Abstract
Most species of the protozoan phylum Apicomplexa harbor an endosymbiotic organelle—the apicoplast—acquired when an ancestral parasite engulfed a eukaryotic plastid-containing alga. Several hundred proteins are encoded in the parasite nucleus and are posttranslationally targeted to the apicoplast by a distinctive bipartite signal. The N-terminal 20 to 30 amino acids of nucleus-encoded apicoplast targeted proteins function as a classical signal sequence, mediating entry into the secretory pathway. Cleavage of the signal sequence exposes a transit peptide of variable length (50 to 200 amino acids) that is required for directing proteins to the apicoplast. Although these peptides are enriched in basic amino acids, their structural and functional characteristics are not well understood, which hampers the identification of apicoplast proteins that may constitute novel chemotherapeutic targets. To identify functional domains for a model apicoplast transit peptide, we generated more than 80 deletions and mutations throughout the transit peptide of Toxoplasma gondii ferredoxin NADP+ reductase (TgFNR) and examined the ability of these altered transit peptides to mediate proper targeting and processing of a fluorescent protein reporter. These studies revealed the presence of numerous functional domains. Processing can take place at multiple sites in the protein sequence and may occur outside of the apicoplast lumen. The TgFNR transit peptide contains at least two independent and functionally redundant targeting signals, each of which contains a subdomain that is required for release from or proper sorting within the endoplasmic reticulum. Certain deletion constructs traffic to multiple locations, including the apicoplast periphery, the rhoptries, and the parasitophorous vacuole, suggesting a common thread for targeting to these specialized compartments.
Plastid organelles are found in a diverse array of eukaryotic organisms, including the chloroplasts of green algae and plants, the rhodoplasts associated with red algae, and a variety of plastids in other taxa. For example, apicomplexan parasites, including Plasmodium species (which cause ∼500 million cases of human malaria annually, resulting in 2 to 3 million deaths [36, 58]), Toxoplasma gondii (which can lead to severe manifestations in immunocompromised hosts or unborn fetuses [50]), and Eimeria species (which are responsible for coccidiosis in poultry and other farm animals [19]), contain a plastid organelle called the apicoplast. In retrospect, the apicoplast was first noted nearly 50 years ago in morphological studies of these parasites (34, 44) and was variously described as the Golgi adjunct, the spherical body, the organelle plurimembranaire, and the Hohlzylinder (45). It has recently become clear that this organelle is a plastid (13, 21, 26, 30) that is essential for parasite survival, raising the possibility that the apicoplast provides a parasite-specific target for chemotherapy (4, 14, 17).
All plastids are thought to be monophyletic in origin (31, 35), tracing their ancestry to a single endosymbiotic event involving the colonization of a eukaryote by a photosynthetic cyanobacterium. Subsequent divergence led to the establishment of red and green algal lineages. Secondary endosymbiotic colonizations of diverse lineages by eukaryotic red or green algae have given rise to plastids in the brown algae, euglenoids, cryptomonads, chlorarachniophytes, apicomplexans, etc. (51). All of these plastids share several common features. First, they retain a vestigial genome derived from the cyanobacterial ancestor. The apicoplast genome of T. gondii and Plasmodium falciparum is ∼35 kb long and encodes fewer than 30 proteins (21, 28, 56, 57; http://www.sas.upenn.edu/∼jkissing/toxomap.html). Second, these organelles are surrounded by multiple membranes, typically two for primary plastids, but three or four for secondary endosymbionts, betraying the complex origins of the latter (13, 21, 41). Third, the vast majority of proteins responsible for plastid metabolic functions have been transferred to the nuclear genome during the course of eukaryotic evolution; these proteins are translated on cytoplasmic ribosomes and must find their way back into the plastid, using targeting information acquired during transfer to the nucleus (10, 21, 27, 41, 43).
In plants and green algae and in glaucocystophyte algae, the cytoplasmic synthesis of proteins destined for the plastid includes an N-terminal domain responsible for targeting mature proteins to the plastid import machinery and transport across both chloroplast or cyanelle membranes, respectively (reviewed in references 9 and 46). Targeting of proteins into complex plastids occurs by a variety of mechanisms, all of which require a bipartite N-terminal extension (reviewed in reference 51). The plastids of Euglena and dinoflagellates are enclosed within three membranes and acquire their proteins from the endoplasmic reticulum (ER) via the Golgi complex (49). In contrast, diatom and cryptomonad plastids are enclosed by four membranes, the outermost of which is studded with ribosomes, suggesting that the first step of protein import is cotranslational translocation across the outer membrane of these plastids (6).
Apicomplexan plastids are also enclosed by four membranes, and proteins destined for the apicoplast possess a bipartite leader sequence (53). The first 20 to 30 amino acids (aa) of nucleus-encoded apicoplast-targeted (NEAT) proteins function as a classical secretory signal sequence (SS). Cleavage by a signal peptidase within the ER exposes a 50- to 200-aa transit peptide (TP) that is responsible for targeting the protein to the organelle (41, 54). A detailed characterization of NEAT protein targeting signals is likely to enhance our understanding of plastid evolution and organellar targeting and to facilitate the identification of apicoplast proteins, many of which are attractive as parasiticidal drug targets that are unlikely to be found in mammalian host species.
Previous studies of the TP of T. gondii RPS9 (ribosomal small subunit protein 9) suggested that targeting information is located within the N-terminal portion of this peptide and that the TP may be processed multiple times during import into the apicoplast (11, 59). More recent evidence suggests that TgRPS9 contains redundant targeting domains (60). Analyses of NEAT proteins from P. falciparum have revealed the importance of a charged amino acid distribution in the TP (15), permitting the development of computational tools for the detection of candidate apicoplast proteins in malaria parasites (15, 61; http://plasmoDB.org). These tools fail to detect many NEAT proteins in T. gondii, however, despite proper targeting of P. falciparum NEAT proteins in transgenic T. gondii and vice versa (unpublished observations). In summary, precisely which elements of the TP are necessary and sufficient for targeting and translocation into the apicoplast remains unclear.
We have undertaken a systematic study of TP function, focusing on the NEAT protein ferredoxin NADP+ reductase (FNR) from T. gondii because the leader of this protein (SS plus TP) is relatively short, easily manipulated, and functions in both T. gondii and P. falciparum. Our results indicate that the TgFNR TP is processed at least twice and that processing is required for translocation into the lumen of the apicoplast. Furthermore, this peptide includes two functionally redundant TP domains, each containing subdomains that facilitate exit from the ER. Several constructs target proteins to multiple locations—the apicoplast periphery, rhoptries, and parasitophorous vacuoles—suggesting a possible link for targeting to these different subcellular compartments.
MATERIALS AND METHODS
Plasmids and deletion constructs.
