Abstract
The yeast Candida albicans is a major opportunistic pathogen of immunocompromised individuals. It can grow in several distinct morphological states, including budded and hyphal forms, and the ability to make the dynamic transition between these forms is strongly correlated with virulence. Recent studies implicating the Cdc42p GTPase in hypha formation relied on cdc42 mutations that affected the mitotic functions of the protein, thereby precluding any substantive conclusions about the specific role of Cdc42p in the budded-to-hypha-form transition and virulence. Therefore, we took advantage of several Saccharomyces cerevisiae cdc42 mutants that separated Cdc42p's mitotic functions away from its role in filamentous growth. The homologous cdc42-S26I, cdc42-E100G, and cdc42-S158T mutations in C. albicans Cdc42p caused a dramatic defect in the budded-to-hypha-form transition in response to various hypha-inducing signals without affecting normal budded growth, strongly supporting the conclusion that Cdc42p has an integral function in orchestrating the morphological transition in C. albicans. In addition, the cdc42-S26I and cdc42-E100G mutants demonstrated a reduced ability to damage endothelial cells, a process that is strongly correlated to virulence. The three mutants also had reduced expression of several hypha-specific genes, including those under the regulation of the Efg1p transcription factor. These data indicate that Cdc42p-dependent signaling pathways regulate the budded-to-hypha-form transition and the expression of hypha-specific genes.
Candida albicans is an opportunistic fungal pathogen, causing significant morbidity as well as life-threatening systemic disease in immunocompromised individuals. C. albicans cells can exist in a range of morphological states and possess the ability to radically alter their growth pattern from budded to hyphal growth in the presence of inducing signals, such as serum, N-acetylglucosamine, or numerous different formulations of laboratory media that induce a starvation response (5, 12). The starvation response appears to be physiologically relevant, since a starvation response can be induced in C. albicans cells when they are incubated in the presence of neutrophils (43). The ability to switch between the two distinct morphological states is an aspect of C. albicans biology that is strongly correlated to virulence, and mutants that are unable to make the transition are avirulent in a mouse model of disseminated candidiasis (9, 18, 24-26, 29, 39, 47, 57). The timing of hyphal production in the infection process is also crucial (45), and both budded and hyphal cells are seen in infected tissues, emphasizing the importance of this dynamic switch between morphological forms in the infection process.
Expression of virulence determinants, such as adhesins (Hwp1p), cell wall proteins (Ece1p), secreted aspartyl proteases (Sap6p), and phospholipases (Plb1p), is up-regulated in the presence of hypha-inducing signals (4, 23, 48). Several transcriptional activators and repressors regulate this transcriptional induction. Efg1p and Cph1p are the primary transcriptional activators, since strains with both of these genes deleted are unable to form hyphae and are avirulent in mice (29). Additionally, gene array analyses suggest that Efg1p and Cph1p together are responsible for inducing transcription of many of the described virulence factors in C. albicans (4, 23, 48). Recent studies suggest that the Tup1p transcriptional repressor may act in concert with other repressor proteins, such as Mig1p, Nrg1p, and Rfg1p, to negatively regulate transcription of genes involved in the budded-to-hyphal-form transition (11, 34). Altogether, these data suggest there are several overlapping signaling cascades that regulate the expression of hypha-specific genes in C. albicans.
Cdc42p is a Rho-type GTPase responsible for establishing and maintaining polarized growth in many, if not all, eukaryotic cell types (21). Rho-type GTPases act as molecular switches that are inactive when GDP bound and active when GTP bound, thereby enabling them to interact with downstream effector proteins. Three domains within Cdc42p, comprised of residues 5 to 20, 53 to 62, and 115 to 118, are primarily responsible for GTP binding and hydrolysis (53). Other defined regions of Cdc42p include the Rho insert domain (residues 122 to 135) and the effector domain (residues 26 to 50), both of which have been implicated in the interactions with various downstream effectors (31, 54, 55). Effector domain mutations, which have differential effects on interactions with subsets of effector proteins (40, 41), have been used to separate different Cdc42p-dependent signaling pathways in Saccharomyces cerevisiae, indicating that the myriad of Cdc42p functions can be mutationally separated.
Cdc42p has recently been shown to be necessary for the establishment and maintenance of polarized growth in C. albicans (52). Additionally, regulation of the nucleotide-bound state of Cdc42p is imperative for proper budded and hyphal cell formation. Consistent with the differences in growth patterns between budded and hyphal cells, green fluorescent protein (GFP)-tagged Cdc42p has differential localization to sites of polarized growth in these cell types (16). Hyphal cells continuously localize GFP-Cdc42p to the growing apical tip, whereas budded cells localize GFP-Cdc42p to growing bud tips, where the GFP-Cdc42p signal diminishes as the bud enlarges, until GFP-Cdc42p reappears at the mother-bud neck before cytokinesis. GFP-Cdc42p localization correlates temporally and spatially to actin polarization at these sites.
A recent study in which Cdc42p was expressed under the control of the MET3 repressible promoter suggested that Cdc42p, probably because of the reduced levels of expressed protein, was important for C. albicans virulence (2). None of the experiments to date, however, has specifically examined the function of Cdc42p in the budded-to-hyphal-form transition without altering the mitotic functions of the protein, which are necessary for cell viability and growth. Therefore, we took advantage of S. cerevisiae cdc42 mutants that had defects in pseudohyphal formation while retaining the normal mitotic functions of Cdc42p in budded cells (33). The cdc42-S26I, cdc42-E100G, and cdc42-S158T mutations lay outside of the Cdc42p domains involved in interactions with effectors or in GTP binding and hydrolysis (see Fig. 1A). These mutations in C. albicans resulted in serious defects in hyphal formation on a variety of hypha-inducing media but did not have noticeable defects in nuclear division, chitin deposition, septin structures, or actin polarity under budded growth conditions, suggesting that the normal mitotic roles of Cdc42p had been preserved. The cdc42-S26I and cdc42-E100G mutants had a reduced ability to damage human endothelial cells (ECs), supporting the notion that Cdc42p-dependent regulation of the budded-to-hyphal-form transition was important in the infection process. Additionally, several Efg1p-dependent, hypha-specific transcripts had lower expression levels in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants, suggesting that the Cdc42p signaling cascade unexpectedly affected Efg1p transcription factor activity. However, the levels of a Cph1p-dependent transcript remained unchanged, suggesting that the cdc42 mutants did not affect the recognized Cdc42p-dependent signaling pathway leading to activation of Cph1p.
