Abstract
Blastomyces dermatitidis is a dimorphic fungal pathogen that converts from mycelia or conidia to a host-adapted yeast morphotype upon infection. Conversion to the yeast form is accompanied by the production of the virulence factor BAD1. Yeast-phase-specific expression of BAD1 is transcriptionally regulated, and its promoter shares homology with that of the yeast-phase-specific gene YPS3 of Histoplasma capsulatum. Serial truncations of the BAD1 upstream region were fused to the lacZ reporter to define functional areas in the promoter. Examination of PBAD1-lacZ fusions in B. dermatitidis indicated that BAD1 transcription is upregulated in the yeast phase. The 63-nucleotide box A region conserved in the YPS3 upstream region was shown to be an essential component of the minimal BAD1 promoter. A matched PYPS3-lacZ construct indicated that this same region was needed for minimal YPS3 promoter activity in B. dermatitidis transformants. Reporter activity in H. capsulatum transformants similarly showed a requirement for box A in the minimal BAD1 promoter. Several putative transcription factor binding sites were identified within box A of BAD1. Replacement of two of these predicted sites within box A—a cAMP responsive element and a Myb binding site—sharply reduced transcriptional activity, indicating that these regions are critical in dictating the yeast-phase-specific expression of this crucial virulence determinant of B. dermatitidis.
Most systemic fungal infections of immunocompetent humans are caused by dimorphic fungi. This group of pathogens includes Blastomyces dermatitidis, Histoplasma capsulatum, Paracoccidioides brasiliensis, Coccidioides immitis, and Sporothrix schenkii. These fungi share the unique ability to undergo reversible cellular differentiation, existing as an infectious form or morphotype in the environment but converting to a second, pathogenic form upon entry into the host. This morphological switch promotes host invasion and evasion (40). A change in fungal gene expression is one possible mechanism by which morphogenesis affects the outcome of infection. Factors required for colonization of the host or modulation of the host immune response may be upregulated, while other factors only necessary for growth in the environment may be downregulated. Identification of products whose expression is linked only to the pathogenic phase may uncover virulence factors of these fungi. Also, since prevention of this essential developmental event may hold the key to control of infections by these fungi, identification of molecules necessary for the morphological switch could lead to new drug or vaccine targets that block the earliest events in colonization or infection.
Several genes have been identified whose expression is specific to one morphotype, a subset of which have been shown to be required for virulence. For example, expression of SOWgp58, an extracellular matrix-binding protein of C. immitis, is limited to the endosporulating spherule form, which is the parasitic phase of this fungus (22). Deletion of SOWgp resulted in a reduction in virulence, and its product is presumed to play a role as an adhesin (23). In H. capsulatum, expression of the calcium-binding protein CBP1 was limited to the yeast phase, and a knockout strain lacking CBP1 was unable to destroy macrophages in vitro or colonize in a mouse model of pulmonary histoplasmosis (45). BAD1 of B. dermatitidis, like CBP1, was also shown to be a yeast-phase-specific gene and a virulence factor (41, 12). BYS1 of B. dermatitidis is a yeast-phase-specific gene of unknown function that contains 3′ signals found in message with regulated stability (13), but no such signals were found in the 3′ untranslated region of BAD1 (unpublished observation). Promoter analysis of CBP1 revealed that this gene is transcriptionally upregulated in the yeast and identified a stretch of DNA required for yeast-phase expression of this gene (35).
B. dermatitidis exists as a mold in the acidic soil found along the Mississippi and Ohio River valleys. Upon inhalation of conidia or mycelial fragments, this fungus converts to the yeast morphotype and colonizes the host alveoli (10). Infection can lead to an acute pulmonary disease or can disseminate to other sites, most notably bone and skin. Conversion to the yeast form induces the expression of BAD1, which becomes arrayed on the surface of the yeast and is released into the extracellular milieu (27, 41). A knockout strain lacking BAD1 was avirulent in a mouse model of infection, while a strain in which BAD1 expression was reconstituted was restored to virulence matching that of the parental strain (12). BAD1 has been shown to bind to complement receptor 3 and may serve as an adhesin, although recently it was shown that this property is dispensable for virulence (31, 11). Both cell-associated and secreted forms of the protein modulate the host inflammatory response in favor of the pathogen by downregulating production of the proinflammatory cytokine tumor necrosis factor alpha (17, 18).
Expression of BAD1 is limited to the yeast phase and regulated transcriptionally. Analysis of the promoter region identified two regions of homology with that of another yeast-phase-specific gene, YPS3 of H. capsulatum (26). These regions were termed boxes B and A and shared 86 and 80% identity between the BAD1 gene of B. dermatitidis strain 26199 and the YPS3 gene of H. capsulatum strain G217B, respectively (see Fig. 1). An additional stretch of nucleotides upstream of boxes B and A in the YPS3 upstream region, which is able to bind protein present in a yeast nuclear extract via electrophoretic mobility shift assay (24), was also identified in the BAD1 upstream region. When the BAD1 promoter was expressed heterologously in H. capsulatum, its transcription was phase regulated, indicating that the regulatory machinery is conserved between these two fungi (41).
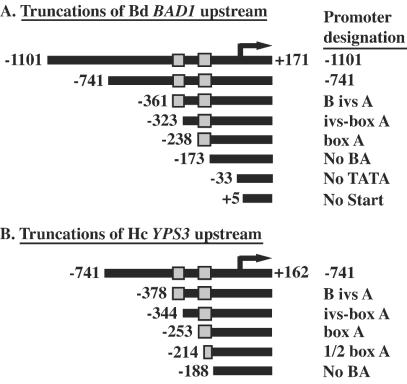
FIG. 1.
Serial truncations of the upstream regions of BAD1 and YPS3. Shown are schematic representations of amplified fragments that were fused to lacZ to assess the contributions of promoter regions to B. dermatitidis (Bd) BAD1 and H. capsulatum (Hc) YPS3 transcription (A and B, respectively). Transcriptional start sites are indicated by arrows. Numbering of the 5′ and 3′ (at the translation-initiating ATG) ends of the promoter fragments are indicated relative to the transcriptional start sites (+1). At the right are promoter fragment designations, based on either the distance upstream of the transcriptional start site or homology between the BAD1 and YPS3 promoters at boxes B and A, shown as shaded boxes.
Herein, we explored the sequences required for regulating expression of BAD1. Serial truncations of upstream sequences fused to a β-galactosidase reporter were used to functionally analyze homologous regions of the BAD1 and YPS3 promoters in both B. dermatitidis and H. capsulatum. We observed that the conserved box A region is an essential component of the minimal BAD1 promoter. This same region is needed for minimal YPS3 promoter activity in B. dermatitidis, whereas additional upstream sequence 5′ to box A is required for YPS3 transcription in H. capsulatum. We also pinpointed two putative transcription factor binding sites within box A that are essential for upregulating BAD1 expression in the yeast phase.
MATERIALS AND METHODS
Strains and growth.
B. dermatitidis strain 26199 and Saccharomyces cerevisiae strain 204509 were obtained from the American Type Culture Collection (Rockville, Md.). H. capsulatum G217Bura5-23 (39) was provided by J. Woods (University of Wisconsin). Agrobacterium tumefaciens strain LBA1100 (4) was provided by C. van den Hondel (Leiden University, Leiden, The Netherlands) and grown at 28°C on Luria broth supplemented with 0.1% glucose. S. cerevisiae was grown on yeast extract-peptone-dextrose at 28°C. B. dermatitidis yeast was maintained on Middlebrook 7H10 medium containing oleic acid-albumin complex prior to transformation. For reporter assays, B. dermatitidis was grown as yeast at 37°C or as mycelia at 22°C in Histoplasma macrophage medium (HMM) (58). Conversion from yeast to mold was accomplished by transferring log-phase yeast cultures to 22 to 25°C. For studies of mycelia, conversion was allowed to proceed for approximately 8 weeks of growth at 22 to 25°C on an orbital shaker. H. capsulatum was grown on an orbital shaker in HMM or HMM supplemented with 100 μg of uracil/ml as yeast at 37°C in 5% CO2.
