been less clearly demonstrated in a more physiologic in vitro system such as human hepatocytes. As an expansion of earlier work (Subramanian et al., 2010), in which recombinant CYP2C9 activity decreased with increasing levels of CYP3A4, the current study modulated CYP3A4 content in human hepatocytes to determine the impact on CYP2C9. Modulation of CYP3A4 levels in situ was enabled by the use of a long-term human hepatocyte culture model (HepatoPac) shown to retain phenotypic hepatocyte function over a number of weeks. The extended period of culture allowed time for knockdown of CYP3A4 protein by small interfering RNA (siRNA) with subsequent recovery, as well as upregulation through induction with a recovery period. CYP3A4 gene silencing resulted in a 60% decrease in CYP3A4 activity and protein levels with a concomitant 74% increase in CYP2C9 activity, with no change in CYP2C9 mRNA levels. Upon removal of siRNA, both CYP2C9 and CYP3A4 activities returned to pre-knockdown levels. Importantly, modulation of CYP3A4 protein levels had no impact on cytochrome P450 reductase activities or levels. However, the possibility for competition for limiting reductase cannot be ruled out. Interestingly, lowering CYP3A4 levels also increased UDP-glucuronosyltransferase 2B7 activity. These studies clearly demonstrate that alterations in CYP3A4 levels can modulate CYP2C9 activity in situ and suggest that further studies are warranted to evaluate the possible clinical consequences of these findings.
Introduction
Cytochrome P450 (P450) protein levels are regulated by nuclear receptor activation (Honkakoski and Negishi, 2000) and by endobiotic endocrine (e.g., cytokine) (Morgan, 1997; Morgan et al., 2008) or autocrine (e.g., oxidative stress and hypoxia) factors (Zangar et al., 2004; Legendre et al., 2009). Protein-protein interactions may also modulate P450 activity. The ability of one P450 protein to modulate the activity of another P450 was first suggested in rat studies evaluating the effect of enzyme induction on warfarin metabolism (Kaminsky and Guengerich, 1985). The authors observed that metabolite production was not necessarily commensurate with the enzymes that were induced, suggesting that increases of one P450 isoform were correlated with modulation of the activity of other P450 isoforms that were not induced. This led to the hypothesis that such interactions might not involve a change in P450 content, but rather a decrease in the specific P450 activity. Possible mechanisms for this change in enzyme activity include allosteric alteration of the active site, substrate-dependent ionic interactions between each protein, and obstruction of, or competition for, cytochrome P450 reductase (CPR) (Hazai et al., 2005; Kelley et al., 2005). Interactions between a number of different P450s have been evaluated, including CYP1A2 and CYP3A4 (Yamazaki et al., 1997), CYP2D6 and CYP3A4 (Li et al., 1999), CYP2E1 and CYP2A6 (Tan et al., 1997), CYP2D6 and CYP2C9 (Subramanian et al., 2009), and CYP2C9 and CYP3A4 (Subramanian et al., 2010). For example, previous studies from our laboratory demonstrated that protein-protein interactions occurring between CYP2D6 and CYP3A4 with CYP2C9 result in reduction in the rate of catalysis (Subramanian et al., 2009, 2010). These interactions were not due to competition for limited CPR, because reduction of CYP2C9 activity was still observed at saturating concentrations of CPR. Additionally, it has been reported that both CYP3A4 and CYP2C9 have similar Kd values for CPR, suggesting that the proteins are not simply competing for CPR, although this aspect cannot be completely ruled out (Shimada et al., 2005; Locuson et al., 2007; Subramanian et al., 2010; Reed and Backes, 2012).
To date, P450 protein-protein interactions have largely been characterized using reconstituted, purified P450s or recombinantly expressed P450 systems (Reed and Backes, 2012). However, in these systems, the ratios of P450 to CPR or lipids are artificial, and as such it is not clear whether these findings would translate to clinically meaningful changes in enzyme activities. To overcome these issues, the current study employed a long-term human hepatocyte culture system, HepatoPac (Hepregen, Medford, MA). HepatoPac is a primary hepatocyte coculture model with extended viability of hepatocytes and drug-metabolizing enzyme (DME) activity for several weeks after seeding. This is in contrast to standard hepatocyte models in which differential loss of DME and liver function over culture time is observed (Khetani and Bhatia, 2008). It is well accepted that human hepatocytes are the closest in vitro model to human liver (Gomez-Lechon et al., 2003, 2007), containing the full array of DME and necessary cofactors. The HepatoPac model is a useful tool for studies requiring long-term and stable enzyme expression such as characterization of low-clearance drugs and quantitative metabolite profiling (Wang et al., 2010; Chan et al., 2013). Maintenance of P450, UDP-glucuronosyltransferase (UGT), and transporter proteins has been reproducibly observed over an extended period of culture with this model, enabling mechanistic studies to study transporter and enzyme interplay (Ramsden et.al, 2014a). HepatoPac was successfully applied to bridge in vitro to in vivo data for faldaprevir, in which rates of metabolism were replicated and functional regulation of P450 through induction by rifampicin resulted in in vitro changes in faldaprevir exposure that were comparable to clinical drug-drug interaction (DDI) findings (Ramsden et al., 2014a, 2014b). These studies provide a level of confidence that the HepatoPac model is a physiologically relevant model for investigating the impact of protein-protein interactions on end points of metabolism.
In the current study, protein-protein interactions between CYP3A4 and CYP2C9 and between CYP3A4 and other DMEs were evaluated by altering CYP3A4 enzyme expression. Modulation of enzyme activity was achieved through both downregulation [(small interfering RNA (siRNA)] and induction (pregnane X receptor agonist rifampicin) of CYP3A4. Taking advantage of the long-term cell survival of the HepatoPac model, it was also possible to permit enzyme recovery after removal of the modulators (siRNA and rifampicin). These experiments demonstrated that CYP3A4 protein levels alter CYP2C9 activity in a native environment, suggesting that CYP2C9:CYP3A4 protein-protein interactions can affect enzyme activity in situ, and as such there is a need to consider possible clinical implications.
Materials and Methods
Human HepatoPac cultures, proprietary maintenance, and probing media were supplied by Hepregen (Medford, MA), and cultures were prepared from cryoplateable human hepatocytes purchased from Invitrogen Life Technologies (Grand Island, NY) or CellzDirect (Research Triangle Park, NC). Azidothymidine (AZT), AZT glucuronide, estradiol, estradiol 3-glucuronide, dextromethorphan hydrobromide monohydrate, dextrorphan d-tartrate, rifampicin, and diclofenac sodium salt were purchased from Sigma-Aldrich (St. Louis, MO). Midazolam, 1′OH-midazolam, 4′OH-diclofenac, and isotopically labeled internal standards ([13C6]hydroxydiclofenac, [13C3]1′-hydroxymidazolam, dextrorphan-d3) were purchased from BD Biosciences/Gentest (Woburn, MA). MagMax total RNA recovery bead kits, reverse transcription polymerase chain reaction (PCR), and TaqMan low-density arrays were purchased from Invitrogen Life Technologies (Grand Island, NY). Enzyme-linked immunosorbent assay (ELISA) kits were purchased for quantifying protein levels of CPR, CYP2C9, and CYP3A4 from MyBiosource (San Diego, CA). Dharmacon Smart Pool Accell RNAi, nontargeting pools, glyceraldehyde-3-phosphate dehydrogenase pools, and delivery media were purchased from Thermo Scientific (Lafayette, CO). Nevirapine, as an internal standard, was synthesized at Boehringer Ingelheim Pharmaceuticals (Ridgefield, CT).
Cell Culture.
