Abstract
Single-nucleotide polymorphisms (SNPs) are essential tools for studying a variety of organismal properties and processes, such as recombination, chromosomal dynamics, and genome rearrangement. This paper describes the development of a genome-wide SNP map for Candida albicans to study mitotic recombination and chromosome loss. C. albicans is a diploid yeast which propagates primarily by clonal mitotic division. It is the leading fungal pathogen that causes infections in humans, ranging from mild superficial lesions in healthy individuals to severe, life-threatening diseases in patients with suppressed immune systems. The SNP map contains 150 marker sequences comprising 561 SNPs and 9 insertions-deletions. Of the 561 SNPs, 437 were transition events while 126 were transversion events, yielding a transition-to-transversion ratio of 3:1, as expected for a neutral accumulation of mutations. The average SNP frequency for our data set was 1 SNP per 83 bp. The map has one marker placed every 111 kb, on average, across the 16-Mb genome. For marker sequences located partially or completely within coding regions, most contained one or more nonsynonymous substitutions. Using the SNP markers, we identified a loss of heterozygosity over large chromosomal fragments in strains of C. albicans that are frequently used for gene manipulation experiments. The SNP map will be useful for understanding the role of heterozygosity and genome rearrangement in the response of C. albicans to host environments.
Single nucleotide markers are essential tools for studying a variety of organismal properties and processes, such as recombination, chromosomal dynamics, genome rearrangement, and genetic relatedness between individuals. If one compares the same stretch of DNA for two or more individuals, nucleotide polymorphisms are the most frequently observed differences at the nucleotide level within diploid organisms. Single-nucleotide polymorphisms (SNPs) can be located in coding regions of genes or in intergenic regions, where they are most abundant (6, 25). In coding regions, SNPs can alter the function and structure of encoded proteins, e.g., proteins involved in drug metabolism (27, 44). In humans, single-nucleotide substitutions are the cause for most of the known recessively or dominantly inherited monogenic disorders, and missense SNPs also often contribute to common diseases (31, 50). SNPs are estimated to occur once every 1 kb throughout the human genome and are being targeted for association mapping of disease susceptibility genes (42) and used to study traits of diseases, such as cancers, which are often accompanied by a loss of heterozygosity at SNP loci (12). High-density SNP maps were developed for the model organism Caenorhabditis elegans to facilitate rapid gene mapping, to characterize natural isolates, and to compare different strains of C. elegans (23, 25, 51, 60). To make large-scale studies of SNPs possible, high-throughput methods such as SNP microarrays have been developed to analyze regions of interest in humans (14, 38, 54) and for genetic mapping projects and recombination studies in plants (7, 46).
This paper describes the development of a SNP map for Candida albicans to study recombination as a genome-wide process. C. albicans is a diploid yeast which propagates primarily by clonal mitotic division (18, 20, 40, 62) and demonstrates high levels of heterozygosity (41, 53, 56, 57). It is the leading fungal pathogen that causes infections in humans, ranging from mild superficial lesions in healthy individuals to severe, life-threatening diseases in patients with suppressed immune systems (5). To date, the causes for the transition from a commensal to a pathogenic organism are not entirely clear. Certainly, factors such as host susceptibility play an important role, as does the developmental transition from a budding yeast form to a filamentous form, which is essential for virulence (11, 15, 22). High levels of heterozygosity and genome rearrangement remain relatively unexplored sources of variation for adaptive responses of C. albicans populations to host environments.
Clinical isolates of C. albicans exhibiting high levels of variation among strains have been studied in great detail by such techniques as randomly amplified polymorphic DNA analysis (35), restriction fragment length polymorphism (RFLP) analysis (62), karyotyping (32, 33, 43), and DNA fingerprinting (39). Correlations between multilocus genotypes and the evolution of drug resistance have been examined in populations of C. albicans with the help of heterozygous genomic loci (10). In addition, SNPs have been used for population genetic studies with C. albicans to demonstrate that some form of recombination must also occur, albeit at a low level (18, 20).
SNP loci, as well as larger heterozygous genomic regions, can be used to study the mechanism and rates of mitotic recombination by an analysis of loss of heterozygosity (LOH). Forche et al. (17) constructed strains that were heterozygous at the GAL1 locus and determined the in vivo frequencies of mitotic recombination at that locus through the loss of the second copy of the GAL1 gene. Others have examined the rate of loop-out in transformation cassettes used for gene disruption in C. albicans and have reported mitotic recombination rates ranging from 6 × 10−6 to 6 × 10−8/cell/generation (13, 16). In a recent study, Yesland and Fonzi found that the integration of a disruption cassette is allele, and thus sequence, dependent. The failure to disrupt the second copy of a gene was due not to its essential nature but to allelic variation in the flanking regions of that gene (63). Mitotic recombination and LOH occur at observable rates and frequencies in vitro and in vivo, respectively, suggesting that C. albicans is a tractable system for studying the role of recombination in the evolution of virulence.
Our goal was to use SNP loci to study genome-wide mitotic recombination in C. albicans in vitro and in vivo and to understand the role of mitotic recombination and genome rearrangement in the evolution of virulence. With the complete genome sequence of strain SC5314 of C. albicans now available (http://www-sequence.stanford.edu:8080/haploid19.html), abundant heterozygosity is apparent and is represented by approximately 62,000 transitions and transversions across the 32-Mb diploid genome (24). Using these data for strain SC5314, we developed a SNP map representing the entire genome of C. albicans and show here that we can detect LOH by using these markers. We address the concern that LOH, through chromosome loss and reduplication or mitotic recombination, might confound the interpretation of results for targeted manipulations by assessing LOH at a number of SNPs flanking targeted loci (13, 16, 17, 36, 61).
MATERIALS AND METHODS
Strains and DNA extraction.
The strains used for this study are listed in Table 1. DNA extraction was performed by a quick prep method that is described elsewhere (1).
TABLE 1.