Plasmid constructions.
A base vector to allow fusion of promoter fragments to the E. coli lacZ coding region was made by deletion of the BamHI site downstream of lacZ to create pWLTΔ3′Bam. Promoter fragments were amplified from BAD1 and YPS3 using the vectors pWLT (41) and pY1 (YPS3 of H. capsulatum strain G217B in the vector pGEM-Teasy, provided by J. Woods, University of Wisconsin) as templates, respectively. A fragment of the S. cerevisiae ACT1 locus, representing bases 1940 to 2002 (GenBank accession number L00026.1), was amplified from genomic DNA. The URA5 promoter of H. capsulatum strain G184AS (from −311 to +87) was amplified from pWU20 (57), using primers N-XURAFOR (5′-ATGCGGCCGCTCTAGAAAGCTTTATATAAACTTGAGAG) and URABAMREV (5′-TAGGATCCATTGTAGCAATTCCCGCTGGGTC). Amplification of BAD1 promoter fragments was performed using forward primers in Table 1, with added 5′ NotI and XbaI sites (5′-AAGCGGCCGCTCTAGA-) and reverse primer DELTAREV (5′-TAGGATCCATTCTCCCGGTAGGTAGCTC). Amplification of YPS3 promoter fragments was performed using the forward primers in Table 1, including 5′ NotI, XbaI, BsrGI, and StuI sites (5′-AAGCGGCCGCTCTAGATGTACAAGGCCT-) and reverse primer YPSREV (5′-TAGGATCCATTTTTGCGGTTGATTATTT). Modified BAD1 and URA5 promoter fragments were created by amplification, either via splicing by overlap extension (SOE) or primer-induced sequence alterations (Table 2). Creation of the −741 AΔ::ACT1 and B ivs AΔ ACT1 promoter modifications required two-step SOE, in which two products were fused, and these fusion products were then spliced to a third fragment prior to cloning. Amplifications were performed using Elongase (Invitrogen Corp., Carlsbad, Calif.), and the primers used are listed in Table 2. Those creating the 5′ end of the promoter included additional 5′ NotI and XbaI sites.
TABLE 1.
Primers for amplification of BAD1 and YPS3 promoter truncation fragments
| Gene | Promotera | Forward primer sequenceb |
|---|---|---|
| BAD1 | −1101 | ACCCTGGTCGAATATTCCAGA |
| BAD1 | −741 | GAGAGTACGGGTCGTATATTT |
| BAD1 | B ivs A | CAGATGTTTCGATAAGAGGCC |
| BAD1 | ivs-box A | TACTACCCCTATCCGATAATG |
| BAD1 | box A | ATTGTGTGGATAGACGTCATA |
| BAD1 | No BA | AGGTATCAGCTAGCCGGCTTG |
| BAD1 | No TATA | GCTCTCGTCTTGCTCCTTTTT |
| BAD1 | No Start | CGCCCAGTCATTCGTTCATTT |
| YPS3 | −741 | AAGCTTTATTCAGCCGCAAGA |
| YPS3 | B ivs A | TAGATGTTTCGATAAGTCAAG |
| YPS3 | ivs-box A | ACAGCGCAGTGATGCAATAAT |
| YPS3 | box A | GTTGTTTAGACAGACGTCATA |
| YPS3 | 1/2 box A | TTGACTCTTGTCCATGTTTCT |
| YPS3 | No BA | TAGCATCAGTTAGCCTGCCCA |
Nomenclature based on areas of homology between the two promoters contained within the PCR product or the 5′ end of PCR product numbered relative to the transcriptional start site.
Primers had 5′ extensions (see Materials and Methods). The YPS3 − 741 product was obtained by binding to a site containing a partial primer match.
TABLE 2.
Primers for amplification of BAD1 and YPS3 modified promoter fragments
| Promotera | Primersb |
|---|---|
| −741 AΔ::ACT1 | GAGAGTACGGGTCGTATATTT*/ |
| GAAAGAAAGTACTCCGTCTGGATGTGCCGAATCAATAATGGAC; | |
| GTCCATTATTGATTCGGCACATCCAGACGGAGTACTTTCTTTC/ | |
| CAAGCCGGCTAGCTGATACCTATGGCTCCATCTTCCATGAAGGT; | |
| ACCTTCATGGAAGATGGAGCCATAGGTATCAGCTAGCCGGGCTTG/ | |
| TAGGATCCATTCTCCCGGTAGGTAGCTC | |
| B ivs AΔ::ACT1 | CAGATGTTTCGATAAGAGGCC*/ |
| GAAAGAAAGTACTCCGTCTGGATGTGCCGAATCAATAATGGAC; | |
| GTCCATTATTGATTCGGCACATCCAGACGGAGTACTTTCTTTC/ | |
| CAAGCCGGCTAGCTGATACCTATGGCTCCATCTTCCATGAAGGT; | |
| ACCTTCATGGAAGATGGAGCCATAGGTATCAGCTAGCCGGGCTTG/ | |
| TAGGATCCATTCTCCCGGTAGGTAGCTC | |
| Box A-PURA5 | TTGTGTGGATAGACGTCATATATTGTTCCGTTTTGGCTTA*/ |
| CATCACGGAGGGCATTTCCATCAACAGAAACATTGGCAAGAG; | |
| CTCTTGCCAATGTTTCTGTTGATGGAAATGCCCTCCGTGATG/ | |
| TAGGATCCATTGTAGCAATTCCCGCTGGGTC | |
| ACT1-PURA5 | TCCAGACGGAGTACTTTCTTTC*/ |
| CATCACGGAGGGCATTTCCATTGGCTCCATCTTCCATGAAGGT; | |
| ACCTTCATGGAAGATGGAGCCAATGGAAATGCCCTCCGTGATG/ | |
| TAGGATCCATTGTAGCAATTCCCGCTGGGTC | |
| A-CREΔ::sp | TTGTGTGGAGACTACGTGCATATTGTTCCGTTTTGGCTTA* |
| A-Sox-5Δ::sp | TTGTGTGGATAGACGTCATAGACTACGTGCTTTTGGCTTAGGCTTCTTGCCA* |
| A-MybΔ::sp | TTGTGTGGATAGACGTCATATATTGACTACGTGCGGCTTAGGCTCTTGCCAATGT* |
| A-NF-1Δ::sp | TTGTGTGGATAGACGTCATATATTGTTCCGTTTTGGCTTA*/ |
| ACAACAGAAACGCACGTAGTCGCCTAAGCCAAAACGGAACAA; | |
| TTGTTCCGTTTTGGCTTAGGCGACTACGTGCGTTTCTGTTGT/ | |
| TAGGATCCATTCTCCCGGTAGGTAGCTC | |
| A-AREB6Δ::sp | TTGTGTGGATAGACGTCATATATTGTTCCGTTTTGGCTTA*/ |
| ACAGCACGTAGTCTGGCAAGAGCCTAAGCCAAAAC; | |
| GTTTTGGCTTAGGCTCTTGCCAGACTACGTGCTGT/ | |
| TAGGATCCATTCTCCCGGTAGGTAGCTC | |
| A-CRE/MybΔ::sp | TTGTGTGGAGACTACGTGCATATTGACTACGTGCGGCTTAGGCTCTTGC* |
All vectors contain modified PBAD1 fragments, unless otherwise indicated. Replacements of predicted transcription factor binding sites are indicated by the site (CRE) or the protein (Sox-5, Myb, NF-1, and AREB6) that may bind it. ACT1 represents a 63-bp fragment of the S. cerevisiae actin locus (see Materials and Methods). Sp, 10-bp spacer.
Primers indicated by an asterisk had 5′ extensions (see Materials and Methods). Paired SOE primers are separated by a slash, and sets used together to generate the final promoter fragment are separated by a semicolon. The 10-base replacement sequences are underlined.
