Abstract
Fenofibrate is a peroxisome proliferator-activated receptor (PPAR) α ligand that has been widely used as a lipid-lowering agent in the treatment of hypertriglyceridemia. ABCD2 (D2) is a peroxisomal long-chain acyl-CoA transporter that is highly induced by fenofibrate in the livers of mice. To determine whether D2 is a modifier of fibrate responses, wild-type and D2-deficient mice were treated with fenofibrate for 14 days. The absence of D2 altered expression of gene clusters associated with lipid metabolism, including PPARα signaling. Using 3T3-L1 adipocytes, which express high levels of D2, we confirmed that knockdown of D2 modified genomic responses to fibrate treatment. We next evaluated the impact of D2 on effects of fibrates in a mouse model of diet-induced obesity. Fenofibrate treatment opposed the development of obesity, hypertriglyceridemia, and insulin resistance. However, these effects were unaffected by D2 genotype. We concluded that D2 can modulate genomic responses to fibrates, but that these effects are not sufficiently robust to alter the effects of fibrates on diet-induced obesity phenotypes.
Introduction
Fenofibrate has been widely used clinically to treat dyslipidemia. As an agonist of peroxisome proliferator-activated receptor (PPAR) α, fenofibrate has been shown to significantly reduce triglyceride (TG) levels and raise high density lipoprotein cholesterol levels in plasma (Schoonjans et al., 1996b; Staels et al., 1998). However, the clinical response to fenofibrate therapy is highly variable (Farnier, 2008). It has been proposed that response variability may be partially due to gene-drug interactions (Lai et al., 2007). Polymorphisms at several genes have been reported to interact with the lipid-lowering effects of fenofibrate during the postprandial period (Liu et al., 2008; Shen et al., 2008; Irvin et al., 2010; Wojczynski et al., 2010; Aslibekyan et al., 2012a; Frazier-Wood et al., 2013; Kraja et al., 2013). However, only a limited number of genes have been investigated to date, and the effects of other candidate genes, especially those that play important roles in lipid metabolism, remain unexplored.
ABCD2 (D2) is a mammalian peroxisomal ABC transporter thought to facilitate very long-chain acyl-CoA import into peroxisomes for β-oxidation (Ferrer et al., 2005; Fourcade et al., 2009; Weber et al., 2014). Impaired peroxisome function leads to very long-chain fatty acid (VLCFA) and branched-chain fatty acid accumulation in plasma and tissues, resulting in severe metabolic diseases (Wanders and Waterham, 2006). X-linked adrenoleukodystrophy is a peroxisomal disorder caused by inactivation of the ABCD1 (D1) gene (Mosser et al., 1993). D2 shares 66% amino acid identity with D1 and is presumed to have overlapping substrate specificity based on complementation studies in yeast (Lombard-Platet et al., 1996; van Roermund et al., 2011). Additionally, a ubiquitous promoter-controlled D2 transgene corrected both biochemical and neurologic phenotypes in D1-deficient mice, indicating that they share overlapping functions (Lu et al., 1997; Pujol et al., 2004).
D2 protein has been detected in adrenal, brain, kidney, liver, and skeletal muscle, but is most abundant in adipose (Liu et al., 2010). Mice lacking D2 are characterized by late-onset cerebellar and sensory ataxia with loss of cerebellar Purkinje cells and accumulation of VLCFA in dorsal root ganglia cells (Ferrer et al., 2005). D2-deficient mice also showed evidence of oxidative stress with spontaneous and premature ceroid deposition in adrenal medulla cells (Lu et al., 2007), reduced rates of VLCFA (C26:0) and polyunsaturated fatty acid (PUFA) (C24:6ω3) oxidation, as well as altered long-chain fatty acid (LCFA) and VLCFA levels (C20:0, 22:1ω9, 22:6ω3, and 22:5ω6) in the cerebral cortex (Fourcade et al., 2009). D1 and D2 appear to cooperate in monounsaturated C26:1ω9 import (Fourcade et al., 2010) as serum levels of C22:1ω9, a fatty acid associated with disruptions in lipid metabolism in rats and other species (Bremer and Norum, 1982), were elevated in fed and fasting states (Fourcade et al., 2009; Liu et al., 2010). The absence of D2 sensitized mice to dietary erucic acid, resulting in the rapid onset of obesity and insulin resistance (Liu et al., 2012).
The expression of D2 is dynamically regulated by several transcription factors, including PPARα, sterol regulatory element-binding proteins 1a and 1c, retinoid X receptor, thyroid hormone receptor, liver X receptor (LXR) α, and T-cell factor (TCF) 4/β-catenin (Fourcade et al., 2001; Weinhofer et al., 2005, 2008; Park et al., 2013; Gondcaille et al., 2014). D2 expression levels in liver are near the lower limit of reverse transcription-polymerase chain reaction (rtPCR) and immunoblotting detection, but are dramatically upregulated by PPARα agonists and in genetic and high-fat feeding obesity models (Berger et al., 1999; Liu et al., 2012). Although no functional peroxisome proliferator response element has been identified in the D2 promoter, the expression of D2 is induced by fibrate treatment and is PPARα dependent (Fourcade et al., 2001). Prolonged fasting for 48 hours was also shown to increase both hepatic D2 expression and the amount of some peroxisomal metabolites (C20:0 and C22:1) in mouse serum (Fourcade et al., 2009). Collectively, these data demonstrate a number of key functions for D2 as a lipid homeostasis regulator. However, the precise physiologic role of D2 in lipid metabolism remains largely unknown.
Our initial studies revealed that the absence of D2 altered the genomic response to fenofibrate treatment in mouse liver and confirmed that D2 protein levels altered responses to fibrate treatment in vitro. This prompted us to propose that D2 may play a role in the regulation of PPARα signaling and response to fenofibrate therapy. We next tested the hypothesis that D2 would alter the effect of fibrate in a model of diet-induced obesity. Fibrates decreased weight gain, adiposity, hyperlipidemia, and insulin resistance, but these effects were not altered by the D2 genotype.
Materials and Methods
Analysis of Hepatic Gene Expression and Sample Preparation
Animals.
D2-deficient mice are maintained on C57Bl/6J background and strain refreshed every five generations to C57Bl/6J (strain 000664). Age-matched (3-month, n = 6) wild-type (WT) and Abcd2 (D2) knockout (KO) male mice on the C57Bl/6J background were gavaged (200 µl/d) with fenofibrate (100 mg/kg per day) dispersed in water containing 3% gum arabic for 14 days, which is sufficient time for fenofibrate treatment to alter the transcriptome and significantly decrease plasma triglycerides (Schoonjans et al., 1996a; Guillou et al., 2002; Lu et al., 2011). A shorter-term treatment period was not used because the effects of fenofibrate on hepatic lipid metabolism, peroxisome proliferation, and plasma lipids are not well established at less than 14 days. Control mice were gavaged with vehicle alone (3% gum arabic). Livers were dissected from the mice, and total RNA was extracted using RNeasy kit (Qiagen, Valencia, CA ).
Microarray.