The TP of TgFNR (52) was subcloned into the BglII and AvrII sites of the T. gondii expression vector ptubYFP/sagCAT (47) to produce ptubFNR-YFP/sagCAT. High-fidelity PCR (with Herculase enhanced DNA polymerase; Stratagene) was utilized to generate the various deletion constructs presented in this study, using the primers listed in Table 1; the resulting constructs were named according to the predicted FNR peptide and are referred to by their corresponding numbers in Table 1. To generate sequential 5-aa deletions from the C terminus of the TP, we subcloned PCR products, using sense primer 1 and one of the antisense primers (2 to 25), between the BglII and AvrII sites of ptubYFP/sagCAT. FNR-34 was generated by using primers 1 and 26. Constructs designed to define the FNR SS were generated by using primers 1 and 27, 28, or 29. Deletion of the FNR SS was achieved by utilizing primers 30 (containing a BglII site and an ATG initiation codon) and 31. For the generation of FNRΔ26-50, FNRΔ26-82, and FNRΔ26-86, primer 33 was utilized in combination with primers 34, 35, and 36, respectively. The 5′ end of primer 33 was designed to contain nucleotide substitutions that generated a unique restriction (BsrGI) site without affecting the amino acid sequence. Primers 34 and 35 each contained a BsrGI site at their 5′ ends to facilitate cloning. Deletions designed to define the second FNR targeting domain were generated by utilizing primer 1 and primes 12 to 17, with either FNRΔ26-50 or FNRΔ26-82 as a template (see Results). The deletion of aa 27 to 30 was achieved with primers 37 and 38, while the deletion of aa 83 to 86 was achieved with primers 39 and 40. In order to assess the number of 25-aa targeting subdomains present within the FNR TP, we used primer 41 in conjunction with each of the primers 42 to 61, followed by blunt-end ligation. The template for each of these reactions varied based on which 25-aa stretch was being generated. For example, to generate the construct FNRΔ26-30;56-150, we combined primer 41 with primer 42 in a PCR, with the FNR55 construct as the template.
TABLE 1.
PCR primers used for this study
| Primer no. | Deletion | Sequence |
|---|---|---|
| 1 | Tub-for | CAGTCTCGTAGAGAACAAGCACTC |
| 2 | FNR-145 | GCGCCTAGGCGGGGTCGCGAAAAGGCC |
| 3 | FNR-144 | GCGCCTAGGGGTCGCGAAAAGGCCTGTTG |
| 4 | FNR-143 | GCGCCTAGGCGCGAAAAGGCCTGTTGG |
| 5 | FNR-140 | GCGCCTAGGGCCTGTTGGCGTATCAGAACGC |
| 6 | FNR-135 | GCGCCTAGGAGAACGCTTGCGCGTACTACTG |
| 7 | FNR-130 | GCGCCTAGGACTACTGCGGTCTAGACGCACTG |
| 8 | FNR-125 | GCGCCTAGGACGCACTGAAAGAGGTCTTTCGAGT |
| 9 | FNR-120 | GCGCCTAGGTCTTTCGAGTGTGCCCCACTGTGGT |
| 10 | FNR-115 | GCGCCTAGGCCACTGTGGTAGGAAACCTTCAGACGA |
| 11 | FNR-110 | GCGCCTAGGACCTTCAGACGACTTGGGGAGACAA |
| 12 | FNR-105 | GCGCCTAGGGGGGAGACAATGCGTCCTCCTCCT |
| 13 | FNR-100 | GCGCCTAGGCCTCCTCCTGTCACTGCACTGTCT |
| 14 | FNR-95 | GCGCCTAGGGCACTGTCCCGTTGCACCCGGCAG |
| 15 | FNR-90 | GCGCCTAGGACCCGGCAGAAGGAAGCTGGAGAC |
| 16 | FNR-85 | GCGCCTAGGGCTGGAGACGCCACGGAATCGACC |
| 17 | FNR-80 | GCGCCTAGGGAATCGACCCTGGTAGCCTAAAACCGAG |
| 18 | FNR-75 | GCGCCTAGGGCCTAAAACCGAGTTAGCTGTAGC |
| 19 | FNR-70 | GCGCCTAGGAGCTGTAGCCGCGGCTCGCAGCGG |
| 20 | FNR-65 | GCGCCTAGGTCGCAGCGGTCCCAAATATGCGGT |
| 21 | FNR-60 | GCGCCTAGGATATGCGGTGGAGGAAGAGTCAAA |
| 22 | FNR-55 | GCGCCTAGGAGAGTCAAGCACTCGCACACCGCA |
| 23 | FNR-50 | GCGCCTAGGCACACCGCACGAGGCTGTATCGCC |
| 24 | FNR-45 | GCGCCTAGGTGTATCGCCATGATCAATCACATG |
| 25 | FNR-40 | GCGCCTAGGAATCACATGCGGTCCTCTCTTTGC |
| 26 | FNR-34 | GCGCCTAGGCTTTGCGACGCGGAAGCTCACGAC |
| 27 | FNR-24 | GCGCCTAGGAACGGATATAGCCACAGAGGCTAC |
| 28 | FNR-22 | GCGCCTAGGTATAGCCACAGAGACTACAACCGC |
| 29 | FNR-20 | GCGCCTAGGCACAGAGGCTACAACCGCAACAAA |
| 30 | FNRΔ1-24 | GCGAGATCTACAATGCAAGAGGTCGTGAGCTTCCGCGTCGCAAAGAGAGGA |
| 31 | FNR-rev | GCGCCTAGGGGATGTTTGGTCGGTCGGGGT |
| 32 | YFP-rev | GCTGTTCACCGGGGTGGTGCC |
| 33 | BsrGrev25 | GCGTTGTACAGATATAGCCACAGAGGCTACAACCGCAAC |
| 34 | FNRΔ26-50 | GCGTCTGTACAAGTGCGAGTGCTTGACTCTTCCTCCAC |
| 35 | FNRΔ26-82 | GCGTCTGTACAAGTCTCCAGCTTCCTTCTGCCGGGTGC |
| 36 | FNRΔ26-86 | GCGTCTGTACAACTTCTGCCGGGTGCAACGAGACAGTG |
| 37 | FNRΔ27-30-for | CGCGTCGCAAAGAGAGGACCGCATGTG |
| 38 | FNRΔ27-30-rev | CTCTTGAACGGATATAGCCACAGAGGC |
| 39 | FNRΔ83-86-for | CTTCTGCCGGGTGCAACGAGACAGTGC |
| 40 | FNRΔ83-86-rev | GCCACGGAATCGACCCTGGTAGCCTAA |
| 41 | FNR-SS-rev | AACGGATATAGCCACAGAGGCTACAACCGCAAC |
| 42 | FNRΔ26-30, 56-150 | TTCCGCGTCGCAAAGAGAGGACCGCATGTGATTG |
| 43 | FNRΔ26-35, 61-150 | AGAGGACCGCATGTGATTGATCATGGCGATACAG |
| 44 | FNRΔ26-40, 66-150 | ATTGATCATGGCGATACAGCCTCGTGCGGTGTGC |
| 45 | FNRΔ26-45, 71-150 | ACAGCCTCGTGCGGTGTGCGAGTGCTTGACTCTT |
| 46 | FNRΔ26-50, 76-150 | GTGCGAGTGCTTGACTCTTCCTCCACCGCATATT |
| 47 | FNRΔ26-55, 81-150 | TCTTCCTCCACCGCATATTTGGGACCGCTGCGAG |
| 48 | FNRΔ26-60, 86-150 | TATTTGGGACCGCTGCGAGCCGCGGCTACAGCTA |
| 49 | FNRΔ26-65, 91-150 | CGAGCCGCGGCTACAGCTAACTCGGTTTTAGGCT |
| 50 | FNRΔ26-70, 96-150 | GCTAACTCGGTTTTAGGCTACCAGGGTCGATTCC |
| 51 | FNRΔ26-75, 101-150 | GGCTACCAGGGTCGATTCCGTGGCGTCTCCAGCT |
| 52 | FNRΔ26-80, 106-150 | TTCCGTGGCGTCTCCAGCTTCCTTCTGCCGGGTG |
| 53 | FNRΔ26-85, 111-150 | AGCTTCCTTCTGCCGGGTGCAACGAGACAGTGCA |
| 54 | FNRΔ26-90, 116-150 | GGTGCAACGAGACAGTGCAGTGACAGGAGGAGGA |