Plateable cryopreserved primary human hepatocytes were purchased from commercial vendors permitted to sell products derived from human organs procured in the United States by federally designated Organ Procurement Organizations. Donor information can be found in Table 1. HepatoPac plates were prepared by the manufacturer, as previously described (Khetani and Bhatia, 2008), prior to shipment. Cryopreserved human hepatocytes were seeded with 32,000 (in a 24-well plate) or 5000 (in a 96-well plate) hepatocytes per well and cultured for 9 days postseeding before initiating experiments. The maintenance medium was supplied by Hepregen and contains 10% bovine serum [human HepatoPac culture medium (hHCM)]. For incubations, proprietary serum-free probing medium supplied by Hepregen was used (human HepatoPac probing medium). The plates were cultured in an incubator with 10% CO2 and 99% relative humidity at 37°C. Culture medium was replaced every 2 days (400 µl per 24 well or 64 µl per 96 well) prior to incubation with siRNA and probe substrates. HepatoPac cultures were maintained in hHCM with 10% serum until after day 9 postseeding prior to siRNA treatment.
TABLE 1.
Demographics for hepatocyte donors used in the HepatoPac model to study protein interactions
| Demographics |
Donor 1 |
Donor 2 |
Donor 3a |
Donor 4b |
|---|---|---|---|---|
| Gender | Female | Male | Female | Female |
| Age | 19 | 56 | 54 | 61 |
| Race/Ethnicity | Caucasian | African American | Caucasian | Not provided |
| Cause of Death | Intracerebral bleed secondary to ICH/stroke | CVA | Anoxia secondary to CVA | Not provided |
| Medical History | Lupus | Diabetes, hypertension, kidney disease, insulin | Diabetes and hypertension, 3 years, medicine compliant | Not provided |
| Social History | No alcohol, tobacco, or drug use | No alcohol, tobacco, or drug use | Nonsmoker, social drinker | Not provided |
CVA, cerebrovascular accident; ICH, intracerebral hemorrhage.
This donor was determined through genotyping analysis to be organic anion-transporting polypeptide (OATP) wild type and CYP3A5 *3*3
Donor ID “RTM” from the manufacturer.
siRNA Treatment.
Dharmacon Smart Pool Accell RNAi was used at a concentration of 1 µM, as described in the product kit insert. siRNA sequences were chosen because they were previously shown by the vendor to be effective at knocking down CYP3A4. A pool of sequences was used because pools have shown better selectivity toward the target while decreasing off-target effects. Initial studies were performed to optimize the conditions for maximal knockdown (data not shown). Seventy-two or ninety-six hours after siRNA treatment, cells were washed and enzyme activity was assessed under linear time conditions. Ninety-six hours was used, in some experiments, to assess whether CYP3A4 activity could be further decreased after longer exposure to siRNA. Because there was not much difference between the 72- and 96-hour data, with regard to decreases in CYP3A4 activity, data from each experimental condition were pooled to determine the average decrease in CYP3A4 activity from multiple experiments.
Enzyme Activity Assessment.
Metabolite production was used to monitor P450 activity and was accomplished by in situ incubation of probe substrates with hepatocyte cultures at 37°C with 10% CO2. Probe substrate solutions [midazolam (CYP3A4), diclofenac (CYP2C9), dextromethorphan (CYP2D6), estradiol (UGT1A1), and AZT (UGT2B7)] were prepared at 200-fold the desired final concentration and added to serum-free medium. Total concentration of solvent did not exceed 1%. Wells containing the hepatocyte cultures were washed with serum-free medium (64 µl for a 96-well plate or 400 µl for a 24-well plate) prior to substrate incubation. After washing, medium was aspirated and reactions were initiated by adding the same volume of medium containing probe substrate. Substrates were generally incubated at concentrations achieving maximum reaction velocity [diclofenac (40 µM), dextromethorphan (50 µM), AZT (1 mM), and estradiol (40 µM)]. Midazolam was incubated at a concentration of 15 µM, because midazolam can exhibit substrate inhibition kinetics at higher concentrations in vitro (Giragossian et al., 2009). Initial studies were performed using 24-well plates, and the methods were transferred to 96-well plates. There was no difference in enzyme activities determined between plating formats (data not shown). During the recovery phase, siRNA treatments were removed and replaced with proprietary human HepatoPac culture medium containing 10% serum (hHCM). Substrate incubations (CYP2C9 and CYP3A4 only) were performed daily or every other day, as described, and until recovery was observed. For the induction study, samples were pretreated with siRNA or vector control following the same protocol described above. After siRNA pretreatment, additional siRNA or vector control was added to wells with or without 25 µM rifampicin for an additional 48 hours. After the 48-hour induction period, the activities of CYP2C9 and CYP3A4 were determined, as described above. Utilizing a 24-well plate format, multiple time points were evaluated by removing a 50 µl aliquot at designated time points between 2.5 and 120 minutes and quenching the aliquot with 100 µl reaction termination solutions (described below). When 96-well plates were used, each well represented an individual time point and reactions were terminated at linear time points, two or three per substrate, by adding 130 µl quench solution directly into the well. Reaction termination solutions for P450s (60% acetonitrile and 0.1% acetic acid in water) contained internal standards, either isotopically labeled authentic standards or nevirapine (0.1 µM). Reaction termination solutions for reactions monitoring UGT1A1 and UGT2B7 activity contained 40% acetonitrile, 5% acetic acid, and 0.5 μM napthylglucuronide (internal standard) in water.
Cytochrome P450 Reductase Activity Measurements (Human Liver Microsomes, Suspended Hepatocytes, Hepatocytes).
The activity levels of CPR were determined using a modification of the methods described (Guengerich et al., 2009). Data were compared across human liver microsomes (HLMs), suspended hepatocytes, and human HepatoPac samples (incubated for up to 22 days). For hepatocyte incubations, an additional lysis step was included. Hepatocyte samples were diluted with ice-cold phosphate buffered saline and centrifuged at 14,000g for 5 minutes. Multiple wells (>12 wells/plate) were pooled to create one sample for analysis of HepatoPac CPR activity due to the low cell number present in each well of ∼5000. The pellet was lysed with lysis buffer [10 mM Tris-HCL (pH 7.5), 10 mM NaH2PO4/NaHP04, 130 mM NaCl, 1% Triton X-100, and 10 mM PPi]. Protein levels from samples were quantitated in phosphate buffer using a modified Bradford assay and measuring absorbance at 595 nm. The level of total protein was determined from 1 million suspended hepatocytes from two donors to correlate total HepatoPac protein absent of fibroblast protein to a cell concentration. The total protein in hepatocytes from this method was 1.15 mg/ml in 1 × 106 hepatocytes. The hepatocellularity of 2.67 × 106 hepatocytes/1 mg microsomal protein was used as a scaling factor to compare across activity levels from HLMs and hepatocyte samples. CPR activity was determined with the equation below and is reported as nmol/min in 1 mg microsomal protein.
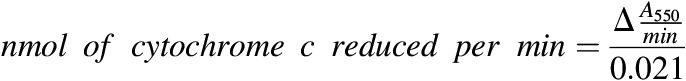 |
Quantitation of Protein Levels from HLMs and HepatoPac.
The levels of protein for CPR, CYP2C9, and CYP3A4 were quantitated from HLM and HepatoPac samples through ELISA kits and following the specific protocol details provided by the vendor. The assays were confirmed to have high sensitivity and excellent specificity for detection of their respective target proteins. Concentrations of these proteins present in the HepatoPac samples were determined through linear regression extrapolation of the optical density at 450 nm against known concentrations of protein. Multiple samples from vector control and siRNA treatment groups were pooled to ensure adequate detection signal. The concentration of cells and protein present in the sample was quantitated using a modified Bradford assay, as above. The amount of protein present in the HepatoPac samples was scaled similarly to the activity assay by using the scaling factors noted. Concentrations of protein present in one donor of HLMs were also determined for comparison with literature data.
