Abstract
The Cryptococcus neoformans LYS9 gene (encoding saccharopine dehydrogenase) was cloned and found to be part of an evolutionarily conserved chimera with SPE3 (encoding spermidine synthase). spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants were constructed, and these were auxotrophic for lysine and spermidine, spermidine, and lysine, respectively. Thus, SPE3-LYS9 encodes functional spermidine synthase and saccharopine dehydrogenase gene products. In contrast to Saccharomyces cerevisiae spe3 mutants, the polyamine auxotrophy of C. neoformans spe3-LYS9 mutants was not satisfied by spermine. In vitro phenotypes of spe3-LYS9 mutants included reduced capsule and melanin production and growth rate, while SPE3-lys9 mutants grew slowly at 30°C, were temperature sensitive in rich medium, and died upon lysine starvation. Consistent with the importance of saccharopine dehydrogenase and spermidine synthase in vitro, spe3-lys9 mutants were avirulent and unable to survive in vivo and both functions individually contributed to virulence. SPE3-LYS9 mRNA levels showed little evidence of being influenced by exogenous spermidine or lysine or starvation for spermidine or lysine; thus, any regulation is likely to be posttranscriptional. Expression in S. cerevisiae of the full-length C. neoformans SPE3-LYS9 cDNA complemented a lys9 mutant but not a spe3 mutant. However, expression in S. cerevisiae of a truncated gene product, consisting of only C. neoformans SPE3, complemented a spe3 mutant, suggesting possible modes of regulation. Therefore, we identified and describe a novel chimeric SPE3-LYS9 gene, which may link spermidine and lysine biosynthesis in C. neoformans.
To better understand the metabolism, gene regulation, and gene structure of Cryptococcus neoformans, an important opportunistic pathogenic fungus (6, 30), we are studying biosynthetic genes and pathways. One such pathway is the lysine biosynthetic pathway. The length, complexity, and fungus-specific nature (reviewed in reference 55) make this pathway interesting from the perspective of both potential antifungal drug targets and gene regulation. Polyamine biosynthesis comprises another interesting pathway due to the pivotal role of polyamines in cell growth and development and the highly regulated and ubiquitous nature of the pathway (8, 44, 45, 51). Our initial focus was on the C. neoformans gene encoding saccharopine dehydrogenase (saccharopine reductase; NADP+, l-glutamate forming; EC:1.5.1.10), catalyzing the penultimate step in the lysine biosynthetic pathway. Unexpectedly, we found that the C. neoformans saccharopine dehydrogenase gene (LYS9) is part of a chimera with SPE3, encoding spermidine synthase (putrescine aminopropyltransferase; EC:2.5.1.16). Both saccharopine dehydrogenase and spermidine synthase activities contributed to the virulence and survival of C. neoformans in vivo. While both activities were fully functional in C. neoformans and the SPE3-LYS9 cDNA complemented Saccharomyces cerevisiae lys9 lysine auxotrophy, S. cerevisiae spe3 spermidine and pantothenate auxotrophy was complemented only by a truncated cDNA containing only the SPE3 sequence, suggesting different modes of regulation operating in the two species. In contrast to S. cerevisiae LYS9 transcription, exogenous lysine or lysine starvation barely affected SPE3-LYS9 transcription, nor did spermidine presence or starvation; thus, any regulation in response to spermidine or lysine is likely to be posttranscriptional. This novel gene structure suggests that there is an unexpected link between lysine and polyamine biosynthesis in C. neoformans.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All S. cerevisiae and C. neoformans strains used in this study are listed in Table 1. Standard yeast culture media were prepared as described previously (42). Medium lacking pantothenate contained 1.7% (wt/vol) yeast nitrogen base (YNB) without amino acids, ammonium sulfate, and vitamins (American Biorganics, Inc.), to which biotin, folic acid, inositol, niacin, p-aminobenzoic acid, pyridoxine HCl, riboflavin, and thiamine HCl were added at amounts described previously (42). Where specified, the medium was supplemented with nourseothricin (Nat; 100 μg ml−1; Hans Knöll Institute für Naturstoff-Forschung, Jena, Germany), geneticin (200 μg ml−1; Life Technologies), proline (43 mM), lysine (0.16 mM), sorbitol (1 M), H-Ala-Lys-OH · HCl (Ala-Lys; 0.26 mM; Bachem), H-Lys-Ala-OH · HBr (Lys-Ala; 0.31 mM; Bachem), spermidine (33 μM), spermine (33 μM), glycerol (20 g liter−1), or ethanol (25 ml liter−1). Melanin production was compared on Niger seed agar (24) as described previously (23), except that the medium was supplemented with spermidine and lysine. Capsule production was induced in Dulbecco's modified Eagle's medium (with 22 mM NaHCO3 and 25 mM Na-morpholinepropanesulfonic acid) supplemented with spermidine, and capsule thickness was determined as described previously (23). One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen) was used for most cloning purposes, and DH10B was used for other plasmid propagation. Bacterial culture media were prepared according to standard protocols (39). S. cerevisiae and C. neoformans cultures were incubated at 30°C and E. coli cultures were incubated at 37°C, unless specified otherwise.
TABLE 1.
Strain list
| Strain | Genotype | Source |
|---|---|---|
| S. cerevisiae | ||
| S157 | MATα ura3Δ | 54 |
| S313 | MATα ura3Δ lys9Δ::natMX4 | This study |
| S3545 | MATα ura3Δ spe3Δ::kanMX4 | This study |
| C. neoformans | ||
| CDC1, CDC2 | Serotype A | J. Xu, personal communication |
| B4495, B4496 | Serotype B | J. Xu, personal communication |
| ATCC 34880, ATCC 34883 | MATα serotype C | 4, 40 |
| JEC20 | MATa serotype D | 18, 25 |
| JEC21 | MATα serotype D | 18, 25 |
| H99 | MATα serotype A | 35 |
| H99-24, H99-26 | MATα serotype A, spe3-lys9::NAT1 | This study |
| H99-25, H99-44, H99-49 | MATα serotype A, SPE3-LYS9 | This study |
| H99-29, H99-30 | MATα serotype A, spe3-LYS9 | This study |
| H99-41, H99-43 | MATα serotype A, SPE3-lys9 | This study |
Manipulation of nucleic acids.
Plasmid DNA was recovered from E. coli by using the QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer's instructions. Plasmid DNA from S. cerevisiae and genomic DNA from C. neoformans and S. cerevisiae for PCR analyses were isolated as described previously (19). C. neoformans genomic DNA for Southern analyses was isolated as described previously (54). DNA (2 μg) was digested with various restriction enzymes, separated by gel electrophoresis on a 0.7% (wt/vol) agarose gel, denatured, and transferred to a nylon membrane (Roche) as described previously (39).
To isolate total RNA, C. neoformans cells were grown overnight in yeast extract-peptone-dextrose (YPD) medium, harvested by centrifugation, and washed twice in water. Cells were inoculated into 50 ml of YNB plus proline medium, with or without lysine and/or spermidine, to a concentration of approximately 2 × 107 cells ml−1. After incubation for 5 h, the cells were harvested by centrifugation. RNA was isolated as described previously (54). Approximately 10 μg of total RNA was separated in a 1.0% agarose-formaldehyde gel and transferred to a nylon membrane (Roche).
Probes for Southern and Northern analyses were prepared from either PCR products or restriction digestion fragments. Probes for Southern analyses were labeled with a digoxigenin nonradioactive labeling kit (Roche), and those for Northern analyses were labeled with [α-32P]dCTP (Perkin-Elmer), using the RediprimeII Random Prime labeling system (Amersham Biosciences) according to the manufacturer's instructions. Blots were prehybridized in ULTRAhyb hybridization buffer (Ambion) for 1 h and hybridized overnight in the same buffer following addition of the denatured probe. Washing and detection of hybridized DNA bands were performed according to the manufacturer's instructions (Roche). Northern blots were similarly washed, bands were visualized with a Typhoon 9200 Variable Mode Imager (Molecular Dynamics), and expression was quantified by using ImageQuaNT 5.2 software (Molecular Dynamics).