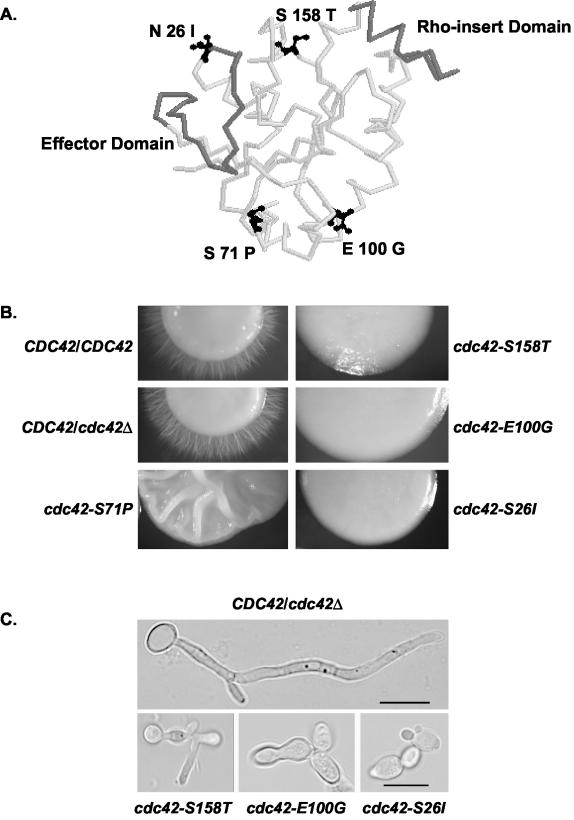
FIG. 1.
(A) Location of C. albicans hypha-defective Cdc42p point mutations modeled onto the crystal structure of human Cdc42p. Side chains of mutated amino acids are depicted in black; the Rho insert domain and effector domain are highlighted in grey. The image was generated from PDB file 1AN0 of the Cdc42Hs-GDP complex using RasMol version 2.6. (B) Cdc42p mutants were defective in hypha formation on solid Spider medium. Strains AV05 (cdc42-S158T), AV03 (cdc42-E100G), AV02 (cdc42-S71P), AV06 (cdc42-S26I), CaDH50, and Sc5314 were grown for 3 to 4 days at 30°C. (C) Cdc42p mutants had defects in hypha formation in 10% FBS. Strains (see Fig. 2B) were grown in minimal medium plus 10% FBS for 4 h at 37°C. Bar, 10 μm. The scale bar applies to all three images in the bottom panel.
MATERIALS AND METHODS
Media and growth conditions.
The C. albicans strains used are listed in Table 1. All cells used for microscopic observations, protein preparation, or mRNA analysis were taken from saturated overnight cultures inoculated into fresh medium, usually minimal medium (0.8% yeast nitrogen base plus 2% dextrose) or YEPD (1% yeast extract, 2% peptone, and 2% dextrose). Routine hypha induction utilized 10% fetal bovine serum (FBS; Gibco-BRL, Grand Isle, N.Y.) in minimal medium unless otherwise indicated. Other hypha-inducing media, including medium 199 buffered with sodium bicarbonate (Sigma Chemical, St. Louis, Mo.), Lee's (27), Spider (28), and N-acetylglucosamine in salt base (47) were prepared as described elsewhere. For GAP1 induction, peptone and K2HPO4-based medium was used with the addition of 0.5% glucose in GPK medium and N-acetyl-d-glucosamine in NPK medium (3). To test strains for growth under embedded conditions, ∼50 cells were added to YPS (1% yeast extract, 2% peptone, 2% sucrose) plus 1% agar and grown at 25°C for ∼5 days, as previously described (6).
TABLE 1.
C. albicans strains used
| Strain | Relevant genotype | Reference |
|---|---|---|
| Sc5314 | URA3/URA3 | 15 |
| CAI-12 | ura3Δ::imm434/URA3 | 51 |
| CaDH50 | ura3/ura3 CDC42/cdc42Δ::hisG-URA3-hisG | 52 |
| CaDH85 | ura3/ura3 CDC42/cdc42Δ::hisG | 52 |
| AV06 | ura3/ura3 cdc42Δ::hisG/cdc42-S26I::URA3 | This study |
| AV02 | ura3/ura3 cdc42Δ::hisG/cdc42-S71P::URA3 | This study |
| AV03 | ura3/ura3 cdc42Δ::hisG/cdc42-E100G::URA3 | This study |
| AV05 | ura3/ura3 cdc42Δ::hisG/cdc42-S158T::URA3 | This study |
| AV08 | ura3/ura3 cdc42Δ::hisG/cdc42-S26I ARG4/ ARG4::URA3 | This study |
| AV09 | ura3/ura3 cdc42Δ::hisG/cdc42-E100G ARG4/ ARG4::URA3 | This study |
| AV15 | ura3/ura3 CDC42/cdc42Δ::hisG ARG4/ ARG4::URA3 | This study |
Generation of mutant strains.
A 3.77-kb fragment containing the CaCDC42 gene and ∼1.5-kb 5′ and ∼1.7-kb 3′ sequences were amplified from the BWP17 genome and cloned into the pGEM-T Easy vector (Promega, Madison, Wis.). A PvuII fragment of the URA3 gene (from pGEM-URA3) was inserted 691 nucleotides downstream from the CDC42 stop codon using a SmaI site engineered with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The S26I, S71P, E100G, and S158T mutations were introduced into CDC42 by site-directed mutagenesis using primers 605 and 606, 601 and 602, 607 and 608, and 638 and 639, respectively (Table 2). A linear fragment liberated by cutting the CDC42::URA3 vector with AflII and SnaBI was transformed into the C. albicans CDC42/cdc42Δ CaDH85 strain (generously donated by M. Whiteway) (Table 1), using the spheroplasting method (22). Resulting transformants were screened by PCR using the following strategy. Primary screening utilized primers 616 and 641 (Table 2) to screen for strains containing URA3 at the CDC42 locus. Secondary screening utilizing primers 499 and 646 generated a product if the Salmonella hisG region present in the cdc42Δ allele was at the CDC42 locus. Isolates that contained both the cdc42Δ allele and mutant cdc42 at the correct locus then had the region just outside of the CDC42 open reading frame amplified using primers 465 and 466; this fragment was sequenced to screen for isolates solely containing a mutant copy of cdc42.
TABLE 2.
Primers used
| Primer | Sequencea | Sequence targetedb |
|---|---|---|
| 465 | CGTCCTAAATCTATTTTTATTG | 5′ CDC42 |
| 466 | GCCCAACACATCAAACGAGATC | 3′ CDC42 |
| 641 | CGAACTTGGTAGTGGGACCGAATGG | 3′ CDC42 |
| 616 | GCAACAACCCCATACACACTGG | URA3 |
| 646 | CCTCTTTATCCTGTCTGAACCGG | HisG |
| 499 | GCATGGATATTCCATGTCAAGG | 5′ CDC42 |
| 601 | GATTACGACAGATTACGGCCGTTGCCATATCCATCGACTG | cdc42-S71P |
| 602 | CAGTCGATGGATATGGCAACGGCCGTAATCTGTCGTAATC | cdc42-S71P |
| 605 | CGTATACCACTATTAAATTTCCAGC | cdc42-S26I |
| 606 | GCTGGAAATTTAATAGTGGTATACG | cdc42-S26I |
| 607 | GAAAAATGGTTCCCCGGGGTTCATCACCATTGTCC | cdc42-E100G |
| 608 | GGACAATGGTGATGAACCCCGGGGAACCATTTTTC | cdc42-E100G |
| 638 | GTATGTTGAGTGCACTGCATTGACTCAAAG | cdc42-S158T |
| 639 | CTTTGAGTCAATGCAGTGCACTCAACATAC | cdc42-S158T |
Underlined nucleotides are those changed from wild type to alter amino acids.