| 55 | FNRΔ26-95, 121-150 | TGCAGTGACAGGAGGAGGACGCATTGTCTCCCCA |
| 56 | FNRΔ26-100, 126-150 | AGGACGCATTGTCTCCCCAAGTCGTCTGAAGGTT |
| 57 | FNRΔ26-105, 131-150 | CCCAAGTCGTCTGAAGGTTTCCTACCACAGT |
| 58 | FNRΔ26-110, 136-150 | GGTTTCCTACCACAGTGGGGCACACTCGAAAGAC |
| 59 | FNRΔ26-115, 141-150 | TGGGGCACACTCGAAAGACCTCTTTCAGTGCGTC |
| 60 | FNRΔ26-120, 146-150 | AGACCTCTTTCAGTGCGTCTAGACCGCAGTAGTA |
| 61 | FNRΔ26-126 | CGTCTAGACCGCAGTAGTACGCGCAAGCGGTCTG |
Cell culture and fluorescence microscopy.
T. gondii tachyzoites (RH strain) were maintained by serial passaging in human foreskin fibroblast (HFF) cells as previously described (42). Uninfected HFF monolayers were maintained exactly as previously described (42). The transfection of T. gondii was achieved by electroporation of 50 μg of DNA into 107 freshly harvested parasite tachyzoites as previously described (42). Parasites were inoculated onto confluent HFF monolayers on 22-mm-diameter glass coverslips in six-well plates and incubated for 24 or 48 h prior to processing.
For direct visualization of yellow fluorescent protein (YFP), infected coverslips were inverted onto glass slides and examined immediately. For indirect immunofluorescence, coverslips were fixed for 10 min in 4% paraformaldehyde, permeabilized for 10 min in 0.25% Triton X-100, blocked for 1 h in phosphate-buffered saline (PBS) (pH 7.4) plus 3% bovine serum albumin fraction V (Fisher), incubated for 1 h with a primary antibody (in blocking solution), washed, and incubated for 1 h with a secondary antibody. The primary antibodies used were as follows: rabbit polyclonal anti-acyl carrier protein (ACP; diluted 1:1,000) (53), mouse monoclonal anti-green fluorescent protein (GFP) (Clontech; 1:1,000), and mouse monoclonal anti-ROP2/3/4 (1:3,000) (3, 24). The following were used as secondary antibodies: Alexa 488-conjugated goat anti-mouse (Molecular Probes; 1:500) and Alexa 594-conjugated goat anti-rabbit (Molecular Probes; 1:500). Finally, apicoplast and nuclear DNAs were stained with 4′,6′-diamidino-2-phenylindole (DAPI) for 5 min (in PBS), and samples were washed with PBS and mounted on glass slides by the use of Fluoromount-G (Southern Biotechnology Associates, Inc.) for examination under either a Zeiss Axiovert 35 microscope equipped with a heated stage (Biopteks) or a Leica DM IRBE microscope equipped with a motorized filter wheel. Both inverted microscopes were equipped with a 100-W Hg-vapor lamp and an Orca-ER digital camera (Hamamatsu). Images were captured with Openlab 3.1 software (Improvision), and serial 0.1-μm-thick sections were subjected to iterative deconvolution and three-dimensional reconstruction with Volocity 2.0 software (Improvision).
Protein analysis.
Parasites were harvested shortly after monolayer lysis, filtered through 3-μm-pore-size polycarbonate filters (Nuclepore), pelleted at 900 × g, and resuspended in PBS at 106 parasites/μl. NuPAGE LDS sample buffer (Invitrogen) and 0.5 M dithiothreitol were added to each sample, followed by denaturation at 70°C for 10 min. Samples of ∼107 parasites were loaded into precast bis-Tris-4 to 12% polyacrylamide gels (Invitrogen) and were run in NuPAGE morpholineethanesulfonic acid-sodium dodecyl sulfate running buffer at 200 V for 45 min. The transfer of proteins to nitrocellulose was performed by using a Trans-Blot SemiDry apparatus (Bio-Rad) according to the manufacturer's instructions. After the transfer, the membrane was blocked for 1 h in PBS plus 5% nonfat dry milk, incubated for 1 h with a mouse anti-GFP antibody (1:1,000 in blocking solution) plus 0.2% Tween 20 (Sigma), washed twice in PBS plus Tween 20, and then incubated for 1 h with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2,500) (Sigma) plus Tween 20. After further washes in PBS plus Tween 20, a chemiluminescence reaction was performed by using ECL Western blotting detection reagents according to the manufacturer's (Amersham Biosciences) instructions, and blots were exposed on Kodak BioMax film.
RESULTS
Defining the boundaries of the bipartite FNR leader sequence (SS plus TP).
Based on multiple sequence alignments of FNR proteins from various species (52), the T. gondii FNR leader sequence is predicted to be ∼150 aa long. The fusion of either the full-length TgFNR gene (18) or the complete TgFNR leader (TgFNR-150, including both the presumed SS and the TP) (Fig. 1a) to a YFP reporter targets YFP to the apicoplast in T. gondii (Fig. 1b). The processing profile for this fusion construct revealed that the mature form (Fig. 2b, lane 1) was slightly larger than the ∼27-kDa molecular mass expected for YFP alone (arrowhead), suggesting that cleavage occurred upstream of the reporter protein. The removal of five or six additional amino acids from the C terminus of the FNR peptide yielded a processed product of close to 27 kDa (TgFNR-145 [Fig. 2b, lane 2] and TgFNR-144 [data not shown]), but further deletions resulted in alternative processing (TgFNR-140 [Fig. 2b, lane 3]; similar results were obtained for TgFNR-141 to -143 [not shown]). N-terminal sequencing of the purified protein revealed that the mature N terminus of FNR is Ala143.