Liquid Chromatography–Tandem Mass Spectrometry Conditions.
Samples were analyzed for metabolite production on a 4000 Qtrap (AB Sciex, Thornhill, Ontario, Canada) attached to either a CTC PAL autosampler (Leap Technologies, Carborboro, NC) with Shimadzu or Perkin Elmer pumps or a Waters Acquity UPLC system (Milford, MA). The aqueous mobile phase (A) and organic mobile phase (B) consisted of 95:5 (v/v) water/acetonitrile and 95:5 (v/v) acetonitrile/water, respectively. Both mobile phases contained 0.1% acetic acid. Samples were eluted through Acquity UPLC columns BEH C18 1.7µm (for CYP2C9, CYP2D6, and CYP3A4), HSS T3 1.7µm (2.1 mm × 50 mm), for UGT1A1 and UGT2B7 or when the Acquity UPLC system was not used a Phenomenex Synergi Max RP (150 × 2 mm, 4 µm) column (for CYP2C9, CYP2D6, and CYP3A4), using validated probe substrate analysis methods. The multiple reaction monitoring transitions used were 312.0→230.0 (4′-hydroxydiclofenac); 318.0→236.0 ([13C6]4′hydroxydiclofenac); 258.0→157.0 (dextrorphan); 261.0→160.0 (dextrorphan-d3); 342.0→324.0 (1′-hydroxymidazolam); 347.0→329.0 ([13C6]1′-hydroxymidazola); 267.2→226.1 (neviripine); 447.4→271.5 (estradiol-3-glucuronide); 466.0→290.0 (AZT glucuronide); and 319.0→142.9 (naphthylglucuronide), in positive ion mode.
Relative Quantitation of mRNA Levels.
Following incubations, cell lysates were treated with RNAlater solution (Ambion, Life Technologies, Grand Island, NY) and stored at −20°C until isolation of RNA. Total RNA was isolated from cell lysates using the MAGmax bead-based system. The relative mRNA levels for specific gene targets were determined by TaqMan real-time reverse transcription PCR methods, under a two-step assay. The first step was preparation of cDNA by reverse transcription of 200 ng total RNA. The RNA concentration of each sample was measured in triplicate using a Nanodrop2000 spectrophotometer (Thermo). Comparative (quantitative) PCR assay was conducted using a 7900 real-time PCR system (Applied Biosystems, Foster City, CA) and a custom-designed TaqMan low-density array (Table 2). Relative mRNA levels were determined from four different donors by combining triplicate samples and analyzing in duplicate (intra-assay variability).
TABLE 2.
Details of the TaqMan low-density array gene targets analyzed
| Number | Gene Target | Assay ID (Applied Biosystems) |
|---|---|---|
| 1 | HMOX1 | Hs01110250_m1 |
| 2 | CYP1A2 | Hs00167927_m1 |
| 3 | UGT1A4 | Hs01655285_s1 |
| 4 | UGT1A1 | Hs02511055_s1 |
| 5 | UGT1A3 | Hs04194492_g1 |
| 6 | UGT1A6 | Hs01592477_m1 |
| 7 | ABCG2 (BCRP) | Hs01053790_m1 |
| 8 | SLCO1B1(OATP1B1) | Hs00272374_m1 |
| 9 | GAPDH (endogenous control) | Hs99999905_m1 |
| 10 | FMO5 | Hs00356233_m1 |
| 11 | CYP2B6 | Hs00167937_g1 |
| 12 | CYP3A4 | Hs00604506_m1 |
| 13 | CYP2C8 | Hs02383390_s1 |
| 14 | CYP2C9 | Hs00426397_m1 |
| 15 | UGT2B7 | Hs00426592_m1 |
| 16 | ABCC2 (MRP2) | Hs00166123_m1 |
| 17 | UGT1A8 | Hs01592482_m1 |
| 18 | ALDH1A1 | Hs00946916_m1 |
| 19 | ABCC3 (MRP3) | Hs00978473_m1 |
| 20 | ABCB1 (MDR1) | Hs00184500_m1 |
| 21 | SLCO2B1 (OATP2B1) | Hs00200670_m1 |
| 22 | CYP2D6 | Hs02576167_m1 |
| 23 | SLCO1B3 (OATP1B3) | Hs00251986_m1 |
| 24 | GAPDH | Hs02758991_g1 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
Results
Impact of siRNA-Mediated Downregulation of CYP3A4.
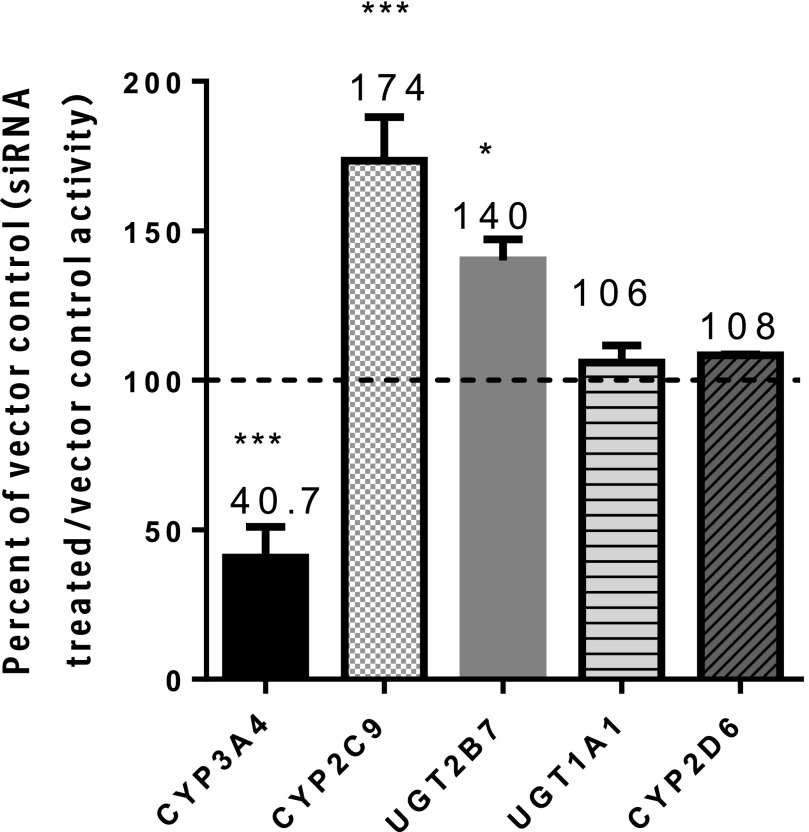
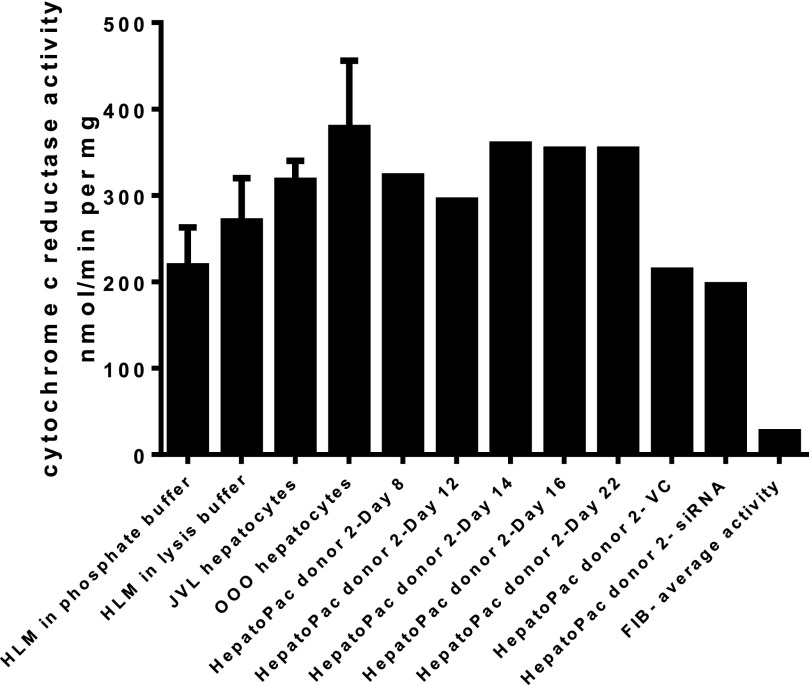
To determine whether downregulation of CYP3A4 expression might impact the activity of CYP2C9, a pool of siRNA sequences specific for CYP3A4 was used to alter CYP3A4 expression. Preincubation of HepatoPac cultures for 72 or 96 hours with CYP3A4 siRNA led to an average 60% decrease in CYP3A4 activity with a concomitant mean increase in CYP2C9 activity of 74% (Fig. 1). The mean changes in activity were pooled from data derived from four donors and included repeat experiments in two donors. The CYP2C9 and CYP3A4 activity levels in vector control wells were 85–115% of the transfection medium (Accell)-only wells, confirming that the vector had no impact on activity. Knockdown of CYP3A4 levels was reproducible between donors (n = 4) and experimental dates (n = 6). CYP3A4 siRNA treatment did not affect UGT1A1, CYP2D6, or CPR activities (Figs. 1 and 2). In contrast, lowering of CYP3A4 levels also resulted in a 40% increase in UGT2B7 activity (Fig. 1). CPR activity was stable across the incubation time course (Fig. 2), and application of the lysis solution had no impact on measured activity. It is of note that CPR activity in the HepatoPac samples was also comparable to activity measured from two lots of suspended human hepatocytes and one lot of HLM (Fig. 2).