This represents either the region of the gene where the primer anneals or the introduced mutation.
For strains used in EC invasion assays, URA3 was disrupted at the cdc42 locus and reintroduced at the ARG4 locus in order to ensure an adequate level of URA3 expression. Plasmid pGEM-URA3 (56) had an EcoRI site added by site-directed mutagenesis 487 nucleotides downstream of the only other EcoRI site. The EcoRI fragment was removed; the vector was religated to generate a disrupted ura3Δ allele on the pGEM-ura3Δ plasmid. A PvuII fragment from the pGEM-ura3Δ plasmid was transformed into AV05, AV06, and CaDH50 strains, and transformants were selected on 5-fluoroorotic acid-containing plates. Resulting ura3Δ strains were transformed with NotI-linearized pRS-ARG-URA-BN plasmid (10) to generate strains AV08, AV09, and AV15, which have URA3 reintroduced at the ARG4 locus.
Immunofluorescence microscopy.
To observe chitin localization and DNA content, cells were fixed for 30 min in 4% formaldehyde, Calcofluor White (1 mg/ml) was added at a dilution of 1/1,000, and 4′,6-diamidino-2-phenyl-indole (DAPI; 1 mg/ml) was added at a 1/3,000 dilution. Septins were stained by the method of Sudbery (49) using an anti-Cdc11p antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) and an Alexa-488-conjugated secondary antibody (Molecular Probes, Eugene, Oreg.). Actin staining with rhodamine phalloidin (Molecular Probes) diluted 1/250 to 1/500 was performed using a protocol for stationary-phase yeast cells (38), modified with the addition of a 0.1% Triton X-100 soak for 15 min after zymolyase treatment. Phase-contrast optics and Omega filter cubes (Omega Optical, Brattleboro, Vt.) were used to visualize cells on a Nikon E400 fluorescence microscope, using filters XF106 for DAPI and Calcofluor White, XF100 for Alexa-488, and XF102 for rhodamine phalloidin. Digital images were obtained using a SPOT RT monochrome camera driven by SPOT version 3.5.8. Pictures were processed using Adobe Photoshop 7.0. Photomicrographs of wild-type and cdc42-E100G cells were taken at the same magnification and measured to determine the lengths of the long and short cellular axes.
Immunoblotting.
Cells were grown from saturated overnight cultures, inoculated into fresh YEPD with or without 10% FBS, and grown for 4 h. Protein preparation and immunoblot assays were done as previously described, with affinity-purified polyclonal anti-Cdc42p antibody used to detect Cdc42p (58). The blot was also probed with goat anti-yeast actin antibody (1:3,000 dilution) as a loading control with horseradish peroxidase-conjugated rabbit anti-goat secondary antibody (1:2,000 dilution). Densitometric analysis using NIH Image version 1.63 normalized Cdc42p levels to actin levels in each sample.
Endothelial cell invasion assay.
The assay was performed as described previously (37), using human umbilical vein ECs incubated for 3 h in the presence of 5 × 105 of C. albicans cells/ml. Endothelial cells were obtained following appropriate guidelines approved by the Institutional Review Board. Results shown were normalized for spontaneous 51Cr release and are the average of two independent experiments performed in triplicate.
Northern analysis.
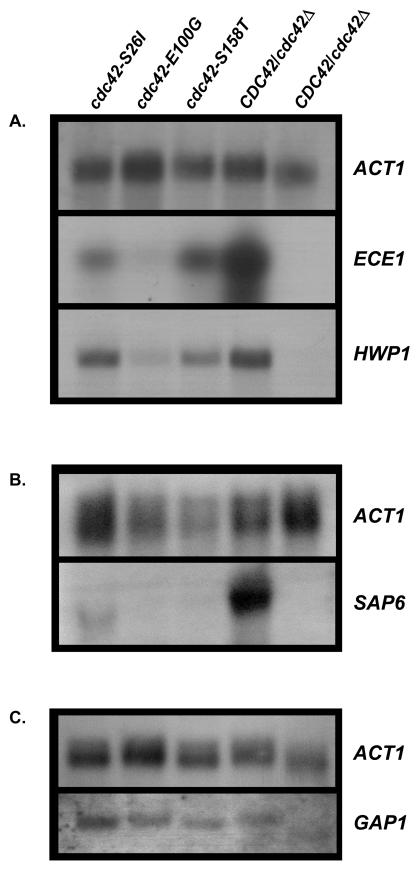
RNA extractions were performed as previously described (8) using a hot acid phenol method. The DIG High Prime labeling kit (Roche, Indianapolis, Ind.) using digoxigenin (DIG)-labeled PCR probes was used to detect mRNA, following the manufacturer's instructions. In order to control for variance in RNA loading, densitometry was performed as above. The ratios of ECE1, HWP1, SAP6, and GAP1 densitometric values to the actin value were used to compare the fold change between samples. Densitometric ratios presented are representative of results from two experiments.
RESULTS
Mutant Cdc42-S26Ip, Cdc42-S71Pp, Cdc42-E100Gp, and Cdc42-S158Tp could function as the sole copy of Cdc42p in the cell.
A heterozygous CDC42/cdc42Δ disruption strain, CaDH85 (Table 1), was transformed with the cdc42-S26I, cdc42-E100G, cdc42-S71P, and cdc42-S158T mutant alleles and marked by URA3, and transformants that contained both mutant cdc42 and the cdc42Δ allele were isolated (see Materials and Methods). These four cdc42 mutations (Fig. 1A) had the most striking defects in pseudohypha formation in S. cerevisiae. The C. albicans cdc42-S26I, cdc42-E100G, cdc42-S71P, and cdc42-S158T mutants were viable at 30°C under budded growth conditions, suggesting that these mutant alleles did not disrupt the normal mitotic functions of Cdc42p (see below). The CDC42/cdc42Δ starting strain was also transformed with a wild-type CDC42 allele; the resulting CDC42::URA3/cdc42Δ strain was indistinguishable from the starting CDC42/cdc42Δ strain CaDH85 in morphology and growth characteristics (data not shown), indicating that integration of a URA3-marked allele at the CDC42 endogenous locus does not adversely affect these properties.
The cdc42 mutants displayed defects in the budded-to-hyphal-form transition on various hypha-inducing media.
Although the cdc42-S26I, cdc42-E100G, cdc42-S71P, and cdc42-S158T mutants were viable on budded growth media, all four mutants had smooth colony borders when grown on hypha-inducing Spider medium (Fig. 1B), indicating that they were impaired in the budded-to-hyphal-form transition and/or hyphal growth. The cdc42-S71P mutant had an unusual crinkly colony morphology, reminiscent of other C. albicans mutants that had a mixture of yeast and pseudohyphal cells (1, 47). On Lee's medium, the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants likewise had smooth colony borders (data not shown). However, growth on minimal media plus 10% FBS stimulated hypha formation in the cdc42-S71P mutant but not in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants (data not shown). These results suggested that the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants had defects in the budded-to-hyphal-form transition in response to several hypha-inducing signals and may have a general hypha formation defect, whereas the cdc42-S71P mutant only had a defect on Spider plates, a weak inducer of hypha formation (this mutant was not examined further).