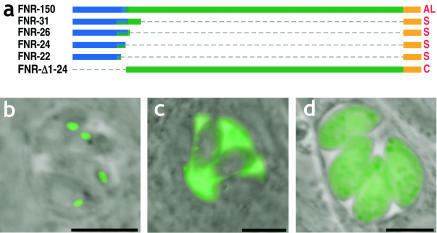
FIG. 1.
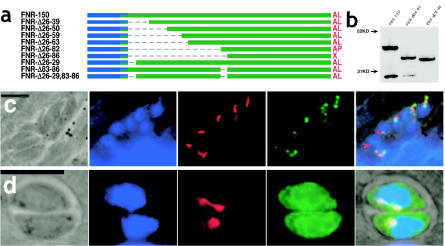
The bipartite apicoplast targeting signal. (a) Diagram representing the constructs used for the experiments shown in panels b, c, and d. The secretory signal is shown in blue, the plastid TP is in green, and YFP is in orange. (b) The fusion of the first 150 aa of FNR to YFP results in its targeting to the apicoplast, seen as oval structures in the apical juxtanuclear region of transfected T. gondii tachyzoites. (c) Deletion constructs containing only the FNR SS fused to YFP (FNR-31, −26, −24, and −22) result in secretion via dense granules (small dots within the parasites) into the parasitophorous vacuole surrounding the parasites. (d) Deletion of the SS (FNR-Δ1-24) results in YFP expression within the parasite cytoplasm only. FNR constructs diagrammed at the top are shown to scale, with summaries of the targeting results shown to the right. AL, apicoplast lumen; AP, apicoplast periphery; C, cytoplasmic; ER, endoplasmic reticulum; R, rhoptry; S, secreted; X, complex localization. Panels B, C, and D are representative images showing direct YFP fluorescence in live parasites. Bars = 5 μm.
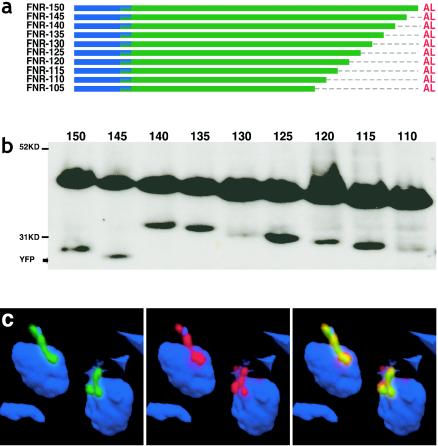
FIG. 2.
C-terminal deletions of the TP. (a) Diagram of FNR constructs depicting sequential 5-aa deletions between aa 105 and 150. (b) Western blot of deletion constructs FNR-150 through FNR-110, illustrating mature and processed forms of each fusion construct; note the alternative processing site in constructs shorter than TgFNR-145. Inefficient processing has been previously described (53). (c) Deconvolved fluorescence image of TgFNR-105 (representative of all constructs shown in panel a) illustrating targeting into the apicoplast lumen. The abbreviations are as described in the legend to Fig. 1. For microscopic images, green = YFP (anti-GFP), red = apicoplast (anti-ACP [53]), blue = nuclear and apicoplast DNA (DAPI [42]). A video of the images shown in panel c is available for download (http://roos.bio.upenn.edu/∼oharb/Harb2c.mov).
In order to define the beginning (N terminus) of the TP domain, we identified potential SSs by using a variety of computational algorithms. The neural network-based algorithm SignalP-NN (33; http://www.cbs.dtu.dk/services/SignalP/) predicted a eukaryotic signal cleavage site between aa 29 and 30; the SignalP hidden Markov model predicted cleavage between aa 21 and 22, while PSORT (32; http://psort.ims.u-tokyo.ac.jp/) predicted cleavage between aa 23 and 24. To determine which prediction was accurate, we generated several deletion constructs (Fig. 1a) and examined their ability to direct YFP to the secretory pathway. TgFNR-31, TgFNR-26, TgFNR-24, and TgFNR-22 were all sufficient for YFP secretion (Fig. 1c), while shorter fragments (e.g., TgFNR-20) were unable to direct secretion (not shown). The sizes of the proteins observed on Western blots also suggested that cleavage occurs close to aa 21, although we have not been able to purify sufficient amounts of incompletely processed protein to directly examine the signal peptidase cleavage site. Amino acids 1 to 21 constitute a typical eukaryotic SS (33), containing a positively charged N-terminal region, a 10-aa h-region, and small residues at the −1 and −3 positions relative to the cleavage site.
The deletion of aa 1 to 24 (Fig. 1a) abolished entry into the secretory pathway, resulting in the cytosolic expression of YFP (Fig. 1d), as has previously been observed for the NEAT protein ACP in both P. falciparum and T. gondii (54). Studies of the NEAT protein TgRPS9 have shown that deletion of the SS can result in the mistargeting of GFP to the mitochondria (11, 59), but this was not observed for TgFNR. The replacement of the TgFNR SS with an alternative SS derived from the secreted protein TgGRA8 (8) restored the targeting of YFP to the apicoplast (data not shown).
The C-terminal 45 aa of the TgFNR TP are not required for apicoplast targeting or translocation.
Previous deletion studies of the TPs of T. gondii NEAT proteins have been unsuccessful in clearly delineating specific domains involved in targeting to, or translocation into, the apicoplast (11, 59), because truncated proteins are often expressed poorly or are retained within the ER (presumably due to misfolding). Fortunately, this appears not to be the case for FNR protein deletions. In order to identify regions of the TgFNR TP that affect processing, targeting, and/or translocation, we generated nested 5-aa deletions from the C terminus of the 150-aa leader domain, as shown in Fig. 2.
Progressive deletions, from TgFNR-150 to TgFNR-105 (Fig. 2a), resulted in progressively smaller unprocessed preproteins (Fig. 2b, upper bands). Alternative processing was observed beyond TgFNR-144, however, as indicated by the increased size of the processed (lower) bands in Fig. 2b (note that the processing of the TP was inefficient, as has previously been described [53]). This observation is consistent with an intermediate or cryptic processing site between aa 100 and 105 (multiple processing sites have also been reported for TgRPS9 [59]). Despite the alternative processing of the TP domain, deletions down to TgFNR-105 had no apparent effect on the targeting of YFP to the apicoplast (Fig. 2c), consistent with previous results suggesting that the N-terminal portion of the TP is more important than the C-terminal portion for apicoplast targeting (11, 59). These results also demonstrate that processing at the native TP cleavage site is not necessary for import.
Separating targeting to the apicoplast from translocation into the organellar lumen.