Fig. 1.
Enzyme activity of multiple enzymes after CYP3A4 siRNA treatment compared with respective vector controls. CYP3A4 (midazolam), CYP2C9 (diclofenac), CYP2D6 (dextromethorphan), UGT1A1 (estradiol), and UGT2B7 (AZT). Statistical analysis was performed to evaluate whether there was any significant difference between groups using a two-tailed unpaired [two-sample equal variance (homoscedastic)] Student t test in Microsoft Excel. The levels of significance are defined as follows: *P < 0.05, ***P < 0.001.
Fig. 2.
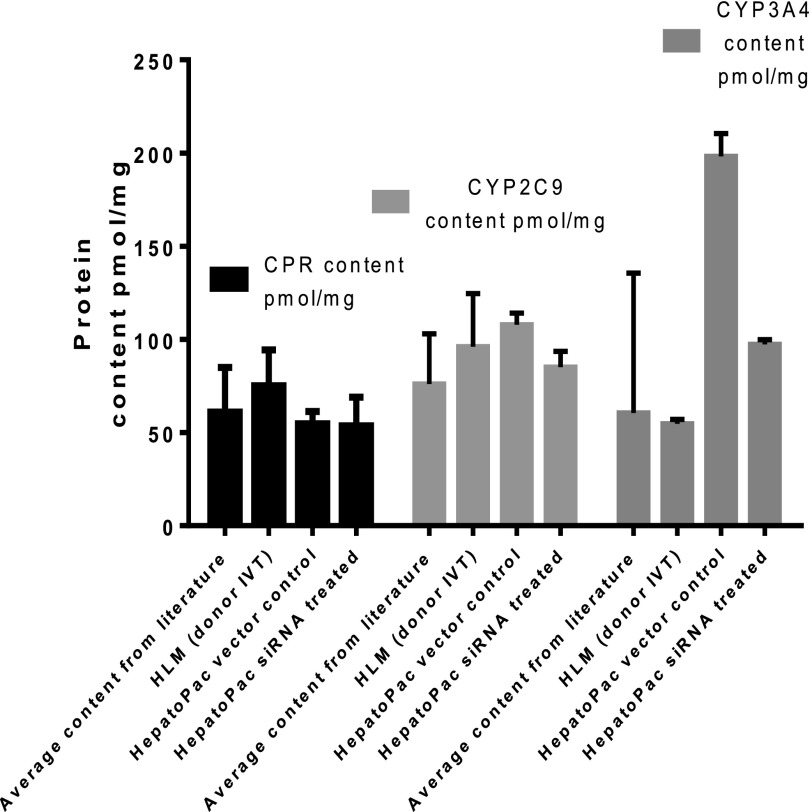
Activity of cytochrome P450 reductase in the HepatoPac model was shown to be stable in the HepatoPac model up through 22 days in culture. CPR activity was measured by pooling n = 12 samples at each time point or condition (vector control or siRNA treated) using donor 2. Activity levels were shown to be similar to the activity levels observed from two separate donor preparations of human hepatocytes and HLM samples. Although there was slightly lower activity observed after treatment of vector control or siRNA, most likely due to the difference in media supplementation for the delivery media, there was no difference between vector control and siRNA treatments, demonstrating that CPR activity is not impacted by reduction in CYP3A4 levels.
Recovery of CYP3A4 after siRNA Treatment.
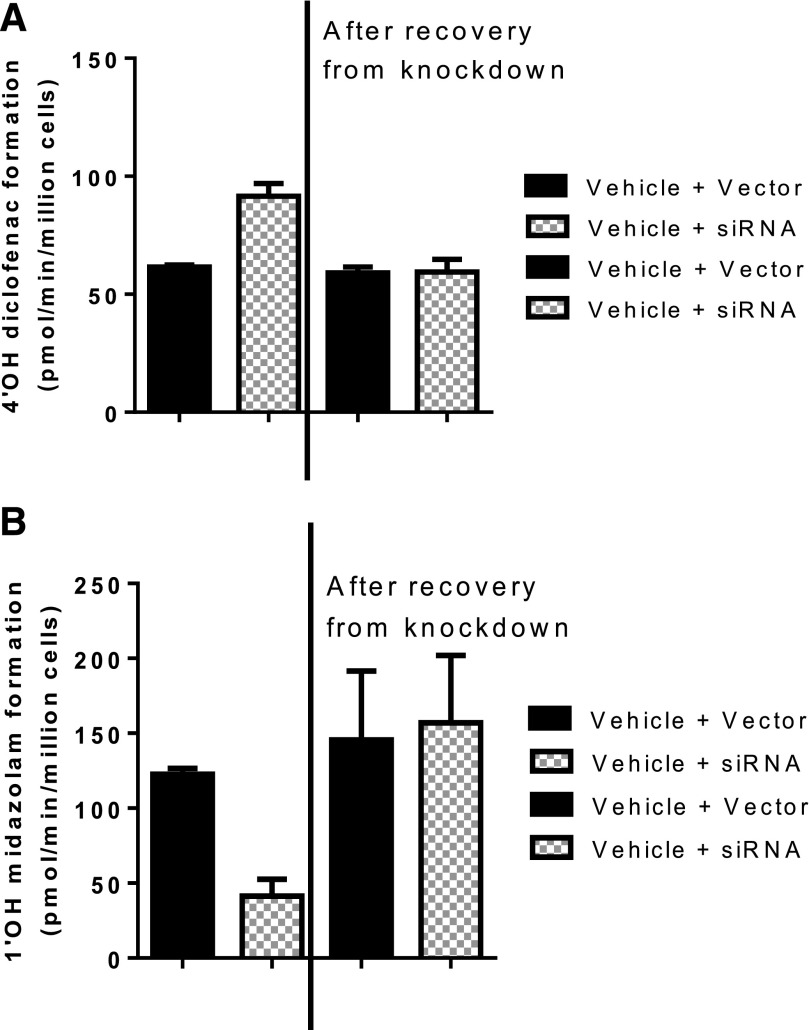
Because downregulation of CYP3A4 expression resulted in concomitant increases in CYP2C9 and UGT2B7 activity, it was important to evaluate whether activity of these two enzymes returned to baseline when the same preparation was allowed to recover to baseline CYP3A4 activity levels. Thus, hepatocytes treated with siRNA were allowed to recover by treating the hepatocyte cultures with standard culture medium (hHCM) and monitoring the activity intermittently (daily or every other day). After 8 days, CYP2C9 activity returned to vector control levels when CYP3A4 activity concomitantly reached baseline levels, suggesting that the modulation of activity was reversible (Fig. 3).