PCR amplification reactions were performed using Expand High Fidelity PCR system polymerase (Roche) when high-fidelity DNA amplification was required; otherwise, Taq polymerase (Invitrogen BRL) was used. Unless specified otherwise, amplification incubations consisted of 94°C for 3 min, 55°C for 30 s, and 72°C (when using Taq) or 68°C (when using Expand) for 1 min per 1 kb of DNA to be amplified. The cycle was repeated 29 times except that the initial 94°C incubation was reduced to 30 s in subsequent cycles. The final cycle included a 10-min extension step at 72°C. See Table S2 in the supplemental material for a list of primers used.
S. cerevisiae strain construction.
The LYS9 and SPE3 open reading frames (ORFs) in S. cerevisiae strain S157 (MATα ura3Δ) were replaced with a natMX4 or kanMX4 cassette by PCR-mediated gene deletion (15, 50). Primers for the disruption of LYS9 included ZY021 and ZY036, and SPE3 disruption primers included ZY047 and ZY048. Deletion of LYS9 in strain S313 (lys9Δ::natMX4) was confirmed by colony PCR using the primer pair JM37 and ZY051 and by lysine auxotrophy. The spe3Δ mutation in strain S3545 (spe3Δ::kanMX4) was confirmed by colony PCR using primers JM37 and ZY049.
Isolation of SPE3-LYS9 cDNA.
The SPE3-LYS9 cDNA from C. neoformans serotype A strain H99 was isolated by complementation of an S. cerevisiae lys9Δ mutation. A C. neoformans H99 cDNA library, with inserts placed under control of the S. cerevisiae GAL1 promoter of the vector pYES2.0 (Invitrogen) (43), was a generous gift from Brian Wong (Yale University School of Medicine, New Haven, Conn.) to the Duke University Mycology Research Unit (B. Wong and J. Perfect, personal communication). For complementation, library plasmid DNA was used to transform the S. cerevisiae ura3 lys9Δ strain S313, using a lithium acetate transformation protocol (14). Transformants were selected on uracil dropout medium and were screened for lysine prototrophy on lysine dropout medium containing galactose as the sole carbon source. Plasmid DNA was isolated from the Lys+ transformants, propagated in E. coli DH10B, and purified DNA was sequenced by the Duke University Cancer Center Sequencing Facility.
Identification of the 5′ region of the SPE3-LYS9 mRNA.
A library of adaptor-ligated, double-stranded cDNA, prepared using the Marathon cDNA Amplification kit (Clontech) from RNA isolated from C. neoformans H99 that had been cultured in YPD medium, was a generous gift from the laboratory of J. A. Alspaugh. The library was used as the template for PCR amplification of the 5′ end of the SPE3-LYS9 mRNA (5′-RACE), using primer ZY152 and the Marathon cDNA Amplification kit Adaptor Primer 1 according to the manufacturer's instructions. The 5′-RACE products obtained were gel purified and cloned into pCR2.1-TOPO according to the manufacturer's manual (Invitrogen). The inserts within five resulting plasmids were sequenced.
Plasmid construction.
Plasmids were constructed that contained derivatives of C. neoformans SPE3-LYS9 or S. cerevisiae SPE3 under galactose-regulated expression. The plasmid containing the longest SPE3-LYS9 cDNA, pCnSPE3-LYS9, was engineered to contain an ATGG start sequence (pCnATGG-SPE3-LYS9). pCnSPE3-LYS9 was restriction digested with EcoRI, resulting in the removal of approximately 200 bp of 5′ SPE3 sequence. A PCR fragment was amplified that overlapped the sequence deleted by restriction digestion and contained an ATGG sequence at the 5′ end of the SPE3 sequence, using primers JO213 and JO221 and pCnSPE3-LYS9 as the template. The purified PCR and restriction-digested products were introduced into S. cerevisiae S3545 (ura3Δ spe3Δ), and strains in which the plasmid had been gap repaired (33) by the PCR product were selected by acquisition of Ura prototrophy.
Plasmids containing C. neoformans SPE3-homologous cDNA with an engineered ATGG start site and a TAA termination sequence (pCnSPE3) or S. cerevisiae SPE3 (pScSPE3) were constructed in a manner similar to that used for pCnSPE3-LYS9. A PCR product consisting of C. neoformans SPE3 containing the ATGG start and TAA termination codon and flanked by pYES2.0 sequence was amplified from pCnSPE3-LYS9 by using primers JO221 and ZY135. The S. cerevisiae SPE3 ORF, flanked by pYES2.0 sequence, was amplified by using primers JO244 and JO245. The PCR products, together with pYES2.0 DNA, created from restriction digestion of pCnSPE3-LYS9 with EcoRI and NotI to excise the SPE3-LYS9 sequence, were used to transform S. cerevisiae S3545. Strains containing gap-repaired plasmids were selected as described above. All plasmids were recovered from yeast cells and propagated in E. coli, and following isolation, constructs were verified by restriction digestion and by PCR analyses with primers used for the plasmid construction.
C. neoformans strain construction.
A C. neoformans spe3-lys9 mutant was constructed in which a 0.46-kb region consisting of the 3′ end of the SPE3-homologous region and the 5′ end of the LYS9-homologous region was replaced with the NAT1 gene cassette (29). First, an in vitro spe3-lys9::NAT1 targeting allele was constructed by a modified PCR overlap technique (11). The first round of PCRs amplified a 0.87-kb fragment containing the 5′ end of SPE3 (primers JO269 and JO215) and a 1.19-kb fragment consisting of the 3′ end of LYS9 (primers JO267 and JO220) from C. neoformans H99 chromosomal DNA. Also, a 1.70-kb product containing the NAT1 marker cassette was amplified from template pGMC200 (29) by using primers JO266 and JO268. Primers JO266 and JO267 were complementary to JO269 and JO268, respectively; thus, the ends of the three amplified products overlapped. The purified products were used as the template for the final reaction, using primers JO220 and JO215, to yield the 3.68-kb spe3-lys9::NAT1 targeting allele. The purified PCR product was cloned into pCR2.1-TOPO, from which the spe3-lys9::NAT1 allele was amplified by using primers JO215 and JO220, and introduced into C. neoformans H99 by biolistic transformation using a Bio-Rad model PDS-1000/He biolistic particle delivery system (46). Transformation was performed on YPD plates that were supplemented with sorbitol. Plates were incubated at 30°C for 4 h, after which time cells were scraped off and replated on YPD plus Nat medium. Nat-resistant transformants were purified and screened for inability to grow on synthetic medium lacking amino acids (SD medium). Replacement of the SPE3-LYS9 gene by the targeting allele was confirmed by PCR (primer pairs JO89 and ZY100, JO187 and ZY100, and JO217 and JO206) and Southern hybridization analysis (Fig. 1). Two spe3-lys9::NAT1 mutants, H99-24 and H99-26, were selected for use in further experiments.