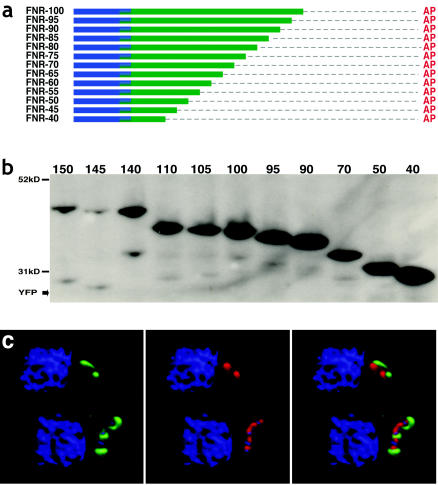
Further nested deletions down to TgFNR-40 (Fig. 3a) permitted the targeting of YFP to the region of the apicoplast, but not into the apicoplast lumen (Fig. 3c). This phenotype coincides with the elimination of the second cleavage site (Fig. 3b, lanes 6 to 11), suggesting that cleavage of the TP may be required for import into the apicoplast lumen but not for targeting to the organelle periphery. In summary, the region from aa ∼23 to 40 contains sufficient information for apicoplast targeting (when combined with a SS).
FIG. 3.
C-terminal deletions of the TP delineate between targeting and translocation. (a) Diagram of FNR constructs depicting sequential 5-aa deletions between aa 40 and 105. (b) Western blot of deletion constructs down to TgFNR-40. (c) Deconvolved fluorescence image of TgFNR-50 (representative of all constructs shown in panel a) illustrating targeting to, but not into, the apicoplast. The abbreviations are as described in the legend to Fig. 1. For microscopic images, green = YFP (anti-GFP), red = apicoplast (anti-ACP [53]), blue = nuclear and apicoplast DNA (DAPI [42]). Bars = 5 μm. A video of the images shown in panel c is available for download (http://roos.bio.upenn.edu/∼oharb/Harb3c.mov).
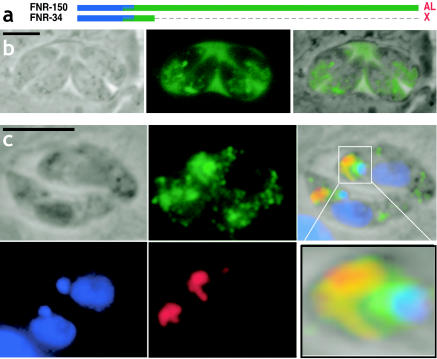
Reducing this minimal TP by 6 aa (TgFNR-34) (Fig. 4a) resulted in the simultaneous targeting of YFP to the apicoplast periphery, the rhoptries, and the parasitophorous vacuole (Fig. 4b and c). Moreover, YFP fluorescence never colocalized with dense granule markers (not shown). These results suggest that proteins destined for the apicoplast and rhoptries may share a common targeting pathway beyond the obvious fact that both pass through the ER. However, further experiments to confirm this observation are required. Interestingly, a similar phenotype was observed for several other deletion constructs (see below).
FIG. 4.
TgFNR-34 targets YFP to multiple compartments. (a) Diagram of TgFNR-34 and -150. (b) Direct YFP fluorescence image of TgFNR-34, showing complex targeting pattern within the parasite and secretion into the parasitophorous vacuole. (c) Fluorescence image of TgFNR-34 (fixed specimen), showing complex targeting pattern within the parasite, including colocalization with rhoptries. The insert (bottom right) is an enlarged view of the apical region of one of the parasites showing green fluorescence at the periphery of the apicoplast and colocalizing with the rhoptries. The abbreviations are as described in the legend to Fig. 1. For microscopic images, green = YFP (anti-GFP or native fluorescence in panel b), red = rhoptries (anti-ROP2/3/4 [3]), blue = nuclear and apicoplast DNA (DAPI [42]). Bars = 5 μm.
A second apicoplast targeting domain in the TgFNR TP.
In order to assess the importance of the minimal TP defined above for apicoplast targeting, a deletion construct lacking aa 26 to 50 of the FNR leader was generated, as shown in Fig. 5a (TgFNR-Δ26-50). This construct remained able to traffic to the apicoplast, indicating the presence of redundant targeting signals (data not shown). A series of further deletions from the N terminus of the TP were therefore engineered (Fig. 5a); of these, only TgFNR-Δ26-82 and TgFNR-Δ26-86 failed to fully translocate into the apicoplast lumen. A microscopic evaluation indicated that TgFNR-Δ26-82 was targeted to the apicoplast periphery only (Fig. 5c), but this protein was processed nevertheless (Fig. 5b, lane 2), suggesting that TP maturation may not require complete translocation into the organellar lumen (see Discussion). Further deletion (TgFNR-Δ26-86) resulted in a combination of targeting to the periphery of the apicoplast and retention within the ER (Fig. 5d), and Western blots showed no evidence of processing beyond SS cleavage (Fig. 5b, lane 3).
FIG. 5.
Redundant motif within apicoplast targeting signal. (a) Diagram depicting deletions that define a redundant domain with the FNR TP. (b) Western blot demonstrating processing of TgFNR-Δ26-82, but not TgFNR-Δ26-86. (c) Despite deletion of one apicoplast targeting signal, TgFNR-Δ26-82 still localizes to the organelle periphery. (d) Deletion of an additional 4 aa results in targeting to both the ER and apicoplast periphery (TgFNR-Δ26-86). The abbreviations are as described in the legend to Fig. 1. For microscopic images, green = YFP (anti-GFP), red = apicoplast (anti-ACP [53]), blue = nuclear and apicoplast DNA (DAPI [42]). Bars = 5 μm.
To identify potential motifs that may be associated with apicoplast targeting signals, we compared patterns detected by the Teiresias algorithm (http://cbcsrv.watson.ibm.com/Tspd.html) with the observed targeting patterns. Val-(Val/Ser)-Ser-Phe occurs at both aa 27 to 30 and 83 to 86, which are closely linked to the redundant apicoplast targeting signals mapped above. The deletion of either or both motifs (Fig. 5a) had minor effects on the processing intermediates (not shown), but these mutations failed to affect apicoplast targeting. However, these motifs may be associated with release from the ER (see below).
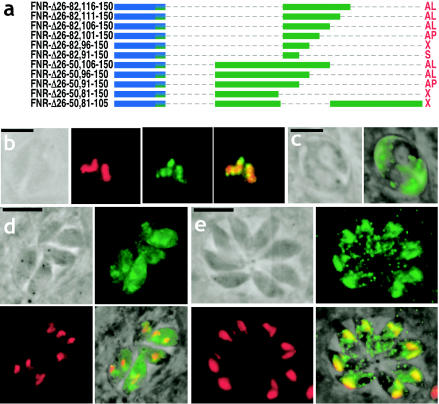
The second apicoplast targeting signal was mapped more precisely via C-terminal deletions of the TP domain in the TgFNR-Δ26-50 and TgFNR-Δ26-82 backgrounds, as shown in Fig. 6a. The deletion of aa 106 to 150 had no effect on proper protein trafficking (in either background), but further deletions resulted in increasingly compromised targeting to the apicoplast, as follows. A Δ101-150 mutation in the TgFNR-Δ26-82 background produced targeting to the apicoplast periphery, a Δ96-150 mutation produced staining in the apicoplast periphery and the ER, and a Δ91-150 mutation yielded a secretory protein (Fig. 6c). Similarly, in the TgFNR-Δ26-50 background, Δ96-150 and Δ91-150 mutations yielded localization to the apicoplast periphery (Fig. 6b), while a Δ81-150 mutation yielded a complex pattern of localization to the apicoplast periphery, the rhoptries, and the parasitophorous vacuole, as noted previously for extreme deletions of the upstream apicoplast targeting signal (TgFNR-34; Fig. 4b and c).