PCR products were digested with NotI and BamHI and cloned into pWLTΔ3′Bam in replacement of the large (∼2.7-kb) BAD1 upstream region, and their sequences were confirmed. The 2.5-kb BAD1 upstream region fused to lacZ in T-DNA vector pBTS92 (41) was modified to receive promoter fragments. The ClaI site outside of the lacZ coding region, and the XbaI site not immediately 5′ of the BAD1 promoter, were removed to create vector pBPR25. Promoters and a fused 5′ fragment of lacZ were removed by digestion of the pWLTΔ3′Bam-based vectors with XbaI and ClaI and cloned into the E. coli-A. tumefaciens-B. dermatitidis shuttle vector pBPR25.
Construction of H. capsulatum telomeric vectors necessitated reamplification of BAD1 promoter fragments using the forward primers listed in Table 1, with additional 5′ NotI, XbaI, BsrGI, and StuI sites (see above) and reverse primer DELTAREV. PCR products were cloned into the NotI and BamHI sites of pWLTΔ3′Bam and confirmed by sequencing. To prevent read-through transcription previously noted in telomeric reporter constructs in H. capsulatum (data not shown), the transcriptional terminator region from the Aspergillus nidulans trpC gene was amplified from pAN7-1 (37) with primers B-TRPCF (5′-AATGTACAACTTAACGTTACTGAAATCATC) and S-TRPCR (5′-AAAGGCCTAAGAAGGATTACCTCTAAACA), cloned and sequenced (pPJ56), and subcloned into the BsrGI-StuI sites upstream of the BAD1 and YPS3 promoters fused to lacZ. Generation of a vector for selection in H. capsulatum began with deletion of a PmeI fragment from pWU56, an EcoRI deletion derivative of pWU55 (57) (provided by J. Woods, University of Wisconsin). The Podospora anserina URA5 (PaURA5) marker from pWU55 was added into the EcoRI site, and a plasmid in which the marker would be transcribed towards the nearest PmeI site was selected for further modification (pPJ11). A kanamycin resistance (kanr) marker amplified from pBIN19 (5) using primers ESKAN-U (5′-GATATCACTAGTCGATAAACCCAGCGAACCATT) and ESKAN-L (5′-GATATCACTAGTCATCTAGGTACTAAAACAATT), with EcoRV and SpeI sites added at each end, was cloned into the EcoRV site to form pPJ11Kan. Terminator-promoter-lacZ-containing fragments were removed by digestion with XbaI and cloned into the SpeI site of pPJ11Kan in replacement of the kanr cassette. Clones were selected in which lacZ would be transcribed opposite the PaURA5 selectable marker. Telomeric repeats able to be exposed by PmeI digestion were isolated from pWU55 and cloned into the NotI site of each H. capsulatum-ready reporter vector.
Fungal transformation.
Transformation of B. dermatitidis using A. tumefaciens strain LBA1100 derivatives was performed as previously described (41, 48). Transformants were selected on HMM containing 100 μg of hygromycin B/ml. As the resulting plates were crowded with dense growth, transformant colonies were first patched and then streaked out for single-colony isolation on HMM containing 100 μg of hygromycin B/ml prior to inoculating liquid cultures for reporter analysis. This method of DNA delivery results in random insertion in the genome (48). Since the position of insertion can affect transgene expression, to allow comparison of transformant promoter activity among the vectors, eight transformants of each deletion vector were analyzed for β-galactosidase (LacZ) activity and the results were averaged. To reduce the effects of passage number, constructs to be compared were transformed simultaneously into the same recipient culture. H. capsulatum transformation with PmeI linearized plasmid DNA was accomplished using electroporation conditions described previously (57). Transformants were selected on HMM at 37°C with 5% CO2.
DNA analysis.
Alignment and sequence comparison of the BAD1 and YPS3 upstream regions was performed using ClustalW, v1.4, of the MacVector nucleotide analysis software package (Accelrys, Burlington, Mass.). Determinations of sequence identity were performed using short sequence files of the regions being compared. The reported percentage identity between these promoters at box B and the region containing a probe used in gel shift assays (24) required use of sequence files containing additional sequence beyond the alignment of these regions highlighted in Fig. 1. This was necessary to eliminate gaps created at the ends of these regions when the approximately 1.1 kb of upstream sequence was aligned.
As a test of reporter transfer, selected B. dermatitidis transformants were analyzed by Southern hybridization. Genomic DNA from B. dermatitidis was isolated using a liquid nitrogen grinding technique for cellular disruption, followed by organic extraction (21). For each sample, 2.5 μg of genomic DNA was separately digested with EcoRI, isolating a fragment containing the PBAD1- or PYPS3-lacZ fusion, and XbaI, which digests the T-DNA reporter vectors at a single site. Transforming plasmid DNAs digested with the same enzymes were included as controls. DNA analysis by Southern blot hybridization was performed using standard techniques (43). Blots were probed for lacZ using a BamHI fragment of pWLT (41). Probes for hybridizations were gel purified using the QIAquick gel extraction kit (QIAGEN, Inc., Valencia, Calif.) and radiolabeled using the Prime-a-Gene labeling system (Promega Corp., Madison, Wis.). Four transformants each for plasmids containing the No BA-lacZ and No Start-lacZ reporters (Table 1) were tested in this fashion to ensure that their relatively low level of β-galactosidase (LacZ) activity was not due to rearrangement of the promoter-reporter fusion. Results indicated that the fate of transforming DNA was integration at a single site, with multimerization of the input DNA. Hybridizing fusion-containing EcoRI fragments were of the expected sizes (data not shown).
Five other B. dermatitidis transformants whose β-galactosidase (LacZ) activity levels were dramatically lower than those of other transformants of the same vector were similarly analyzed (data not shown). Results indicated the partial transfer of a single copy of the T-DNA in three transformants, resulting in the truncation of the lacZ fusion. A fourth transformant with low LacZ activity gave a hybridizing PBAD1-lacZ fragment of the expected size, but only a single (nonmultimerized) copy of the transforming DNA was present in the genome. An additional transformant with low LacZ activity showed that transforming DNA was multimerized and the PYPS3-lacZ hybridizing fragment was of the expected size. In this case, the lack of LacZ activity may have resulted from point mutations or rearrangements not detected by Southern hybridization. In transformations of vectors containing partial BAD1 promoter replacements, three transformants showed low LacZ activity but were not analyzed by Southern hybridization. Since the fate of transforming DNA was not determined for all transformants, LacZ data from these eight aberrant transformants was included in Fig. 2 and 4 and in statistical analyses.
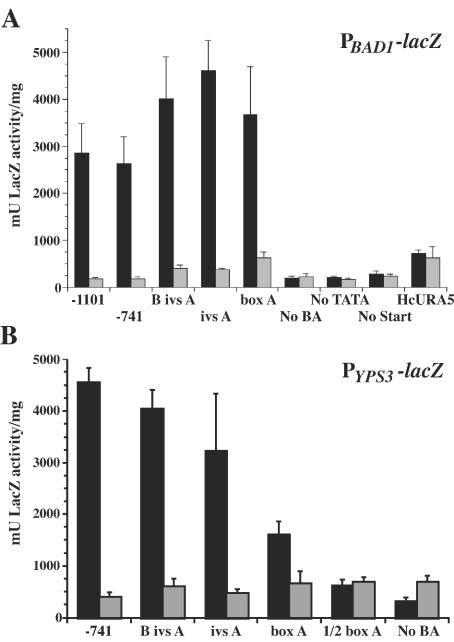
FIG. 2.