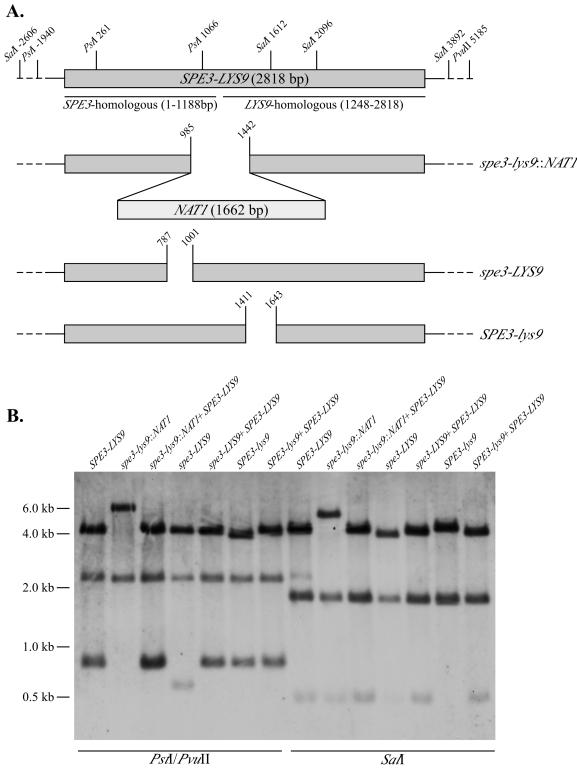
FIG. 1.
(A) Schematic representation of the SPE3-LYS9, spe3-lys9::NAT1, spe3-LYS9, and SPE3-lys9 alleles. (B) Southern blot analysis confirming the spe3-lys9::NAT1, spe3-LYS9, and SPE3-lys9 disruptions and subsequent reconstitution by SPE3-LYS9. Genomic DNA from strains H99 (SPE3-LYS9), H99-24 (spe3-lys9), H99-25 (spe3-lys9 SPE3-LYS9), H99-29 (spe3-LYS9), H99-44 (spe3-LYS9 SPE3-LYS9), H99-41 (SPE3-lys9), and H99-49 (SPE3-lys9 SPE3-LYS9) was digested with the restriction enzymes indicated and probed with genomic SPE3-LYS9 DNA.
Strains were constructed that contained either a 213-bp in-frame deletion within the SPE3-homologous region (spe3-LYS9) or a 231-bp in-frame deletion within the LYS9-homologous region (SPE3-lys9). First, spe3-LYS9 and SPE3-lys9 constructs were made in vitro by using the PCR overlap procedure. For the spe3-LYS9 construct, a 0.77-kb fragment containing the 5′ end of SPE3 and a 1.71-kb fragment containing LYS9 and the 3′ end of the SPE3 sequence were amplified from H99 chromosomal DNA, using primer pair JO214 and JO293 and primer pair JO219 and JO294, respectively. Primers JO293 and JO294 were complementary and homologous to bp 768 to 787 and bp 1001 to 1020 of the SPE3-LYS9 sequence. The purified PCR products were used as the template for a fusion PCR using primers JO214 and JO219. The resulting product was purified and cloned into pCR2.1-TOPO.
For the SPE3-lys9 construct, a 1.40-kb fragment containing SPE3 and 3′ LYS9 sequence and a 1.07-kb fragment containing 5′ LYS9 sequence were amplified by using primer pair JO214 and JO295 and primer pair JO219 and JO296, respectively. Primers JO295 and JO296 were complementary and were homologous to bp 1392 to 1411 and bp 1643 to 1662 of the SPE3-LYS9 sequence. The SPE3-lys9 targeting construct was then created by a PCR overlap reaction, using the previous two PCR products as the template and primers JO214 and JO219. The purified construct was cloned into pCR2.1-TOPO.
The cloned spe3-LYS9 and SPE3-lys9 constructs were amplified by using primers JO214 and JO219 and introduced into C. neoformans H99-24 (spe3-lys9::NAT1) by biolistic transformation. Transformants were selected on SD medium supplemented with sorbitol and spermidine when the transforming DNA was the spe3-LYS9 construct and on YNB medium containing proline, lysine, and sorbitol when the transforming DNA was the SPE3-lys9 construct. Purified prototrophic transformants were screened for the loss of Nat resistance. Replacement of the spe3-lys9::NAT1 allele by spe3-LYS9 and SPE3-lys9 was verified by PCR (primers included pair JO186 and JO206 and pair JO186 and JO205 for the spe3-LYS9 allele and pair JO187 and JO219 and pair JO188 and JO219 for the SPE3-lys9 allele) and Southern analysis (Fig. 1). spe3-LYS9 strains included H99-29 and H99-30, and SPE3-lys9 strains included H99-41 and H99-43.
Strains were also constructed in which the spe3-lys9::NAT1, SPE3-lys9, and spe3-LYS9 alleles were replaced by wild-type SPE3-LYS9. A 2.66-kb region of C. neoformans H99 SPE3-LYS9 DNA was PCR amplified (primers JO214 and JO219) and introduced into H99-24, H99-26, H99-29, and H99-41 mutants by biolistic transformation, and transformants were selected by reversion to prototrophy on SD medium containing sorbitol. Purified transformants from transformation of H99-24 were screened for the loss of the NAT1 marker. Replacement of the spe3-lys9::NAT1, SPE3-lys9, and spe3-LYS9 alleles by the wild-type allele was confirmed by PCR and Southern analyses (Fig. 1), using the same conditions that discriminated the wild-type SPE3-LYS9 and disrupted spe3-lys9, spe3-LYS9, and SPE3-lys9 alleles. Reconstituted strains H99-25, H99-44, and H99-49 were derived from the parent strains H99-24, H99-29, and H99-41, respectively.
C. neoformans virulence tests.
The virulence phenotypes of mutant and wild-type strains were compared by using the murine inhalation model of cryptococcal infection as described previously (10). Results for mouse survival were analyzed by the Mann-Whitney test. The mice infected with the spe3-lys9::NAT1 (H99-24) and spe3-LYS9 (H99-29) strains were sacrificed at 70 days postinfection, and their brains and lungs were removed and homogenized in 1 ml of phosphate-buffered saline. Entire homogenates were plated (in aliquots) onto YPD medium containing chloramphenicol and cultured. All animal experiments met with institutional guidelines and were approved by the institutional animal care and use committee.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the C. neoformans serotype A SPE3-LYS9 cDNA and genomic DNA sequence are AF368901 and AY486138, respectively.
RESULTS
Complementation of an S. cerevisiae lys9Δ mutant with C. neoformans cDNA.
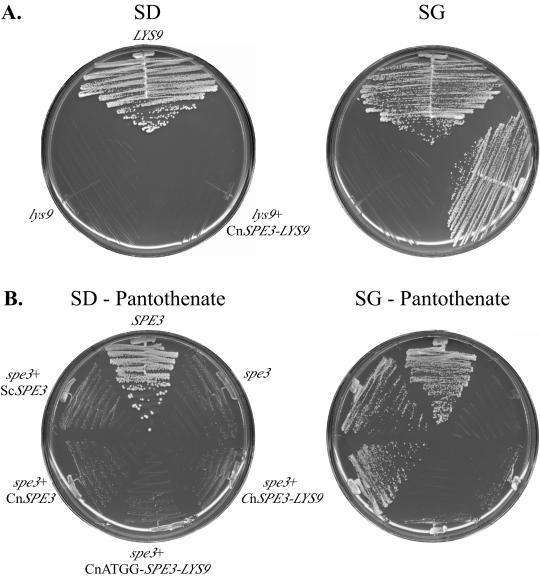
A C. neoformans cDNA library, with cDNAs inserted downstream of an S. cerevisiae GAL1 promoter (43), was used to transform the S. cerevisiae ura3 lys9Δ strain S313 to uracil (Ura) prototrophy. From approximately 47,000 Ura+ transformants, 9 independent Lys+ colonies were obtained. Complementation of lys9Δ was galactose dependent, as judged by growth on lysine-deficient medium containing galactose as the sole carbon source but not on the equivalent glucose-containing medium (Fig. 2). In addition, all nine plasmids showed galactose-dependent complementation of the lys9Δ mutation when the purified plasmids were reintroduced into the original ura3 lys9Δ strain, S313.