The cdc42 mutants displayed budded and pseudohyphal morphologies on hypha-inducing media.
The cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants caused dramatic defects in hypha formation in liquid minimal medium supplemented with 10% FBS, a strong inducer of hyphal formation (Fig. 1C). The CDC42/cdc42Δ parental CaDH50 strain had 86% hyphal cells after 4 h of incubation, with only 12% budded cells (Table 3). The cdc42-S158T mutant had 17% hyphal cells, cdc42-E100G had 2% hyphal cells, and cdc42-S26I had 3% hyphal cells, illustrating a drastic reduction of hypha formation in 10% FBS for each of these mutants. A small proportion of cells (<8% in cdc42-E100G, <4% in cdc42-S158T, and <2% in cdc42-S26I) had an abnormal cell shape with predominantly peanut-, pear-, or L-shaped cells, which may contribute to the slightly slower growth rates observed for these strains (see below). Similar results were observed after 2 h of growth in 10% FBS, and defects in hyphal cell formation were observed out to 8 h of growth (data not shown), indicating that the cdc42 mutations were not merely causing a delay but also a true defect in the budded-to-hyphal-form transition. These data do not rule out the possibility that these mutants may also have an effect on subsequent hyphal elongation processes.
TABLE 3.
Hyphal, pseudohyphal, and budded cells in cdc42 mutants
| Strain | CDC42 allele | Growth condition | % of totala
|
||
|---|---|---|---|---|---|
| Hyphae | Pseudohyphae | Budded | |||
| Sc5314b | CDC42 | 10% FBS | 99 | 1 | 0 |
| CaDH50b | CDC42 | 10% FBS | 86 | 2 | 12 |
| AV05 | S158T | 10% FBS | 17 | 51 | 32 |
| AV09 | E100G | 10% FBS | 2 | 13 | 85 |
| AV06 | S26I | 10% FBS | 3 | 38 | 59 |
| Sc5314 | CDC42 | Spider | 60 | 16 | 24 |
| CaDH50 | CDC42 | Spider | 46 | 23 | 31 |
| AV05 | S158T | Spider | 0 | 17 | 83 |
| AV09 | E100G | Spider | 0 | 8 | 92 |
| AV06 | S26I | Spider | 0 | 5 | 95 |
Cells were counted for the proportion of buds versus hyphae using DAPI to stain DNA and Calcofluor White to stain the cell wall, in order to ensure that counts reflected the true number of individual cells and that pseudohyphal constrictions would be accentuated by the Calcofluor stain. After 4 h of growth, categories of cells counted were as follows; hyphae were long, filamentous cells with parallel sides and no constrictions visible between cells; pseudohyphae were at least twice the length of a budded cell, with constrictions visible at the cell junctions, or were long filamentous cells with nonparallel sides. In all categories, only nucleated cells (n = 200) were counted.
Sc5314 is a clinical isolate, while CaDH50 is the parental laboratory strain. Both are CDC42/CDC42.
Hypha formation was similarly impeded in liquid Spider medium (Table 3). CaDH50 cells formed 46% hyphal cells, but none of the mutants formed hyphal cells, with the majority of cells growing as budded cells. Similar reductions in hypha formation were also observed in the presence of other hypha-inducing signals, including medium 199 and N-acetyl-d-glucosamine in salt base (data not shown). These results suggested that Cdc42p-dependent signaling was responsive to a variety of hypha-inducing signals in solid and liquid media.
The cdc42 mutants maintained normal mitotic functions.
S. cerevisiae Cdc42p functions in many cellular processes, including initiation and maintenance of polarized growth, switching from apical to isotropic growth in G2/M phase, cytokinesis, and septation (21). It was therefore necessary to ensure that these mitotic cellular processes were not perturbed in C. albicans mutants grown under budding conditions. Additionally, observations were made at both 30 and 37°C to ensure that these cdc42 mutations were not conferring a temperature-sensitive phenotype under any of the conditions tested. Strains were grown in minimal medium instead of YEPD to reduce the amount of pseudohyphal cells that would be present in cultures after 4 h of growth.
DAPI staining for DNA content and Calcofluor White staining of the chitin cell wall were performed on fixed cells. The cdc42 mutants had normal DNA content, with normally partitioned nuclei visible (Fig. 2A), indicating that cellular division was occurring normally in each of the mutants. Cell wall staining revealed normal chitin deposition (Fig. 2A) and a normal pattern of bud site selection (17). The CDC42/cdc42Δ strain had 55% of cells with a bipolar budding pattern, 22% with an axial pattern, and 3% with a random pattern. In comparison, the cdc42-E100G mutant had 59% bipolar, 33% axial, and 9% random buds. Overall, the percentage of axial to bipolar budding varied no more than 18%. The percentage of random buds in each of the mutants was no higher than the 9% observed in the cdc42-E100G mutant. Together, these results indicated that nuclear division, cell wall deposition, and bud site selection occurred normally during budding growth in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants.
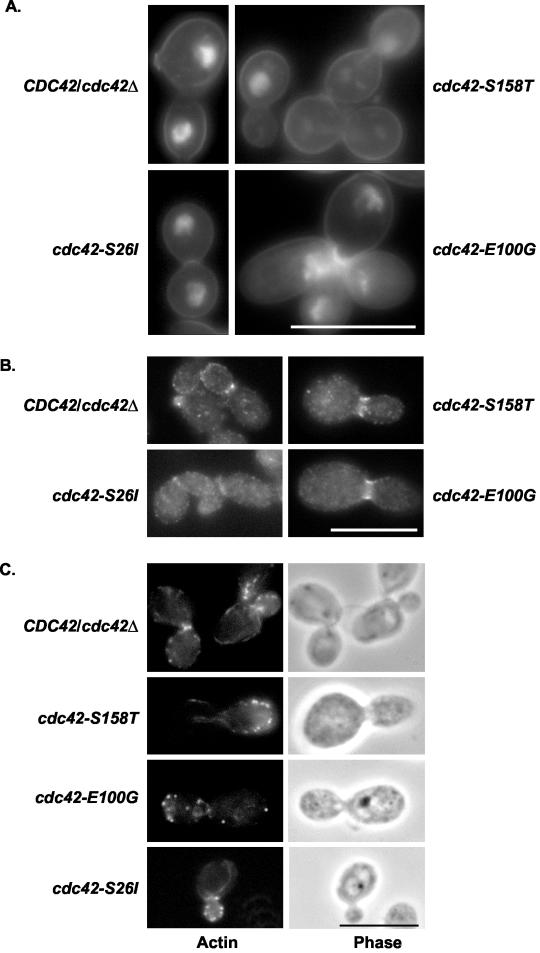
FIG. 2.