β-Galactosidase activity in B. dermatitidis PBAD1-lacZ, PYPS3-lacZ, and control PURA5-lacZ transformants. B. dermatitidis 26199 transformed with lacZ fusion vectors was analyzed in both the yeast and mycelial phase for β-galactosidase activity using an o-nitrophenyl β-d-galactopyranoside assay. Activity levels for yeast are shown as black bars, and the levels for mold are shown as shaded bars. Test promoters driving lacZ expression are indicated on the x axis according to the designations indicated in Table 1 and/or Fig. 2. Eight independent transformants of each vector were analyzed in three experiments, and the results were averaged. Values are reported in milliunits of β-galactosidase activity per milligram of protein and represent the mean ± standard error. (A) β-Galactosidase activity of transformants of BAD1 promoter truncations fused to lacZ. (B) β-Galactosidase activity of transformants of YPS3 promoter truncations fused to lacZ.
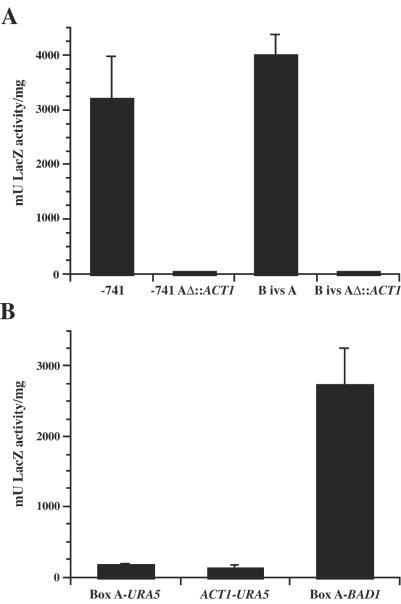
FIG. 4.
β-Galactosidase activity in B. dermatitidis transformants of reporter fusions containing modified BAD1 or URA5 promoters. B. dermatitidis 26199 transformed with lacZ fusion vectors was analyzed in the yeast phase for β-galactosidase activity using an o-nitrophenyl β-d-galactopyranoside assay. Test promoters driving lacZ expression are indicated on the x axis according to the designations indicated in the Results section. Eight independent transformants of each vector were analyzed in three experiments, and the results were averaged. Values are reported as milliunits of β-galactosidase activity per milligram of protein and represent the mean ± standard error. (A) β-Galactosidase activities of transformants of constructs in which the BAD1 promoter is fused to lacZ, comparing activities from box A-containing vectors versus those in which box A was replaced with sequence from the actin gene (ACT1) of S. cerevisiae. (B) β-Galactosidase activities of transformants harboring the URA5 promoter fused to lacZ, with the upstream addition of either the box A region of the BAD1 promoter or a similarly sized fragment of the ACT1 coding region. The activity of the BAD1 box A-lacZ construct is included for comparison.
H. capsulatum transformants were screened by Southern hybridization to select clones in which the transforming DNA was not integrated into the genome. H. capsulatum genomic DNA was isolated according to the published protocol of Woods and Goldman (55). Undigested genomic DNA samples were probed with an EcoRI fragment of pWU56 containing the PaURA5 gene. Episomal vectors were noted as appearing in multiple sizes, including the size of the linearized transforming DNA, a larger form that corresponds to the size of a linear, dimerized vector, and sometimes appearing as both forms in the same transformant. Transformants selected for analysis contained dimerized vectors, the predominant fate of the transforming DNA. To determine the relative copy number of episomes in H. capsulatum transformants, genomic DNA was serially diluted in round-bottomed wells of a microtiter plate. DNA was denatured by NaOH (final concentration, 0.1 N), with incubation at 37°C for 5 min, followed by neutralization by addition of SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a final concentration of 6×. DNA was transferred to nytran using a 96-well vacuum blotting manifold (Schleicher & Schuell, Keene, N.H.). Samples to be compared (e.g., all PBAD1-lacZ transformants) were included on the same membrane. Membranes were first hybridized with the PaURA5 probe to determine the relative amounts of transforming DNA per well. Blots were then stripped and reprobed with a 700-bp XhoI-SmaI fragment of the H. capsulatum URA5 (HcURA5) gene (isolated from pWU20) to determine the relative amounts of genomic DNA/well. Hybridization signals were quantified using a Storm blot imaging system and ImageQuant for Macintosh software (Amersham Biosciences, Piscataway, N.J.). The PaURA5/HcURA5 hybridization signal was determined for each well. For each clone, this derived signal was averaged across wells of the dilution series within linearity. The average for each clone was divided by the lowest average value of a clone in its comparison group (PBAD1-lacZ or PYPS3-lacZ transformants) to give the relative copy number factor, which varied from 1 to 3.5 (data not shown).
Quantitation of β-galactosidase activity.
Histoplasma yeast and B. dermatitidis yeast and mycelial cultures were grown in HMM and assayed for β-galactosidase activity as previously described (41). β-Galactosidase activity was normalized to the protein content of extracts, which were quantified using 10 μl of supernatant in the BCA microtiter plate assay (Pierce Chemical Co., Rockford, Ill.). Activity from H. capsulatum transformants was then also normalized to the relative copy number of vector by dividing the protein-normalized value by the relative copy number factor. Pairwise comparisons of β-galactosidase activity values were performed using the two-sided Wilcoxon test.
RESULTS
Activity of PBAD1-lacZ fusions in B. dermatitidis.
To map functional regions of the BAD1 promoter, a series of vectors containing nested deletions of the BAD1 promoter fused to the E. coli lacZ coding region were introduced into B. dermatitidis by Agrobacterium-mediated transformation. Constructs were designed to test the contributions of boxes B and A, as well as upstream sequences, to BAD1 transcription (Fig. 1A). Transformants were first analyzed for yeast-phase LacZ activity (Fig. 2A). Those harboring constructs containing BAD1 promoter sequence down to and including the 63-nucleotide box A sequence possessed robust yeast-phase reporter activity. Within this set, there were no significant differences in LacZ expression. In contrast, transformants with fusions lacking box A yielded LacZ activity consistent with those of control PBAD1-lacZ vectors that lacked the TATA signal (No TATA-lacZ) or transcriptional start site (No Start-lacZ). This represents the background transcriptional activity in transformants of promoterless lacZ vectors. The sharp drop in LacZ activity between vectors containing box A and those lacking this region is statistically significant (P < 0.01).
Reporter activity was also quantified for each transformant grown as a mold. The level of mold-phase LacZ activity detected was similar for transformants of all constructs, including the No TATA-lacZ and No Start-lacZ controls (Fig. 2A). These data point to the presence of a positively acting region contributing to yeast-phase activity. Transformants of a control vector in which the H. capsulatum URA5 promoter was fused to lacZ showed similar levels of activity in yeast and mold phases.
Activities of PYPS3-lacZ fusions in B. dermatitidis.
The upstream region of the YPS3 gene of H. capsulatum was also analyzed in B. dermatitidis using a set of constructs containing nested YPS3 promoter deletions fused to lacZ. To determine if the areas of homology between the BAD1 and YPS3 promoters were functionally conserved, these constructs were designed to match the set of BAD1 promoter deletions (Fig. 1B). Deletions of the YPS3 promoter showed a slightly different trend than that seen for the BAD1 promoter. While transformants of the PBAD1-lacZ fusions showed consistent LacZ activity from all constructs containing box A, those of the PYPS3-lacZ fusions displayed a stepwise decline in activity as the YPS3 promoter driving lacZ transcription was successively deleted from the 5′ end (Fig. 2B). For example, reporter activity was significantly greater from the construct containing both boxes B and A (B ivs A-lacZ) than from that containing box A alone (box A-lacZ; P < 0.001). Though less robust than the reporter activity of longer YPS3 upstream regions fused to lacZ, the activity of box A-lacZ transformants was still much greater than that of No BA-lacZ transformants (P < 0.001). As was the case with the BAD1 promoter, the loss of box A from the YPS3 promoter reduced reporter transcription to background levels. An additional similarity between transformants of the BAD1 and YPS3 reporter constructs was the lack of mold-phase LacZ activity. Thus, as with BAD1, a positively acting region is involved in the yeast-phase transcription of YPS3 in B. dermatitidis transformants.