FIG. 6.
Mapping the second redundant targeting motif within the apicoplast targeting signal. (a) Diagram depicting various deletions generated to map the second redundant targeting motif within the FNR TP. (b) TgFNR-Δ26-50,91-150 is targeted to the apicoplast periphery. (c) TgFNR-Δ26-82,91-150 is secreted into the parasitophorous vacuole. (d and e) Deleting both of the mapped apicoplast targeting signals results in a complex pattern of protein secretion targeting TgFNR-Δ26-50,83-105 to the apicoplast periphery (d), rhoptries (e), and the parasitophorous vacuole (not shown). The abbreviations are as described in the legend to Fig. 1. For microscopic images, green = YFP (anti-GFP or native fluorescence [c]), red = apicoplast (anti-ACP) or rhoptries (anti-ROP2/3/4 [e]). Bars = 5 μm.
In summary, aa 20 to 40 and 82 to 105 can each function as an independent signal for targeting to (but not into) the apicoplast. Remarkably, a deletion construct lacking both of these domains (TgFNR-Δ26-50,Δ83-105) was still able to target YFP to the apicoplast periphery (Fig. 6d) in addition to the rhoptries (Fig. 6e) and the parasitophorous vacuole (not shown). This protein appeared unable to reach the apicoplast lumen, however, despite the retention of the native protein-processing site, and neither aa 51 to 82 nor aa 106 to 150 alone were able to function as an apicoplast targeting signal.
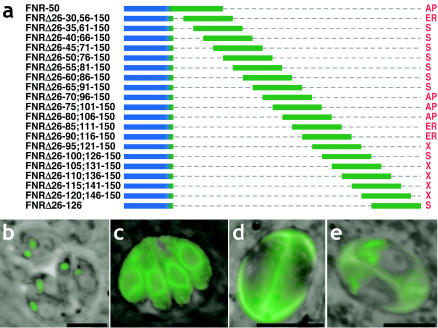
To more directly test whether any other linear spans are capable of mediating apicoplast targeting, we generated peptide sequences of 25 aa in length (sufficient to encompass either of the plastid targeting domains defined above) every 5 aa throughout the TP and sandwiched them between the TgFNR SS and the YFP reporter (Fig. 7a). As shown in Fig. 7, four phenotypic groups were defined based on the subcellular localization of YFP: (i) targeting to the apicoplast, associated with the two linear domains identified above (Fig. 7b); (ii) retention within the ER, correlated with the deletion of the V[V/S]SF motif from both targeting domains (Fig. 7c); (iii) secretion into the parasitophorous vacuole via dense granules (Fig. 7d); and (iv) complex localization within multiple secretory pathway compartments, including the apicoplast periphery, the rhoptries, and the parasitophorous vacuole (Fig. 7e).
FIG. 7.
Scanning the apicoplast TP. (a) Diagram of overlapping sequential 25-aa segments scanning through the TgFNR TP inserted downstream of the SS and upstream of a YFP reporter. Four targeting patterns were observed, labeling the apicoplast (b), ER (c), parasitophorous vacuole (d), or rhoptries and parasitophorous vacuole (e). The abbreviations are as described in the legend to Fig. 1. All micrographs are direct fluorescence images of living parasites. Bars = 5 μm.
DISCUSSION
The identification of nucleus-encoded apicoplast proteins offers the potential for understanding the function and evolution of this organelle, and secondary endosymbiotic plastids in general (40). In addition, the ability to recognize NEAT proteins in the genomes of apicomplexan parasites suggests possible targets for parasite-specific chemotherapy (29). Signal peptide recognition algorithms perform acceptably in recognizing the first part of the bipartite apicoplast targeting signal. However, despite advances that facilitated the identification of NEAT proteins in the genome of P. falciparum (15, 61), these algorithms remain imperfect, particularly across species boundaries, in contrast to the sequences themselves, which function surprisingly well across large phylogenetic distances.
In order to facilitate the identification of apicoplast targeting signals, we performed a careful analysis of the T. gondii FNR TP, expressing a large series of deletion mutants in transgenic parasites and using fluorescent protein reporters to resolve subcellular localization. The results of these studies are summarized in Fig. 8. Deconvolution of high-resolution wide-field microscopic images (Fig. 2C and 3C) enabled targeting to the apicoplast periphery (designated AP) to be clearly distinguished from translocation into the organellar lumen (designated AL). Further analysis will be required to determine if these deletion constructs constitute targeting and/or translocation intermediates.
FIG. 8.
Summary of signal features within the TgFNR leader. The secretory SS is shown in magenta, and redundant apicoplast targeting domains are shown in green. Computationally predicted and experimentally determined cleavage sites are indicated by black and blue text, respectively. Red highlighting indicates the V-(V/S)-S-F motif that may be associated with release from the ER. The mature FNR gene is indicated, with conserved amino acids shown in red.
The elimination of the native TP processing site had no effect on luminal targeting, perhaps because of the presence of additional (or cryptic) processing sites (Fig. 2B), but further deletion of this second processing site (Fig. 3B) resulted in targeting to the periphery only. Targeting to the apicoplast periphery does not preclude TP processing, however (Fig. 5B). These results suggest that TP processing may be necessary for release into the apicoplast lumen, but translocation is not essential for processing.
The processing of chloroplast TPs is carried out by metal-binding stromal endopeptidases, and multiple processing sites have previously been described in plants and algae (20, 39, 48), but a consensus cleavage motif remains elusive, possibly because several chloroplast peptidases may recognize different cleavage sites (12, 22). The role of TP cleavage in plastid import is not clearly understood, but the down-regulation of a chloroplast-processing enzyme by antisense technology has been correlated with an inhibition of protein import into the chloroplast (55), consistent with our observation that the abolition of processing is correlated with protein localization to the apicoplast periphery.
Two functionally redundant apicoplast targeting signals have been identified; both are ∼20 aa in length, and a series of scanning mutations (Fig. 7) argues that these are the only two linear epitopes within the FNR TP that are capable of mediating targeting to the apicoplast. The TPs of plant and algal chloroplasts are composed of multiple domains (2, 37, 38), each of which may be involved in distinct steps of the targeting and import process. An additive effect of redundant targeting sequences has been reported for the mitochondrial F1-ATPase beta subunit precursor (5). Functional redundancy in the TP may also reflect the evolutionary history of these sequences, as domain shuffling has been shown to facilitate the acquisition of targeting information (7, 16, 25).
Each of the two apicoplast targeting domains appears to contain functional microdomains. The N-terminal region is implicated in release from the ER (perhaps attributable to the consensus motif V[V/S]SF). C-terminal truncations appear to lower the fidelity of apicoplast targeting, resulting in mislocalization to the rhoptries and the parasitophorous vacuole in addition to apicoplast targeting. Other mutations within the TP also produce this complex targeting pattern (designated “X”), suggesting that targeting to these organelles branches off of a common pathway.