Strains of C. albicans used for this study
| Strain | Parent | Genotype or description | Reference |
|---|---|---|---|
| SC5314 | Clinical isolate | 24a | |
| CAI4 | SC5314 | ura3Δ::λimm434/ura3Δ::λimm434 | 16 |
| RM1000 | CA14 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG | 36 |
| BWP17 | RM1000 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 61 |
| AF6 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 gal1::URA3/GAL1 | 17 |
| AF14 | AF6 | ura3Δ::λimm434/ura3Δ::λimm434 gal1::URA3/gal1::URA3 | 17 |
| AF12 | RM1000 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG gal1::URA3/GAL1 | 17 |
| AF27 | AF12 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG gal1::URA3/gal1::HIS1 | 17 |
Development of SNP markers.
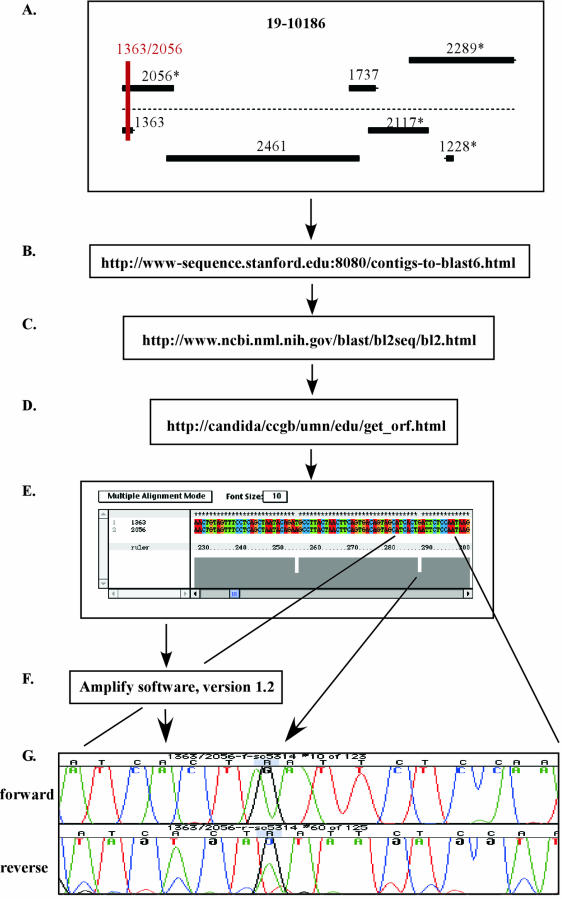
The goal for the development of the SNP map was to design one SNP marker for every 100 to 200 kb of sequence and to space the SNP markers as evenly as possible across the genome. For this study, we defined a SNP marker as a small PCR product ranging in size between 100 and 400 bp and containing at least one SNP. To design SNP markers, we took advantage of the available genome sequence represented as contigs-6 (http://www-sequence.stanford.edu:8080/contigs-to-blast6.html) and the contig-19 assembly of sequencing contigs-6 (http://www-sequence.stanford.edu:8080/haploid19.html) at the Stanford C. albicans genome web site (Fig. 1). Our method was as follows. First, two overlapping sequence contigs from a 10× genome coverage, representing the two potentially heterozygous alleles for each genomic region, were selected from the map option on the contig-19 sequence web page for C. albicans (http://www-sequence.stanford.edu:8080/haploid19.html) (Fig. 1A). Combinations of such pairs were further examined only when at least one of the two contigs-6 sequences was mapped to the corresponding chromosome. Complete sequences for both contigs were then obtained from the web site http://www-sequence.stanford.edu:8080/contigs-to-blast6.html (Fig. 1B). The contig sequences were compared by using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) to get the exact start-to-end base positions of the overlapping sequence (Fig. 1C). This step was followed by obtaining the actual sequences for the overlapping parts of the contigs at the web site http://candida/ccgb/umn/edu/get_orf.html (Fig. 1D). The two sequences were then aligned with ClustalX software, version 1.81 (Fig. 1E), to locate heterozygous positions. Forward and reverse primers flanking apparent SNPs were designed. To test their compatibility and to ensure the amplification of a single product, we performed PCR simulations with the software Amplify, version 1.2. Primer pairs that gave a single, strong amplification product in Amplify were synthesized by IDTDNA (Iowa City, Iowa). To confirm the heterozygous base pair positions, we amplified regions by PCRs, using the same primer pairs and the genomic DNA of strain SC5314, and conducted a sequence analysis (Fig. 1G; also see below). Thirty-one polymorphic markers from previous studies (9, 18, 20, 62) were obtained and evaluated for polymorphisms in strain SC5314 as described below.
FIG. 1.
Flow chart for web-based development of SNP markers in C. albicans, with marker 1363/2056 used as an example. Two overlapping sequence contigs representing the two potentially heterozygous alleles for each genomic region were selected (A). Complete sequences for both contigs were then obtained (B) and compared by using the BLAST algorithm (C), followed by obtaining the actual sequence for overlapping parts of the contigs (D). The two sequences were aligned (E), heterozygous positions were identified, and forward and reverse primers flanking apparent SNPs were designed and synthesized. PCR simulations were performed (F), primer pairs were synthesized, and SNPs were confirmed by sequencing (G).
PCRs were carried out in a final volume of 25 μl containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, a 100 μM concentration (each) of dATP, dCTP, dGTP, and dTTP, 2.5 U of Taq polymerase, 0.5 μl of a 10 μM stock solution of each primer, and 30 ng of quick prep genomic DNA under the following conditions: initial denaturation for 3 min at 94°C, denaturation step for 1 min at 94°C, primer annealing step for 30 s at 55°C, extension step for 1 min at 72°C, and a final extension step for 5 min at 72°C. Five microliters of the PCR product was run in a 3% agarose gel (0.5× Tris-acetate-EDTA) to verify the amplification of a single fragment of the appropriate size. Primer pairs failing to give a single strong band were subjected to PCR optimization by adjusting the annealing temperature or MgCl2 concentration, and those that still failed to amplify single bands were discarded.
Confirmation of heterozygosity of SNP markers by sequencing.
To confirm the heterozygous polymorphisms obtained from the genome sequence and reported in other studies, we again sequenced each marker for strain SC5314. Internal primers were designed to sequence through larger PCR products (e.g., RFLP markers from reference 62). PCRs were performed as described above, PCR products were purified by using a PCR purification kit (Qiagen Inc., Valencia, Calif.), and 50 to 100 ng of purified DNA was used as a sequencing template. Sequencing reactions were performed with both forward and reverse primers according to the manufacturer's instructions (ABI Big Dye Chemistry; Perkin-Elmer, Boston, Mass.) and were run on an ABI377 automated sequencer. Sequences were edited and aligned with Sequencer, version 3.1.1, software (Perkin-Elmer).