Cdc42p mutants did not have defects in cell wall components, cell division, or septin localization under budded growth conditions. Strains AV05 (cdc42-S158T), AV03 (cdc42-E100G), AV06 (cdc42-S26I), and CaDH50 were grown in minimal medium at 37°C for 4 h and then stained with DAPI and Calcofluor White (A), by immunofluorescence with anti-Cdc11p antibodies (B), or rhodamine-phalloidin (C). Bar, 10 μm. The scale bar applies to all four figures in panels A, B, and C.
As septin proteins are important for the structural integrity of the mother-bud junction and have been proposed to interact with Cdc42p (30), localization of one of the septin proteins was carried out by immunofluorescent antibody staining. Immunofluorescent antibody staining using antibodies specific for the Cdc11p septin protein demonstrated normal septin localization in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants, with discrete spots of fluorescence visible at incipient bud sites and single and double septin bands visible at mother-bud necks (Fig. 2B). These data suggested that the septin structures were assembled properly during budding growth.
In addition, the cdc42-S26I and cdc42-S158T mutants had no observable differences in the amount of polarized actin patches or cables at either 30 or 37°C, as assayed by rhodamine phalloidin staining (Fig. 2C). The cdc42-E100G mutant, however, reproducibly had decreased numbers of cells with polarized cortical actin patches and cables and had decreased levels of polarized actin patches with very few actin cables visible at 37°C (Table 4). These results suggested that the cdc42-E100G mutant had a mild defect in actin polarity at 37°C.
TABLE 4.
Actin localization in cdc42 mutant cells
| Strain | CDC42 allele | Growth conditionsa | Actin localization (% of cells)b
|
||
|---|---|---|---|---|---|
| Random | Polar patches | Polar and cables | |||
| AV03 | E100G | 30°C | 67 | 33 | <1 |
| CaDH50 | CDC42 | 30°C | 25 | 65 | 10 |
| AV03 | E100G | 37°C | 82 | 17 | <1 |
| CaDH50 | CDC42 | 37°C | 7 | 84 | 9 |
| AV03 | E100G | 37°C + serum | 82 | 17 | 1 |
| CaDH50 | CDC42 | 37°C + serum | 0 | 55 | 45 |
| AV06 | S26I | 37°C + serum | 31 | 69 | <1 |
| AV05 | S158T | 37°C + serum | 47 | 49 | 4 |
A total of 300 rhodamine-phalloidin-stained cells were counted after 4 h of growth at the temperature indicated in minimal medium (+/− 10% FBS).
Random, no polarized actin patches; polar patches, polarized actin patches with no actin cables visible; polar and cables, polarized actin patches and actin cables visible. For details, see text.
Mutants with defects in vacuolar morphogenesis also have demonstrated defects in hypha formation (7, 36). Therefore, mutant cells were stained with FM4-64, a lipophilic dye that labels membranes and can be used as a pulse-chase reagent to observe the internalization of endocytic vesicles. Observations of each of the mutants were made at 30 min and 1 h, and no appreciable differences in FM4-64 staining were detectable between Sc5314, CaDH50, and any of the mutants (data not shown). This result suggested that the mutants did not have defects in endocytosis or vacuolar morphogenesis.
Strains containing mutant Cdc42p were also characterized under hypha-inducing conditions for structural and polarity defects. Several possible mechanisms could be responsible for the defect in the budded-to-hyphal-form transition seen in the mutants, including defects in polarized growth, cellular division, or cytokinesis. The cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants were grown in minimal medium plus 10% FBS, fixed, and stained with DAPI and Calcofluor (Fig. 3A). Each mutant had bud scar patterns similar to those of budded cells and normal chitin deposition. Occasionally, a cell was observed that had a thick band of chitin in the middle of a pseudohyphal cell, but this was observed in less than 1% of cells. DAPI staining showed normal DNA content, with one nucleus per Calcofluor-stained cell body. These results indicated that cellular division and chitin deposition occurred normally during growth in the presence of hypha-inducing signals. Normal septin staining was observed in all mutants (Fig. 3B), with less than 1% of cells having a wide band of septin staining, suggesting that improper septin organization was not the cause of the defect in hypha formation in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants.
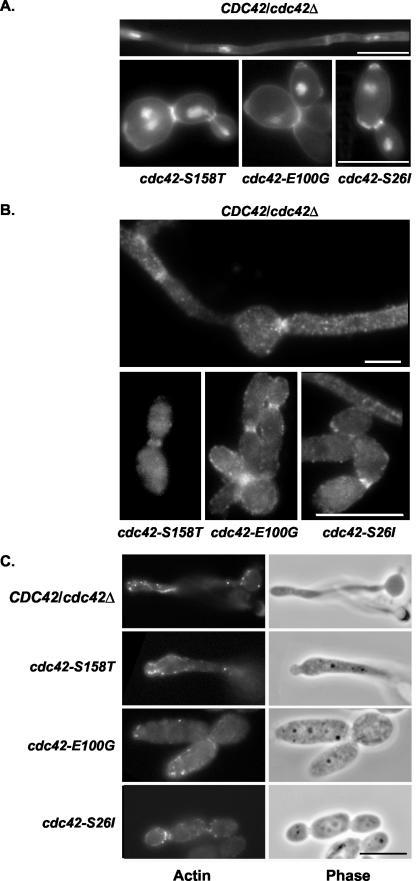
FIG. 3.
Cdc42p mutants grown in the presence of 10% FBS did not have major defects in cell wall composition, cell division, septin localization, or actin polarity. Strains AV05 (cdc42-S158T), AV03 (cdc42-E100G), AV06 (cdc42-S26I), and CaDH50 were grown in minimal medium plus 10% FBS for 4 h at 37°C and stained as described in the legend for Fig. 2. Bar, 10 μm. The scale bar applies to all three images in the bottom panel.
The cdc42-S26I and cdc42-S158T mutants grown in minimal medium plus 10% FBS appeared to have properly polarized actin patches and cables at bud and pseudohypha tips (Fig. 3C). However, quantification of the percentage of cells showing no polarized actin patches in serum-induced cells grown at 37°C was 7% in CDC42/cdc42Δ cells, 31% in cdc42-S26I cells, and 47% in cdc42-S158T cells (Table 4), suggesting that the cdc42-S26I and cdc42-S158T mutants had a mild defect in polarizing actin when grown in the presence of serum. The cdc42-E100G mutant had 67% of cells with no polarized actin patches at 30°C (Table 4), and this phenotype was exacerbated at 37°C, with 82% of cdc42-E100G mutant cells having no polarized actin patches either with or without the addition of 10% FBS. These results suggested that the cdc42-E100G mutant had a mild defect in polarizing actin at 30°C which was exacerbated at 37°C, and this defect was independent of hypha-inducing signals. It is interesting that the cdc42-E100G mutant had virtually no actin cables visible under any of the conditions tested.