A cDNA microarray was manufactured at Institut de Génétique et de Biologie Moléculaire et Cellulaire (Illkirch-Graffenstaden, France), and the experiments were carried out as previously described (Schluter et al., 2012). The clones were selected from public cDNA libraries, the NIA 15K (http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html) and the IMAGE collection (http://image.llnl.gov/), whose length and sequence specificity were optimized to reduce the cross-hybridization problems encountered with cDNA-based microarrays. Selection of the unique gene set was done by all-against-all similarity searches with BLAST algorithm with a cutoff score of 800 to group expressed sequence tags (ESTs) into unique genes, as previously described (Tanaka et al., 2000). The array consisted of 20,736 mouse cDNA clones representing approximately a total of 15,000 unique genes or UniGene clusters. Some clones were also acquired from a private cDNA library collection (C. Benoist, Joslin Diabetes Center, Boston, MA). The cDNAs were amplified by polymerase chain reaction (PCR) using vector-specific primers, purified using MultiScreen FB filter plates (Millipore, Billerica, MA), and verified by gel electrophoresis. The length of cDNA inserts was in the range of 300–1200 bp, with a mean value of ∼750 bp. cDNA library constructions were based on oligo-dT–primed reverse transcription of total RNA (Tanaka et al., 2000). After dehydration, PCR products were resuspended in the spotting buffer (75% formamide and 25% water) and spotted in duplicate onto Ultra-GAPSTM amino-silane–coated slides (Corning BV, Schiphol-Rik, The Netherlands) using a MicroGrid II Arrayer (BioRobotics, Cambridge, UK). The printed slides were dried for 48 hours and cross-linked by UV at 600 mJ/cm2. For probe synthesis in the dye swap experiments, total RNA (200 ng) was amplified by linear PCR and the amplification products were labeled with dUTP-Cy3 and dUTP-Cy5 by random priming (Bioprime; Invitrogen, Grand Island, NY). Labeled cDNAs were purified using Nucleospin Extract II columns (Macherey Nagel, Düren, Germany) and hybridized in a Discovery station using ChipHybe 80 hybridization buffer at 42°C for 12 hours without any final stringency washes (hybridization automate, reagents, and microarray hybridization procedure are from Ventana Medical System, Tucson, AZ). Arrays were scanned using ScanArray 4000 (Packard Biochips, Billerica, MA), and images were quantified using Imagene 5.0 (BioDiscovery Microarray Bioinformatics Software, Marina Del Rey, CA). The microarray experiment has been deposited in the Array Express Database under accession number E-MTAB-2006.
Hybridization and Data Processing.
All samples were hybridized in duplicate and analyzed using dye swap to avoid label-specific bias. Preprocessing of raw data and statistical analyses were performed using Bioconductor packages in R programming environment (Gentleman et al., 2004). For mouse custom cDNA arrays, background correction was performed using the “normexp” method implemented in the Bioconductor LIMMA package to adjust local median background estimates (Gentleman et al., 2005; Smyth, 2005). Background-corrected intensity data were normalized using the Print-tip loess method to remove the bias within each array and the A-values quantile normalization (Aquantile) to remove the bias between arrays.
Functional Enrichment Analysis.
To evaluate which pathways or functional categories were enriched in differentially expressed genes, we computed both the gene set enrichment analysis (G) and hypergeometric distribution function (H). We used P < 0.05 as the cutoff point to determine whether the metabolic and cell-signaling pathways extracted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) (Kanehisa and Goto, 2000) were significantly enriched. GeneSetTest function from LIMMA package tests whether a set of genes is enriched for differential expression. Its principle is the same as for Gene Set Enrichment Analysis introduced by Mootha et al. (2003), but the statistical tests used are different. It is based on a set of probe-wise t-statistics arising for microarray analysis. We have computed three different tests, as follows: 1) upregulated genes, with positive t-statistics, as indicated by red spots; 2) downregulated genes, with negative t test statistics, as indicated by green spots; and 3) genes differentially expressed regardless of the direction, as indicated by black spots. The sizes of spots are scaled positively with the probability values in a semiquantitative pattern. We used the “phyper” hypergeometric distribution function from stats package to determine enriched Kyoto Encyclopedia of Genes and Genomes pathways in the subset of genes differentially expressed at P < 0.05.
Impact of D2 on Fibrate Response In Vitro
Generation of D2-Deficient 3T3-L1 Cell Line.
The 3T3-L1 cells were propagated as fibroblasts in subconfluent cultures in Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with antibiotics and 10% newborn calf serum. One day postconfluence, cells were infected with lentiviral particle expressing short hairpin RNA (shRNA) directed against D2 [sequence: 5′-CCGGCCTCGGACTTTCATCATCAAACTCGAGTTTGATGATGAAAGTCCGAGGTTTTTG-3′; Sigma-Aldrich (St. Louis, MO) MISSION shRNA Lentiviral Transduction Particles TRCN0000105346] and a control shRNA in the presence of hexadimethrine bromide (polybrene, 8 μg/ml). Cell-viral particle mixtures were incubated in 37°C overnight. Medium was replaced with DMEM/F12 containing antibiotics and 10% newborn calf serum. Puromycin (2 μg/ml) was added to select successfully transduced cells. Medium was replaced every 3 days until resistant clones could be identified. Several puromycin-resistant colonies were selected by limiting dilution, expanded separately in medium containing reduced amount of puromycin (1 μg/ml), and frozen in liquid nitrogen. Cell lysates of each expanded colony were prepared and subjected to immunoblot analysis to confirm knockdown of D2.
Adipocyte Differentiation and Fenofibrate Treatment.
D2-deficient 3T3-L1 and control cells were propagated as fibroblasts in subconfluent cultures in DMEM/F12 supplemented with antibiotics and 10% newborn calf serum. Two days postconfluence (day 2), cells were treated with 1.7 µM insulin, 0.5 µM dexamethasone, 0.5 mM isobutylmethylxanthine, and 1 µM rosiglitizone in DMEM/F12 supplemented with 10% fetal calf serum and antibiotics for 48 hours to promote differentiation to adipocytes, as previously described (Liu et al., 2010). Thereafter, cells were cultured in DMEM/F12 supplemented with 10% fetal calf serum. Four days postconfluence (day 4), the medium was replaced with DMEM/F12 supplemented with antibiotics, 10% fetal calf serum, and 1 μg/ml insulin. Fresh medium was replaced every 2 days. The 3T3-L1 adipocytes were analyzed between 8 and 12 days postconfluence. Eight days postconfluence (day 8), adipocytes were treated with carrier (dimethylsulfoxide) or fenofibrate (500 mM) for 16 hours. Cells were washed with phosphate-buffered saline (4°C), and total RNA was extracted using RNeasy kit (Qiagen).
Protein Sample Preparation and Immunoblotting.
Cell lysates were prepared by incubating the cells in Triton lysis buffer (80 mM NaCl, 50 mM Tris, 2 mM CaCl2, 1% Triton, pH 8.0) at 4°C for 30 minutes, scraped off the dish, and homogenized with polytron homogenizer. Samples were added with protein sample buffer (30 mM Tris base, 10 mM EDTA, pH 6.8, 3% SDS, 20% glycerol, and 0.00625% bromphenol blue) to achieve uniform concentrations. β-mercaptoethanol was added to a final concentration of 1.2% (v/v). Samples were heated to 95°C for 5 minutes before loading. Proteins were size-fractionated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were incubated in buffer A (20 mM Tris, 137 mM NaCl, 0.2% Tween 20, and 5% nonfat milk, pH 7.6) for 30 minutes at room temperature. Anti-ABCD2 antibody (Liu et al., 2010) was diluted in buffer A and incubated with membranes for 60 minutes at room temperature. Membranes were washed three times for 5 minutes in buffer B (20 mM Tris, 137 mM NaCl, and 0.2% Tween 20, pH 7.6). Horseradish peroxidase–conjugated goat anti-rabbit IgG were diluted in buffer B and incubated with membranes for 30 minutes at room temperature. Membranes were washed three times for 5 minutes in buffer B and visualized by enhanced chemiluminescence (SuperSignal West Pico; Thermo Scientific, Waltham, MA).
Quantitative Real-Time PCR.
Total RNA was reverse transcribed using SuperScript First Strand Synthesis system (Invitrogen). Quantitative rtPCR was performed on an Applied Biosystems 7900HT sequence detection system. Standard reaction volume was 30 μl containing 1× SYBR Green PCR master mix (Applied Biosystems, Grand Island, NY), 1 μl cDNA template, and 150 nM concentrations of each oligonucleotide. Initial steps of rtPCR were 10 minutes at 95°C. Cycles (n = 40) consisted of a 15-second melt at 95°C, followed by a 1-minute annealing/extension at 60°C. All reactions were performed in triplicate. Means of the differences in threshold cycle (CT) values from cyclophilin and their S.D. were calculated for each treatment group (ΔCT). The relative abundance of each transcript within treatment groups was determined by subtracting the control group mean difference from the remaining treatment groups (ΔΔCT) and calculated according to the expression 2−ΔΔCT. The S.D. of the difference between control and each treatment group was calculated as the square root of the sums of squares for the S.D. of the ΔCT means.