Inter- and intragenic locations of SNP markers and changes in predicted amino acid sequence.
For all SNP markers, we determined whether the SNPs were located within open reading frames (ORFs) or in intergenic regions. The sequence of each marker was compared against the entire genome sequence of C. albicans (http://www-sequence.stanford.edu:8080/bncontigs6.html) to obtain the exact nucleotide positions within the sequence contigs. The ORF information available at http://www-sequence.stanford.edu:8080/contigs-to-blast6.html was used to determine whether the SNP marker sequence was located inside, outside, or partially within an ORF. For SNPs located within an ORF, we assessed alterations of the predicted amino acid sequence for the entire ORF.
LOH in laboratory strains of C. albicans.
We were interested in whether processes such as transformation or growth on selective media, such as 5-fluoroorotic acid or 2-deoxygalactose (2DG), would lead to LOH in C. albicans. Strains that had undergone one or multiple rounds of transformation and passaging on selective media were analyzed (Table 1). These strains were derived in previous transformation experiments from the wild-type strain SC5314. SNP markers flanking altered genomic regions (URA3, HIS1, ARG4, and GAL1) were used to assess LOH (Table 2; also see the supplemental material) and then were mapped onto the SNP map (see Fig. 3).
TABLE 2.
Detection of LOH in strains of C. albicans
| Marker no.a | Marker nameb | Flanking gene | Chromosome | Genotype of strainc
|
||||
|---|---|---|---|---|---|---|---|---|
| CAI4 | RM1000 | BWP17 | AF14 | AF27 | ||||
| 76 | 2195/2207d | URA3 | 3P | Het | Het | Het | Het | Het |
| 118 | 1341/2493e | HIS1 | 5I | Het | Het | Het | Het | Het |
| 120 | 2340/2493e | HIS1 | 5I | Het | Hom | Hom | Het | Hom |
| 141 | 1883/2139f | ARG4 | 7G | Het | Het | Het | Het | Het |
| 143 | 1530/2473 (1 to 3)f | ARG4 | 7G | Het | Het | Het | Het | Het |
| 22 | 1322/2294 | NA | 1S | Het | Het | Het | Hom | Het |
| 23 | 1799/2450 | NA | IS | Het | Het | Het | Hom | Het |
| 24 | MNS1 | NA | 1S | Het | Het | Het | Hom | Het |
| 25 | 2036/2375 | NA | 1S | Het | Het | Het | Hom | Het |
| 26 | 2281/2302 | NA | 1S | Het | Het | Het | Hom | Het |
| 27 | 2032/2371 | NA | 1S | Het | Het | Het | Hom | Het |
| 28 | 1449/2362 | NA | 1S | Het | Het | Het | Hom | Het |
| 29 | 1363/2056g | GAL1 | 1S | Het | Het | Het | Hom | Het |
| 30 | 1622/2428g | GAL1 | 1E | Het | Het | Het | Het | Het |
| 42 | 2106/2441 | NA | 1L | Het | Hom | Het | Het | Hom |
| 43 | 1729/2211 | NA | 1L | Het | Hom | Het | Het | Hom |
| 46 | 1584/2265 | NA | 1 | Het | Het | Het | Hom | Het |
| 48 | 1353/2363 | NA | 2U | Het | Hom | Hom | Het | Hom |
| 103 | 1718/2417 (HST3) | NA | 5M | Het | Het | Het | Hom | Hom |
| 113 | 1969/2162 | NA | 5M | Het | Het | Het | Het | Hom |
| 137 | 2397/2498 | NA | 7F | Hom | Hom | Hom | Hom | Hom |
| 147 | 2080/2297 (DLH1) | NA | Het | Hom | Hom | Het | Hom | |
Numbering corresponds to the supplemental material and Fig. 2.
The names of markers correspond either to the two contig-6 sequence numbers from which they were generated or to the corresponding gene.
Het, heterozygous; hom, homozygous; NA, not applicable.
URA3 is at the end of contig-19-10123 (see Fig. 2); only SNP marker 2195/2207 was available for sequencing.
Flanks HIS1.
Flanks ARG4.
Flanks GAL1.
FIG. 3.
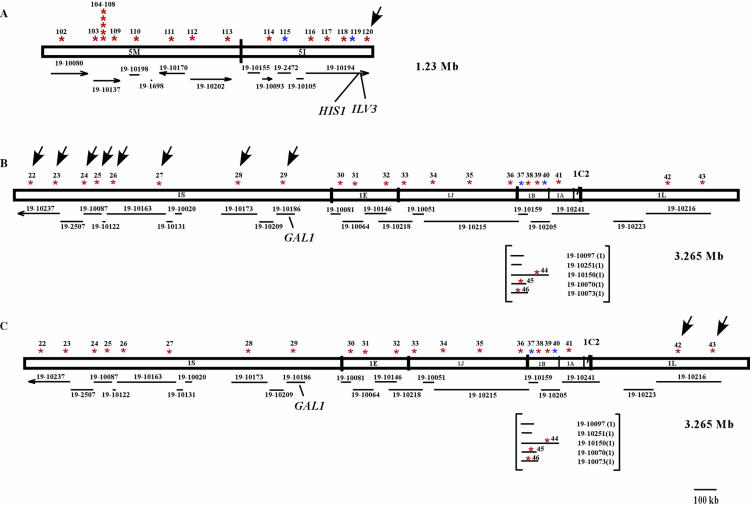
LOH detected in selected strains of C. albicans. (A) LOH in strain AF14 on chromosome fragment 1S. (B) LOH in strains RM1000, BWP17, AF12, and AF27 on chromosome fragment 1L. (C) LOH in strains RM1000, BWP17, AF12, and AF27 on chromosome fragment 5I. Arrows indicate LOH of SNP markers. Red stars, SNP markers that are heterozygous in strain SC5314; blue stars, SNP markers that are homozygous in strain SC5314.
RESULTS
Development and characterization of SNP markers.