These actin observations in the cdc42-E100G mutant correlated with a 30% increase in doubling time at 37°C (data not shown). To examine this increase in more detail, the budding indices of cdc42-E100G and CaDH50 cells grown to mid-log phase were determined in both minimal medium and in YEPD. The percentage of unbudded to budded cells varied by only 1% between the CaDH50 parental strain and cdc42-E100G in either growth medium. However, repeated observations indicated that cdc42-E100G cells were enlarged compared to CaDH50 cells, with the average short and long axes of cdc42-E100G cells being 28 and 21% longer, respectively, than CaDH50 cells. This observation could potentially correlate to an actin cable defect seen in the cdc42-E100G mutants.
Defects in hyphal formation were not due to decreased levels of Cdc42p.
Immunoblot analysis was performed on total protein extracts of cells grown under hypha-inducing conditions (YEPD plus 10% FBS at 37°C) to address the question of whether mutant Cdc42p was being expressed at levels similar to wild type. Densitometric analysis that normalized Cdc42p levels to actin protein levels indicated that Cdc42-S26Ip levels were 129% of wild-type levels, Cdc42-E100Gp levels were 234% of wild-type levels, and Cdc42-S158Tp levels were 99% of wild-type levels. This analysis demonstrated that there was no significant reduction in the levels of mutant Cdc42p compared to wild-type Cdc42p. The reason for elevated levels of Cdc42-E100Gp is unknown.
The cdc42-S26I and cdc42-E100G mutants caused a decreased level of EC damage.
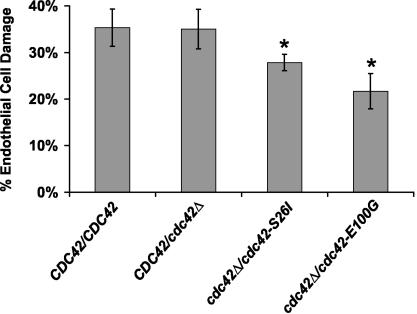
The cdc42 mutants showed a slight increase (20 to 30%) in their doubling times compared to CaDH50, which precluded the use of these mutants in a meaningful analysis of virulence in a mouse model of disseminated candidiasis. Therefore, an EC invasion assay was performed. The EC invasion assay is an excellent predictor of virulence, as strains with a reduction in EC invasion also have reduced virulence in a mouse model of disseminated candidiasis (19, 20). The assay measures the combined effects of induced phagocytosis, hyphal penetration and damage, and the release of cellular damaging agents such as phospholipases and secreted aspartyl proteases. The cdc42-S26I and cdc42-E100G mutants, which had URA3 integrated at the ARG4 locus to eliminate the positional effects sometimes seen with the URA3 marker (50), were tested for their ability to cause EC damage in an in vitro model system using primary cultured human EC. Briefly, cdc42-S26I, cdc42-E100G, and wild-type C. albicans cells were incubated with 51Cr-labeled ECs, where after 3 h the amount of 51Cr released into the supernatant was measured, reflecting the amount of EC damage that had occurred. The cdc42-E100G and cdc42-S26I mutants showed a statistically significant decrease in EC damage, dropping from 35% in the AV15 positive control to 22 and 28%, respectively, in the mutants (Fig. 4). These results demonstrated that the cdc42-S26I and cdc42-E100G mutants caused reduced damage in an EC model for invasion.
FIG. 4.
cdc42-S26I and cdc42-E100G mutants had a reduced ability to cause EC damage. Strains CAI-12, AV15 (CDC42/cdc42), AV08 (cdc42-S26I), and AV09 (cdc42-E100G) were incubated for 3 h with human ECs. The percentage of EC damage was measured with a chromium release assay. Results are the averages ± standard deviations of triplicate measurements. Asterisks indicate a P value of <0.005 compared to that for cells infected with CAI-12, as determined by the Wilcoxon rank sum test.
The cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants had decreased expression of hypha-induced, Efg1p-regulated mRNA transcripts.
C. albicans Cdc42p has been proposed to signal through the putative effector protein Cst20p to the Cst11p-Hst7p-Cek1p mitogen-activated protein kinase cascade, culminating in the activation of the Cph1p transcription factor (9). The GAP1 gene is solely dependent upon Cph1p for its induction (3), while expression of other virulence determinants, such as SAP6 (14, 23), the putative cell wall protein ECE1 (4, 46), and HWP1 (4, 46, 48), depends upon Efg1p. Northern analysis was performed on total RNA prepared from the parental CDC42/cdc42Δ strain and each of the mutants grown in 10% FBS, with a budded control grown in YEPD. The budded control had no detectable expression of the HWP1, ECE1, or SAP6 hypha-specific genes, whereas the CDC42/cdc42Δ serum-induced cells showed strong expression of all three genes (Fig. 5). Densitometric analysis confirmed that there was a reduction in ECE1 expression of approximately twofold for the cdc42-S158T mutant and threefold for the cdc42-S26I mutant, with ECE1 expression being all but eliminated in the cdc42-E100G mutant compared to wild type (Fig. 5A). Expression of HWP1 was reduced sixfold in the cdc42-E100G mutant (Fig. 5A). SAP6 expression was dramatically reduced in each of the three mutants (Fig. 5B), with the cdc42-S26I mutant having a greater-than-twofold reduction in SAP6 expression, and expression was all but eliminated in the cdc42-S158T and cdc42-E100G mutants. These data supported the conclusion that HWP1, ECE1, and SAP6 mRNA levels were decreased in the cdc42 mutants. This result was surprising in light of the fact that Cdc42p was not known to signal through the cyclic AMP/protein kinase A pathway leading to the activation of the Efg1p transcription factor. It should be noted that the activity of an Efg1p-dependent promoter fused to a heterologous reporter gene was not assayed in these mutants.
FIG. 5.
(A) cdc42 mutants have reduced expression of Efg1p-dependent hypha-specific transcripts. Total RNA was prepared from the following strains grown for 2 h in minimal medium with 10% FBS and probed with ACT1, HWP1, and ECE1. Lane 1, AV06 (cdc42-S26I); lane 2, AV03 (cdc42-E100G); lane 3, AV05 (cdc42-S158T); lane 4, CaDH50 (CDC42/cdc42Δ); lane 5, control CaDH50 grown in YEPD (budded growth). (B) Total RNA was prepared and probed for ACT1 and SAP6; lanes were loaded as for panel A. (C) cdc42 mutants did not have reduced expression of Cph1p-dependent hypha-specific transcripts. Total RNA was isolated from N-acetyl-d-glucosamine-induced strains. Lane 1, AV06 (cdc42-S26I); lane 2, AV03 (cdc42-E100G); lane 3, AV05 (cdc42-S158T); lane 4, CaDH50 (CDC42/cdc42Δ); lane 5, control CaDH50 grown in noninducing GPK medium.
GAP1 expression was unaltered relative to the actin loading control in the cdc42 mutants (Fig. 5C). This result suggested that the signaling pathway leading to Cph1p activation was intact in these cdc42 mutants. To further address this point, the ability of cdc42 mutants to undergo filamentation was examined under embedded conditions in which Cph1p, but not Efg1p, is required (6, 42). The CDC42/cdc42Δ starting strain and all four cdc42 mutants could produce filaments under embedded conditions (data not shown), indicating that the Cph1p signaling pathway was functional in the cdc42 mutants. Under the growth conditions used to assay GAP1 expression, there was no expression of Efg1p-dependent genes (i.e., similar to budded conditions for Efg1p-dependent transcription).