Our findings on FNR are consistent with previous studies on the T. gondii NEAT protein TgRPS9, which suggested the presence of multiple processing sites and more than one apicoplast targeting peptide (59, 60). These results are also consistent with indications that the N-terminal portion of the TP is particularly important (59), although this region is not particularly basic or enriched in lysine or asparagine, as found in P. falciparum (15). Protein targeting into the apicoplast shares with mitochondria the requirement for an N-terminal protein extension, but neither the FNR TP nor the ACP TP mediates mitochondrial targeting (Fig. 1). The observation that TgRPS9 SS deletions target to the mitochondria (11, 59) may be related to the high frequency with which randomly generated sequences function as mitochondrial targeting domains (1, 23).
Overall, NEAT protein sequences appear to be organized in a remarkably linear fashion. The extreme N terminus provides the SS (magenta segment in Fig. 8) required for entry into the secretory pathway itself (S in Fig. 1, 6, and 7); the removal of this domain results in cytosolic localization (Fig. 1, C). The penultimate domain (green) is required for targeting to the apicoplast periphery (AP); the N-terminal portion (red) is associated with release from the ER, while the C-terminal portion is required for efficient apicoplast targeting since truncations in this region result in complex targeting (X). Finally, proteolytic cleavage sites (blue) are required for proper entry into the apicoplast lumen (AL). The ability to recognize apicoplast targeting sequences, combined with the rapidly expanding genome and expressed sequence tag databases for apicomplexan parasites, should facilitate studies of the evolution of complex plastids, the relationships between apicomplexan parasites, the biological function(s) of the apicoplast, and the potential of this organelle as a parasiticidal drug target.
Acknowledgments
We acknowledge Boris Striepen and other past and present members of the Roos lab for helpful comments and Paul Warfel for editing the supplemental videos. We also thank Frank Seeber for the original FNR construct, Con Beckers for the anti-ROP2/3/4 antibody, and Geoff McFadden for the anti-ACP antibody.
O.S.H. and M.J.C. are supported by National Research Service Awards F32 AI10654 and F32 AI10482, and this research was supported by grants from the National Institutes of Health to D.S.R., who is also a Burroughs-Wellcome Scholar in Molecular Parasitology and an Ellison Foundation Senior Scholar in Global Infectious Diseases.
REFERENCES
- 1.Allison, D. S., and G. Schatz. 1986. Artificial mitochondrial presequences. Proc. Natl. Acad. Sci. USA 83:9011-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apt, K. E., L. Zaslavkaia, J. C. Lippmeier, M. Lang, O. Kilian, R. Wetherbee, A. R. Grossman, and P. G. Kroth. 2002. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 115:4061-4069. [DOI] [PubMed] [Google Scholar]
- 3.Beckers, C. J., J. F. Dubremetz, O. Mercereau-Puijalon, and K. A. Joiner. 1994. The Toxoplasma gondii rhoptry protein ROP 2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host cell cytoplasm. J. Cell Biol. 127:947-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckers, C. J., D. S. Roos, R. G. Donald, B. J. Luft, J. C. Schwab, Y. Cao, and K. A. Joiner. 1995. Inhibition of cytoplasmic and organellar protein synthesis in Toxoplasma gondii. Implications for the target of macrolide antibiotics. J. Clin. Investig. 95:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedwell, D. M., D. J. Klionsky, and S. D. Emr. 1987. The yeast F1-ATPase beta subunit precursor contains functionally redundant mitochondrial protein import information. Mol. Cell. Biol. 7:4038-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya, D., and A. Grossman. 1991. Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol. Gen. Genet. 229:400-404. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, B. D. 2001. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541:2-21. [DOI] [PubMed] [Google Scholar]
- 8.Carey, K. L., C. G. Donahue, and G. E. Ward. 2000. Identification and molecular characterization of GRA8, a novel, proline-rich, dense granule protein of Toxoplasma gondii. Mol. Biochem. Parasitol. 105:25-37. [DOI] [PubMed] [Google Scholar]
- 9.Chen, K., X. Chen, and D. J. Schnell. 2000. Mechanism of protein import across the chloroplast envelope. Biochem. Soc. Trans. 28:485-491. [PubMed] [Google Scholar]
- 10.Delwiche, C. F. 1999. Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154:S164-S177. [DOI] [PubMed] [Google Scholar]
- 11.DeRocher, A., C. B. Hagen, J. E. Froehlich, J. E. Feagin, and M. Parsons. 2000. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J. Cell Sci. 113:3969-3977. [DOI] [PubMed] [Google Scholar]
- 12.Emanuelsson, O., H. Nielsen, and G. von Heijne. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fast, N. M., J. C. Kissinger, D. S. Roos, and P. J. Keeling. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18:418-426. [DOI] [PubMed] [Google Scholar]
- 14.Fichera, M. E., and D. S. Roos. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390:407-409. [DOI] [PubMed] [Google Scholar]
- 15.Foth, B. J., S. A. Ralph, C. J. Tonkin, N. S. Struck, M. Fraunholz, D. S. Roos, A. F. Cowman, and G. I. McFadden. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299:705-708. [DOI] [PubMed] [Google Scholar]
- 16.Gantt, J. S., S. L. Baldauf, P. J. Calie, N. F. Weeden, and J. D. Palmer. 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, C. Y., M. K. Shaw, C. H. Pletcher, B. Striepen, L. G. Tilney, and D. S. Roos. 2001. A plastid segregation defect in the protozoan parasite Toxoplasma gondii. EMBO J. 20:330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, C. Y., B. Striepen, C. H. Pletcher, J. M. Murray, and D. S. Roos. 2001. Targeting and processing of nuclear-encoded apicoplast proteins in plastid segregation mutants of Toxoplasma gondii. J. Biol. Chem. 276:28436-28442. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, M. C. 2001. Advances and prospects for subunit vaccines against protozoa of veterinary importance. Vet. Parasitol. 101:291-310. [DOI] [PubMed] [Google Scholar]
- 20.Keegstra, K., and K. Cline. 1999. Protein import and routing systems of chloroplasts. Plant Cell 11:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler, S., C. F. Delwiche, P. W. Denny, L. G. Tilney, P. Webster, R. J. Wilson, J. D. Palmer, and D. S. Roos. 1997. A plastid of probable green algal origin in apicomplexan parasites. Science 275:1485-1489. [DOI] [PubMed] [Google Scholar]
- 22.Koussevitzky, S., E. Ne'eman, A. Sommer, J. C. Steffens, and E. Harel. 1998. Purification and properties of a novel chloroplast stromal peptidase. Processing of polyphenol oxidase and other imported precursors. J. Biol. Chem. 273:27064-27069. [DOI] [PubMed] [Google Scholar]