An extra construct was included in the analysis of the YPS3 promoter in which the box A region was truncated. This vector lacked the 5′ half of box A, leaving only the 3′ half and downstream sequence fused to lacZ (1/2 box A-lacZ) (Fig. 1B). Strains harboring this fusion gave yeast-phase LacZ activity between that of box A-lacZ and that of No BA-lacZ transformants (Fig. 2B). The decrease in activity of transformants of box A-lacZ vectors versus that of 1/2 box A-lacZ vectors was highly significant (P < 0.001). Although the level of yeast-phase LacZ activity of 1/2 box A-lacZ was greater than that of the No BA-lacZ fusion (P < 0.05), it had returned to the background levels seen in mold-phase transformants. This suggests that box A is an essential component of the minimal promoter for YPS3.
β-Galactosidase activities of BAD1 and YPS3 promoter truncations in H. capsulatum.
A subset of the BAD1 and YPS3-nested promoter deletions fused to lacZ was tested in H. capsulatum to determine if the upstream regions of these genes function in a manner similar to that seen in B. dermatitidis. In H. capsulatum, positional effects necessitating the screening of multiple transformants for each reporter construct can be avoided by the use of vectors that can remain episomal (56). To this end, reporter constructs were cloned into telomeric vectors. Yeast-phase H. capsulatum transformants in which the transforming vectors remained episomal were analyzed for LacZ activity. Although quality genomic DNA could not be isolated from mycelium-phase cultures, cells which had undergone transition to mold were converted back to yeast and reexamined to ensure that the transforming DNA remained episomal. In nearly all samples tested, the transforming DNA had integrated into the genome after conversion, potentially affecting the level of reporter transcription (data not shown). As such, LacZ activity was quantified exclusively from yeast-phase cultures.
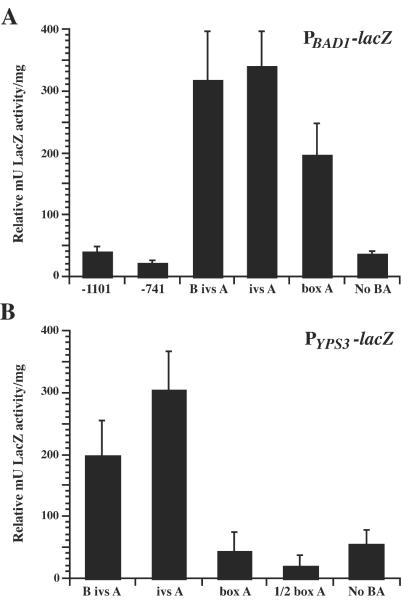
Overall, transcriptional activity from PBAD1-lacZ fusions demonstrated a similar requirement for box A in yeast-phase H. capsulatum, but some fusions gave unexpected results. Transformants containing the longest BAD1 upstream regions (−1101-lacZ and −741-lacZ) expressed background levels of LacZ activity, equivalent to that in untransformed H. capsulatum (Fig. 3A; data not shown). Those with BAD1 upstream regions from box B (B ivs A-lacZ) to box A (box A-lacZ) possessed LacZ activity, though at a much lower level than that of matched B. dermatitidis transformants. As with the matched integrated constructs in B. dermatitidis, however, there was no significant difference in LacZ activity between these constructs. Similarly, the No BA-lacZ transformant did not express LacZ activity above background levels, indicating that the minimal promoter region for BAD1 in H. capsulatum is the same as that in B. dermatitidis and requires box A.
FIG. 3.
β-Galactosidase activity in H. capsulatum PBAD1-lacZ and PYPS3-lacZ transformants. Selected yeast-phase H. capsulatum strain G217B transformants in which the lacZ fusion vectors were episomal were analyzed for β-galactosidase activity using an o-nitrophenyl β-d-galactopyranoside assay. Independent transformants analyzed are indicated along the x axis according to the promoter fragment driving lacZ expression (see Fig. 2). Transformants were analyzed in three experiments, and the results were averaged. Values, scored in milliunits of β-galactosidase activity per milligram of protein, were normalized to the relative vector copy number per genome (see Materials and Methods) and are reported as relative milliunits of galactosidase activity per milligram of protein, representing the mean ± standard error. (A) β-Galactosidase activities of transformants of BAD1 promoter truncations fused to lacZ. (B) β-Galactosidase activities of transformants of YPS3 promoter truncations fused to lacZ.
H. capsulatum PYPS3-lacZ activity differed in some aspects from PBAD1-lacZ activity in this fungus. The longest PYPS3-lacZ construct (−741-lacZ) produced a high level of LacZ activity (2,200 relative mU/mg). Also, while transformants of vectors containing B ivs A-lacZ and ivs A-lacZ expressed LacZ activity, LacZ activity was reduced to background levels in the box A-lacZ transformant (Fig. 3B).
Box A is necessary but not sufficient for transcriptional activation in B. dermatitidis.
Serial truncations of the BAD1 promoter fused to lacZ demonstrated that inclusion of box A was necessary for minimal BAD1 promoter activity. To determine whether sequences upstream of box A could compensate for a loss of this region, the 63-bp box A sequence was replaced within constructs containing 741 or 361 bp of BAD1 promoter upstream of the transcriptional start site, fused to lacZ. Box A (−175 to −237) was replaced with a 63-bp fragment amplified from the coding region of the S. cerevisiae actin gene. Resulting constructs containing −741 AΔ::ACT1-lacZ and B ivs AΔ::ACT1-lacZ, and −741-lacZ and B ivs A-lacZ matched controls, were introduced into B. dermatitidis, and transformants were analyzed for yeast-phase LacZ activity. Replacement of box A in either promoter background resulted in a loss of LacZ activity, indicating that this region is necessary for transcription from the BAD1 promoter (Fig. 4A).
Since box A is required for activating transcription from the BAD1 promoter in the yeast phase, this sequence was analyzed for its ability to confer transcriptional activation on a foreign promoter in B. dermatitidis. For this purpose, we employed the H. capsulatum URA5 promoter, which produces a low level of activity in B. dermatitidis (Fig. 2A). Box A, or a 63-bp fragment of the S. cerevisiae actin gene, was fused upstream of a URA5 promoter fragment containing 175 bp upstream of the transcriptional start site, driving lacZ transcription. The placement of box A, or the actin fragment, relative to the transcriptional start site was conserved between these constructs and the BAD1 box A-lacZ vector. No statistically significant increase in reporter activity was observed from yeast-phase B. dermatitidis transformants of the box A-PURA5-lacZ vector over that for those harboring the ACT1-PURA5-lacZ vector (Fig. 4B). Both groups possessed relatively low levels of promoter activity compared to that of the BAD1 box A-lacZ transformants. These data indicate that the box A region of the BAD1 promoter is not sufficient for transcriptional activation of the H. capsulatum URA5 promoter.
Analysis of BAD1 upstream region for transcription factor binding sites.