DISCUSSION
Recent studies have implicated the C. albicans Cdc42p GTPase in the morphological transition from budded to hyphal growth. In this study, the Cdc42-S26Ip, Cdc42-E100Gp, and Cdc42-S158Tp mutant proteins preserved their normal mitotic functions while exhibiting a specific defect in forming hyphae, even in a potent inducer of hyphal cells, 10% FBS. The defect in the budded-to-hyphal-form transition was apparent in numerous different hypha-inducing media, suggesting that Cdc42p is a convergence point for the regulation of a variety of upstream signals that induce hyphal growth. Not only were cells containing these mutant Cdc42p morphologically normal, but also septin proteins and actin polarity were normal in budded cells grown at 30°C. Since Cdc42p is responsible for modulating rearrangements of the actin cytoskeleton in numerous cell types, it is not surprising that Cdc42p was required to specifically regulate the proper formation of hyphal cells.
A recent study indicated that when CDC42 was expressed under the control of the MET promoter (repressed by methionine and cysteine), cells could not initiate hyphal agar invasion on certain types of media (2). This result suggested that the MET promoter allowed adequate expression of the Cdc42p to allow growth but not agar invasion on the type of hypha-inducing medium used. Similarly, in a mouse model of systemic candidiasis, cells containing Cdc42p expressed from the MET promoter were avirulent under repressing conditions. A caveat in interpreting these results is that the growth rate of these strains and the levels of Cdc42p were not reported, so it is unknown what contribution the level of Cdc42p expression had on the avirulence of the strains. Altering the levels of Cdc42p has been shown to ameliorate or eliminate a growth defect or even lethality in S. cerevisiae (32). Therefore, it was with the aim of maintaining the endogenous regulation of Cdc42p while specifically affecting the role of Cdc42p in the budded-to-hyphal-form transition that these specific mutant cdc42 strains were generated.
The reduction in the number of actin cables observed in the cdc42-E100G mutant was not a lethal defect, since strains do grow at 37°C in various liquid media and form colonies when plated on 10% FBS-containing solid medium (data not shown). In S. cerevisiae, Cdc42-E100Gp has lost its ability to interact with the known Cdc42p effector Bni1p in a two-hybrid assay (33). Bni1p is an actin scaffold protein recently demonstrated to be necessary for the proper orientation of actin cables from the mother into the growing tip of daughter cells, where the actin cables serve as a track for the delivery of secretory vesicles (13, 35, 44). It is possible that the actin cable defect seen with the cdc42-E100G mutant and the increase in cell size are reflective of the loss of Cdc42p-Bni1p interactions.
An EC invasion assay demonstrated a decrease in the amount of EC damage caused by the cdc42-S26I and cdc42-E100G mutants compared to wild-type strains. This result suggested that the Cdc42p-mediated role in promoting the budded-to-hyphal-form transition is important in a more physiologically relevant setting than what can be produced using laboratory media. A cla4Δ/cla4Δ strain had a similar decrease in the amount of EC damage as the cdc42-E100G mutant and, concomitantly, had reduced virulence in a mouse model of disseminated candidiasis (26, 37). These data support a model in which a hypha-specific effector function(s) of Cdc42p may have been disrupted in the mutants.
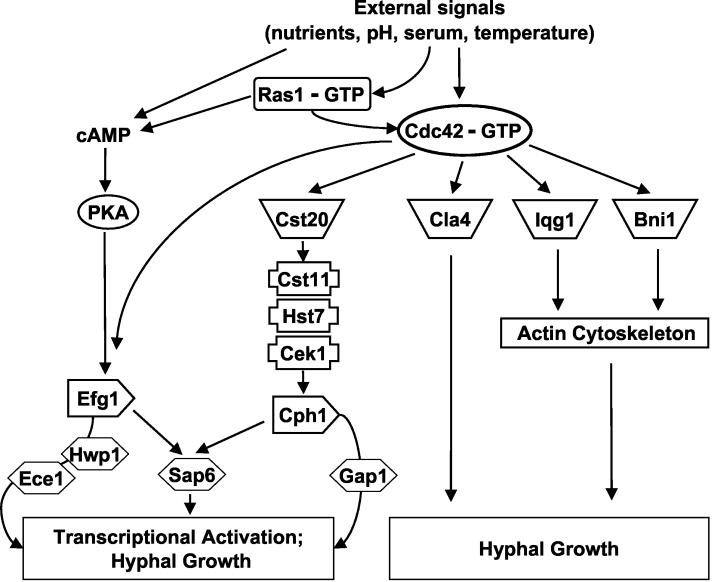
Recent gene array studies suggested that C. albicans cells that are grown under hypha-inducing conditions up-regulate a set of hypha-specific transcripts, including some demonstrated virulence factors. Northern blotting results showed a reduction in expression of ECE1, HWP1, and SAP6 in the cdc42 mutants, indicating that Cdc42p has an important role in regulating the expression of hypha-specific genes. This result also suggested that Cdc42p had an additional signaling function that impinges upon the Efg1p transcription factor (Fig. 6), since ECE1, HWP1 and SAP6 are under the control of Efg1p. Previous studies have suggested that Cdc42p-depleted strains (where CDC42 is under control of a repressible promoter) are still able to express HWP1 and ECE1 when grown in the presence of 10% FBS (52). These studies did not utilize mutations that separated the mitotic functions of Cdc42p away from the protein's role in the budded-to-hyphal-form transition and may have had a reduction, but not a complete elimination, of Efg1p-dependent transcription under these conditions. C. albicans Cdc42p has been proposed to signal through Cst20p to the Cst11p-Hst7p-Cek1p mitogen-activated protein kinase cascade, leading to the activation of the Cph1p transcription factor (9, 52). It was recently shown that the expression of GAP1 is solely dependent upon Cph1p for its expression. However, the levels of GAP1 transcript were not altered in the cdc42 mutants, suggesting that the signaling pathway leading to Cph1p activation was still intact (Fig. 6). This is not entirely surprising, as Cst20p is predicted to interact with a portion of the Cdc42p effector domain that is not altered in the cdc42-S26I, cdc42-E100G, and cdc42-S158T mutants. Taken together, these data raise the interesting and unsuspected possibility that Cdc42-S26Ip, Cdc42-E100Gp, and Cdc42-S158Tp have lost an interaction with a specific downstream effector that impinges upon the Efg1p-dependent pathway but not the Cph1p-dependent pathway. In this sense, these mutations would be analogous to S. cerevisiae Cdc42p effector domain mutations that have differential effects on interactions with subsets of effector proteins (40, 41).
FIG. 6.
Potential Cdc42p-dependent signaling pathways regulating hyphal growth and hypha-specific gene expression. All known Cdc42p effector proteins identified through the C. albicans sequencing project are shown in the model. See text for details.