Fig. 3.
The ability of the cells to recover back to vector control treatment levels of enzyme activity was monitored after removal of siRNA treatment of both CYP2C9 (diclofenac, n = 3) and CYP3A4 (midazolam, n = 3). After 72 hours of siRNA treatment and confirmation of reduced CYP3A4 activity (48 hours post-siRNA treatment removal), treated cells were allowed to recover by incubating the cells in hHCM and monitoring the activity of CYP2C9 and CYP3A4 until activity levels rebounded to the vector control treatment data. The activity of CYP3A4 completely recovered after 8 days. (A) Results shown for CYP2C9 activity before and after recovery of siRNA treatment. On the left-hand side of the graph, in the presence of CYP3A4 siRNA (hatched bars), the activity of CYP2C9 (diclofenac metabolism to 4′OH diclofenac) increased above the vector control treatment (solid bars), which is in line with observations from Fig. 1. The right-hand side of the figure demonstrates that, upon increased CYP3A4 activity through recovery from siRNA treatment, the activity of CYP2C9 decreased back to the vector control level. (B) Similarly, upon siRNA treatment, CYP3A4 activity (midazolam metabolism to 1′OH midazolam) was markedly reduced by siRNA treatment (left-hand side hatched bar) compared with vector control (solid bar). The reduction of CYP3A4 activity by siRNA was completely reversible (right-hand side), confirmed by rebound in activity to vector control treatment levels.
Impact of siRNA Treatment on Multiple DME Gene Targets.
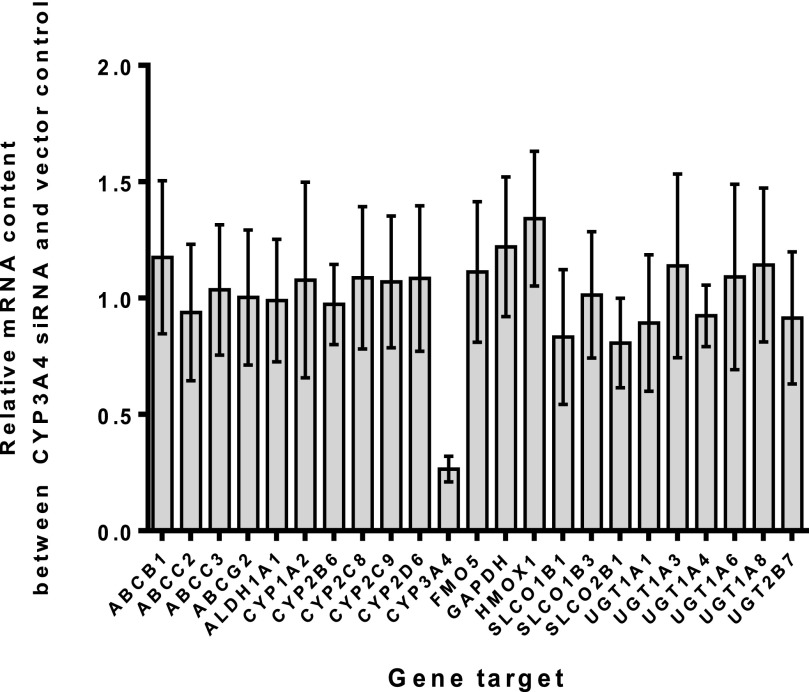
To determine whether this effect of reduced CYP3A4 expression, leading to increased CYP2C9 and UGT2B7 activity, was due to changes in mRNA levels of these enzymes, and to also confirm specificity of siRNA toward CYP3A4, an analysis of mRNA levels was conducted. Only CYP3A4 mRNA was impacted by the siRNA treatment, demonstrating the specificity of the siRNA construct (Fig. 4). Neither CYP2C9 nor UGT2B7 mRNA levels were increased, confirming that the observed increased activity of these enzymes was not due to increased transcription (i.e., higher mRNA levels).
Fig. 4.
Relative quantitation analysis of multiple gene targets was performed to evaluate the specificity of CYP3A4 siRNA treatment on multiple drug-metabolizing enzymes. The data displayed represent the average changes observed from four donors after treatment of CYP3A4 siRNA for 72 hours. Triplicate samples of both vector control and siRNA-treated cells were combined and analyzed in duplicate (siRNA-treated/vector control treatment); mean values displayed from n = 4 donor treatments, each containing n = 3 interassay pooled replicates, n = 2 intra-assay replicates. The data demonstrate that CYP3A4 siRNA was highly specific toward reducing mRNA levels of CYP3A4 (∼90%) when compared with the vector control mRNA levels. No other drug-metabolizing enzyme (including CYP2C9 and UGT2B7) was affected by CYP3A4 siRNA treatment.
Impact of siRNA Treatment on Protein Levels of CPR, CYP2C9, and CYP3A4.
Levels of protein were quantitated by ELISA methodology from vector control and siRNA-treated hepatocytes from donor 2 following the combining of 24 wells of samples (to attain enough protein for analysis) (Fig. 5). Levels of protein were compared with one donor of HLM. There was no impact of siRNA treatment on the level of CPR or CYP2C9 protein. As expected, siRNA targeting CYP3A4 markedly reduced the protein content of CYP3A4 after 72 hours.
Fig. 5.
Quantitation of protein levels of CPR, CYP2C9, and CYP3A4 in HepatoPac donor 2 was achieved using ELISA by pooling n = 24 samples, of donor 2, and measuring n = 2 intra-assay replicates. Concentrations were compared with literature reported (Gan et al., 2009; Ohtsuki et al., 2012) and those generated in the same assay for a single HLM donor. The levels of CPR, CYP2C9, and CYP3A4 protein present in the HepatoPac model were consistent with those present in a single donor of HLM and in literature reports. There was no change in CPR or CYP2C9 content from treatment of the HepatoPac model with siRNA-targeting CYP3A4. The content of CYP3A4 protein decreased by 2.1-fold after treatment with siRNA-targeting CYP3A4.
Impact of CYP3A4 siRNA Treatment on Magnitude of Rifampicin Induction of CYP2C9 and CYP3A4.