FIG. 2.
(A) Complementation of S. cerevisiae lys9 auxotrophy by C. neoformans SPE3-LYS9 cDNA. S. cerevisiae S157 transformed with pYES2.0 (LYS9) and S. cerevisiae S313 transformed with plasmid pYES2.0 (lys9) or pCnSPE3-LYS9 (lys9 + CnSPE3-LYS9) were plated on minimal medium with dextrose (SD) or galactose (SG) as the sole carbon source for 3 days. (B) Complementation of S. cerevisiae spe3 auxotrophy by C. neoformans SPE3-LYS9 cDNA was tested on minimal medium without pantothenate and with dextrose (SD-pantothenate) or galactose (SG-pantothenate) as the sole carbon source, with incubation for 4 days. Strains (clockwise from the top) included S. cerevisiae S157 transformed with pYES2.0 (SPE3) and S3545 transformed with plasmids pYES2.0 (spe3), pCnSPE3-LYS9 (spe3 + CnSPE3-LYS9), pCnATGG-SPE3-LYS9 (spe3 + CnATGG-SPE3-LYS9), pCnSPE3 (spe3 + CnSPE3), or pScSPE3 (spe3 + ScSPE3). Cn, C. neoformans; Sc, S. cerevisiae.
The nine plasmids contained three different-size inserts of approximately 2.3, 2, and 1.8 kb. Sequencing revealed that the 3′ ends of plasmid inserts from each of the size classes were identical, while the smaller plasmids contained cDNA inserts that were truncated at the 5′ end. The complete sequence of the largest insert (GenBank accession number AF368901) revealed that this cDNA contained a single ORF, the 5′ end (∼900 bp) of which was homologous to SPE3 (encoding spermidine synthase) and the 3′ end (∼1,400 bp) of which was homologous to LYS9 (encoding saccharopine dehydrogenase). The plasmid containing the largest cDNA was designated pCnSPE3-LYS9.
The deduced amino acid sequence from both the SPE3- and the LYS9-homologous regions of the SPE3-LYS9 ORF showed high sequence identity with other Spe3ps and Lys9ps. The deduced amino acid sequence of Spe3p shared over 60% sequence identity with Spe3ps from Neurospora crassa, S. cerevisiae, and Schizosaccharomyces pombe, and the deduced amino acid sequence of Lys9p shared over 45% identity with other Lys9ps from Magnaporthe grisea, N. crassa, S. cerevisiae, and S. pombe. There was a 20-amino-acid putative linker sequence located between the Spe3p- and Lys9p-homologous regions (DGVNVLPKFSGARPTPTTTK) which showed no similarity with either Spe3ps or Lys9ps from the fungal species listed above.
The chimeric SPE3-LYS9 gene structure is present as a single copy and is evolutionarily conserved.
Two criteria were used to show that the chimeric SPE3-LYS9 gene structure was not a cloning artifact and was, in fact, present in the genome. First, we identified a contig (cneo 010512.contig 7427) from the Stanford Genome Technology Center serotype D strain JEC21sequence database (http://sequence-www.stanford.edu/group/C.neoformans) that was highly similar (95% sequence identity) to the serotype A SPE3-LYS9 cDNA sequence. Second, the primer pair ZY099 and ZY100, designed from the 5′ and 3′ ends, respectively, of the largest insert, was used to amplify genomic DNA from the serotype A strains H99, CDC1, and CDC2, the serotype D strains JEC20 and JEC21, the serotype B strains B4495 and B4496, and the serotype C strains ATCC 34883 and ATCC 34880. Only one band of approximately 2.7 kb was amplified from the four different serotypes (data not shown), the size differences between the cDNA and genomic DNA being due to the presence of 10 introns noted in the serotype D genome sequence. Therefore, the SPE3-LYS9 gene structure in C. neoformans was evolutionarily conserved.
To determine if there were other sequences with significant similarity to SPE3 or LYS9 in the genome or other copies of the chimeric SPE3-LYS9 gene, genomic DNA was isolated from strain H99 and Southern blot hybridization was performed with probes containing either 700 bp of the SPE3 region or ∼1,600 bp of the predominantly LYS9 portion of the SPE3-LYS9 cDNA. The same hybridization pattern was observed with both probes (data not shown), and single bands were detected with three different restriction enzyme digests under relatively low stringency conditions. These results suggested that SPE3-LYS9 was the only copy of the coding region for both SPE3 and LYS9 in the C. neoformans genome.
Serotype A chromosomal SPE3-LYS9 sequence.
No ATG start codon was identified in the SPE3 coding region of even the longest SPE3-LYS9 cDNA clone isolated. Therefore, the SPE3-LYS9 cDNA sequence was extended by 5′-RACE, using a primer homologous to nucleotides 385 to 409 downstream of the 5′ end of the most complete SPE3 cDNA sequence (ZY152). Three sequenced 5′-RACE products contained additional SPE3 sequence than had previously been obtained. This additional sequence contained an ATG codon four nucleotides upstream of the 5′ end of the previously longest sequence. We predict that this is the start codon for SPE3, as there were no other ATG codons between it and an upstream termination codon. Also, the start position was consistent with Spe3p sequences from other fungi.
Using primers designed from the H99 SPE3-LYS9 cDNA and the serotype D sequence, we amplified and sequenced the chromosomal H99 SPE3-LYS9 sequence (GenBank accession number AY486138). A comparison between this and the cDNA sequence revealed the presence of 11 introns, none of which were near the junction between the Spe3p- and Lys9p-homologous regions. Six introns with sizes of 68, 54, 51, 51, 53, and 53 bp resided in the 5′ coding region of SPE3, four introns with sizes of 52, 48, 58, and 80 bp were spaced nearer the 3′ end of LYS9, and a 55-bp intron was present upstream of the putative ATG start site of SPE3. The last intron was present in one of the sequenced 5′-RACE cDNA clones but absent from another.
Complementation of S. cerevisiae spe3Δ by C. neoformans SPE3-LYS9.
To test whether the SPE3-LYS9 gene chimera encoded a functional spermidine synthase capable of complementing an S. cerevisiae spe3 mutant, an S. cerevisiae spe3Δ strain (S3545) was transformed with the plasmid containing the largest SPE3-LYS9 cDNA (pCnSPE3-LYS9) or the empty vector (pYES2.0). Since spermidine is a precursor of pantothenic acid biosynthesis in S. cerevisiae (52), the auxotrophic phenotype of S. cerevisiae spe3Δ mutants was scored on minimal medium that lacked polyamines and pantothenic acid. The spe3Δ mutant containing pCnSPE3-LYS9 was not able to grow better than the strain transformed with pYES2.0 when plated on medium that induced SPE3-LYS9 expression (minimal medium without pantothenate and with galactose [SG-pantothenate]) (Fig. 2). In contrast, when transformed with pYES2.0 that contained S. cerevisiae SPE3 under galactose-regulated expression (pScSPE3), the spe3Δ mutant was able to grow under these conditions. Therefore, the C. neoformans SPE3-LYS9 cDNA did not complement the S. cerevisiae spe3Δ mutation.
We investigated whether the absence of the proper amino terminus, including the start codon, or the presence of Lys9p attached to Spe3p might contribute to the lack of C. neoformans Spe3p function in S. cerevisiae. Complementation of the S. cerevisiae spe3 mutant pantothenate and spermidine auxotrophy was tested using plasmids that contained the full-length SPE3-LYS9 cDNA with an engineered ATGG start sequence (pCnATGG-SPE3-LYS9) or the SPE3-homologous cDNA region only with an engineered ATGG start sequence and a TAA stop codon inserted directly after the SPE3-homologous sequence. As seen in Fig. 2, pCnSPE3, but not pCnATGG-SPE3-LYS9, could complement the spe3 pantothenate auxotrophy when growth was under galactose-inducing conditions. Therefore, the lack of complementation of S. cerevisiae spe3Δ mutant phenotypes by pCnATGG-SPE3-LYS9 was due to the presence of Lys9p in the chimeric gene product.