Impact of D2 on Fibrate Response In Vivo
D2-deficient mice are maintained on C57Bl/6J background and strain refreshed every five generations (The Jackson Laboratory, Bar Harbor, ME). The C57Bl/6J strain is a commonly used mouse model that is sensitive to high-fat feeding (Collins et al., 2004) and readily responds to fenofibrate treatment (Chaput et al., 2000; Guerre-Millo et al., 2000).
At 8 weeks of age, male Abcd2 KO and their WT littermates were housed four per cage with both genotypes randomly allocated and provided free access to a high-fat diet (control, 60% kCal; Research Diets, New Brunswick, NJ) or the same diet formulated with fenofibrate (Fibrate, 60% kCal plus 0.05% w/w fenofibrate; Cayman Chemical, Ann Arbor, Michigan). Mice were housed in a 10-hour dark/14-hour light cycle. Every week, mice were weighed and food was replaced. Fasting blood glucose levels and body composition (analyzed by magnetic resonance imaging; EchoMRI, Houston, TX) were measured at the initiation (8 weeks of age) and every 4 weeks of the study. A glucose tolerance test was conducted at the midpoint (8 weeks on diet) and the end (16 weeks on diet) of the study.
At 17 weeks, a subgroup of mice was placed in the metabolic cages to record metabolic parameters. Each mouse was housed separately and provided with free access to diet and water. Mice were allowed to acclimatize for 48 hours. Metabolic data were collected for the next 72 hours. Airflow rate (flow, l/ml), oxygen percentage (dO2, %), carbon dioxide percentage (dCO2, %), food and water consumption, and locomotor activity were measured. Mean values of energy expenditure (H) in the active (dark), resting (light) phase and whole day average were calculated for each mouse. Because the data displayed the same pattern for both active and resting phases, we only show the whole day average data in this work.
Blood samples of these mice were taken using heparinized capillary tubes before and 0.5, 1, 2, and 3 hours after tail vein injection of 650 mg/kg body weight of Triton WR-1339 (Sigma-Aldrich). Total plasma TG concentrations of all time points were determined, and TG secretion rates were calculated using linear regression analysis.
At 16 weeks, mice were anesthetized with ketamine/xylazine (9/1, w/w) for tissue collection. Tissues were snap-frozen in liquid nitrogen, embedded in Tissue-Tek O.C.T. (Sakura Finetek USA, Torrance, CA), formalin-fixed (10% in phosphate-buffered saline), and stored at −80°C until subsequent analysis. All animal procedures were performed with the approval of the Institutional Animal Care and Use Committee.
Statistical Analysis of Data.
Data were analyzed by two-way analysis of variance and presented as mean with S.E.M. Bonferroni post hoc tests were employed, where indicated. All statistical analyses were conducted using GraphPad Prism statistical analysis software (GraphPad Software, La Jolla, CA).
Impact of D2 Polymorphisms on Fibrate Responses in Human Subjects
Study Population.
The Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study, described in detail in previous publications (Corella et al., 2007; Aslibekyan et al., 2012b), was designed to identify genetic markers associated with response to fenofibrate therapy in a population of related metabolically healthy individuals of European American ancestry. Recruitment was restricted to European Americans to minimize potential confounding by population admixture (Aslibekyan et al., 2012b). Participants were asked to discontinue use of lipid-lowering medications, fast for at least 8 hours, and abstain from alcohol prior to study visits. Circulating plasma lipids were measured at baseline and after 3 weeks of daily treatment with 160 mg micronized fenofibrate. Genotype information was collected on 906,600 single-nucleotide polymorphisms (SNPs) using the Affymetrix Genome-Wide Human 6.0 array, and Human Genome Build 36 was used as the reference to impute untyped SNPs. After imputation and quality control, a hybrid set of 2,543,887 SNPs was created, with complete genotype and covariate information available on 861 participants.
SNP Selection.
Of the three ABCD2 variants previously implicated in affecting gene expression in humans (Matsson et al., 2012), the GOLDN data set had genotyped variants that were in perfect linkage disequilibrium (R2 = 1), with two of them, rs7309234 and rs11172592, used as proxies for rs4285917 and rs4284427, respectively. The third SNP, rs10783969, was neither typed nor in linkage disequilibrium with any of the GOLDN variants and was excluded from the analysis.
Statistical Analysis of Data.
Linear mixed models were used to assess the association of each ABCD2 variant with the fenofibrate-induced change in plasma triglycerides, defined as area under the curve calculated using the trapezoidal rule. The models were adjusted for age, sex, and study site as fixed effects, and pedigree as a random effect. Sensitivity analyses were conducted using the alternative phenotype definition of log-transformed plasma triglyceride concentration ratios comparing post- to prefenofibrate treatment levels. The study was adequately powered to detect the causal SNPs (power = 99% for two loci at the significance level of 0.025). To address the multiple testing problem, we implemented a stringent Bonferroni correction, setting the α-level at 0.05/number of SNPs tested = 0.05/2 = 0.025.
Results
Identification of ABCD2-Regulated Pathways with and without Fenofibrate Treatment.
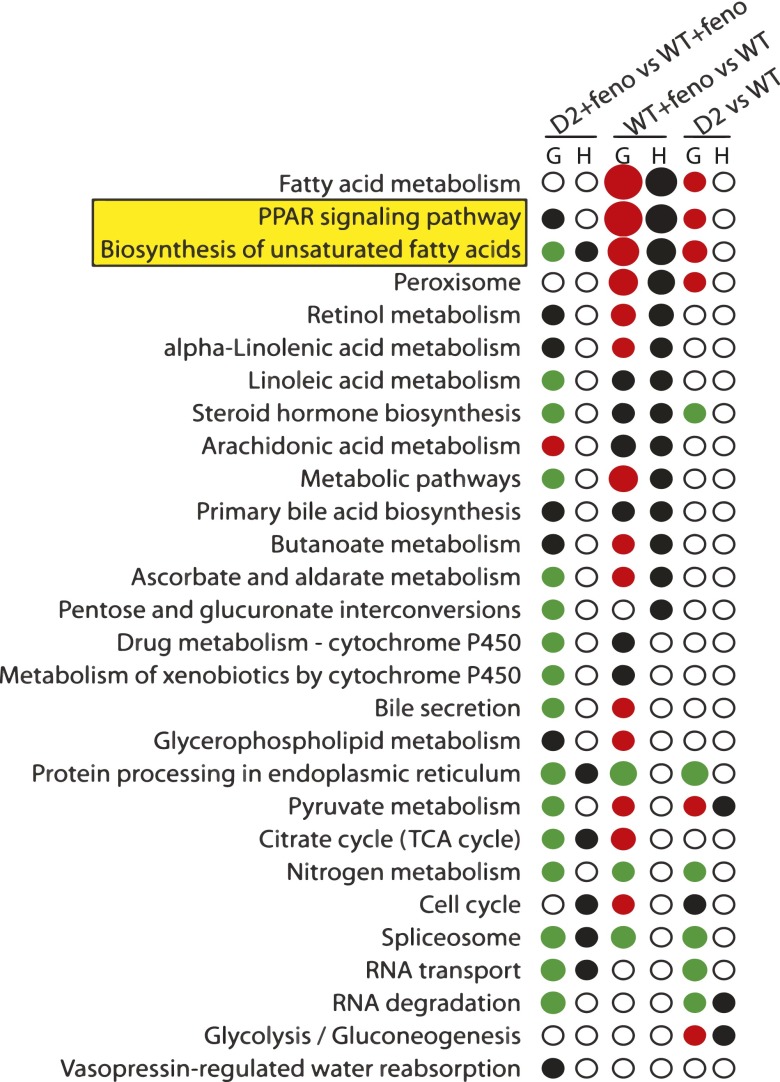
To evaluate the influence of D2 on genomic responses to fibrate treatment, age-matched WT and D2 KO C57Bl/6J mice (n = 6) were administered fenofibrate for 14 days. Total RNA was extracted from liver and subjected to microarray analysis (Supplemental Table 2). To determine which pathways or functional categories were enriched among differentially expressed genes, we computed both the Gene Set Enrichment analysis (G) and Hypergeometric distribution function (H) (Fig. 1). Results were presented as KO + feno compared with WT + feno, WT + feno compared with WT, and KO compared with WT. Red indicates significance with a preponderance for upregulated genes within the pathway or gene cluster. Green indicates significant downregulation. Black spots indicate significance within the cluster, but without overall up or down trend. Open circles indicate no significant differences. The size of the symbols is scaled to the magnitude of the P value. The influence of genotype alone was assessed by comparing WT and KO mice without fenofibrate treatment (D2 versus WT). The effect of fibrate treatment was determined by comparing WT mice with or without fibrate (WT + feno versus WT). The interaction between genotype and treatment was evaluated by comparing WT and KO mice treated with fenofibrate (D2 + feno versus WT + feno).