Using a web-based strategy to develop SNP markers (PCR fragments that contain one or more SNPs), we generated 119 potential marker sequences that are heterozygous in strain SC5314 (Fig. 1 and Fig. 2). By adding 31 markers from previous studies (10, 18, 20, 62), we obtained a total of 150 potential SNP markers for strain SC5314 (see the supplemental material). The SNP marker sequences in the set of 150 ranged in size from 120 to 1,821 bp, with an average of 322 bp. We confirmed most of the SNPs reported in the genome sequence of strain SC5314 (http://www-sequence.stanford.edu:8080/contigs-to-blast6.html)by resequencing. However, for four markers developed in our study, not all of the SNPs could be confirmed. For markers 26 and 63, only half of the reported polymorphisms were detected by sequencing, and for markers 12 and 52, one SNP could not be confirmed for each (data not shown). We found that 19 of the markers from previous studies were homozygous once they were examined for heterozygosity in SC5314 by resequencing. On the other hand, for six markers from previous studies (9, 18, 62), the genome sequence of SC5314 indicated these loci to be homozygous, but our sequences for strain SC5314 indeed revealed the presence of SNPs (see the supplemental material). In total, 131 of the SNP marker sequences used for this study were heterozygous and revealed 561 SNPs and 9 insertions-deletions (indels), for a total of 570 polymorphisms (Table 3).
FIG. 2.
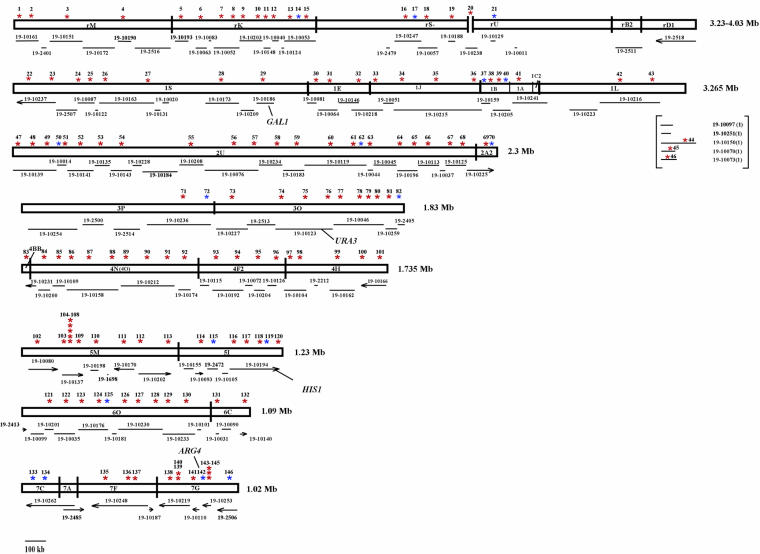
Whole-genome SNP map of C. albicans. Red stars, SNP markers that are heterozygous in strain SC5314; blue stars, SNP markers that are homozygous in strain SC5314. The numbers above the stars correspond to the numbering in Table S1 in the supplemental material. Based on the estimated sizes for C. albicans strain 1006 (obtained by pulsed-field gel electrophoresis), a map was created for the SfiI restriction sites of each chromosome (8). SfiI sites are indicated by vertical bars, and the names assigned by Chu et al. (8) are indicated within the boxes representing the chromosomes (e.g., 7A). The corresponding contig-19 sequences are shown along the lower side of each chromosome. Mapped genes from the fosmid map (http://alces.med.umn.edu/candida/) were used to order contig-19 sequences onto the map. When known, the directions (5′ to 3′) of contig-19 sequences are marked by arrows. Five contig-19 sequences on chromosome 1 have not been assigned to their corresponding SfiI fragments. They are located under chromosome 1 as a separate group (in brackets).
TABLE 3.
Summary of SNP marker characterizationa
| Chromosome | No. of markersb
|
Total no. of polymorphisms | No. of base pairs sequenced | SNP frequency (1 SNP/no. of bp) | No. of SNPs | No. of indels | No. of transitions | No. of transversions
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Heterozygous | Homozygous | G/T (C/A) | C/G | A/T | |||||||
| R | 18 | 3 | 56 | 5,507 | 1/98 | 55 | 1 | 47 | 4 | 2 | 2 |
| 1 | 23 | 2 | 103 | 8,117 | 1/79 | 101 | 2 | 76 | 15 | 1 | 9 |
| 2 | 21 | 3 | 74 | 5,690 | 1/77 | 74 | 0 | 54 | 11 | 2 | 7 |
| 3 | 10 | 2 | 38 | 3,739 | 1/98 | 36 | 2 | 29 | 3 | 2 | 4 |
| 4 | 19 | 0 | 101 | 4,785 | 1/47 | 99 | 2 | 76 | 6 | 6 | 11 |
| 5 | 17 | 2 | 75 | 8,103 | 1/108 | 74 | 1 | 60 | 5 | 6 | 3 |
| 6 | 11 | 1 | 60 | 2,894 | 1/48 | 60 | 0 | 44 | 4 | 6 | 6 |
| 7 | 10 | 4 | 53 | 6,288 | 1/119 | 53 | 0 | 42 | 5 | 3 | 3 |
| Unknown | 2 | 2 | 10 | 1,959 | 1/196 | 9 | 1 | 9 | 0 | 0 | 0 |
| Total | 131 | 19 | 570 | 47,190 | 1/83 | 561 | 9 | 437 | 53 | 28 | 45 |
Markers are ordered and summarized by chromosomal location. For each chromosome, the number of heterozygous and homozygous SNP markers is given followed by the total number of SNP markers. The number of bp sequenced is listed followed by the frequency of SNPs. The number of SNPs and indels is given and the total number of SNPs is categorized into transition and transversion events.
Heterozygous and homozygous markers were based on sequences obtained for C. albicans strain SC5314.
The frequency of SNPs for our data set varied between 1 per 47 bp on chromosome 4 and 1 per 119 bp on chromosome 7. For all of the chromosomes, the average SNP frequency was 1 SNP per 83 bp (Table 3). Disregarding SNP markers 104 to 107 and markers 143 and 144 because of their very close proximities to each other (Fig. 2), on average our map contains 1 SNP marker every 111 kb in the 16-Mb haploid genome. Of the 561 SNPs, 437 were transition events while 126 were transversion events (Table 3), yielding a transition-to-transversion ratio of 3:1, as expected for a neutral accumulation of mutations.