Auxotrophic phenotypes of spe3-lys9, SPE3-lys9, and spe3-LYS9 mutants.
To better understand the role of the SPE3-LYS9 gene product(s), we constructed spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants of the C. neoformans serotype A strain H99. In addition, to ensure that the disruption of SPE3-LYS9 was responsible for mutant phenotypes, we replaced the spe3-lys9::NAT1, spe3-LYS9, and SPE3-lys9 alleles with wild-type SPE3-LYS9 sequence. The spe3-lys9 mutants (H99-24 and H99-26) had 0.46 kb of SPE3- and LYS9-homologous sequence replaced by NAT1. The spe3-LYS9 mutants (H99-29 and H99-30) had bases 788 to 1000 within the SPE3-homologous region of the SPE3-LYS9 ORF deleted. The SPE3-lys9 mutants (H99-41 and H99-43) had bases 1412 to 1642 within the LYS9-homologous region of the SPE3-LYS9 ORF deleted. H99-25, H99-44, and H99-49 were prototrophic transformants in which the spe3-lys9, spe3-LYS9, and SPE3-lys9 alleles, respectively, were replaced with SPE3-LYS9.
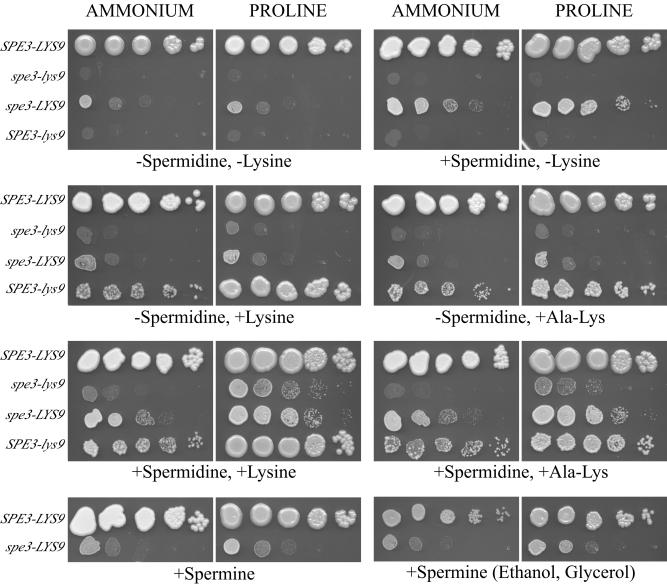
The auxotrophic requirements of the spe3-lys9, SPE3-lys9, and spe3-LYS9 mutants were assayed on SD or YNB plus proline medium that was supplemented individually or with combinations of spermidine, lysine, and Ala-Lys and Lys-Ala dipeptides (Fig. 3). The spe3-lys9 mutants failed to grow on SD medium regardless of the supplement(s), even after 5 days of incubation at 30°C. However, spe3-lys9 mutants grew on YNB plus proline medium supplemented with (i) spermidine and lysine and (ii) spermidine and lysine dipeptides but not on plates that contained only spermidine, only lysine, or only lysine dipeptides. SPE3-lys9 mutants required lysine but not spermidine, and spe3-LYS9 mutants required spermidine but not lysine. Also, satisfaction of the lysine auxotrophy of SPE3-lys9 mutants and the spermidine auxotrophy of spe3-LYS9 mutants by lysine and spermidine, respectively, was enhanced when proline was the nitrogen source instead of ammonium. Therefore, results indicate that (i) SPE3-LYS9 encodes both functional saccharopine dehydrogenase and spermidine synthase products, (ii) these activities are separable, and (iii) uptake of spermidine and lysine are regulated by the quality of the nitrogen source.
FIG. 3.
Auxotrophic satisfaction of C. neoformans spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants. Strains included C. neoformans H99 (SPE3-LYS9), H99-24 (spe3-lys9), H99-29 (spe3-LYS9), and H99-41 (SPE3-lys9). The medium was SD (ammonium as the nitrogen source) or YNB plus proline (proline as the nitrogen source), except when ethanol and glycerol were the carbon sources, in which case the medium was synthetic ethanol-glycerol medium (ammonium as the nitrogen source) or YNB-ethanol-glycerol plus proline medium (proline as the nitrogen source). Media were supplemented, as indicated, with various combinations of the following: spermidine, lysine, Ala-Lys dipeptide, and spermine. Plates were incubated for 4 days at 30°C.
Finally, since spermine can satisfy the spermidine auxotrophy of S. cerevisiae spe3 mutants (17), we investigated whether this was also true for C. neoformans spe3-LYS9 mutants. However, growth of spe3-LYS9 mutants was indistinguishable between media supplemented with spermine or lacking polyamines, regardless of whether ammonium or proline was the nitrogen source, or when a nonfermentable carbon source was present (Fig. 3). Therefore, under the growth conditions tested, spermine did not satisfy the spermidine auxotrophy of C. neoformans spe3-LYS9 mutants.
Reduced growth rate of spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants at 30°C.
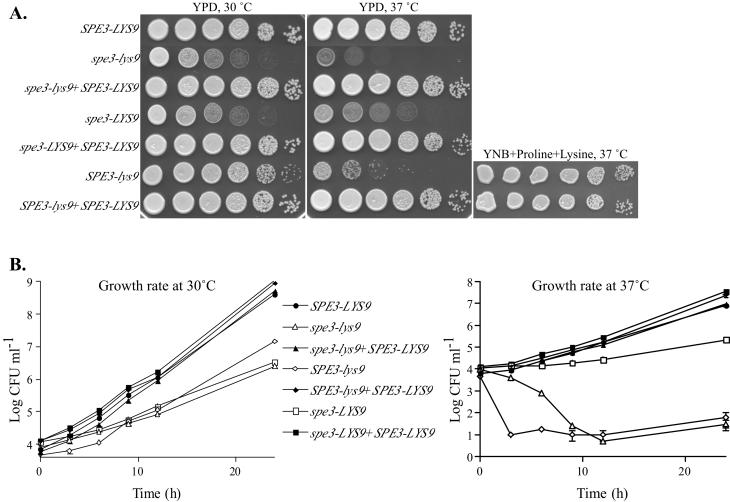
A substantial difference in growth was observed between spe3-lys9, spe3-LYS9, and SPE3-lys9, and wild-type strains when they were plated on YPD at 30°C (Fig. 4A). We therefore compared the growth rates of log-phase cultures in liquid YPD medium at 30°C of the wild-type (H99), spe3-lys9 (H99-24 and H99-26), spe3-LYS9 (H99-29 and H99-30), SPE3-lys9 (H99-41 and H99-43), and SPE3-LYS9-reconstituted (H99-25, H99-44, and H99-49) strains (Fig. 4B).
FIG. 4.
Temperature sensitivity of C. neoformans spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants. Strains included C. neoformans H99 (SPE3-LYS9), H99-24 (spe3-lys9), H99-25 (spe3-lys9 + SPE3-LYS9), H99-29 (spe3-LYS9), H99-44 (spe3-LYS9 + SPE3-LYS9), H99-41 (SPE3-lys9), and H99-49 (SPE3-lys9 + SPE3-LYS9). (A) Five-microliter volumes of 10-fold dilutions of strains were spotted onto YPD or YNB plus proline medium and incubated for 2 days at 30 or 37°C, as indicated. (B) Growth rates of strains incubated in YPD broth at 30 or 37°C.