Fig. 1.
Pathways altered by fibrate treatment (WT + feno vs WT), D2 genotype (D2 vs WT), and the effect of D2 genotype on fibrate response (D2 + feno versus WT + feno) in mouse liver. Significantly deregulated KEGG pathways were identified by using the statistics Gene Set Enrichment Analysis (GSEA) and hypergeometric distribution function (H). Black circles in column H denote significant deregulation by the hypergeometric test (P < 0.05), but do not indicate direction. Circles in column G denote significance deregulation by (P < 0.05) by GSEA. Red indicates upregulation, green indicates downregulation, and black indicates significance within the pathway, but without directional trend (i.e., both up- and downregulated genes in the pathway). Circle size is scaled to the probability value.
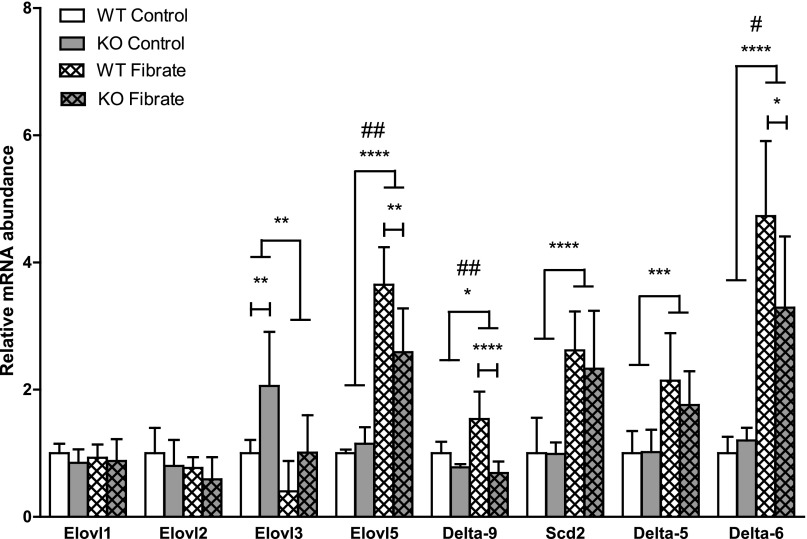
In the absence of fibrate, most pathways were either not affected or only modestly influenced by the loss of D2 (D2 versus WT) in both G and H tests. Fibrate treatment in WT mice resulted in altered gene expression in a collection of pathways, including fatty acid metabolism, PPAR signaling, and biosynthesis of polyunsaturated fatty acids (WT + feno versus WT). The PPAR signaling pathway was the most significantly influenced by fenofibrate, as indicated by the size of spots denoting significance in both statistical tests (P < 3.27E-21; Supplemental Table 1). Notably, the PPAR signaling pathway was altered in KO mice in the absence of fenofibrate treatment (G, D2 versus WT, P < 0.002; Supplemental Table 1). With fibrate treatment, the change of gene expression in KO mice was still maintained, although the pattern of dysregulation was different (G, D2 + feno versus WT + feno, P < 0.001; Supplemental Table 1). This observation indicates that loss of D2 not only influenced PPAR signaling itself, but also modulated the genomic response to fibrates. Similar results were observed for biosynthesis of polyunsaturated fatty acids (Supplemental Table 1). Alterations in selected PPARα target genes (Elovl5, Delta-9, Scd2, Delta-6) (Rakhshandehroo et al., 2010), in the biosynthesis of PUFAs pathway, were confirmed by rtPCR (Fig. 2).
Fig. 2.
D2 modulates fenofibrate effects on hepatic fatty acid biosynthesis. Relative mRNA abundance for fatty acid elongases and desaturases was determined by rtPCR (n = 6, mean ± S.D.). Data were analyzed by two-way analysis of variance with genotype and treatment as factors. #, Over lines terminating in horizontal bars indicates significant genotype by treatment interaction. Bonferroni post-tests were conducted within fibrate-treated groups and control groups. Asterisks over lines terminating in horizontal bars indicate a significant overall effect of fibrate treatment regardless of genotype. Asterisks over bars terminating in vertical lines indicate a significant difference between genotypes within either fibrate-treated or control groups. #P < 0.05, ##P < 0.01, *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
Cellular Response to Fenofibrate Treatment Is Dependent on ABCD2 in 3T3-L1 Cells.
Based on alterations in gene expression in mice, we hypothesized that the lack of ABCD2 may impact PPAR signaling and responses to fibrate treatment. First, we evaluated the influence of D2 on these responses in vitro. We selected NIH3T3-L1 adipocytes because they are the only cell line known to express D2 at high levels and respond to fibrate treatment (Brun et al., 1996; Liu et al., 2010).
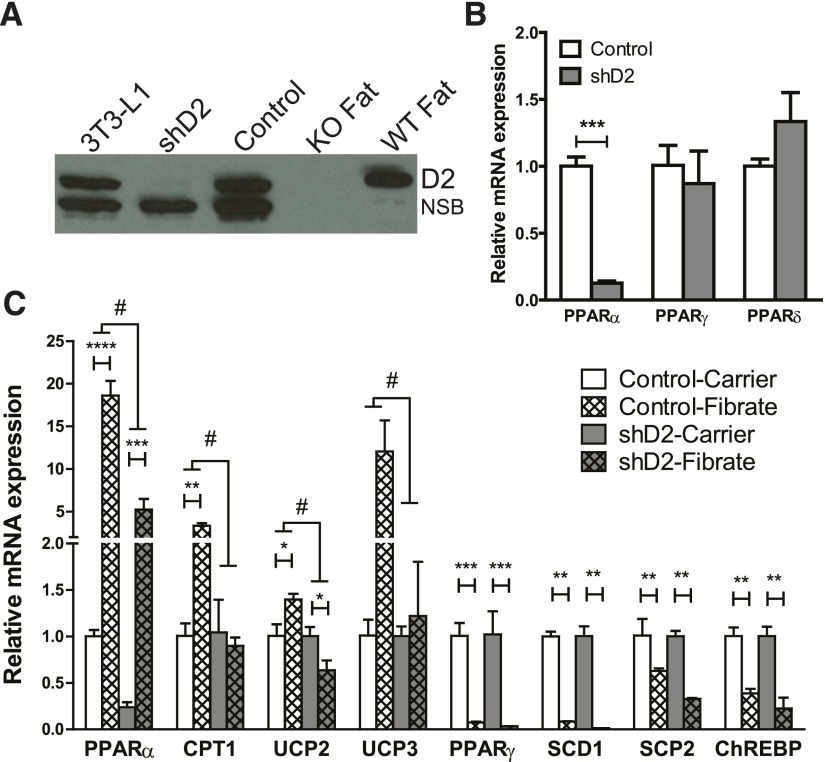
We developed a stable clone harboring a shRNA directed against D2 to repress expression. Parent 3T3-L1 cells, stable clones with D2 shRNA, and control shRNA were differentiated to adipocytes. Levels of D2 were confirmed by immunoblotting. Fat from WT and KO mice were used as positive (+) and negative (−) controls (Fig. 3A). Our D2 antibody cross-reacts with a protein with similar size to D2 in a number of tissues and in 3T3-L1 adipocytes, but this protein is absent in fat (Liu et al., 2010). Total RNAs of shD2 and control cells were extracted and analyzed by quantitative real-time PCR to determine the expression levels of PPAR family members. Interestingly, the lack of D2 specifically suppressed the expression of PPARα, but the other two members of PPAR family, β and γ, were unchanged (Fig. 3B).
