Abstract
Summary: Circleator is a Perl application that generates circular figures of genome-associated data. It leverages BioPerl to support standard annotation and sequence file formats and produces publication-quality SVG output. It is designed to be both flexible and easy to use. It includes a library of circular track types and predefined configuration files for common use-cases, including. (i) visualizing gene annotation and DNA sequence data from a GenBank flat file, (ii) displaying patterns of gene conservation in related microbial strains, (iii) showing Single Nucleotide Polymorphisms (SNPs) and indels relative to a reference genome and gene set and (iv) viewing RNA-Seq plots.
Availability and implementation: Circleator is freely available under the Artistic License 2.0 from http://jonathancrabtree.github.io/Circleator/ and is integrated with the CloVR cloud-based sequence analysis Virtual Machine (VM), which can be downloaded from http://clovr.org or run on Amazon EC2.
Contact: jcrabtree@som.umaryland.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
There are numerous circular genome visualization tools, with varying degrees of interactivity, usability and utility. They differ in the data types and formats they accept, the types of output they produce and the ease of customizing the mapping from input data to graphical display. Flexibility and ease-of-use are frequently at odds, with the most flexible tools often being the hardest to customize, particularly for non-programmers. To address this issue, Circleator provides multiple configuration options, allowing each user to choose the one that best suits his/her needs.
Similar tools include GenomePlot (Gibson and Smith, 2003), a Perl/Tk application and DNAPlotter (Carver et al., 2008), a Java application, both of which have graphical user interfaces and also support linear displays. GenoMap (Sato and Ehira, 2003) is a Tcl/Tk microarray data viewer, and the GeneWiz browser (Hallin et al., 2009) is an interactive Java applet. Some combine analysis and visualization: BRIG (Alikhan et al., 2011) incorporates a BLAST-based prokaryotic genome comparison algorithm, and CGView Server (Grant and Stothard, 2008) is a CGView-based (Stothard and Wishart, 2005) web service that runs on-the-fly BLAST comparisons. At the other extreme, some tools display only predefined datasets: The Microbial Genome Viewer (Kerkhoven et al., 2004) and Genome Projector (Arakawa et al., 2009) are web-based tools in this category.
GenomeDiagram (Pritchard et al., 2006) supports linear and circular displays, but requires Python programming expertise. Circos (Krzywinski et al., 2009) is a popular and powerful stand-alone tool but its use of complex hierarchical configuration files may put it out of reach of some users. The D3.js toolkit (Bostock et al., 2011) has a Circos-like ‘chord diagram’, but does not, to our knowledge, accept standard bioinformatic file formats. Circster (Goecks et al., 2013), which uses D3.js, adds circular drawing capabilities to Galaxy.
Circleator follows the computer design principle of making the common case fast: if a researcher has data in a standard format and needs a routine visualization then he/she should not have to reformat the data or experiment with parameter values. Conversely, with sufficient time, it should be possible to create novel and intricately detailed figures. Circleator users who wish to accomplish the former can choose a predefined configuration file, whereas those seeking more flexibility can write their own. Circleator’s configuration file format supports several novel high-level abstractions (e.g. loops, symbolic track references, feature-based coordinate scaling) and reuses existing standards e.g. SVG, CSS (Cascading Style Sheets), where possible.
2 IMPLEMENTATION
Circleator is a stand-alone Perl application that has also been incorporated into CloVR (Angiuoli et al., 2011). It uses BioPerl (Stajich et al., 2002) for internal data representation and produces SVG (Dahlström et al., 2011) output, from which PDF, PNG and JPEG may be generated.
2.1 Input data
Circleator accepts reference sequence(s) and annotation in any BioPerl-supported format, including GenBank format; Sequence Alignment/Map and BGZF-compressed SAM (SAM/BAM) alignment files; output from Cufflinks (Trapnell et al., 2010), Tandem Repeats Finder (TRF) (Benson, 1999) and the BLAST Score Ratio (Rasko et al., 2005) utility; SNPs in Variant Call Format (VCF) and tab-delimited quantitative data, such as gene expression data.