The average generation time (calculated using CFU values from 0 and 12 h) for the wild type, 1.63 h, was approximately twice as fast as the spe3-lys9 mutant generation times of 3.58 h (H99-24) and 3.45 h (H99-26). The spe3-LYS9 mutants had generation times similar to those of the spe3-lys9 mutants, namely, 3.29 h (H99-29) and 3.27 h (H99-30). The growth rates of SPE3-lys9 mutants were also reduced compared with that of the wild type, although they were greater than those of the spe3-lys9 and spe3-LYS9 mutants, with generation times of 2.65 h (H99-41) and 2.45 h (H99-43). Replacement of each of the mutant alleles with the wild-type SPE3-LYS9 gene chimera restored generation times to approximately wild-type levels, with average doubling times of 1.65 h (H99-25), 1.85 h (H99-44), and 1.71 h (H99-49). Therefore, while saccharopine dehydrogenase mutants have a reduced growth rate at 30°C, the loss of spermidine synthase activity results in a more significant contribution than the loss of saccharopine reductase activity to the slow growth rate of the spe3-lys9 mutants at 30°C.
Conditional temperature sensitivity of spe3-lys9 and SPE3-lys9 mutants.
Since the ability to survive and proliferate at 37°C is important for cryptococcal virulence (32), we investigated whether the spe3-lys9 and SPE3-lys9 mutants also had a growth defect at 37°C. The ability to grow at 37°C was initially scored for 10-fold dilutions of strains spotted onto YPD plates. Almost no growth of spe3-lys9 mutants was observed following incubation for up to 5 days at 37°C, compared with the significant amount of growth observed for the wild type (Fig. 4A). Similar temperature-sensitive growth was observed for the SPE3-lys9 mutants. Furthermore, spe3-LYS9 mutants grew extremely poorly at this temperature when compared with the wild type. Wild-type levels of growth were restored for all mutants by the reintroduction of the wild-type SPE3-LYS9 allele. Therefore, results indicate that spermidine synthase is important, and saccharopine dehydrogenase is essential, for growth at 37°C on YPD.
The changes in CFU of strains grown in liquid YPD medium at 37°C were measured to determine the growth rate of spe3-LYS9 mutants and to establish whether incubation is cytocidal or cytostatic for spe3-lys9 and SPE3-lys9 mutants under these conditions (Fig. 4B). The average generation times of the spe3-LYS9 mutants were almost four times longer than wild-type generation times, namely, 9.60 h (H99-29) and 9.41 h (H99-30), compared with 2.54 h for the wild type. However, both spe3-lys9 and SPE3-lys9 mutants rapidly lost viability at 37°C. Following 12 h of incubation, the average CFU for spe3-lys9 mutants were 0.05% (H99-24) and 0.15% (H99-26) of initial levels, and the average CFU for SPE3-lys9 mutants were 0.27% (H99-41) and 1.56% (H99-43) of initial levels. Loss of spermidine synthase or saccharopine dehydrogenase activity was responsible for the growth defects observed in YPD medium at 37°C, as generation times of all mutants in which the wild-type SPE3-LYS9 allele had been reintroduced were similar to wild-type generation times, namely, 2.75 h (H99-25), 3.05 h (H99-44), and 2.68 h (H99-49).
A possible explanation for the cytocidal effect of incubation at 37°C in YPD medium observed for C. neoformans saccharopine dehydrogenase mutants is that cells may be starving for lysine under these conditions. This model assumes that starvation for lysine is cytocidal to the saccharopine dehydrogenase mutants; thus, we followed the survival of SPE3-lys9 mutants in minimal medium lacking lysine (SD medium) at 30°C. Like the cytocidal effect observed when saccharopine dehydrogenase mutants were grown at 37°C, SPE3-lys9 mutants lost viability when starved for lysine. Following 24 h of incubation in SD medium, the CFU of SPE3-lys9 mutants were reduced to 0.67% (H99-41) and 0.69% (H99-43) of initial levels. Therefore, the cytocidal effect of lysine starvation on C. neoformans saccharopine dehydrogenase mutants is consistent with the cytocidal effect at 37°C being due to lysine starvation under these conditions.
Possible lysine starvation of C. neoformans saccharopine dehydrogenase mutants on YPD medium at 37°C may be a consequence of reduced lysine uptake under these conditions. As C. neoformans lysine transport is likely influenced by the nitrogen source of the culture medium, we investigated whether saccharopine dehydrogenase mutants remained temperature sensitive when plated on a different medium type (YNB plus proline plus lysine medium). SPE3-lys9 mutants were able to grow on this medium at 37°C (Fig. 4A); thus, the temperature-sensitive phenotype of saccharopine dehydrogenase mutants is medium dependent.
Reduced capsule and melanin production by spe3-lys9 and spe3-LYS9 mutants.
The production of a polysaccharide capsule and melanin are important for the virulence of C. neoformans (5, 7, 13, 26, 47, 53). Capsule production was compared for strains grown for 2 days in Dulbecco's modified Eagle's medium plus 22 mM NaHCO3 plus 25 mM Na-morpholinepropanesulfonic acid plus spermidine Average capsule thicknesses were 4.33 ± 1.25 μm (H99, wild type), 1.46 ± 0.46 μm (H99-24, spe3-lys9), 1.84 ± 0.67 μm (H99-26, spe3-lys9), 4.38 ± 1.02 μm (H99-25, spe3-lys9 + SPE3-LYS9), 2.12 ± 0.77 μm (H99-29, spe3-LYS9), 1.67 ± 0.72 μm (H99-30, spe3-LYS9), 3.24 ± 0.95 μm (H99-44, spe3-LYS9 + SPE3-LYS9), 3.33 ± 0.86 μm (H99-41, SPE3-lys9), 4.14 ± 1.59 μm (H99-43, SPE3-lys9), and 4.06 ± 1.03 μm (H99-49, SPE3-lys9 + SPE3-LYS9). Therefore, loss of spermidine synthase activity, but not loss of saccharopine dehydrogenase activity, has an effect on capsule production.
Melanin production was assayed by spotting 106 cells of each strain on Niger seed agar that was supplemented with spermidine and lysine. After approximately 24 h of incubation at 30°C, pigment production was readily discernible for the wild-type (H99), SPE3-lys9 (H99-41 and H99-43), and SPE3-LYS9-reconstituted (H99-25, H99-44, and H99-49) strains but not for the spe3-lys9 (H99-24 and H99-26) or spe3-LYS9 (H99-29 and H99-30) mutants (data not shown). After 48 h of incubation, pigmentation of spe3-LYS9 mutants was slightly darker than that of spe3-lys9 mutants, although levels for both mutants were still decreased compared with that of the wild type. Coloration of all strains was similar after 72 h. Therefore, the delay in melanin production by spe3-lys9 mutants is mostly attributable to the loss of spermidine synthase activity.
Virulence phenotypes of the spe3-lys9, SPE3-lys9, and spe3-LYS9 mutants.
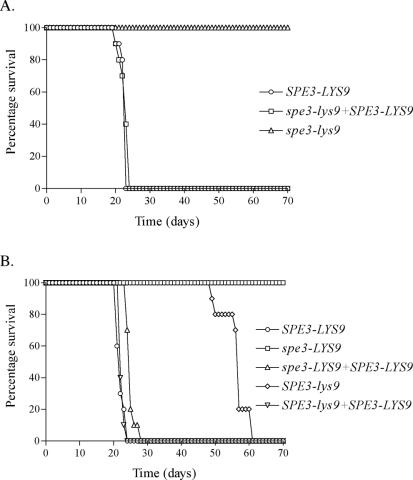
The virulence of the wild-type (H99), spe3-lys9 (H99-24), and SPE3-LYS9-reconstituted (H99-25) strains were compared using the murine inhalation infection model (10). As shown in Fig. 5, mice infected with the wild type and SPE3-LYS9-reconstituted strain survived for averages of 21.6 ± 0.97 and 21.8 ± 1.40 days postinfection (P < 0.001 for both strains compared with the spe3-lys9 mutant), and all were dead after 23 and 24 days, respectively. In contrast, mice that were infected with the spe3-lys9 mutant still remained healthy 70 days after infection. Following this time, no spe3-lys9 mutants were recovered from brain and lung homogenates of the sacrificed mice. Therefore, the spe3-lys9 mutants were avirulent and unable to survive in mice.