Fig. 3.
D2 modulates PPARα and fibrate responses in 3T3-L1 adipocytes. (A) 3T3-L1 cells and two stable subclones infected with lentiviral particles expressing shRNAs directed against D2 (shD2) or a control shRNA (Control) were differentiated into adipocytes that express endogenous D2. Cell lysates were analyzed for D2 expression by immunoblotting. Membrane preparations from epididymal fat dissected from KO and WT mice were used as negative and positive controls. NSB indicates nonspecific band in 3T3-L1 cells. (B) Expression levels of PPAR family members were determined in control and 3T3-L1 adipocytes by rtPCR. Data were analyzed by unpaired t test (n = 3 independent wells). (C) Control and shD2 3T3-L1 adipocytes were treated with fenofibrate (500 mM) or carrier (dimethylsulfoxide) for 16 hours. Gene expression was determined by rtPCR (n = 3, mean ± S.E.M.). Data were analyzed by two-way analysis of variance with genotype and treatment as factors. #, Over lines terminating in horizontal bars indicates significant genotype by treatment interaction. Bonferroni post-tests were conducted within fibrate-treated groups and control groups. Asterisks over lines terminating in horizontal bars indicate a significant overall effect of genotype regardless of fibrate treatment. Asterisks over bars terminating in vertical lines indicate a significant difference between treatment within either control or D2-deficient cells. #P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
Control and shD2 cells were differentiated to adipocytes and treated overnight with either carrier or fenofibrate. Expression of PPARα and its target genes was determined using real-time PCR (Fig. 3C). The mRNA levels of PPARα, CPT1, uncoupling protein 2, and uncoupling protein 3 were significantly increased in response to fenofibrate treatment in control cells, but the elevation of gene expression was blunted in the shD2 cells. Conversely, fenofibrate treatment decreased the expression levels of stearoyl-CoA desaturase 1, sterol carrier protein 2, PPARγ, and carbohydrate-responsive element-binding protein in control cells, but that effect was not influenced by the loss of D2. This suggests D2 modulates genomic response to fibrate by influencing the expression of some, but not all the genes, in the PPAR signaling pathway.
Triglyceride Secretion from Liver Is Accelerated by Loss of ABCD2.
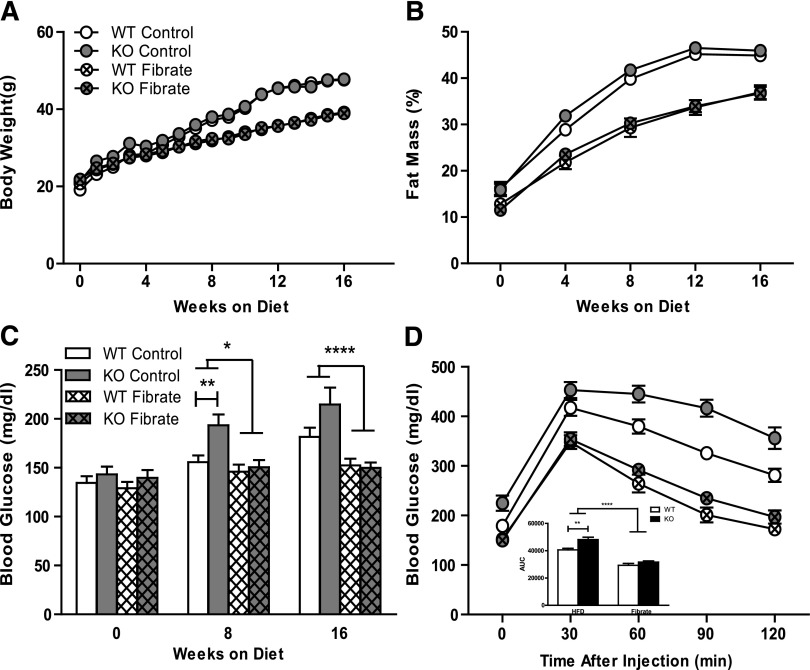
Fibrate therapy has been shown to successfully oppose diet-induced obesity in mice (Chaput et al., 2000; Guerre-Millo et al., 2000). To investigate the impact of D2 on fibrate response in vivo, we challenged KO and WT mice with high-fat diet (control, 60% kcal) or the same diet formulated with 0.05% (w/w) fenofibrate (Fibrate) for 16 weeks starting at 8 weeks of age. No significant differences in body weight, body composition, insulin sensitivity, or other measures of lipid metabolism between genotypes were observed at 8 weeks. After high-fat diet feeding, both KO and WT mice in untreated (control) group developed obesity and hyperglycemia with elevated body weight, fat mass percentage, and blood glucose levels (Fig. 4). Fenofibrate treatment mitigated the increase in body weight and adiposity, but these effects were not influenced by loss of D2 (Fig. 4, A and B). Blood glucose levels were measured at the initiation (0 weeks on diet), midpoint (8 weeks on diet), and end of the study (16 weeks on diet) (Fig. 4C). Control KO mice tended to have higher blood glucose levels compared with their WT littermates at 8 and 16 weeks of age. However, the difference between genotypes was lost in fibrate-treated animals whose blood glucose levels were normalized. A glucose tolerance test was done at the endpoint of the study (Fig. 4D). Area under the curve was calculated using data from 0 to 120 minutes based on trapezoidal rule. Glucose tolerance was compromised by the absence of D2. Fibrate lowered blood glucose and improved glucose tolerance, but this effect was not altered by the absence of D2.
Fig. 4.
Measurement of physiologic parameters. Body weights (A) were recorded for wild-type mice on high-fat diet (WT control, n = 12) and high-fat diet containing 0.05% fenofibrate (WT Fibrate, n = 17), D2 knockout mice on high-fat diet (KO Control, n = 8), and high-fat diet containing 0.05% fenofibrate (KO Fibrate, n = 17). Fat mass percentage (B) of these mice was measured by MRI every 4 weeks (mean ± S.E.M., n = 8∼17). Blood glucose concentrations (C) were measured every 4 weeks (mean ± S.E.M., n = 8∼17). Glucose tolerance tests (D) were performed by intraperitoneal injection of glucose (1 g/kg) and measurement of blood glucose concentration. Area under the curve for the GTT measurement was also listed within (D). Statistical comparison was made between fibrate-treated goups and control groups using two-way analysis of variance. Statistical comparison between genotypes within each group was made using Student test. Asterisks over lines terminating in horizontal bars indicate a significant overall effect of fibrate treatment regardless of genotype. Asterisks over bars terminating in vertical lines indicate a significant difference between genotypes within either fibrate-treated or control groups. *P < 0.05, **P < 0.01, ****P < 0.0001.