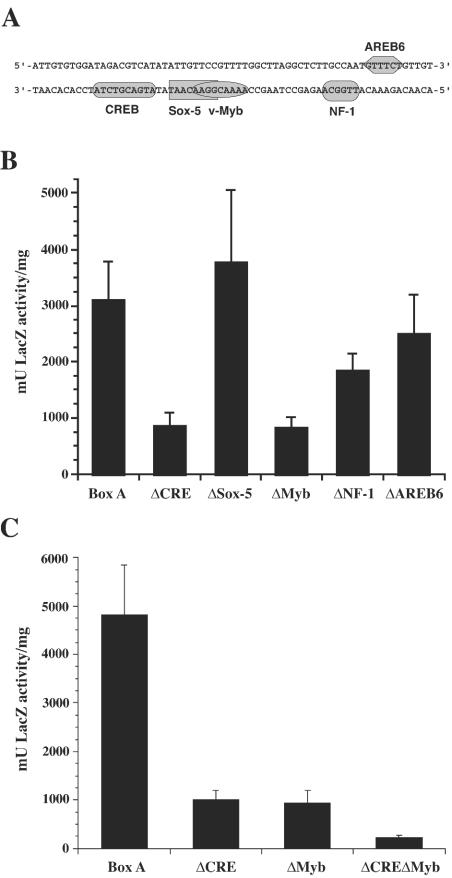
Since the minimal BAD1 promoter region in B. dermatitidis requires box A, we sought to determine if this region contained any potential binding sites for known transcription factors. To this end, the box A sequence was subjected to analysis by MatInspector (v3.0) (http://www.genomatix.de) (38), comparing it to both the fungal and vertebrate transcription factor binding site libraries. The most probable matches for transcription factors binding this region include a cAMP-responsive element (CRE) and binding sites for the mammalian transcription factors Sox-5, v-Myb, nuclear factor 1 (NF-1), and AREB6 (Fig. 5A). Although fungal homologs to a subset of these factors have been identified, the presence of consensus binding sites may simply suggest that transcription factors with similar DNA binding domains could interact at these locations. In S. cerevisiae, three ATF/CREB proteins that associate with CRE have been identified, including both repressors and activators of transcription (51, 19). Sox-5 is a member of a protein family involved in governing cell fate decisions during embryogenesis. These proteins contain a high-mobility group (HMG)-type DNA binding domain that associates with the minor groove and bends DNA, allowing establishment of a local chromatin structure favorable to transcription (36). HMG-box proteins have been identified in fungi, including some factors regulating mating in Schizosaccharomyces pombe (20). Overlapping the Sox-5 binding site was a match with the consensus v-Myb binding site (6). The Myb family of proteins includes transcriptional activators found in many eukaryotes, including plants, mammals, fungi, and Drosophila. Myb family members share a repeated helix-turn-helix DNA binding domain (50). Although the ubiquitous nuclear factor 1 has not been identified in any fungi, its binding site has been noted in another fungal promoter (34, 47). AREB6 is a transcription factor possessing two zinc-finger DNA binding domains and is important for tissue-specific expression and development in vertebrates (25). It can act as either a positive or a negative regulator of gene expression, depending on the concentration and promoter context. A match to the consensus binding site for the N-terminal zinc finger is present in box A, but AREB6 is proposed to only bind this sequence when in conjunction with a second consensus site (CACCTGT) and in such context act as a negative regulator. This second consensus site was not identified in or adjacent to the box A region of the BAD1 promoter. The box A region of YPS3 contains several identical binding elements, including the CRE and a partial AREB6 site.
FIG. 5.
Possible transcription factor binding sites within the box A region and their effect on transcription from the BAD1 promoter. (A) Transcription factor binding sites in which the four core bases of the consensus were found at 100% identity and the entire consensus was 90% or greater by MatInspector matrix similarity. Sites are named for the proteins recruited, shown in shaded boxes over the box A sequence. (B and C) β-Galactosidase activities of yeast-phase B. dermatitidis transformants of BAD1 wild-type and modified box A-lacZ vectors. On the x axes, modified box A promoters, in which a 10-bp region is replaced, are indicated by a delta symbol, followed by the predicted transcription factor binding site (CRE), or the protein (Sox-5, Myb, NF-1, and AREB6) that may bind the site found within the altered region. Eight independent transformants of each vector were analyzed in three experiments, and the results were averaged. Values are reported as milliunits of β-galactosidase activity per milligram of protein and represent the mean ± standard error. Panels B and C represent analyses of transformant sets generated in separate experiments.
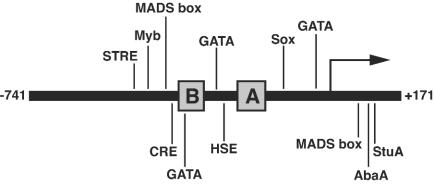
Numerous putative transcription factor binding sites were identified elsewhere within the upstream region of the BAD1 promoter (Fig. 6). From the 5′ end of box B (−361) to 741 bp upstream of the transcriptional start site are additional consensus matches for v-Myb and CREB binding sites. Also of note, a stress response element (STRE) (16) and a MADS box factor binding site similar to those which bind Rlm1 and Mcm1 of S. cerevisiae and the human myocyte enhancer factor 2 (Mef2a) were identified in this same region (2, 32, 53). Box B contains a GATA site (44), and the intervening sequence between boxes B and A contains a second GATA site and a heat shock element (HSE) homologous to those of S. cerevisiae (14). Downstream of box A, additional Sox, GATA, and MADS box factor binding sites were identified, as well as binding sites for the A. nidulans conidiation-regulating transcription factors AbaA and StuA (3, 15).
FIG. 6.
Locations of putative transcription factor binding sites of the BAD1 promoter outside of box A. Sites are labeled for the type of protein or proteins recruited (Myb, MADS box, GATA, Sox, AbaA, and StuA) or the sequence element itself (STRE and HSE) and are shown in their relative positions on the BAD1 promoter map. Binding sites listed share 100% identity with the four core bases of the transcription factor consensus binding site, and the entire consensus was 90% or greater by MatInspector matrix similarity. Note that the MADS box transcription factor binding sites were identified as matching that of the mammalian factor Mef2a (32). The BAD1 transcriptional start site (+1) is shown as an arrow, and boxes B and A are shown as shaded areas. Actual locations of the binding sites are as follows: STRE, −488 to −484; Myb, −463 to −458; CRE, −391 to −384; MADS box, −411 to −402 and +69 to +78; GATA, −352 to −347, −310 to −305, and −54 to −49; HSE, −277 to −264; Sox, −146 to −140; AbaA, +99 to +104; StuA, +114 to +121.
Two regions influence transcriptional activation by box A.
The impact of putative transcription factor binding sites within box A on transcription from the BAD1 promoter was assessed by modification of the BAD1 box A-lacZ construct, in which a 10-bp sequence encompassing each putative binding site was replaced. B. dermatitidis transformants of five modified constructs and the control box A-lacZ vector were analyzed for LacZ activity in the yeast phase (Fig. 5B). Removal of the putative Sox-5, NF-1, or AREB6 binding sites (replacing −217 to −208, −194 to −185, and −186 to −177, respectively) did not significantly affect reporter activity compared to the box A-lacZ control. However, transformants in which −228 to −219 or −213 to −204, encompassing the predicted CRE and the Myb binding site, were replaced displayed a significant reduction in LacZ activity relative to that with box A-lacZ (P < 0.005). Although showing reduced transcriptional activity, neither region alone was essential for the transcriptional activation conferred by box A on the BAD1 promoter in B. dermatitidis. However, replacement of both the CRE and the Myb binding site within box A resulted in a dramatic reduction in transformant LacZ activity compared to that of the wild-type box A-lacZ or either single site replacement reporter (P < 0.001) (Fig. 5C). Thus, the upstream sequences from −228 to −219 and −213 to −204, containing predicted CRE and Myb binding sites, together are critical in transcriptional activation of the BAD1 promoter.
DISCUSSION
Herein, we demonstrate the presence of a region conferring transcriptional activation of both BAD1 and YPS3 in the yeast phase. Transcription of the H. capsulatum yeast-phase-specific factor CBP1 is also activated in the yeast phase, although it does not show sequence similarity to the upstream regions of YPS3 or BAD1. Although both CBP1 and YPS3 are upregulated in the yeast phase, H. capsulatum may possess at least two distinct mechanisms for regulating these phase-specific genes.
We found no evidence for repression of BAD1 transcription in the mold phase. The BAD1 promoter contains p30 M binding sites (also present in the YPS3 promoter) at positions −20, +19, and +47 relative to the transcriptional start site (41). In studies of H. capsulatum, Abidi et al. detected a protein which bound these sites that is present only in nuclear extracts of mycelia, which led them to hypothesize that YPS3 expression is negatively regulated in the mycelial phase (1). None of the BAD1 promoter truncations fused to lacZ yielded activity in mold-phase cells, as one might expect from deletion of a repressor. However, since box A is upstream of the p30 M binding sites and is required for activation of transcription, our approach of serially truncating the BAD1 promoter 5′ to 3′ may not have been suitable for uncovering deregulated, mold-phase reporter expression. Determining whether the TCC motif regulates BAD1 transcription may require deletions of p30 M sites within the context of a longer promoter including box A. Thus, while box A is required for transcriptional activation, p30 M sites could still contribute to regulation of BAD1 production. Activation of transcription in yeast and repression in mold would likely result in tightly regulated expression of this essential virulence factor.