FIG. 5.
Survival of mice infected with C. neoformans SPE3-LYS, spe3-LYS9, SPE3-lys9, and SPE3-LYS9-reconstituted strains. Strains included H99 (SPE3-LYS9), H99-24 (spe3-lys9), H99-25 (spe3-lys9 + SPE3-LYS9), H99-29 (spe3-LYS9), H99-44 (spe3-LYS9 + SPE3-LYS9), H99-41 (SPE3-lys9), and H99-49 (SPE3-lys9 + SPE3-LYS9).
To determine whether the avirulence and inability to survive in vivo of the spe3-lys9 mutant was due to the loss of spermidine synthase or saccharopine dehydrogenase activity, or both activities together, we also determined the virulence of spe3-LYS9 (H99-29) and SPE3-lys9 (H99-41) strains (Fig. 5). Like the spe3-lys9 mutant, all mice infected with the spe3-LYS9 mutant remained healthy at 70 days postinfection. However, in contrast to the spe3-lys9 mutant, several colonies confirmed phenotypically and by PCR analysis as spe3-LYS9 mutants were recovered from the brains of 6 mice and the lungs of all 10 mice sacrificed 70 days postinfection. Mice infected with the SPE3-LYS9-reconstituted derivative of the spe3-LYS9 strain survived for an average of 24.1 ± 1.20 days, and mice infected with the wild type survived for an average of 21.1 ± 1.20 days (P < 0.001 for both strains compared with the spe3-LYS9 mutant). Mice infected with the SPE3-lys9 mutant survived more than twice as long (55.2 ± 3.94 days) as those infected with the SPE3-LYS9-reconstituted derivative of this strain (21.5 ± 0.71 days) or the wild type (P < 0.001 for both strains compared with the SPE3-lys9 mutant). Therefore, both spermidine synthase and saccharopine dehydrogenase are important for virulence, and the complete avirulence and inability to survive in vivo of the spe3-lys9 mutant are likely due to the combined loss of both activities.
SPE3-LYS9 transcript levels.
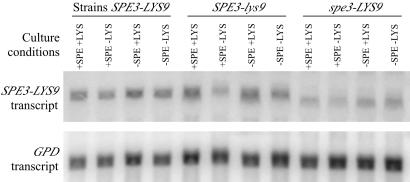
We examined whether the presence of, or starvation for, spermidine and/or lysine influenced SPE3-LYS9 transcription. Northern analysis of the SPE3-LYS9 transcript was performed using RNA isolated from the wild type (H99) and spe3-LYS9 (H99-29) and SPE3-lys9 (H99-41) mutants incubated in the presence or absence of spermidine and/or lysine (Fig. 6). When normalized to levels of the constitutively expressed GPD gene transcript (49), no differences in SPE3-LYS9 transcript levels were observed when the wild type was grown under the various conditions. Likewise, the transcript level for the spe3-LYS9 mutant was not influenced by the presence or absence of spermidine or lysine in the growth medium, although under all conditions tested, SPE3-lys9 transcript levels were only about 50% of wild-type levels. The SPE3-lys9 transcript level was reduced slightly (approximately 50%) when SPE3-lys9 mutants were starved for lysine, compared with levels for conditions where lysine was present, or from wild-type SPE3-LYS9 levels for cells grown under the same conditions. However, a significant reduction in SPE3-lys9 mutant viability was observed when the mutant was grown under lysine starvation conditions (CFU levels were 0.14% of initial levels after 5 h). Therefore, while the impact of lysine starvation on SPE3-LYS9 transcription was difficult to assess due to SPE3-lys9 mutant death, spermidine starvation or lysine and/or spermidine presence in the growth medium had little to no effect on SPE3-LYS9 transcription.
FIG. 6.
Northern analysis of SPE3-LYS9 transcription. RNA was isolated from strains H99 (SPE3-LYS9), H99-29 (spe3-LYS9), and H99-41 (SPE3-lys9) grown in the presence or absence of spermidine (SPE) and/or lysine (LYS). The GPD transcript was a control for RNA loading.
DISCUSSION
The C. neoformans LYS9 serotype A cDNA was isolated by complementation of an S. cerevisiae lys9 mutant. Unexpectedly, while the predicted amino acid sequence encoded by the single ORF within the cDNA was highly similar to other fungal saccharopine dehydrogenases at the 3′ end, the predicted sequence encoded by the 5′ end shared considerable identity with fungal spermidine synthases (encoded by SPE3 genes). The gene chimera was conserved in all four serotypes of C. neoformans and comprised the sole copy of SPE3 and LYS9 present in the genome. The chromosomal SPE3-LYS9 gene was disrupted, and the spe3-lys9 mutant was auxotrophic for both spermidine and lysine, while spe3-LYS9 and SPE3-lys9 mutants required only spermidine and only lysine, respectively. Therefore, LYS9 exists as an evolutionarily conserved chimera with SPE3 in C. neoformans, and this chimera encodes both saccharopine dehydrogenase and spermidine synthase activities.
Unlike the S. cerevisiae lys9 or spe3 mutant auxotrophies, the addition of lysine or lysine dipeptides and spermidine could best satisfy the auxotrophy of C. neoformans spe3-lys9, spe3-LYS9, and SPE3-lys9 mutants in minimal medium when proline, and not ammonium, was the nitrogen source. These results are consistent with the transport of lysine, lysine dipeptides, and spermidine being negatively regulated in the presence of easily utilized nitrogen sources such as ammonium and deregulated when less easily utilized sources are present. Although spermidine transport by S. cerevisiae is active in the presence of ammonium (21), nitrogen-repressible systems regulate amino acid and dipeptide transport in other fungi (28). Also, we have previously observed evidence for nitrogen source-regulated transport of isoleucine and valine amino acids and dipeptides by a C. neoformans ilv2 mutant (23).
While spermine can satisfy the spermidine auxotrophy of S. cerevisiae spe3 mutants due to conversion to spermidine by the polyamine oxidase Fms1p (27), spermine was unable to satisfy the C. neoformans spe3-LYS9 spermidine auxotrophy. The same results have also been observed for Leishmania donovani spermidine synthase mutants (38). A BLAST search of a C. neoformans serotype A database (http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans) identified a sequence with only limited sequence similarity to Fms1p. Therefore, C. neoformans may lack an Fms1p-like polyamine oxidase. Interestingly, a BLAST search of the C. neoformans serotype A sequence for similarity to S. cerevisiae spermine synthase (Spe4p), required for production of spermine from spermidine, detected only a Spe3-Lys9p sequence, and spermine was previously undetected in isolates of C. neoformans (36) and the basidiomycete Ustilago maydis (16). Therefore, C. neoformans and other basidiomycetes may not produce spermine.
Mutants lacking spermidine synthase or saccharopine dehydrogenase activity had pleiotropic phenotypes. First, spe3-LYS9 and SPE3-lys9 mutants had considerably reduced growth rates at 30°C in various conditions, even when spermidine and lysine were present. Spermidine synthase mutants also had reduced production of two known virulence factors, capsule (5, 7, 13, 26) and melanin (47, 53). These defects may be attributable to the reduced growth rate of the spe3 mutants in the assay conditions. The pleiotropic defects of the spermidine synthase mutant are also consistent with the many and varied roles of polyamines in the growth and development of all cells (8, 44, 45).