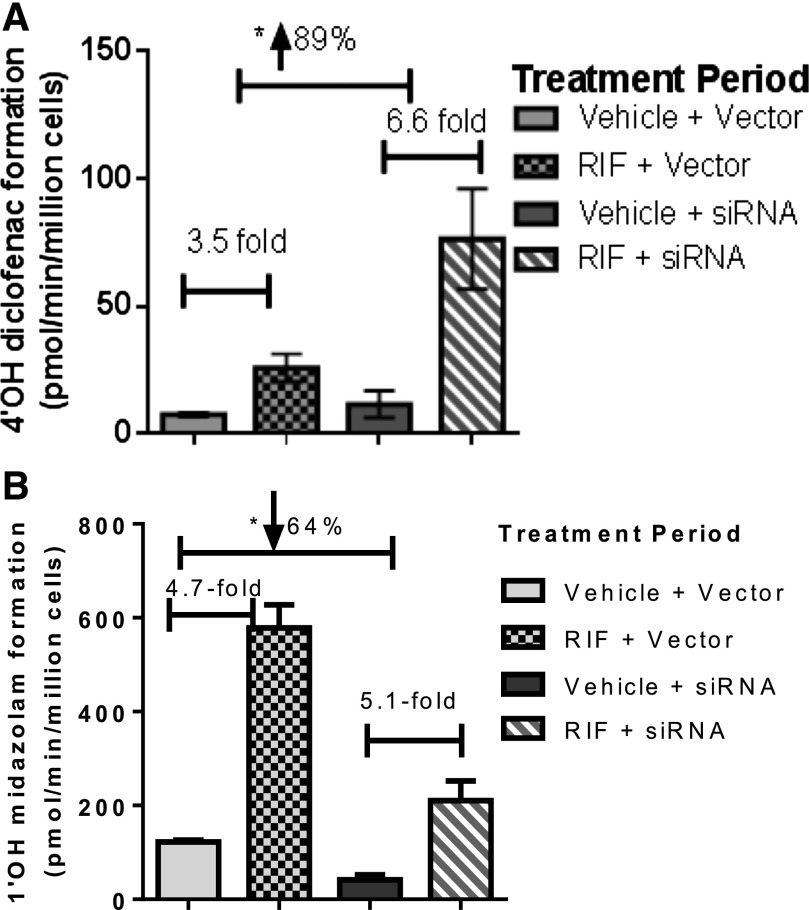
In this set of experiments, CYP3A4 levels were initially reduced by preincubation with siRNA (72 hours), followed by a coincubation including both siRNA and rifampicin (25 µM) (48 hours). The impact of decreased CYP3A4 levels on the extent of observed induction of CYP3A4 was negligible (Fig. 6). Regardless of the absence or presence of siRNA directed against CYP3A4, CYP3A4 was induced approximately fivefold (4.7- and 5.1-fold, respectively) by rifampicin treatment. In contrast, CYP2C9 activity was induced 3.5-fold by rifampicin alone, but in the presence of siRNA against CYP3A4 the extent of induction of CYP2C9 by rifampicin almost doubled to 6.6-fold.
Fig. 6.
Impact of siRNA-mediated CYP3A4 knockdown on induction potentials of CYP2C9 (diclofenac, n = 6) and CYP3A4 (midazolam, n = 6) was determined from duplicate experiments in the same donor of HepatoPac cells (donor 1). Statistical analysis was performed to evaluate whether there was any significant difference between groups using a two-tailed unpaired [two-sample equal variance (homoscedastic)] Student t test in Microsoft Excel. The levels of significance are defined as follows: *P < 0.05. (A) Impact of rifampicin treatment shown on diclofenac activity without (left-hand side) and with (right-hand side) pretreatment of the cells to siRNA-targeting CYP3A4. In the left-hand side of the graph, in which induction by rifampicin was studied in the absence of siRNA, a 3.5-fold increase in CYP2C9 activity was observed. In contrast, when levels of CYP3A4 were selectively reduced prior to induction of CYP2C9 by rifampicin, the magnitude of induction of CYP2C9 activity was significantly increased (89%) to 6.5-fold (right-hand side of the graph). (B) Impact of rifampicin treatment shown on midazolam activity without (left-hand side) and with (right-hand side) pretreatment of the cells to siRNA-targeting CYP3A4. Despite a significant reduction in CYP3A4 activity by siRNA (64%), there was no change in the magnitude of induction observed for CYP3A4 with and without siRNA treatment.
Discussion
Protein-protein interactions can result in changes to activity of specific P450 isoforms (Reed et al., 2012; Reed and Backes, 2012). P450 interacts with other proteins, such as redox transfer proteins that modify P450 activity through provision of electrons from CPR (Lu et al., 1969). Similarly, cytochrome b5 can provide a second electron to P450 (Schenkman and Jansson, 1999, 2003). The disruptive role of detergents (Fisher et al., 2000) also suggests a functional aspect of protein-protein interactions. Early studies with animal P450s (Kaminsky and Guengerich, 1985) confirmed the possibility that protein interactions can alter P450 activity in vivo. Modulating the ratios of different P450 isoforms to study these interactions directly, particularly in humans, is inherently difficult with most studies employing reconstitution systems. To avoid potential artifacts from reconstituted cell systems, such as nonphysiologic P450:CPR ratios, Tan et al. (1997) used differential expression to achieve different molar ratios of enzymes, which demonstrated the usefulness of coexpression systems for mechanistic studies and confirmed that interactions between proteins could occur. Using recombinant P450 and coimmunoprecipitation techniques, Subramanian et al. (2010) provided direct evidence that the N-terminal binding domains of CYP2C9 and CYP3A4 could form quasi-irreversible heteromer complexes (Subramanian et al., 2010). Ionic interactions between P450s were proposed by Kelley et al. (2005), and Takeda et al. (2009) suggested that the J-helix of CYP3A4 contributed to the interaction with UGT2B7 (Takeda et al., 2009). Advancing this concept, hepatocytes represent a more complex and physiologically appropriate environment for evaluating the potential for P450 protein-protein interactions. In particular, the long-term hepatocyte model (HepatoPac) contains relevant concentrations of CPR and P450s over an extended culture time (Fig. 2).
Previous studies demonstrated that increases in CYP3A4 content inhibited CYP2C9 activity by 80% (Subramanian et al., 2010). Similarly, higher ratios of CYP2D6:CYP2C9 resulted in lower activity of CYP2C9 (Subramanian et al., 2009). However, the order of addition of the protein components had differential effects on enzyme activity in these studies, suggesting that the manner in which the reconstituted system was employed could influence the magnitude of the changes, although not the qualitative findings. When CYP2D6 and CPR were initially combined and then CYP2C9 added, higher levels of CPR protein decreased the extent of inhibition. When combining CYP2C9 with CYP2D6 and then adding CPR, higher ratios of CPR had no effect on the decrease in CYP2C9 activity caused by CYP2D6. Thus, a more physiologic system containing relevant ratios of P450s and CPR is more appropriate for evaluating the specificity and magnitude of P450 protein-protein interactions.
Previous experience with HepatoPac confirmed that it represented a physiologic model for studying DMEs (Ramsden et al., 2014a). The use of this long-term hepatocyte culture system allowed the modulation (both decreased and increased) of specific P450 activity in situ through the use of siRNA knockdown of protein levels and induction of P450 proteins, in a system that contains physiologic CPR levels. The long-term culture capability of this system permitted the use of a recovery period for reversal of protein modulation to assess whether the effects of protein-protein interactions on enzyme activity were reversible. In this regard, a 74% increase in CYP2C9 activity was observed upon knockdown of CYP3A4 (Fig. 1), and, upon discontinuation of siRNA treatment and subsequent recovery of expression, both CYP3A4 and CYP2C9 activities returned to pretreatment levels (Fig. 3). These results demonstrate that increases in levels of one protein (e.g., CYP3A4) can modulate the activity of another P450 protein (e.g., CYP2C9) in a system in which the concentration of CPR and baseline ratios of P450s are and remain physiologic. In reconstituted systems, although the exact ratio of the P450s is known, it may not be truly representative of a physiologic ratio, and thus, the magnitude of the interaction observed may be impacted.