In conclusion, these studies have provided evidence that Cdc42p has a pivotal role in promoting the budded-to-hyphal-form transition, which can be mutationally separated from its role in mitotic growth, and that Cdc42p-dependent signaling pathways function in C. albicans virulence as well as induction of hypha-specific gene expression (Fig. 6). At this point, it is unknown whether the hyphal defects observed in the cdc42 mutants were due to altered transcription of hypha-specific genes, altered actin function, or both. Future studies will explore the regulation of Cdc42p-dependent signaling pathways in C. albicans virulence as well as differential interactions between Cdc42p and various C. albicans effector proteins in both budded and hyphal cells.
Acknowledgments
We thank Malcolm Whiteway, Aaron Mitchell, and Paula Sundstrom for valuable reagents and Judy Berman, Eric Bensen, Dana Davis, Peter Sudbery, and members of the Johnson lab for helpful discussions and critical comments on the manuscript.
A.L.V. was supported by the NCI Cancer Biology training grant T32-CAO9286-19 and a predoctoral fellowship from the Vermont Genetics Network through NIH grant number 1 P20 RR16462 from the BRIN Program of the National Center for Research Resources. This work was supported by a grant to D.I.J. from the Vermont Cancer Center and Lake Champlain Cancer Research Organization and Public Health Service grants RO1AI-19990 and PO1AI-37194 awarded to J.E.E.
REFERENCES
- 1.Bahn, Y. S., J. Staab, and P. Sundstrom. 2003. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50:391-409. [DOI] [PubMed] [Google Scholar]
- 2.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, S., M. Roy, and A. Datta. 2003. N-Acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, A. J. P., and N. A. R. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 7.Bruckmann, A., W. Kunkel, K. Augsten, R. Wetzker, and R. Eck. 2001. The deletion of CaVPS34 in the human pathogenic yeast Candida albicans causes defects in vesicle-mediated protein sorting and nuclear segregation. Yeast 18:343-353. [DOI] [PubMed] [Google Scholar]
- 8.Chapon, C., T. R. Cech, and A. J. Zaug. 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3:1337-1351. [PMC free article] [PubMed] [Google Scholar]
- 9.Csank, C., K. Schröppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, N. K., S. Sharma, and G. K. Khuller. 2003. Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit. Rev. Microbiol. 29:259-275. [DOI] [PubMed] [Google Scholar]
- 12.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 13.Evangelista, M., S. Zigmond, and C. Boone. 2003. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603-2611. [DOI] [PubMed] [Google Scholar]
- 14.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schafer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 16.Hazan, I., and H. Liu. 2002. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot. Cell 1:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, A. B., M. C. Lopez, L. Fernandez-Lago, and A. Dominguez. 1999. Candida albicans and Yarrowia lipolytica as alternative models for analysing budding patterns and germ tube formation in dimorphic fungi. Microbiology 145:2727-2737. [DOI] [PubMed] [Google Scholar]
- 18.Hube, B., D. Hess, C. A. Baker, M. Schaller, W. Schafer, and J. W. Dolan. 2001. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147:879-889. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim, A. S., S. G. Filler, D. Sanglard, J. E. Edwards, and B. Hube. 1998. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect. Immun. 66:3003-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim, A. S., F. Mirbod, S. G. Filler, Y. Banno, G. T. Cole, Y. Kitajima, J. E. Edwards, Y. Nozawa, and M. A. Ghannoum. 1995. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect. Immun. 63:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz, M. B., M. W. Cortelyou, and D. R. Kirsch. 1986. Integrative transformation of Candida albicans, using a cloned Candida ADE2 gene. Mol. Cell. Biol. 6:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 24.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. R. Gow, A. J. P. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 26.Leberer, E., K. Ziegelbauer, A. Schmidt, D. Harcus, D. Dignard, J. Ash, L. Johnson, and D. Y. Thomas. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7:539-546. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 28.Liu, H., J. Köhler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 29.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 30.Longtine, M. S., and E. F. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403-409. [DOI] [PubMed] [Google Scholar]
- 31.McCallum, S. J., W. J. Wu, and R. A. Cerione. 1996. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J. Biol. Chem. 271:21732-21737. [DOI] [PubMed] [Google Scholar]
- 32.Miller, P. J., and D. I. Johnson. 1997. Characterization of the S. cerevisiae cdc42-1ts allele and new temperature-conditional-lethal cdc42 alleles. Yeast 13:561-572. [DOI] [PubMed] [Google Scholar]
- 33.Mösch, H.-U., T. Köhler, and G. H. Braus. 2001. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol. Cell. Biol. 21:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murad, A. M. A., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. P. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 35.Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara, K. Irie, and Y. Takai. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer, G. E., A. Cashmore, and J. Sturtevant. 2003. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot. Cell 2:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan, Q. T., P. H. Belanger, and S. G. Filler. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pringle, J. R., R. A. Preston, A. E. M. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 39.Richard, M., S. Ibata-Ombetta, F. Dromer, F. Bordon-Pallier, T. Jouault, and C. Gaillardin. 2002. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 44:841-853. [DOI] [PubMed] [Google Scholar]
- 40.Richman, T. J., and D. I. Johnson. 2000. Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell. Biol. 20:8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 1999. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274:16861-16870. [DOI] [PubMed] [Google Scholar]
- 42.Riggle, P. J., K. A. Andrutis, X. Chen, S. R. Tzipori, and C. A. Kumamoto. 1999. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect. Immun. 67:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and G. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sagot, I., S. K. Klee, and D. Pellman. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42-50. [DOI] [PubMed] [Google Scholar]
- 45.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh, P., S. Ghosh, and A. Datta. 2001. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 69:7898-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 49.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 50.Sundstrom, P., J. E. Cutler, and J. F. Staab. 2002. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 70:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuchimori, N., L. L. Sharkey, W. A. Fonzi, S. W. French, J. E. Edwards, and S. G. Filler. 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ushinsky, S. C., D. Harcus, J. Ash, D. Dignard, A. Marcil, J. Morchhauser, D. Y. Thomas, M. Whiteway, and E. Leberer. 2002. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell 1:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vetter, I. R., and A. Wittinghofer. 2001. Signal transduction—the guanine nucleotide-binding switch in three dimensions. Science 294:1299-1304. [DOI] [PubMed] [Google Scholar]
- 54.Walker, S. J., and H. A. Brown. 2002. Specificity of Rho insert-mediated activation of phospholipase D1. J. Biol. Chem. 277:26260-26267. [DOI] [PubMed] [Google Scholar]
- 55.Walker, S. J., W.-J. Wu, R. A. Cerione, and H. A. Brown. 2000. Activation of phospholipase D1 by Cdc42 requires the Rho insert region. J. Biol. Chem. 275:15665-15668. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yaar, L., M. Mevarech, and Y. Koltin. 1997. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology 143:3033-3044. [DOI] [PubMed] [Google Scholar]
- 58.Ziman, M., and D. I. Johnson. 1994. Genetic evidence for a functional interaction between S. cerevisiae CDC24 and CDC42. Yeast 10:463-474. [DOI] [PubMed] [Google Scholar]