2.2 Features
Circleator outputs SVG natively, rather than using a graphics library that supports only a subset of SVG. It can draw text along circular paths and display semitransparent and overlapping tracks. The scale may vary around the circle, as in Circos, but also along the radius of the circle (i.e. a single figure may combine both global context and local detail, as in Fig. 1B). Regions to scale may be selected with a user-defined filter, e.g. to magnify by 100× all SNP loci at which more than half of the genomes differ from the reference without having to explicitly list the relevant coordinate spans.
Fig. 1.
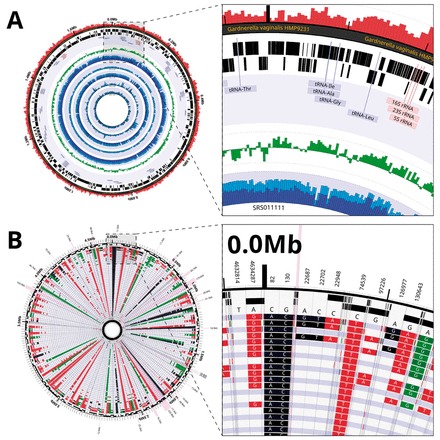
(A) The genome of Gardnerella vaginalis HMP9231 annotated with percent GC content (red), genes, GC-skew (green) and read coverage (blue) from five human metagenomic samples. (B) SNPs from an 80-genome Yersinia pestis SNP panel with the scale in the outer rings expanded to show the affected bases. The reference base and position is shown on the outside and SNPS are color-coded according to their predicted type. Additional details for these figures and others may be found in the supplementary information
2.3 Configuration
Each line in the manually editable Circleator configuration file corresponds to a circular track. The configuration file supports loops, which allow the same set of tracks to be displayed for 80 genomes in a SNP comparison without repeating everything 80 times; pseudo-tracks, which do not appear in the figure but can load data or perform data transformations (e.g. the compute-deserts track, which identifies all regions of a specified length that do not contain any features of a specified type); track references, which allow tracks to reference each other by name, e.g. highlight each of the SNP deserts identified in track SD1 in red; and various feature filters, e.g. to draw only forward-strand genes whose gene product field contains the keyword ‘kinase’. Circleator supports the following configuration options, listed in order of increasing flexibility and decreasing ease-of-use: (i) reuse a predefined configuration file as is; (ii) customize a predefined configuration file; (iii) write a new configuration file using the predefined track types and (iv) define new track types, glyphs and/or filters.
2.4 Documentation and test suite
The predefined configuration files and track types are well documented, and the HTML track documentation is automatically generated from the same Circleator configuration file that defines them. Circleator also has a set of regression tests that help to verify the correctness of the images it produces.
3 CONCLUSIONS
Circleator is a visualization tool that leverages BioPerl and SVG to produce publication-ready circular figures of genome-associated data. It is highly configurable but includes predefined configuration files and a library of well-documented circular track types that allows users to create complex figures without programming expertise.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Hervé Tettelin and Heather Huot Creasy for testing and providing feedback, Hervé Tettelin for the Yersinia pestis SNP data and W. Florian Fricke for assisting with the CloVR integration.
Funding: This project has been funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number HHSN272200900009C and grant U19AI110820.
Conflict of interest: none declared.
REFERENCES
- Alikhan NF, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S, et al. CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa K, et al. A Genome Projector: zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31. doi: 10.1186/1471-2105-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock M, et al. D3: Data-driven documents. IEEE Trans. Vis. Comput. Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- Carver T, et al. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2008;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlström E, et al. 2nd edn. 2011. Scalable Vector Graphics (SVG) 1.1. http://www.w3.org/TR/SVG11/ (30 July 2014, date last accessed) [Google Scholar]
- Gibson R, Smith DR. Genome visualization made fast and simple. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- Goecks J, et al. Web-based visual analysis for high-throughput genomics. BMC Genomics. 2013;14:397. doi: 10.1186/1471-2164-14-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin PF, et al. GeneWiz browser: an interactive tool for visualizing sequenced chromosomes. Stand. Genomic Sci. 2009;1:204–215. doi: 10.4056/sigs.28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven R, et al. Visualization for genomics: the microbial genome viewer. Bioinformatics. 2004;20:1812–1814. doi: 10.1093/bioinformatics/bth159. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L, et al. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics. 2006;22:616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- Rasko D, et al. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–1584. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;10:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