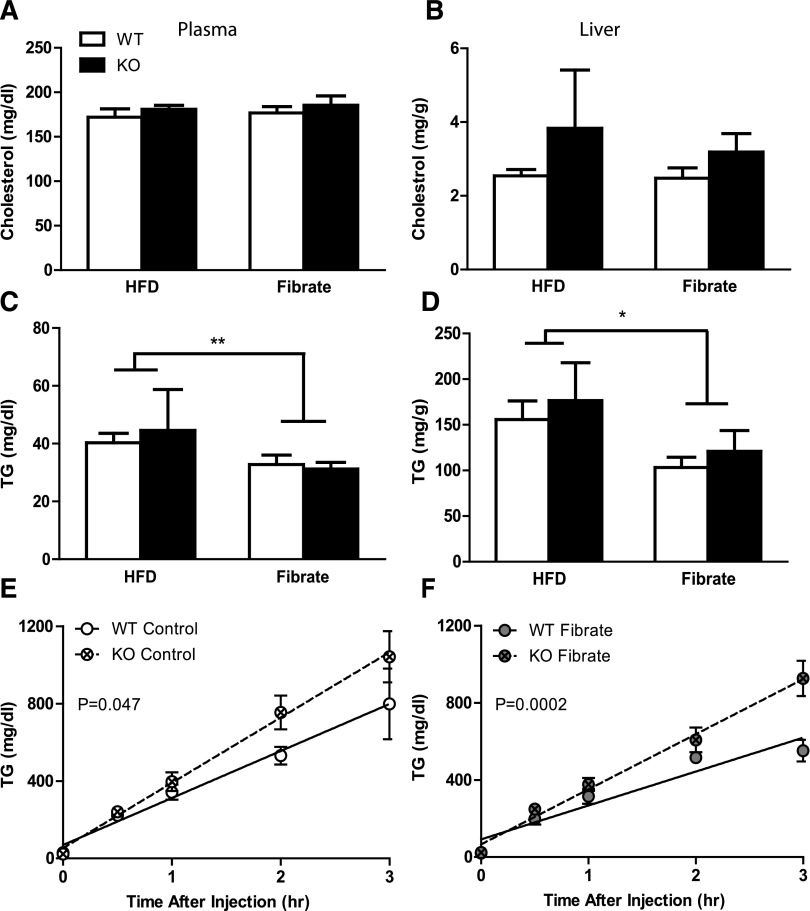
We analyzed plasma and hepatic lipids from both WT and KO mice with and without fenofibrate treatment (Fig. 5, A–D). Neither genotype nor fibrate treatment altered plasma or hepatic cholesterol (Fig. 5, A and B). Fenofibrate reduced the triglyceride levels of plasma and liver, but neither of these effects was altered by the absence of D2 (Fig. 5, C and D). KO mice exhibited significantly higher triglyceride secretion rates than their WT littermates in both control (WT, 243.0 ±33.49 mg/dl per hour; KO, 337.7 ± 27.86 mg/dl per hour) and fibrate (WT, 175.8 ± 17.24 mg/dl per hour; KO, 285.8 ± 21.56 mg/dl per hour) groups (Fig. 5, E and F). However, there was no significant interaction between the genotype and the fibrate treatment. This observation suggests that, although the static plasma triglyceride level was not affected, secretion rate of triglyceride from liver was accelerated by loss of D2 with and without fibrate treatment.
Fig. 5.
Lipid-lowering effect of fibrates dependent on ABCD2. Total cholesterol and triglyceride concentrations in plasma (A and C, respectively, mean ± S.E.M., n = 4∼17) and liver (B and D, respectively, mean ± S.E.M., n = 4∼5) were measured. Statistical analysis was done using two-way ANOVA. Asterisks indicate significant difference between fibrate-treated and control groups regardless of genotype. *P < 0.05, **P < 0.01. (E and F) The TG secretion rate was calculated as the slope, and the significance of difference between two genotypes was demonstrated as P value (control: WT R2 = 0.7453, KO R2 = 0.9187; fibrate: WT R2 = 0.7879, KO R2 = 0.8626).
We placed a subgroup of mice in the metabolic cages by the end of the experiment. Fenofibrate treatment suppressed food intake and promoted energy expenditure, but these effects were almost equal for both genotypes (Supplemental Fig. 1).
SNPs in ABCD2 Do Not Associate with Hypolipidemic Effects of Fenofibrate.
Efficacy of fibrate treatment of dyslipidemia in human subjects is highly variable between individual patients (Farnier, 2008). To evaluate the association of SNPs in ABCD2 to fenofibrate treatment efficacy, we analyzed the data from the GOLDN study in a population of related metabolically healthy individuals of European American ancestry (Table 1). Three SNPs in ABCD2 are associated with alterations in expression levels of D2, suggesting they may alter D2 activity in human subjects (Matsson et al., 2012). Two variants genotyped in GOLDN data set analysis, rs7309234 and rs11172592, were selected and used as proxies for corresponding D2 variants, rs4285917 and rs4284427, as each is in linkage disequilibrium with these expression-associated SNPs, respectively. Neither variant was associated with responses to fibrate among GOLDN participants (P value = 0.59 and 0.50 for rs7309234 and rs11172592, respectively), suggesting D2 expression is unlikely to alter fibrate responses in human subjects.
TABLE 1.
Association of ABCD2 variants and hyolipidemic effects of fenofibrate
Linear mixed models were used to assess the association of two ABCD2 variants, rs7309234 and rs11172592, with the fenofibrate-induced change in plasma triglycerides based on the data from the Genetics of Lipid Lowering Drugs and Diet Network study in a population of related metabolically healthy individuals of European American ancestry.
| SNP | Beta | S.E. | P Value |
|---|---|---|---|
| rs7309234 | 0.009792360 | 0.0183794364 | 0.5943369 |
| rs11172592 | 0.010279890 | 0.0084339715 | 0.4983299 |
SNP, single-nucleotide polymorphism.
Discussion
The major finding of this study is that D2 modulates PPARα signaling and genomic response to fibrate treatment in vitro and in vivo. However, the absence of D2 did not alter the impact of fibrate treatment in a mouse model of diet-induced obesity. Although not a comprehensive assessment, SNPs associated with altered D2 expression levels were not associated with responses to fibrate therapy in human subjects. D2 has long been known as a PPARα target gene that is upregulated in response to fenofibrate. To the best of our knowledge, this is the first report that demonstrates that D2 reciprocally regulates PPARα signaling.
2A number of gene clusters are differentially affected by fibrate in the presence or absence of D2. A limitation of cDNA array analysis is the potential for cross-hybridization of closely related transcripts. Although confounding effects of cross-hybridization cannot be discarded, we interrogated signal intensities of closely related members of the cytochrome P450 family. Cyp4f14 and Cyp4a16 share 66% sequence identity across the 579-bp cDNA probe for Cyp4a16 on our spotted array. Whereas Cyp4f14 is differentially regulated by fibrate treatment, no change in Cyp4a16 was observed, indicating that the extent of cross-hybridization was limited by the stringency of our hybridization conditions. In addition, we verified differential expression of selected genes in the PUFA synthesis pathway by rtPCR in mouse liver. Other clusters affected include pathways associated with lipid, bile acid, and steroid metabolism, but have yet to be confirmed by secondary approaches. In the present study, we elected to focus on PPAR signaling due to the use of PPAR agonists as lipid-modifying agents and the variability in response to fibrate treatment.
In 3T3-L1 cells, suppression of D2 reduced PPARα mRNA levels as well as the upregulation of several PPARα target genes in response to fibrate treatment. Interestingly, the repression of selected genes by fibrates was unaffected. This effect indicates that D2 selectively modulates fibrate responses and perhaps PPARα signaling. The molecular mechanism responsible for differential effects is unknown, but is most likely related to D2-dependent peroxisomal metabolism of fatty acids. LCFAs, VLCFAs, and their coenzyme A thioesters are putative endogenous PPARα ligands with varying affinity for human and mouse PPARs (Lin et al., 1999; Hostetler et al., 2005, 2006; Oswal et al., 2013). We previously demonstrated that the absence of D2 altered endogenous LCFA and VLCFA contents in mouse brain and adipose tissues (Fourcade et al., 2009; Liu et al., 2010). Thus, it is possible that D2 alters the type and availability of PPARα ligands. However, other PPAR (β and γ) were not affected, suggesting that the effect of D2 is related to PPARα abundance. Alternatively, these differences may be due to effects of fenofibrate that are independent of PPARα. The precise molecular mechanism by which D2 alters PPARα expression will require additional studies.
To test the impact of D2 on PPARα signaling and fenofibrate response in vivo, we treated WT and D2 KO mice with fenofibrate during the progression of diet-induced obesity. In the absence of fibrates, D2 deficiency resulted in a modest, but significant increase in fasting glucose at the midpoint and end of the study, as well as reduced glucose tolerance. This is inconsistent with our previously published work in which differences in glycemic control were not observed in D2-deficient mice following high-fat feeding or when crossed onto the ob/ob background (Liu et al., 2012). However, it should be noted that the previous study used a 45% kCal high-fat diet as opposed to the 60% kCal diet. Other phenotypes, such as weight gain and body composition, were as previously reported.