Mapping SNP markers onto the C. albicans genome.
For a better visualization of the locations of the SNP markers, a SNP map was created and is shown in Fig. 2 as a cartoon of the eight chromosomes of strain SC5314. Since all but four of the SNP markers developed in this study were obtained from contigs with known genome locations (chromosome and/or SfiI fragment), the vast majority of marker sequences, whether homozygous or heterozygous, could be mapped to their corresponding chromosomal locations. Markers that are heterozygous in strain SC5314 are shown as red stars. Homozygous markers are indicated by blue stars. Under each chromosome, the corresponding contig-19 sequences (diploid assembly of the C. albicans genome sequence) are mapped. The order of many contig-19 sequences was determined by the association between previously mapped genes and their approximate locations on the physical map (http://alces.med.umn.edu/Candida.html). For contig-19 sequences for which the orientation along the chromosome is known (e.g., see chromosomes 5 and 7 [6a, 6b]) the 5′-to-3′ direction of the sequence is indicated by an arrow (Fig. 2).
The SNP markers are distributed evenly along chromosomes 1, 2, 4, 5, and 6, but there are large chromosomal regions on chromosomes R, 3, and 7 which are apparently highly or entirely homozygous in SC5314. Chromosomal fragments RB2 and RD1 on chromosome R show no or very little heterozygosity. On chromosome 3, a region covering three contig-19 sequences and making up a total of 464 kb of SfiI fragment P is apparently homozygous. On chromosome 7, SfiI fragments 7A and 7C are completely homozygous, and a large portion at the telomere-proximal end of 7G also lacks heterozygosity (Fig. 2). In contrast, there are some genomic locations with abundant polymorphisms. Two examples are the sequences from markers 104 to 108 on chromosome 5 M (near the MTL loci) and from markers 143 to 145 on chromosome 7G.
We were interested in whether the SNPs were located in intergenic or intragenic regions and whether intragenic SNPs changed the amino acid coding sequence (nonsynonymous substitutions). Of the 131 heterozygous marker sequences, 40 were located in intergenic regions and 91 were partially or completely within coding regions. At least one nonsynonymous substitution was found in 81 of the 91 (89%) intragenic SNP marker sequences. Only synonymous substitutions were found in 10 of the intragenic SNP marker sequences (11%).
LOH in selected strains of C. albicans.
To address concerns that various genetic manipulations such as transformation or growth on selective media might increase the rate of LOH, we examined loci that had been targets of knockout experiments for evidence of LOH in flanking regions. We used strains that are often used for gene manipulations as well as some that were grown on selective media during the transformation procedure. Strains CAI4, RM1000, BWP17, AF14, and AF27 were derived from SC5314 by consecutive transformations to knock out both copies of URA3 (CAI4 [16]), HIS1 (RM1000 [36]), and ARG4 (BWP17 [61]), in that order. Furthermore, we examined four strains, AF6, AF14, AF12, and AF27, in which one or both copies of GAL1 had been knocked out (17). One copy of GAL1 was knocked out in AF6, which was derived from CAI4, and in AF12, which was derived from RM1000. The second copy of GAL1 was deleted from strain AF6 to generate AF14 by growth on a 2DG (toxic to Gal+ strains) medium, so AF14 is in a CAI4 background. The second copy of GAL1 was deleted from AF12 to generate AF27 by two sequential rounds of transformation without growth on selective medium, so AF27 is in an RM1000 background (17). The early steps in the construction of most of these knockout strains involved growth on 5-fluoroorotic acid to select for the loss of URA3 due to loop-out events (Table 1).
We sequenced SNP markers adjacent to gene knockouts (URA3, HIS1, ARG4, and GAL1) (see Fig. 2 for genomic locations) from strains CAI4, RM1000, BWP17, AF14, and AF27. We did not observe LOH at URA3 in CAI4 or any of the descendant strains. For HIS1, marker 120, located at the distal side of HIS1, demonstrated LOH in strains RM1000, BWP17, and AF27, but not in CAI4 and its descendant strain AF14 (Fig. 3A, arrow). Thus, the LOH in marker 120 might have accompanied HIS1 mutations in RM1000 such that LOH is observed in RM1000 descendant strains but not in CAI4 descendant strains. SNP marker 118, flanking the proximal side of HIS1, was still heterozygous in all five strains. We did not observe LOH at markers flanking ARG4 in BWP17, the strain from which ARG4 was deleted, or in any of the other strains. Thus, ARG4 deletions were not accompanied by LOH.
To analyze the LOH for strains in which GAL1 manipulations had been done, we initially sequenced markers flanking the GAL1 gene on chromosome fragment 1S for strains AF14 and AF27. Marker 29 was homozygous in strain AF14 but heterozygous in strain AF27. In contrast, marker 30, which flanks GAL1 on the right and is located on the 1E chromosome fragment, was heterozygous in both strains (Fig. 3B). To determine whether the entire chromosome fragment 1S had lost heterozygosity in AF14, we sequenced the remaining SNP markers on fragment 1S for this strain. All SNP markers did indeed show LOH (Fig. 3B). We then sequenced the markers on 1S for strain AF6, the parent of AF14, and found that all were still heterozygous. Together, these results suggest that the homozygosity of marker 29 and all 1S markers distal to marker 29 occurred during continued selection on 2DG in the cell lineage leading from AF6 to AF14.
From the above results, we found that LOH is not directly related to gene manipulations or growth on selective media. To confirm this result, we sequenced all of the SNP markers at other genomic locations for the five strains discussed above, and these results also demonstrated LOH (Table 2). For example, all of the markers sequenced from chromosome fragment 1L demonstrated LOH in strains RM1000, BWP17, AF12, and AF27 (Fig. 3C). Together, our results suggest that transformation per se does not seem to increase LOH but that using a medium such as 2DG to select for a mitotic recombination event can result in the recovery of homozygosity across an entire chromosome region.