- 23.Lemire, B. D., C. Fankhauser, A. Baker, and G. Schatz. 1989. The mitochondrial targeting function of randomly generated peptide sequences correlates with predicted helical amphiphilicity. J. Biol. Chem. 264:20206-20215. [PubMed] [Google Scholar]
- 24.Leriche, M. A., and J. F. Dubremetz. 1991. Characterization of the protein contents of rhoptries and dense granules of Toxoplasma gondii tachyzoites by subcellular fractionation and monoclonal antibodies. Mol. Biochem. Parasitol. 45:249-259. [DOI] [PubMed] [Google Scholar]
- 25.Long, M., S. J. de Souza, C. Rosenberg, and W. Gilbert. 1996. Exon shuffling and the origin of the mitochondrial targeting function in plant cytochrome c1 precursor. Proc. Natl. Acad. Sci. USA 93:7727-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marechal, E., and M. F. Cesbron-Delauw. 2001. The apicoplast: a new member of the plastid family. Trends Plant Sci. 6:200-205. [DOI] [PubMed] [Google Scholar]
- 27.Martin, W., T. Rujan, E. Richly, A. Hansen, S. Cornelsen, T. Lins, D. Leister, B. Stoebe, M. Hasegawa, and D. Penny. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99:12246-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, W., B. Stoebe, V. Goremykin, S. Hapsmann, M. Hasegawa, and K. V. Kowallik. 1998. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162-165. [DOI] [PubMed] [Google Scholar]
- 29.McFadden, G. I., and D. S. Roos. 1999. Apicomplexan plastids as drug targets. Trends Microbiol. 7:328-333. [DOI] [PubMed] [Google Scholar]
- 30.McFadden, G. I., and R. F. Waller. 1997. Plastids in parasites of humans. Bioessays 19:1033-1040. [DOI] [PubMed] [Google Scholar]
- 31.Moreira, D., H. Le Guyader, and H. Philippe. 2000. The origin of red algae and the evolution of chloroplasts. Nature 405:69-72. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Ogino, N., and C. Yoneda. 1966. The fine structure and mode of division of Toxoplasma gondii. Arch. Ophthalmol. 75:218-227. [DOI] [PubMed] [Google Scholar]
- 35.Palmer, J. D. 2000. A single birth of all plastids? Nature 405:32-33. [DOI] [PubMed] [Google Scholar]
- 36.Phillips, R. S. 2001. Current status of malaria and potential for control. Clin. Microbiol. Rev. 14:208-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilon, M., H. Wienk, W. Sips, M. de Swaaf, I. Talboom, R. van't Hof, G. de Korte-Kool, R. Demel, P. Weisbeek, and B. de Kruijff. 1995. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 270:3882-3893. [DOI] [PubMed] [Google Scholar]
- 38.Rensink, W. A., M. Pilon, and P. Weisbeek. 1998. Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 118:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richter, S., and G. K. Lamppa. 1999. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J. Cell Biol. 147:33-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roos, D. S., M. J. Crawford, R. G. Donald, M. Fraunholz, O. S. Harb, C. Y. He, J. C. Kissinger, M. K. Shaw, and B. Striepen. 2002. Mining the Plasmodium genome database to define organellar function: what does the apicoplast do? Philos. Trans. R. Soc. Lond. B 357:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roos, D. S., M. J. Crawford, R. G. Donald, J. C. Kissinger, L. J. Klimczak, and B. Striepen. 1999. Origin, targeting, and function of the apicomplexan plastid. Curr. Opin. Microbiol. 2:426-432. [DOI] [PubMed] [Google Scholar]
- 42.Roos, D. S., R. G. Donald, N. S. Morrissette, and A. L. Moulton. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 43.Schwartzbach, S. D., T. Osafune, and W. Loffelhardt. 1998. Protein import into cyanelles and complex chloroplasts. Plant Mol. Biol. 38:247-263. [PubMed] [Google Scholar]
- 44.Sheffield, H. G., and M. L. Melton. 1968. The fine structure and reproduction of Toxoplasma gondii. J. Parasitol. 54:209-226. [PubMed] [Google Scholar]
- 45.Siddall, M. 1992. Hohlzylinders. Parasitol. Today 8:90-91. [DOI] [PubMed] [Google Scholar]
- 46.Steiner, J. M., and W. Loffelhardt. 2002. Protein import into cyanelles. Trends Plant Sci. 7:72-77. [DOI] [PubMed] [Google Scholar]
- 47.Striepen, B., C. Y. He, M. Matrajt, D. Soldati, and D. S. Roos. 1998. Expression, selection, and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol. 92:325-338. [DOI] [PubMed] [Google Scholar]
- 48.Su, Q., and A. Boschetti. 1993. Partial purification and properties of enzymes involved in the processing of a chloroplast import protein from Chlamydomonas reinhardii. Eur. J. Biochem. 217:1039-1047. [DOI] [PubMed] [Google Scholar]
- 49.Sulli, C., Z. Fang, U. Muchhal, and S. D. Schwartzbach. 1999. Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J. Biol. Chem. 274:457-463. [DOI] [PubMed] [Google Scholar]
- 50.Tenter, A. M., A. R. Heckeroth, and L. M. Weiss. 2000. Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 30:1217-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dooren, G. G., S. D. Schwartzbach, T. Osafune, and G. I. McFadden. 2001. Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta 1541:34-53. [DOI] [PubMed] [Google Scholar]
- 52.Vollmer, M., N. Thomsen, S. Wiek, and F. Seeber. 2001. Apicomplexan parasites possess distinct nuclear-encoded, but apicoplast-localized, plant-type ferredoxin-NADP+ reductase and ferredoxin. J. Biol. Chem. 276:5483-5490. [DOI] [PubMed] [Google Scholar]
- 53.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waller, R. F., M. B. Reed, A. F. Cowman, and G. I. McFadden. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan, J., D. Bringloe, and G. K. Lamppa. 1998. Disruption of chloroplast biogenesis and plant development upon down-regulation of a chloroplast processing enzyme involved in the import pathway. Plant J. 15:459-468. [Google Scholar]
- 56.Wilson, R. J., P. W. Denny, P. R. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
- 57.Wolfe, K. H., C. W. Morden, and J. D. Palmer. 1991. Ins and outs of plastid genome evolution. Curr. Opin. Genet. Dev. 1:523-529. [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization. 1997. World malaria situation in 1994. Wkly. Epidemiol. Res. 72:269-274. [Google Scholar]
- 59.Yung, S., T. R. Unnasch, and N. Lang-Unnasch. 2001. Analysis of apicoplast targeting and transit peptide processing in Toxoplasma gondii by deletional and insertional mutagenesis. Mol. Biochem. Parasitol. 118:11-21. [DOI] [PubMed] [Google Scholar]
- 60.Yung, S. C., T. R. Unnasch, and N. Lang-Unnasch. 2003. Cis and trans factors involved in apicoplast targeting in Toxoplasma gondii. J. Parasitol. 89:767-776. [DOI] [PubMed] [Google Scholar]
- 61.Zuegge, J., S. Ralph, M. Schmuker, G. I. McFadden, and G. Schneider. 2001. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280:19-26. [DOI] [PubMed] [Google Scholar]