Fenofibrate reduced body weight, fat mass, and plasma and hepatic TG in both genotypes. These effects are most likely due to reduced food intake and increased energy expenditure, neither of which was affected by D2 genotype. The absence of D2 also reduced TG secretion rates in both the presence and absence of fibrate treatment. In the absence of D2, TG secretion rates in fibrate-treated mice are indistinguishable from untreated WT mice. However, this difference was insufficient to affect fasting TG levels. microsomal triglyceride transfer protein is known to regulate the TG secretion in mice (Raabe et al., 1999), but its mRNA levels were not influenced by loss of D2 irrespective of fibrate treatment (data not shown). Lipoprotein lipase (LPL) mRNA was elevated in the brown adipose tissue in KO mice compared with WT littermates on high-fat feeding (data not shown). LPL is an enzyme known to hydrolyze triglyceride in lipoprotein to release free fatty acid for uptake into peripheral tissues for metabolism (Mead et al., 2002). Given the role of LPL in TG clearance, the increased LPL expression in the face of elevated TG secretion may account for the absence of differences in steady-state plasma TGs.
A number of genetic and environmental factors influence plasma TGs and responses to lipid-lowering drugs. We took advantage of an ongoing effort to identify genetic variants that influence responses to fibrate therapy to determine whether polymorphisms in the ABCD2 gene might be associated with such differences. Neither of the two proxy SNPs analyzed was statistically significant, suggesting that variants in ABCD2 and alterations in D2 levels are unlikely to contribute to differences in response to fibrate therapy. However, it should be noted that the effects of these SNPs on D2 activity have not been formally characterized. Additional studies are needed to fully elucidate the effect of D2 on TG metabolism and responses to fibrate therapy.
Supplementary Material
Acknowledgments
The authors thank Jeannie Haak for assistance with manuscript preparation.
Abbreviations
- CT
threshold cycle
- DMEM
Dulbecco’s modified Eagle’s medium
- GOLDN
Genetics of Lipid Lowering Drugs and Diet Network
- KO
knockout
- LCFA
long-chain fatty acid
- LPL
lipoprotein lipase
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator-activated receptor
- rtPCR
reverse transcription-PCR
- shRNA
short hairpin RNA
- SNP
single-nucleotide polymorphism
- TG
triglyceride
- VLCFA
very long-chain fatty acid
- PUFA
polyunsaturated fatty acid
- WT
wild-type
Authorship Contributions
Participated in research design: X. Liu, J. Liu, Liang, Schlüter, Fourcade, Aslibekyan, Pujol, Graf.
Conducted experiments: X. Liu, J. Liu, Liang, Schlüter, Fourcade, Aslibekyan.
Contributed new reagents or analytic tools: Aslibekyan.
Performed data analysis: X. Liu, J. Liu, Liang, Schlüter, Fourcade, Aslibekyan, Pujol, Graf.
Wrote or contributed to the writing of the manuscript: X. Liu, Graf.
Footnotes
This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grants DK080874 and DK100892], National Institute of General Medical Sciences [Grant 8 P20 GM103527-05], and National Heart, Lung, and Blood Institute [Grant R01 HL 09135704]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Aslibekyan S, Goodarzi MO, Frazier-Wood AC, Yan X, Irvin MR, Kim E, Tiwari HK, Guo X, Straka RJ, Taylor KD, et al. (2012a) Variants identified in a GWAS meta-analysis for blood lipids are associated with the lipid response to fenofibrate. PLoS One 7:e48663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslibekyan S, Kabagambe EK, Irvin MR, Straka RJ, Borecki IB, Tiwari HK, Tsai MY, Hopkins PN, Shen J, Lai CQ, et al. (2012b) A genome-wide association study of inflammatory biomarker changes in response to fenofibrate treatment in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenet Genomics 22:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Albet S, Bentejac M, Netik A, Holzinger A, Roscher AA, Bugaut M, Forss-Petter S. (1999) The four murine peroxisomal ABC-transporter genes differ in constitutive, inducible and developmental expression. Eur J Biochem 265:719–727 [DOI] [PubMed] [Google Scholar]
- Bremer J, Norum KR. (1982) Metabolism of very long-chain monounsaturated fatty acids (22:1) and the adaptation to their presence in the diet. J Lipid Res 23:243–256 [PubMed] [Google Scholar]
- Brun RP, Tontonoz P, Forman BM, Ellis R, Chen J, Evans RM, Spiegelman BM. (1996) Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev 10:974–984 [DOI] [PubMed] [Google Scholar]
- Chaput E, Saladin R, Silvestre M, Edgar AD. (2000) Fenofibrate and rosiglitazone lower serum triglycerides with opposing effects on body weight. Biochem Biophys Res Commun 271:445–450 [DOI] [PubMed] [Google Scholar]
- Collins S, Martin TL, Surwit RS, Robidoux J. (2004) Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol Behav 81:243–248 [DOI] [PubMed] [Google Scholar]
- Corella D, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, Straka RJ, Province M, Lai CQ, Parnell LD, et al. (2007) The -256T>C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin Chem 53:1144–1152 [DOI] [PubMed] [Google Scholar]
- Farnier M. (2008) Update on the clinical utility of fenofibrate in mixed dyslipidemias: mechanisms of action and rational prescribing. Vasc Health Risk Manag 4:991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Kapfhammer JP, Hindelang C, Kemp S, Troffer-Charlier N, Broccoli V, Callyzot N, Mooyer P, Selhorst J, Vreken P, et al. (2005) Inactivation of the peroxisomal ABCD2 transporter in the mouse leads to late-onset ataxia involving mitochondria, Golgi and endoplasmic reticulum damage. Hum Mol Genet 14:3565–3577 [DOI] [PubMed] [Google Scholar]
- Fourcade S, Ruiz M, Camps C, Schlüter A, Houten SM, Mooyer PA, Pàmpols T, Dacremont G, Wanders RJ, Giròs M, et al. (2009) A key role for the peroxisomal ABCD2 transporter in fatty acid homeostasis. Am J Physiol Endocrinol Metab 296:E211–E221 [DOI] [PubMed] [Google Scholar]
- Fourcade S, Ruiz M, Guilera C, Hahnen E, Brichta L, Naudi A, Portero-Otín M, Dacremont G, Cartier N, Wanders R, et al. (2010) Valproic acid induces antioxidant effects in X-linked adrenoleukodystrophy. Hum Mol Genet 19:2005–2014 [DOI] [PubMed] [Google Scholar]
- Fourcade S, Savary S, Albet S, Gauthe D, Gondcaille C, Pineau T, Bellenger J, Bentejac M, Holzinger A, Berger J, and Bugaut M (2001) Fibrate induction of the adrenoleukodystrophy-related gene (ABCD2): promoter analysis and role of the peroxisome proliferator-activated receptor PPARalpha. Eur. J. Biochem. 268:3490–3500. [DOI] [PubMed] [Google Scholar]
- Frazier-Wood AC, Ordovas JM, Straka RJ, Hixson JE, Borecki IB, Tiwari HK, Arnett DK. (2013) The PPAR alpha gene is associated with triglyceride, low-density cholesterol and inflammation marker response to fenofibrate intervention: the GOLDN study. Pharmacogenomics J 13:312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Huber W, Irizarry RA, Dut S (2005) Bioinformatics and computational biology solutions using R and bioconductor, in Statistics for Biology and Health pp 397-420, Springer, New York, Philadelphia. [Google Scholar]
- Gondcaille C, Genin EC, Lopez TE, Dias AM, Geillon F, Andreoletti P, Cherkaoui-Malki M, Nury T, Lizard G, Weinhofer I, et al. (2014) LXR antagonists induce ABCD2 expression. Biochim Biophys Acta 1841:259–266 [DOI] [PubMed] [Google Scholar]
- Guerre-Millo M, Gervois P, Raspé E, Madsen L, Poulain P, Derudas B, Herbert JM, Winegar DA, Willson TM, Fruchart JC, et al. (2000) Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J Biol Chem 275:16638–16642 [DOI] [PubMed] [Google Scholar]
- Guillou H, Martin P, Jan S, D’Andrea S, Roulet A, Catheline D, Rioux V, Pineau T, Legrand P. (2002) Comparative effect of fenofibrate on hepatic desaturases in wild-type and peroxisome proliferator-activated receptor alpha-deficient mice. Lipids 37:981–989 [DOI] [PubMed] [Google Scholar]