DISCUSSION
This paper describes a web-based approach to developing a genome-wide SNP map for strain SC5314 of the opportunistic human pathogen C. albicans. Heterozygosity in this fungus was demonstrated some 20 years ago (56-58), but only recently have molecular and genomic advances allowed for the exploitation of such variation for the study of the biology of infection. The genome sequence reveals approximately 62,000 SNPs (24), which are a great resource with which to study population genetics, the relationships of strains, and transmission genetics. Heterozygosity can also be exploited to unravel the processes contributing to the diversity and plasticity of the C. albicans genome.
For this study, we took advantage of the publicly available diploid genome sequence (http://www-sequence.stanford.edu:8080/haploid19.html) to develop a genome-wide SNP map for C. albicans. The SNP map contains 131 markers comprising 561 SNPs and 9 indels (see the supplemental material), incorporates 12 markers from previous studies (10, 18, 20, 62), and includes about 1% of the total number of SNPs in the C. albicans genome. The map has one marker every 111 kb, on average.
The frequencies of SNPs per base pair in our data set are quite high and vary over the genome, ranging from 0 in some locations to 1 per 47 bp on chromosome 4 to 1 per 119 bp on chromosome 7, with a genome-wide frequency of 1 per 83 bp. For C. elegans, SNPs have been used to study the relationships among strains and to carry out high-throughput gene mapping (25, 51). In one study using 366 SNPs, the SNP frequencies varied from 1 per 1,445 bp to 1 per 8,750 bp (25). Brumfield et al. (4) examined the utility of SNPs to infer population histories and reviewed SNP frequencies obtained from a variety of different organisms. Studies comparable to ours found SNP frequencies ranging from 1 per 1,001 bp in humans to 1 per 2,119 bp in chickens. Since most organisms covered in that review have complete sexual cycles, the most likely explanation for the high SNP frequency we found in C. albicans is the accumulation of mutations in this largely clonal organism (55). Some form of balancing selection across variable environments might also maintain heterozygosity against the mitotic conversion (LOH) we observed, but it is difficult to demonstrate without genealogical data for specific coding regions.
Recently, multilocus sequence typing (MLST) was recommended as a standard method for determining relationships among strains (2, 3, 52). Sequences of several coding regions (housekeeping genes) are combined into multilocus genotypes to determine the relatedness among strains. Because the SNP markers presented here are distributed across all of the chromosomes, while the MLST markers represent only five of the eight chromosomes, the two systems could be used together in studies of genome regions covered by both. We recommend the use of SNP markers for genome-wide analyses of recombination. A combination of SNP markers and MLST would not only increase the discriminatory power of strain typing, but would unite data from noncoding and coding regions across the entire genome.
To study mitotic recombination, including gene conversion at heterozygous loci, we sequenced several SNP markers for several strains (Table 2; Fig. 3) that were derived sequentially from SC5314 by transformation (see Results and Table 1). We found little evidence that gene knockouts directly increase LOH at flanking regions. No LOH was found for markers flanking URA3 and ARG4 in knockout strains. However, marker 120 (Fig. 2; also see the supplemental material), next to HIS1 and located at the distal end of chromosome fragment 5I, was homozygous for all SNPs tested, while marker 118, approximately 50 kb to the left end of HIS1, was not. Karyotypic analysis has shown that strain RM1000 carries two differently sized chromosome 5 homologues, whereas its progenitor strain, CAI4, carries chromosome 5 homologues of the same size (data not shown). Further analysis showed that the gene ILV3, which is telomeric to HIS1, is missing from the smaller homologue of chromosome fragment 5I in RM1000 (unpublished data). These facts suggest that a partial loss and reconstitution of the telomeric end of one homologue of chromosome fragment 5I occurred in strain RM1000 (Fig. 3A) and may have occurred during the disruption of HIS1. In the second case of large chromosomal fragment changes that we observed, chromosomal fragment 1S (∼1,400 kb) lost heterozygosity during growth on 2DG medium (Fig. 3B and C). Our data indicate that a single mitotic crossover in strain AF6 caused the loss of the second GAL1 allele, generating strain AF14. This event seems to have occurred spontaneously, in line with previous observations of a low but finite rate of mitotic recombination in Candida.
We investigated the genomic locations of the SNP markers with respect to predicted ORFs. More than two-thirds of the heterozygous markers (89%) were located in intragenic regions with the potential to affect the protein sequence or expression level (26). Diverged functions of two alleles at the SAP2 gene were demonstrated by Staib and coworkers (48), who showed that differences in the numbers of pentameric repeats in the promoter region led to differential regulation of the alleles. It was recently shown that allelic variation in gene expression is frequent in the human genome (30). From a SNP array, 326 of 602 (54%) genes expressed in kidney and liver tissues showed two- to fourfold differences between the expression levels of alleles and 170 genes exhibited more than fourfold differences in allelic expression levels. Given the high level of heterozygosity in the C. albicans genome, the differential expression of genes seems likely.
The main goal of this paper was to put together tools to study recombinational and genomic changes in C. albicans under different environmental conditions, such as changing media, growth conditions, exposure to antifungal drugs, and infection in animal hosts. For example, several mechanisms might lead to resistance to the antifungal drug fluconazole in C. albicans. One mechanism involves LOH at the ERG11 locus, a gene in the ergosterol biosynthesis pathway that is targeted by fluconazole. White (59) studied 17 serial isolates of C. albicans and discovered that isolate 13 and all subsequent isolates became resistant to fluconazole. An analysis showed that allelic differences within the ERG11 ORF, in the ERG11 promoter, and in the downstream THR1 gene were lost in these isolates, and a single amino acid substitution in ERG11 was also identified. Another group discovered that multiple mechanisms, including LOH, contribute to a stepwise development of antifungal drug resistance in clinical isolates (19). The fact that LOH at and around the ERG11 gene was discovered by two independent groups suggests that homozygosity at this locus increases fitness in a fluconazole environment. In a second example of the importance of recombination, rates and frequencies of mitotic recombination have been determined for the GAL1 locus in C. albicans in vitro by the use of fluctuation tests and in vivo by the use of a mouse model (17). Strains that were heterozygous at the GAL1 locus were injected into mice, Gal− strains were identified, and the frequencies of mitotic recombination at the GAL1 locus were determined. Although in vitro and in vivo data sets cannot be compared directly, these results suggest that there may be a higher level of mitotic recombination in vivo than in vitro (17).