Notably, the modulation of enzyme activities by CYP3A4 was somewhat selective. Following a reduction of CYP3A4, there was no change in CPR, CYP2D6, or UGT1A1 activities. This selectivity is congruent with previous work that observed that both CYP3A4 and CYP2D6 concentrations affected CYP2C9 activity, but the reverse was not true (Subramanian et al., 2010), demonstrating that the effect is not universal across all P450s or even bidirectional between two P450s. Interestingly, UGT2B7 activity increased 40%. Interactions between CYP3A4 and UGT2B7, resulting in decreased UGT2B7 enzyme catalysis with increasing concentrations of CYP3A4, have also been observed (Fremont et al., 2005; Takeda et al., 2009). This modulation could be physiologically advantageous based on the roles of these enzymes in the sequential metabolism of xenobiotics. Recently, Ishii et al. (2014) examined possible effects of CYP3A4 on UGT1A isoforms and concluded that the presence of CYP3A4 led to increases in UGT1A1 and UGT1A7 activities, while observing differential effects on the activities of allelic variants of these UGT isoforms (Ishii et al., 2014).
Because competition for CPR is a potential mechanism of interaction between CYP2C9 and CYP3A4, the impact of siRNA treatment on activity and protein levels of CPR was determined. The content and activity of CPR measured in multiple preparations of HLM have confirmed that CPR is markedly less variable when compared with P450s (Shephard et al., 1992; Gan et al., 2009; Ohtsuki et al., 2012), and there is no correlation between CPR and CYP3A4 activities. However, it was important to rule out any potential impact of siRNA treatment on the level and activity of CPR. The data indicated that there was no difference in activity or protein levels of CPR, thus confirming that changes in CPR did not contribute to increased CYP2C9 activity observed after reduction of CYP3A4 levels. The human HepatoPac system provides a normal ratio of P450/CPR, 5.5-fold in this study for CYP2C9 (108 pmol/g) and CYP3A4 (198 pmol/mg) to CPR (54.8 pmol/mg) (Fig. 5), consistent with literature reports in which P450/CPR ranges from 3- to 20-fold (Ohtsuki et al., 2012; Reed and Backes, 2012), and CYP2C9 and CYP3A4 represent ∼40–60% of total P450 (Rowland et al., 2004; Boulenc et al., 2011). Although greater access to CPR with lowered CYP3A4 levels cannot be completely excluded as the reason for the increased CYP2C9 activity, it is consistent with the data from a recombinant system in which CPR was not limiting (Subramanian et al., 2010). Additionally, these data confirmed that the level of CPR protein in the HepatoPac model is physiologic, as it is in line with values reported in HLM preparations.
The effect of increasing levels of CYP3A4 through induction on CYP2C9 activity was also evaluated. Without siRNA pretreatment, rifampicin induced CYP2C9 activity 3.5-fold and CYP3A4 activity 5-fold. Upon knockdown of CYP3A4, induction by rifampicin increased CYP2C9 and CYP3A4 to a similar extent (6.6-fold and 5.1-fold, respectively; Fig. 6). Thus, the magnitude of CYP2C9 induction was dependent on the ratio of CYP2C9:CYP3A4 isoforms, suggesting that the degree of induction of a given P450 in humans may be impacted by baseline levels of another P450, adding another level of complexity to predicting the magnitude of P450 induction. These data also provide additional evidence that baseline levels of CYP3A4 could be modulating CYP2C9 activities in vivo in humans.
As stated by Subramanian et al. (2009), the variability of CYP2D6 in the human population, from the extremes of null alleles, where CYP2D6 is absent, to ultra-rapid metabolizers exhibiting multiple gene copies (Rebsamen et al., 2009), should contribute to the overall variability of activity observed with CYP2C9 and emphasizes the need to measure enzyme activities as an end point rather than protein levels (or mRNA levels). Similarly, from the present study, higher CYP2C9 activities (74%) with reduced CYP3A4 levels (60%) could contribute to variability of CYP2C9 activity in the population. However, the extent to which protein-protein interactions contribute to the overall variability, compared with differences in mRNA levels, transcriptional efficiencies, and induction, is unclear.
DDIs have generally been considered to occur through drug-mediated modulation of the activity or expression of DMEs. Although direct inhibition of DMEs remains the most common cause of DDIs (Hollenberg, 2002), induction (Lehmann et al., 1998) or cytokine-mediated suppression of DMEs (Morgan, 2001; Morgan et al., 2008) has also triggered safety concerns. Can protein interactions additionally contribute to DDI through alteration of the activity of a P450 of interest (e.g., CYP2C9) by modulation of another P450 (e.g., CYP3A4)?
The extended period of stability of highly functional enzyme activity offered by the human HepatoPac system (Khetani and Bhatia, 2008) permitted alteration of levels of CYP3A4 by siRNA in situ to clearly demonstrate a change in activity of CYP2C9. Together with previous work (Subramanian et al., 2010), there is a strong suggestion that the change in CYP2C9 activity is a consequence of protein-protein interactions with CYP3A4. Additional analysis on the impact of selective decreases of CYP3A4, mediated by time-dependent inactivators or suppressors (i.e., IL-6), on CYP2C9 activity is ongoing and may provide further insights into the potential clinical consequences of protein-protein interactions to the overall drug-drug interaction potential.
Acknowledgments
The authors thank Dr. Jinling Xie for thoughtful discussions on RNA interference Technologies and training in mRNA isolation and relative quantitation.
Abbreviations
- AZT
azidothymidine
- CPR
cytochrome P450 reductase
- DDI
drug-drug interaction
- DME
drug-metabolizing enzyme
- ELISA
enzyme-linked immunosorbent assay
- hHCM
human HepatoPac culture medium
- HLM
human liver microsome
- P450
cytochrome P450
- siRNA
small interfering RNA
- UGT
UDP-glucuronosyltransferase
Authorship Contributions
Participated in research design: Ramsden, Chan, Tracy.
Conducted experiments: Ramsden.
Performed data analysis: Ramsden.
Wrote or contributed to the writing of the manuscript: Ramsden, Tweedie, Tracy.
Footnotes
This work was supported by Boehringer Ingelheim Pharmaceuticals and National Institutes of Health [Grant GM 086891 to T.S.T.].