Perhaps the most significant finding from these studies is that the box A region conserved between yeast-phase-specific genes BAD1 and YPS3 is a required element of the minimal promoter region for BAD1 expression in B. dermatitidis and its heterologous expression in H. capsulatum. This 63-nucleotide box A region is also essential to the minimal promoter region for YPS3 in B. dermatitidis. Interestingly, the YPS3 box A-lacZ fusion was inactive in H. capsulatum transformants. Since neither gene is shared in the reciprocal fungus, these findings of conserved regulation suggest that the transcriptional machinery is shared in these closely related fungi, though subtle differences in composition or sequence element recognition may exist. This could account for the unexpected silencing and hyperactivation seen only in select H. capsulatum reporter transformants.
DNA sequence analysis identified putative transcription factor binding sites within box A, which were tested functionally, and other sites outside of this region. Replacement of the sequence encompassing the CRE within the BAD1 box A-lacZ vector resulted in a reduction in transcriptional activity in B. dermatitidis transformants. CRE-binding protein (CREB) is ubiquitously expressed. In eukaryotes, protein kinases phosphorylate CREB in response to an increase in cAMP levels, enabling transactivation functions (7). CREB proteins in S. pombe are targets of signal transduction pathways for handling osmotic stress and sexual development (8). Phosphorylated CREB protein recruits CREB-binding protein (CBP), which is a general coactivator molecule able to associate with several transcription factors. One mode by which CBP is thought to mediate CREB-dependent transcriptional activation is by acting as a bridging molecule, linking the DNA-bound CREB to the general transcription factor TFIIB (46). CBP is also an acetyltransferase. This activity can assist in chromatin remodeling to enhance transcriptional activity or can affect the function of other transcription factors by their direct acetylation (7).
Removal of the predicted Myb binding site from the box A-lacZ vector similarly resulted in a loss in transcriptional activation. The Myb protein family members share a DNA binding motif made up of two to three repeats, each forming a helix-turn-helix structure (33). Vertebrate Myb proteins can also associate with the transactivation domain of CBP, using the same region that binds CREB (46). Transcriptional activation by Myb proteins may be mediated in much the same way as CREB-induced transactivation. Although no homolog of CBP has been identified in S. cerevisiae, tests of v-Myb molecules mutated in the activation domain which binds CBP showed an effect on transcriptional activity in a yeast model. This finding suggests the possibility of a functional yeast CBP homolog that is involved in Myb-mediated transactivation (52). Recently, a Myb-like protein was identified in Giardia lamblia. Giardia, like dimorphic fungi, requires cellular differentiation for its success as a pathogen. In this protozoan, gMyb2 was shown to be important for the expression of the encystation-specific gene gbpi-b, encoding glucosamine-6-phosphate isomerase B, the first enzyme in the biosynthetic pathway for synthesis of cyst wall polysaccharides (49). The binding site for gMyb2 was found in the promoters of four encystation-induced genes. The fungus A. nidulans also utilizes a Myb domain containing transcription factor for inducing genes involved in a cellular shape change. In this case, the transcription factor FlbD is required for early conidiophore production (54). Since Myb family members are involved in regulation of differentiation events in diverse systems, from vertebrates (hematopoiesis) (33) and plants (hair cell formation in Arabidopsis) (28) to fungi and protozoans, it is not difficult to envision a role for a Myb-like protein of B. dermatitidis in the modulation of the yeast-phase-specific transcription of BAD1.
Loss of both the CRE and Myb binding site within box A resulted in a substantial reduction in reporter activity compared to replacement of either single site, indicating that these two regions together are critical for transcriptional activation of the BAD1 locus. It is of note that the YPS3 1/2 box A-lacZ construct, which gave equivalent levels of LacZ activity in yeast- and mold-phase transformants (Fig. 2B), is lacking the regions corresponding to the CREB, Sox-5, and v-Myb binding sites within box A of BAD1. Although both CREs and Myb binding sites were found upstream of box A, these sites were unable to compensate for the loss of box A in the −741 AΔ::ACT1-lacZ and B ivs AΔ::ACT1-lacZ vectors.
The binding sites of other transcription factors implicated in governing gene expression during changes in cellular morphology were identified outside of box A (Fig. 6). MADS-box transcription factor binding sites found in the BAD1 promoter shared homology to the consensus binding site for the S. cerevisiae protein Mcm1p, whose C. albicans homolog was shown to be important for morphogenesis (42). Binding sites for the A. nidulans conidiation regulators AbaA and StuA were also identified in the BAD1 upstream. The processes of conidia generation and yeast formation from fungi of a filamentous morphotype share similarities. Both require the coupling of nuclear division with cytokinesis to go from a multicellular mycelium to single cells. Conidiation requires the initial step of generating unicellular forms (metulae and phialides) that give rise to conidia. The formation of these structures is governed by several transcription factors, including AbaA and StuA (30, 29). In the dimorphic fungus Penicillium marneffeii, we see the linkage between conidiation and yeast formation, since AbaA was shown to be required for both processes (9). MADS-box transcription factor, AbaA, and StuA binding sites were found downstream of box A. Since this region was included in the BAD1 box A-lacZ construct, these sites could contribute to the level of transcriptional activity seen in its transformants. The inability of box A alone to stimulate transcriptional activation of the H. capsulatum URA5 promoter suggests that elements downstream of box A may contribute to transcriptional activation from the BAD1 promoter.
The identification of HSE in the BAD1 upstream was surprising, since heat shock responses are generally thought of as reactions to a stress on the cells, rather than a condition for which an organism has a specially adapted morphotype. It is known, however, that the heat shock factor inducing transcription of heat shock proteins in S. cerevisiae is encoded by an essential gene, indicating that heat shock proteins are necessary for survival under normal growth conditions. STRE is found in nearly 200 potential STRE-regulated genes in yeast. Differing patterns of expression of stress-induced genes containing STRE are thought to be due to promoter context. The combination of the HSE with the STRE is found in at least two yeast genes, SSA3, which encodes an Hsp70 involved in prevention of protein aggregation, and HSP104, which has been shown to rescue heat-inactivated proteins (16). The HSE and STRE are both found in the BAD1 upstream region. Although these elements were found upstream of the minimal promoter, they may still play a role in regulation of BAD1 expression. Utilization of these elements may be responsible for the rapid onset of BAD1 gene expression after morphogenesis is stimulated by switching the cells from 25 to 37°C, while other factors may be involved in maintenance of BAD1 transcription in stable yeast-phase cells.
Acknowledgments
This work was supported by NIH grants AI035681 and AI50882 from the USPHS and by a Burroughs Wellcome Fund Scholar Award in Molecular Pathogenic Mycology (B.S.K.).
We thank Jens Eickhoff of the Department of Biostatistics and Medical Informatics, University of Wisconsin, for assistance with statistical analyses.