So far, sequencing has been the method of choice for SNP analysis. However, analyzing large numbers of SNP markers for entire populations of C. albicans by sequencing is very time- and labor-intensive. Recently, SNP microarrays have been developed for a variety of organisms, including humans (14, 21, 37, 45), mice (28), plants (7), and protozoa (47, 49), and they are becoming a fast, accurate, and reproducible method for SNP analysis (7, 12, 29, 34). We have developed a SNP microarray (unpublished data) for C. albicans to study the mitotic recombination of populations of cells in vitro and in vivo to learn more about how and by which genomic processes C. albicans adapts to diverse and constantly changing host environments. This will provide a first glimpse of what makes C. albicans so successful as an opportunistic pathogen of humans.
Supplementary Material
Acknowledgments
We thank Christine Ramos for her excellent technical assistance. Sequence data for C. albicans were obtained from the Stanford DNA Sequencing and Technology Center web site at http://www-sequence.stanford.edu/group/candida.
This work was supported by NIH grant AI46351 awarded to P. T. Magee and G. May and by contract AI054006.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1.Beckerman, J., H. Chibana, J. Turner, and P. T. Magee. 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bougnoux, M.-E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bougnoux, M.-E., A. Tavanti, C. Bouchier, N. A. R. Gow, A. Magnier, A. D. Davidson, M. C. J. Maiden, C. d'Enfert, and F. C. Odds. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumfield, R. T., P. Beerli, D. A. Nickerson, and S. V. Edwards. 2003. The utility of single nucleotide polymorphisms in inference of population history. TREE 18:249-256. [Google Scholar]
- 5.Calderone, R. 2002. Candida and candidiasis. American Society for Microbiology, Washington D.C.
- 6.Chasman, D., and R. M. Adams. 2001. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 307:683-706. [DOI] [PubMed] [Google Scholar]
- 6a.Chibana, H., B. B. Magee, S. Grindle, Y. Ran, S. Scherer, and P. T. Magee. 1998. A physical map of chromosome 7 of Candida albicans. Genetics 149:1739-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b.Chibana, H., J. L. Beckerman, and P. T. Magee. 2000. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10:1865-1877. [DOI] [PubMed] [Google Scholar]
- 7.Cho, R. J., M. Mindrinos, D. R. Richards, R. J. Sapolsky, M. Anderson, E. Drenkard, L. Dewdney, T. L. Reuber, M. Stammers, N. Federspiel, A. Theologis, W. H. Yang, E. Hubbell, M. Au, E. Y. Chung, D. Lashkari, B. Lemieux, C. Dean, R. J. Lipshutz, F. M. Ausubel, R. W. Davis, and P. J. Oefner. 1999. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat. Genet. 23:203-207. [DOI] [PubMed] [Google Scholar]
- 8.Chu, W. S., B. B. Magee, and P. T. Magee. 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J. Bacteriol. 175:6637-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowen, L. E., C. Sirjusingh, R. C. Summerbell, S. Walmsley, S. Richardson, L. M. Kohn, and J. B. Anderson. 1999. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 43:2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumur, C. I., C. Dechsukhum, J. L. Ware, S. S. Cofield, A. M. Best, D. S. Wilkinson, C. T. Garrett, and A. Ferreira-Gonzalez. 2003. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics 81:260-269. [DOI] [PubMed] [Google Scholar]
- 13.Enloe, B., A. Diamond, and A. P. Mitchell. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, J. B., X. Q. Chen, M. K. Halushka, A. Berno, X. H. Huang, T. Ryder, R. J. Lipshutz, D. J. Lockhart, and A. Chakravarti. 2000. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 10:853-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schafer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases SAP4 to SAP6 are required for the invasion of parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forche, A., G. May, J. Beckerman, S. Kauffman, J. Becker, and P. T. Magee. 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39:38-50. [DOI] [PubMed] [Google Scholar]
- 18.Forche, A., G. Schönian, Y. Gräser, R. Vilgalys, and T. G. Mitchell. 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28:107-125. [DOI] [PubMed] [Google Scholar]
- 19.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gräser, Y., M. Volovsek, J. Arrington, G. Schönian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacia, J. G., J.-B. Fan, O. Ryder, L. Jin, K. Edgemon, G. Ghandour, R. A. Mayer, B. Sun, L. Hsie, C. M. Robbins, L. C. Brody, D. Wang, E. S. Lander, R. Lipshutz, S. P. A. Fodor, and F. S. Collins. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22:164-167. [DOI] [PubMed] [Google Scholar]
- 22.Inglis, D. O., and A. D. Johnson. 2002. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Mol. Cell. Biol. 22:8669-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubowski, J., and K. Kornfeld. 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, T., N. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Kelly, R., S. M. Miller, and M. B. Kurtz. 1998. One-step gene disruption by cotransformation to isolate double mutants in Candida albicans. Mol. Gen. Genet. 214:24-31. [DOI] [PubMed] [Google Scholar]
- 25.Koch, R., H. van Luenen, M. van der Horst, K. L. Thijssen, and R. H. A. Plasterk. 2000. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 10:1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landegren, U., M. Nilsson, and P.-Y. Kwok. 1998. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 8:769-776. [DOI] [PubMed] [Google Scholar]
- 27.Levo, A., A. Koski, I. Ojanpera, E. Vuori, and A. Sajantila. 2003. Post-mortem SNP analysis of CYP2D6 gene reveals correlation between genotype and opioid drug (tramadol) metabolite ratios in blood. Forensic Sci. Int. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 28.Lindblad-Toh, K., E. Winchester, M. J. Daly, D. G. Wang, J. N. Hirschorn, J.-P. Laviolette, K. Ardlie, D. E. Reich, E. Robinson, P. Sklar, N. Shah, D. Thomas, J.-B. Fan, T. Gingeras, J. Warrington, N. Patil, T. J. Hudson, and E. S. Lander. 2000. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24:381-386. [DOI] [PubMed] [Google Scholar]