References
- Boulenc X, Schmider W, Barberan O. (2011) Vitro/In Vivo correlation for DrugGÇôDrug interactions, in Drug Discovery and Evaluation: Methods in Clinical Pharmacology (Vogel H, Maas J, Gebauer A. eds) pp 133–160, Springer, Berlin [Google Scholar]
- Chan T, Yu H, Moore A, Khetani S, and Tweedie DJ (2013) Meeting the challenge of predicting hepatic clearance of compounds slowly metabolized by cytochrome P450 using a novel hepatocyte model, HepatoPacTM. Drug Metab Dispos 41:2024–2032. [DOI] [PubMed] [Google Scholar]
- Fisher MB, Campanale K, Ackermann BL, VandenBranden M, Wrighton SA. (2000) In vitro glucuronidation using human liver microsomes and the pore-forming peptide alamethicin. Drug Metab Dispos 28:560–566 [PubMed] [Google Scholar]
- Fremont JJ, Wang RW, King CD. (2005) Coimmunoprecipitation of UDP-glucuronosyltransferase isoforms and cytochrome P450 3A4. Mol Pharmacol 67:260–262 [DOI] [PubMed] [Google Scholar]
- Gan L, von Moltke LL, Trepanier LA, Harmatz JS, Greenblatt DJ, Court MH. (2009) Role of NADPH-cytochrome P450 reductase and cytochrome-b5/NADH-b5 reductase in variability of CYP3A activity in human liver microsomes. Drug Metab Dispos 37:90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giragossian C, LaPerle J, Kosa RE, Gillian S. (2009) Impact of time-dependent inactivation on the estimation of enzyme kinetic parameters for midazolam. Drug Metab Lett 3:45–53 [DOI] [PubMed] [Google Scholar]
- Gómez-Lechón MJ, Castell JV, Donato MT. (2007) Hepatocytes—the choice to investigate drug metabolism and toxicity in man: in vitro variability as a reflection of in vivo. Chem Biol Interact 168:30–50 [DOI] [PubMed] [Google Scholar]
- Gómez-Lechón MJ, Donato T, Ponsoda X, Castell JV. (2003) Human hepatic cell cultures: in vitro and in vivo drug metabolism. Altern Lab Anim 31:257–265 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Martin MV, Sohl CD, Cheng Q. (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4:1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazai E, Bikádi Z, Simonyi M, Kupfer D. (2005) Association of cytochrome P450 enzymes is a determining factor in their catalytic activity. J Comput Aided Mol Des 19:271–285 [DOI] [PubMed] [Google Scholar]
- Hollenberg PF. (2002) Characteristics and common properties of inhibitors, inducers, and activators of CYP enzymes. Drug Metab Rev 34:17–35 [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. (2000) Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J 347:321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Koba H, Kinoshita K, Oizaki T, Iwamoto Y, Takeda S, Miyauchi Y, Nishimura Y, Egoshi N, Taura F, et al. (2014) Alteration of the function of the UDP-glucuronosyltransferase 1A subfamily by cytochrome P450 3A4: different susceptibility for UGT isoforms and UGT1A1/7 variants. Drug Metab Dispos 42:229–238 [DOI] [PubMed] [Google Scholar]
- Kaminsky LS, Guengerich FP. (1985) Cytochrome P-450 isozyme/isozyme functional interactions and NADPH-cytochrome P-450 reductase concentrations as factors in microsomal metabolism of warfarin. Eur J Biochem 149:479–489 [DOI] [PubMed] [Google Scholar]
- Kelley RW, Reed JR, Backes WL. (2005) Effects of ionic strength on the functional interactions between CYP2B4 and CYP1A2. Biochemistry 44:2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. (2008) Microscale culture of human liver cells for drug development. Nat Biotechnol 26:120–126 [DOI] [PubMed] [Google Scholar]
- Legendre C, Hori T, Loyer P, Aninat C, Ishida S, Glaise D, Lucas-Clerc C, Boudjema K, Guguen-Guillouzo C, Corlu A, et al. (2009) Drug-metabolising enzymes are down-regulated by hypoxia in differentiated human hepatoma HepaRG cells: HIF-1alpha involvement in CYP3A4 repression. Eur J Cancer 45:2882–2892 [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DN, Pritchard MP, Hanlon SP, Burchell B, Wolf CR, Friedberg T. (1999) Competition between cytochrome P-450 isozymes for NADPH-cytochrome P-450 oxidoreductase affects drug metabolism. J Pharmacol Exp Ther 289:661–667 [PubMed] [Google Scholar]
- Locuson CW, Wienkers LC, Jones JP, Tracy TS. (2007) CYP2C9 protein interactions with cytochrome b(5): effects on the coupling of catalysis. Drug Metab Dispos 35:1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu AY, Junk KW, Coon MJ. (1969) Resolution of the cytochrome P-450-containing omega-hydroxylation system of liver microsomes into three components. J Biol Chem 244:3714–3721 [PubMed] [Google Scholar]
- Morgan ET. (1997) Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev 29:1129–1188 [DOI] [PubMed] [Google Scholar]
- Morgan ET. (2001) Regulation of cytochrome p450 by inflammatory mediators: why and how? Drug Metab Dispos 29:207–212 [PubMed] [Google Scholar]
- Morgan ET, Goralski KB, Piquette-Miller M, Renton KW, Robertson GR, Chaluvadi MR, Charles KA, Clarke SJ, Kacevska M, Liddle C, et al. (2008) Regulation of drug-metabolizing enzymes and transporters in infection, inflammation, and cancer. Drug Metab Dispos 36:205–216 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92 [DOI] [PubMed] [Google Scholar]
- Ramsden D, Tweedie DJ, Chan TS, Taub ME, Li Y. (2014a) Bridging in vitro and in vivo metabolism and transport of faldaprevir in human using a novel cocultured human hepatocyte system, HepatoPac. Drug Metab Dispos 42:394–406 [DOI] [PubMed] [Google Scholar]
- Ramsden D, Tweedie DJ, St George R, Chen LZ, Li Y. (2014b) Generating an in vitro-in vivo correlation for metabolism and liver enrichment of a hepatitis C virus drug, faldaprevir, using a rat hepatocyte model (HepatoPac). Drug Metab Dispos 42:407–414 [DOI] [PubMed] [Google Scholar]
- Rebsamen MC, Desmeules J, Daali Y, Chiappe A, Diemand A, Rey C, Chabert J, Dayer P, Hochstrasser D, Rossier MF. (2009) The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J 9:34–41 [DOI] [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Connick JP, Cheng D, Cawley GF, Backes WL. (2012) Effect of homomeric P450-P450 complexes on P450 function. Biochem J 446:489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland YK, Rostami-Hodjegan A, Tucker GT. (2004) Abundance of cytochrome P450 in human liver: a meta-analysis. Br J Clin Pharmacol 57:687–688 [Google Scholar]
- Schenkman JB, Jansson I. (1999) Interactions between cytochrome P450 and cytochrome b5. Drug Metab Rev 31:351–364 [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152 [DOI] [PubMed] [Google Scholar]
- Shephard EA, Palmer CN, Segall HJ, Phillips IR. (1992) Quantification of cytochrome P450 reductase gene expression in human tissues. Arch Biochem Biophys 294:168–172 [DOI] [PubMed] [Google Scholar]
- Shimada T, Mernaugh RL, Guengerich FP. (2005) Interactions of mammalian cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b(5) enzymes. Arch Biochem Biophys 435:207–216 [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. (2009) CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos 37:1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam H, Zheng H, Tracy TS. (2010) CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos 38:1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Ishii Y, Iwanaga M, Nurrochmad A, Ito Y, Mackenzie PI, Nagata K, Yamazoe Y, Oguri K, Yamada H. (2009) Interaction of cytochrome P450 3A4 and UDP-glucuronosyltransferase 2B7: evidence for protein-protein association and possible involvement of CYP3A4 J-helix in the interaction. Mol Pharmacol 75:956–964 [DOI] [PubMed] [Google Scholar]
- Tan Y, Patten CJ, Smith T, Yang CS. (1997) Competitive interactions between cytochromes P450 2A6 and 2E1 for NADPH-cytochrome P450 oxidoreductase in the microsomal membranes produced by a baculovirus expression system. Arch Biochem Biophys 342:82–91 [DOI] [PubMed] [Google Scholar]
- Wang WW, Khetani SR, Krzyzewski S, Duignan DB, Obach RS. (2010) Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab Dispos 38:1900–1905 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Gillam EM, Dong MS, Johnson WW, Guengerich FP, Shimada T. (1997) Reconstitution of recombinant cytochrome P450 2C10(2C9) and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450-P450 and cytochrome P450-b5 interactions. Arch Biochem Biophys 342:329–337 [DOI] [PubMed] [Google Scholar]
- Zangar RC, Davydov DR, Verma S. (2004) Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharmacol 199:316–331 [DOI] [PubMed] [Google Scholar]