REFERENCES
- 1.Abidi, F. E., H. Roh, and E. J. Keath. 1998. Identification and characterization of a phase-specific, nuclear DNA binding protein from the dimorphic pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:3867-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianopoulos, A., and W. E. Timerlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beijersbergen, A., A. den Dulk-Ras, R. A. Schilperoort, and P. J. J. Hooykaas. 1992. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324-1327. [DOI] [PubMed] [Google Scholar]
- 5.Bevan, M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12:8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biedenkapp, H., U. Borgmeyer, A. E. Sippel, and K. H. Klempnauer. 1988. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335:835-837. [DOI] [PubMed] [Google Scholar]
- 7.Blobel, G. A. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 8.Borgnes-Walmsley, M. I., and A. R. Walmsley. 2000. cAMP signaling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8:133-141. [DOI] [PubMed] [Google Scholar]
- 9.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2000. The abaA homologue of Penecillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol. Microbiol. 38:1034-1047. [DOI] [PubMed] [Google Scholar]
- 10.Bradsher, R. W. 1996. Histoplasmosis and blastomycosis. Clin. Infect. Dis. 22:S102-S111. [DOI] [PubMed] [Google Scholar]
- 11.Brandhorst, T., M. Wüthrich, B. Finkel-Jimenez, and B. Klein. 2003. A C-terminal EGF-like domain governs BAD1 localization to the yeast surface and fungal adherence to phagocytes, but is dispensable in immune modulation and pathogenicity of Blastomyces dermatitidis. Mol. Microbiol. 48:53-65. [DOI] [PubMed] [Google Scholar]
- 12.Brandhorst, T. T., M. Wüthrich, T. Warner, and B. S. Klein. 1999. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J. Exp. Med. 189:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burg, E. F., III, and L. H. Smith, Jr. 1994. Cloning and characterization of bys1, a temperature-dependent cDNA specific to the yeast phase of the pathogenic dimorphic fungus Blastomyces dermatitidis. Infect. Immun. 62:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J., and D. S. Pederson. 1993. A distal heat shock element promotes the rapid response to heat shock of the HSP26 gene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:7442-7448. [PubMed] [Google Scholar]
- 15.Dutton, J. R., S. Johns, and B. L. Miller. 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16:5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24:469-486. [DOI] [PubMed] [Google Scholar]
- 17.Finkel-Jimenez, B., M. Wüthrich, T. Brandhorst, and B. S. Klein. 2001. The WI-1 adhesin blocks phagocyte TNF-α production, imparting pathogenicity on Blastomyces dermatitidis. J. Immunol. 166:2665-2673. [DOI] [PubMed] [Google Scholar]
- 18.Finkel-Jimenez, B., M. Wüthrich, and B. S. Klein. 2002. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-α production through TGF-β-dependent and -independent mechanisms. J. Immunol. 168:5746-5755. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Gimeno, M. A., and K. Struhl. 2000. Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol. Cell. Biol. 20:4340-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94-100. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, L. H., and B. S. Klein. 1997. Transforming DNA integrates at multiple sites in the dimorphic fungal pathogen Blastomyces dermatitidis. Gene 186:219-226. [DOI] [PubMed] [Google Scholar]
- 22.Hung, C.-Y., N. M. Ampel, L. Christian, K. R. Seshan, and G. T. Cole. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect. Immun. 68:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatov, A., and E. J. Keath. 2002. Gel shift assay of nuclear extracts from Histoplasma capsulatum demonstrates the presence of several DNA binding proteins. Infect. Immun. 70:2238-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda, K., and K. Kawakami. 1995. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur. J. Biochem. 233:73-82. [DOI] [PubMed] [Google Scholar]
- 26.Keath, E. J., and F. E. Abidi. 1994. Molecular cloning and sequence analysis of yps-3, a yeast-phase-specific gene in the dimorphic fungal pathogen Histoplasma capsulatum. Microbiology 140:759-767. [DOI] [PubMed] [Google Scholar]
- 27.Klein, B. S., and S. L. Newman. 1996. Role of cell surface molecules on Blastomyces dermatitidis in host:pathogen interactions. Trends Microbiol. 4:246-251. [DOI] [PubMed] [Google Scholar]
- 28.Martin, C., and J. Paz-Ares. 1997. MYB transcription factors in plants. Trends Genet. 13:67-73. [DOI] [PubMed] [Google Scholar]
- 29.Miller, K. Y., J. Wu, and B. L. Miller. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6:1770-1782. [DOI] [PubMed] [Google Scholar]
- 30.Mirabito, P. M., T. H. Adams, and W. E. Timberlake. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859-868. [DOI] [PubMed] [Google Scholar]
- 31.Newman, S. L., S. Chaturvedi, and B. S. Klein. 1995. The WI-1 antigen on Blastomyces dermatitidis yeasts mediates binding to human macrophage CD18 and CD14 receptors. J. Immunol. 154:753-761. [PubMed] [Google Scholar]
- 32.Nurrish, S., and R. Treisman. 1995. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 15:4076-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, I.-H., and E. P. Reddy. 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017-3033. [DOI] [PubMed] [Google Scholar]
- 34.Paonessa, G., F. Gounari, R. Frank, and R. Cortese. 1988. Purification of a NF1-like DNA-binding protein from rat liver and cloning of the corresponding cDNA. EMBO J. 7:3115-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel, J. B., J. W. Batanghari, and W. E. Goldman. 1998. Probing the yeast phase-specific expression of the CBP1 gene in Histoplasma capsulatum. J. Bacteriol. 180:1786-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penvy, L. H., and R. Lovell-Badge. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7:338-344. [DOI] [PubMed] [Google Scholar]
- 37.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from E. coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 38.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Retallack, D. M., E. L. Heinecke, R. Gibbons, G. S. Deepe, Jr., and J. P. Woods. 1999. The URA5 gene is necessary for Histoplasma capsulatum growth during infection of mouse and human cells. Infect. Immun. 67:624-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney, P. J., and B. S. Klein. 2002. Linking fungal morphogenesis with virulence. Cell. Microbiol. 3:127-138. [DOI] [PubMed] [Google Scholar]
- 41.Rooney, P. J., T. D. Sullivan, and B. S. Klein. 2000. Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism. Mol. Microbiol. 39:875-889. [DOI] [PubMed] [Google Scholar]
- 42.Rottmann, M., S. Dieter, H. Brunner, and S. Rupp. 2003. A screen in Saccharomyces cerevisiae identified CaMCM1, an essential gene in Candida albicans crucial for morphogenesis. Mol. Microbiol. 47:943-959. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Scazzocchio, C. 2000. The fungal GATA factors. Curr. Opin. Microbiol. 3:126-131. [DOI] [PubMed] [Google Scholar]
- 45.Sebghati, T. S., J. T. Engle, and W. E. Goldman. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1368-1372. [DOI] [PubMed] [Google Scholar]
- 46.Shimikama, N., J. Lyon, and N. E. La Thangue. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230-236. [DOI] [PubMed] [Google Scholar]
- 47.Soll, D. R. 1997. Gene regulation during high-frequency switching in Candida albicans. Microbiology 143:279-288. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan, T. D., P. J. Rooney, and B. S. Klein. 2002. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot. Cell 1:895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, C.-H., D. Palm, A. G. McArthur, S. G. Svärd, and F. D. Gillin. 2002. A novel Myb-related protein involved in transcriptional activation of encystations genes in Giardia lamblia. Mol. Microbiol. 46:971-984. [DOI] [PubMed] [Google Scholar]
- 50.Tanikawa, J., T. Yasukawa, M. Enari, K. Ogata, Y. Nishimura, S. Ishii, and A. Sarai. 1993. Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA 90:9320-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent, A. C., and K. Struhl. 1992. ACR1, a yeast ATF/CREB repressor. Mol. Cell. Biol. 12:5394-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, D.-M., and J. S. Lipsick. 2002. Mutational analysis of the transcriptional activation domains of v-Myb. Oncogene 21:1611-1615. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe, Y., K. Irie, and K. Matsumoto. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 15:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wieser, J., and T. H. Adams. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 9:491-502. [DOI] [PubMed] [Google Scholar]
- 55.Woods, J. P., and W. E. Goldman. 1992. In vivo generation of linear plasmids with addition of telomeric sequences by Histoplasma capsulatum. Mol. Microbiol. 6:3603-3610. [DOI] [PubMed] [Google Scholar]
- 56.Woods, J. P., and W. E. Goldman. 1993. Autonomous replication of foreign DNA in Histoplasma capsulatum: role of native telomeric sequences. J. Bacteriol. 175:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods, J. P., E. L. Heinecke, and W. E. Goldman. 1998. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and β-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worsham, P. L., and W. E. Goldman. 1988. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26:137-143. [PubMed] [Google Scholar]