- Hostetler HA, Kier AB, Schroeder F. (2006) Very-long-chain and branched-chain fatty acyl-CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha). Biochemistry 45:7669–7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler HA, Petrescu AD, Kier AB, Schroeder F. (2005) Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem 280:18667–18682 [DOI] [PubMed] [Google Scholar]
- Irvin MR, Kabagambe EK, Tiwari HK, Parnell LD, Straka RJ, Tsai M, Ordovas JM, Arnett DK. (2010) Apolipoprotein E polymorphisms and postprandial triglyceridemia before and after fenofibrate treatment in the Genetics of Lipid Lowering and Diet Network (GOLDN) Study. Circ Cardiovasc Genet 3:462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraja AT, Borecki IB, Tsai MY, Ordovas JM, Hopkins PN, Lai CQ, Frazier-Wood AC, Straka RJ, Hixson JE, Province MA, et al. (2013) Genetic analysis of 16 NMR-lipoprotein fractions in humans, the GOLDN study. Lipids 48:155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CQ, Arnett DK, Corella D, Straka RJ, Tsai MY, Peacock JM, Adiconis X, Parnell LD, Hixson JE, Province MA, et al. (2007) Fenofibrate effect on triglyceride and postprandial response of apolipoprotein A5 variants: the GOLDN study. Arterioscler Thromb Vasc Biol 27:1417–1425 [DOI] [PubMed] [Google Scholar]
- Lin Q, Ruuska SE, Shaw NS, Dong D, Noy N. (1999) Ligand selectivity of the peroxisome proliferator-activated receptor alpha. Biochemistry 38:185–190 [DOI] [PubMed] [Google Scholar]
- Liu J, Liang S, Liu X, Brown JA, Newman KE, Sunkara M, Morris AJ, Bhatnagar S, Li X, Pujol A, et al. (2012) The absence of ABCD2 sensitizes mice to disruptions in lipid metabolism by dietary erucic acid. J Lipid Res 53:1071–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sabeva NS, Bhatnagar S, Li XA, Pujol A, Graf GA. (2010) ABCD2 is abundant in adipose tissue and opposes the accumulation of dietary erucic acid (C22:1) in fat. J Lipid Res 51:162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ordovas JM, Gao G, Province M, Straka RJ, Tsai MY, Lai CQ, Zhang K, Borecki I, Hixson JE, et al. (2008) The SCARB1 gene is associated with lipid response to dietary and pharmacological interventions. J Hum Genet 53:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Platet G, Savary S, Sarde CO, Mandel JL, Chimini G. (1996) A close relative of the adrenoleukodystrophy (ALD) gene codes for a peroxisomal protein with a specific expression pattern. Proc Natl Acad Sci USA 93:1265–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JF, Barron-Casella E, Deering R, Heinzer AK, Moser AB, deMesy Bentley KL, Wand GS, C McGuinness M, Pei Z, Watkins PA, et al. (2007) The role of peroxisomal ABC transporters in the mouse adrenal gland: the loss of Abcd2 (ALDR), not Abcd1 (ALD), causes oxidative damage. Lab Invest 87:261–272 [DOI] [PubMed] [Google Scholar]
- Lu JF, Lawler AM, Watkins PA, Powers JM, Moser AB, Moser HW, Smith KD. (1997) A mouse model for X-linked adrenoleukodystrophy. Proc Natl Acad Sci USA 94:9366–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Boekschoten MV, Wopereis S, Müller M, Kersten S. (2011) Comparative transcriptomic and metabolomic analysis of fenofibrate and fish oil treatments in mice. Physiol Genomics 43:1307–1318 [DOI] [PubMed] [Google Scholar]
- Matsson P, Yee SW, Markova S, Morrissey K, Jenkins G, Xuan J, Jorgenson E, Kroetz DL, Giacomini KM. (2012) Discovery of regulatory elements in human ATP-binding cassette transporters through expression quantitative trait mapping. Pharmacogenomics J 12:214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead JR, Irvine SA, Ramji DP. (2002) Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med 80:753–769 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273 [DOI] [PubMed] [Google Scholar]
- Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. (1993) Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature 361:726–730 [DOI] [PubMed] [Google Scholar]
- Oswal DP, Balanarasimha M, Loyer JK, Bedi S, Soman FL, Rider SD, Jr, Hostetler HA. (2013) Divergence between human and murine peroxisome proliferator-activated receptor alpha ligand specificities. J Lipid Res 54:2354–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Kim HS, Jang J, Lee H, Lee JS, Yoo JE, Lee DR, Kim DW. (2013) ABCD2 is a direct target of β-catenin and TCF-4: implications for X-linked adrenoleukodystrophy therapy. PLoS One 8:e56242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol A, Ferrer I, Camps C, Metzger E, Hindelang C, Callizot N, Ruiz M, Pàmpols T, Giròs M, Mandel JL. (2004) Functional overlap between ABCD1 (ALD) and ABCD2 (ALDR) transporters: a therapeutic target for X-adrenoleukodystrophy. Hum Mol Genet 13:2997–3006 [DOI] [PubMed] [Google Scholar]
- Raabe M, Véniant MM, Sullivan MA, Zlot CH, Björkegren J, Nielsen LB, Wong JS, Hamilton RL, Young SG. (1999) Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Invest 103:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Müller M, Kersten S. (2010) Peroxisome proliferator-activated receptor alpha target genes. PPAR Res 2010:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter A, Espinosa L, Fourcade S, Galino J, Lopez E, Ilieva E, et al. (2012) Functional genomic analysis unravels a metabolic-inflammatory interplay in adrenoleukodystrophy. Hum Mol Genet 21:1062-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, Heyman RA, Briggs M, Deeb S, Staels B, Auwerx J. (1996a) PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J 15:5336–5348 [PMC free article] [PubMed] [Google Scholar]
- Schoonjans K, Staels B, Auwerx J. (1996b) Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res 37:907–925 [PubMed] [Google Scholar]
- Shen J, Arnett DK, Parnell LD, Peacock JM, Lai CQ, Hixson JE, Tsai MY, Province MA, Straka RJ, Ordovas JM. (2008) Association of common C-reactive protein (CRP) gene polymorphisms with baseline plasma CRP levels and fenofibrate response: the GOLDN study. Diabetes Care 31:910–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2005) Limma: linear models for microarray data, in Statistics for Biology and Health pp 397-420, Springer, New York, Philadelphia. [Google Scholar]
- Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. (1998) Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 98:2088–2093 [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. (2000) Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA 97:9127–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund CW, Visser WF, Ijlst L, Waterham HR, Wanders RJ. (2011) Differential substrate specificities of human ABCD1 and ABCD2 in peroxisomal fatty acid β-oxidation. Biochim Biophys Acta 1811:148–152 [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Waterham HR. (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75:295–332 [DOI] [PubMed] [Google Scholar]
- Weber FD, Wiesinger C, Forss-Petter S, Regelsberger G, Einwich A, Weber WH, Kohler W, Stockinger H, and Berger J (2014) X-linked adrenoleukodystrophy: very long-chain fatty acid metabolism is severely impaired in monocytes but not in lymphocytes. Hum Mol Genet 23:2542–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I, Forss-Petter S, Kunze M, Zigman M, Berger J. (2005) X-linked adrenoleukodystrophy mice demonstrate abnormalities in cholesterol metabolism. FEBS Lett 579:5512–5516 [DOI] [PubMed] [Google Scholar]
- Weinhofer I, Kunze M, Rampler H, Forss-Petter S, Samarut J, Plateroti M, Berger J. (2008) Distinct modulatory roles for thyroid hormone receptors TRalpha and TRbeta in SREBP1-activated ABCD2 expression. Eur J Cell Biol 87:933–945 [DOI] [PubMed] [Google Scholar]
- Wojczynski MK, Gao G, Borecki I, Hopkins PN, Parnell L, Lai CQ, Ordovas JM, Chung BH, Arnett DK. (2010) Apolipoprotein B genetic variants modify the response to fenofibrate: a GOLDN study. J Lipid Res 51:3316–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.