- 29.Lindroos, K., S. Sigurdsson, K. Johansson, L. Roennblom, and A. C. Syvanen. 2002. Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res. 30:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, H. S., Z. N. Wang, Y. Hu, H. H. Yang, S. Gere, K. H. Buetow, and M. P. Lee. 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13:1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, H., T. Higashikata, A. Inazu, A. Nohara, W. Yu, M. Shimizu, and H. Mabuchi. 2002. Association of estrogen receptor-α gene polymorphisms With coronary artery disease in patients with familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 22:817-823. [DOI] [PubMed] [Google Scholar]
- 32.Magee, B. B., and P. T. Magee. 1987. Electrophoretic karyotypes and chromosome numbers in Candida species. J. Gen. Microbiol. 133:425-430. [DOI] [PubMed] [Google Scholar]
- 33.Magee, P. T., L. Bowdin, and J. Staudinger. 1992. Comparison of molecular typing methods for Candida albicans. J. Clin. Microbiol. 30:2674-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei, R., P. C. Galipeau, C. Prass, A. Berno, G. Ghandour, N. Patil, R. K. Wolff, M. S. Chee, B. J. Reid, and D. J. Lockhart. 2000. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res. 10:1126-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, W., E. Lieckfeldt, K. Kuhls, E. Z. Freedman, T. Borner, and T. G. Mitchell. 1993. DNA- and PCR-fingerprinting in fungi. EXS 67:311-320. [DOI] [PubMed] [Google Scholar]
- 36.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 37.Pastinen, T., M. Raitio, K. Lindroos, P. Tainola, L. Peltonen, and A.-C. Syvaenen. 2000. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 10:1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Primdahl, H., F. P. Wikman, H. V. D. Maase, X.-G. Zhou, H. Wolf, and T. F. Orntorf. 2002. Allelic imbalances in human bladder cancer: genome-wide detection with high-density single-nucleotide polymorphism arrays. J. Natl. Cancer Inst. 94:216-223. [DOI] [PubMed] [Google Scholar]
- 39.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pujol, C., J. Reynes, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Mallie, and J. M. Bastide. 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggsby, W. S., L. J. Torres-Bauza, J. W. Wills, and T. M. Townes. 1982. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol. Cell. Biol. 2:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risch, N., and K. Merikangas. 1996. The future of genetic studies of complex human diseases. Science 273:1516-1517. [DOI] [PubMed] [Google Scholar]
- 43.Rustchenko-Bulgac, E. P., F. Sherman, and J. B. Hicks. 1990. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J. Bacteriol. 172:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaeda, T., T. Nakamura, and K. Okumura. 2003. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics 4:397-410. [DOI] [PubMed] [Google Scholar]
- 45.Sapolsky, R. J., L. Hsie, A. Berno, G. Ghandour, M. Mittmann, and J. B. Fan. 1999. High-throughput polymorphism screening and genotyping with high-density oligonucleotide arrays. Genet. Anal. 14:187-192. [DOI] [PubMed] [Google Scholar]
- 46.Schmid, K. J., T. R. Sorensen, R. Stracke, O. Torjek, T. Altmann, T. Mitchell-Olds, and B. Weisshaar. 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13:1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibley, L. D., D. G. Mordue, C. Su, R. P. M., and D. K. Howe. 2002. Genetic approaches to studying virulence and pathogenesis in Toxoplasma gondii. Philos. Trans. R. Soc. Lond. B 357:81-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 49.Straub, T. M., D. S. Daly, S. Wunshel, P. A. Rochelle, R. DeLeon, and D. P. Chandler. 2002. Genotyping Cryptosporidium parvum with an hsp70 single-nucleotide polymorphism microarray. Appl. Environ. Microbiol. 68:1817-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su, Z. G., S. Z. Zhang, Y. P. Hou, T. Li, D. W. Nebert, L. Zhang, D. J. Huang, L. C. Liao, and C. Y. Xiao. 2002. Single-nucleotide polymorphisms in the lipoprotein lipase gene associated with coronary heart disease in Chinese. Eur. J. Pharmacol. 454:9-18. [DOI] [PubMed] [Google Scholar]
- 51.Swan, K. A., D. E. Curtis, K. B. McKusick, A. V. Voinov, F. A. Mapa, and M. R. Cancilla. 2002. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 12:1100-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tavanti, A., N. A. R. Gow, S. Senesi, M. C. J. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsang, P. W., B. Cao, P. Y. Siu, and J. Wang. 1999. Loss of heterozygosity, by mitotic gene conversion and crossing over, causes strain-specific adenine mutants in constitutive diploid Candida albicans. Microbiology 145:1623-1629. [DOI] [PubMed] [Google Scholar]
- 54.Wang, D. G., J. B. Fan, C. J. Siao, A. Berno, P. Young, R. Sapolsky, G. Ghandour, N. Perkins, E. Winchester, J. Spencer, L. Kruglyak, L. Stein, L. Hsie, T. Topaloglou, E. Hubbell, E. Robinson, M. Mittmann, M. S. Morris, N. P. Shen, D. Kilburn, J. Rioux, C. Nusbaum, S. Rozen, T. J. Hudson, R. Lipshutz, M. Chee, and E. S. Lander. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077-1082. [DOI] [PubMed] [Google Scholar]
- 55.Welch, D. M., and M. Meselson. 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288:1211-1215. [DOI] [PubMed] [Google Scholar]
- 56.Whelan, W. L., and P. T. Magee. 1981. Natural heterozygosity in Candida albicans. J. Bacteriol. 145:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whelan, W. L., R. M. Partridge, and P. T. Magee. 1980. Heterozygosity and segregation in Candida albicans. Mol. Gen. Genet. 180:107-113. [DOI] [PubMed] [Google Scholar]
- 58.Whelan, W. L., and D. R. Soll. 1982. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol. Gen. Genet. 187:477-485. [DOI] [PubMed] [Google Scholar]
- 59.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston, and R. H. A. Plasterk. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28:160-164. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, J., T. G. Mitchell, and R. Vilgalys. 1999. PCR-restriction fragment length polymorphism (RFLP) analyses reveal both extensive clonality and local genetic differences in Candida albicans. Mol. Ecol. 8:59-73. [DOI] [PubMed] [Google Scholar]
- 63.Yesland, K., and W. A. Fonzi. 2000. Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology 146:2097-2104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.