Importantly, growth rates of both spermidine synthase and saccharopine dehydrogenase mutants were significantly more attenuated at the physiologically relevant temperature of 37°C. The slower growth rate of spermidine synthase mutants at this temperature may be due to an elevated requirement for polyamines at high temperatures to protect cell components against damage, as has been reported for S. cerevisiae (1), or a reduction in spermidine transport at this temperature. The temperature-sensitive phenotype of saccharopine dehydrogenase mutants was complex. While incubation in YPD medium at 37°C was cytocidal to the mutants, minimal proline medium supplemented with lysine was conducive to growth at 37°C. The medium-dependent growth may be due to variations in regulation of lysine transport by C. neoformans in the different media and at different temperatures; thus, the cytocidal effect of incubation in YPD medium at 37°C may be due to starvation for lysine. Consistent with this hypothesis, we observed that lysine starvation by the mutant is cytocidal at lower temperatures. Alternatively, the cytocidal effect could be due to the accumulation of a toxic intermediate that may be either more toxic in certain conditions or accumulated to higher concentrations in certain conditions. Indeed, S. cerevisiae lys9 mutants are known to accumulate the intermediate α-aminoadipate-δ-semialdehyde (3), which may be toxic to cells (56). A possible toxic-intermediate accumulation in certain conditions may also explain why lysine starvation by C. neoformans saccharopine dehydrogenase mutants is cytocidal while starvation for some other amino acids, such as methionine, is cytostatic (23).
Consistent with the pleiotropic defects of C. neoformans spermidine synthase and saccharopine dehydrogenase mutants, the spe3-lys9 mutant lacking both functions was avirulent and unable to survive in vivo. In addition, the independent loss of either function had a substantial effect on virulence. While the virulence of SPE3-lys9 mutants was significantly attenuated, mice eventually succumbed to infection. Therefore, even though incubation at 37°C is cytocidal to SPE3-lys9 mutants in rich medium in vitro, the in vivo environment is conducive to the growth of these mutants, although growth rates at reduced levels are likely compared with that of the wild type, accounting for the attenuated virulence observed. Also, while lysine transport is influenced by the nitrogen source present, lysine is apparently sufficiently abundant and transported in adequate levels to allow the survival and growth of the SPE3-lys9 mutant in vivo. Therefore, saccharopine dehydrogenase, and perhaps other components of the lysine biosynthetic pathway, would not make ideal anticryptococcal drug targets. However, since mice infected with spermidine synthase mutants remained healthy throughout the course of the experiment, spermidine synthase may be a more useful anticryptococcal, and perhaps antifungal, drug target. At least four spermidine synthase inhibitors have been developed (2, 20, 22, 34), at least one of which (cyclohexylamine) is known to inhibit C. neoformans growth (36).
The transcription of S. cerevisiae LYS9 is regulated in response to lysine levels. When lysine is abundant, LYS9 transcription is downregulated (12, 37), while LYS9 transcription is upregulated following starvation for amino acids via the general control of amino acid biosynthesis pathways (48). While we are unaware of any reports of transcriptional regulation of S. cerevisiae SPE3 by spermidine levels, production of other polyamine biosynthetic enzymes is highly regulated in different organisms in response to polyamine levels by a wide variety of mechanisms (9, 41, 51). Therefore, we examined the transcriptional response of SPE3-LYS9 to starvation for lysine and spermidine, as well as to conditions when either or both spermidine and lysine were abundant. However, SPE3-LYS9 transcription was affected little to not at all by changes in spermidine and lysine levels. Therefore, regulation of SPE3-LYS9 transcription differs from S. cerevisiae LYS9 transcriptional regulation.
While the SPE3-LYS9 chimera encodes fully functional spermidine synthase and saccharopine dehydrogenase activities in C. neoformans and the SPE3-LYS9 cDNA complemented the S. cerevisiae lys9 lysine auxotrophy, the polyamine and pantothenate auxotrophy of the S. cerevisiae spe3 mutant was complemented only when the chimera was truncated to contain only the SPE3 region. The difference between the functionality of the full-length SPE3-LYS9 gene products in S. cerevisiae and C. neoformans may be related to differences in regulation between the species. Any differences in regulation are unlikely to occur at the level of transcription since only one size of SPE3-LYS9 mRNA was observed. In principle, such regulation might also occur at the translational level, although no in-frame stop codon was observed directly after the SPE3-homologous sequence. Finally, a posttranslational mechanism might be responsible for SPE3-LYS9 regulation. For example, C. neoformans may contain a protein, not present in S. cerevisiae, which binds Spe3-Lys9p to either modify its function or cleave the polypeptide into functional spermidine synthase and saccharopine dehydrogenase units.
Although an ornithine decarboxylase-S-adenosylmethionine decarboxylase gene chimera has been identified in Plasmodium species (31), the coupling of spermidine synthase with saccharopine dehydrogenase in C. neoformans SPE3-LYS9 is unprecedented and may suggest a link between polyamine and lysine biosynthesis. Since lysine can be decarboxylated to form the polyamine cadaverine, a coupling between lysine and spermidine biosynthesis could potentially control the production of both cadaverine and spermidine. The SPE3-LYS9 gene chimera is present in all four C. neoformans serotypes. In addition, in contrast to ascomycetes, which contain discrete genes encoding saccharopine dehydrogenase and spermidine synthase, we identified SPE3-LYS9 chimera-like sequences in two other basidiomycete sequence databases (Broad Institute Ustilago maydis and Coprinus cinereus databases; http://www.broad.mit.edu/annotation/fungi/ustilago_maydis/index.htmland http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus). Therefore, the SPE3-LYS9 chimera may have arisen in fungi after the basidiomycete and ascomycete lineages diverged.
Supplementary Material
Acknowledgments
We thank J. Xu for the C. neoformans serotype A strains CDC1 and CDC2 and the serotype B and C strains, B. Wong for the C. neoformans cDNA library, and J. A. Alspaugh's laboratory for the library of adaptor-ligated, double-stranded C. neoformans H99 cDNA.
This work was supported by a Public Health Service grant from NIAID (PO1-AI44975).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Balasundaram, D., C. W. Tabor, and H. Tabor. 1996. Sensitivity of polyamine-deficient Saccharomyces cerevisiae to elevated temperatures. J. Bacteriol. 178:2721-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beppu, T., A. Shirahata, N. Takahashi, H. Hosoda, and K. Samejima. 1995. Specific depletion of spermidine and spermine in HTC cells treated with inhibitors of aminopropyltransferases. J. Biochem. 117:339-345. [DOI] [PubMed] [Google Scholar]
- 3.Borell, C. W., L. A. Urrestarazu, and J. K. Bhattacharjee. 1984. Two unlinked lysine genes (LYS9 and LYS14) are required for the synthesis of saccharopine reductase in Saccharomyces cerevisiae. J. Bacteriol. 159:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt, M. E., S. L. Bragg, and R. W. Pinner. 1993. Multilocus enzyme typing of Cryptococcus neoformans. J. Clin. Microbiol. 31:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulmer, G. S., M. D. Sans, and C. M. Gunn. 1967. Cryptococcus neoformans. I. Nonencapsulated mutants. J. Bacteriol. 94:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childs, A. C., D. J. Mehta, and E. W. Gerner. 2003. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 60:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffino, P. 2001. Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie 83:319-323. [DOI] [PubMed] [Google Scholar]
- 10.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 12.Feller, A., E. Dubois, F. Ramos, and A. Pierard. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fromtling, R. A., H. J. Shadomy, and E. S. Jacobson. 1982. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79:23-29. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 16.Guevara-Olvera, L., B. Xoconostle-Cázares, and J. Ruiz-Herrera. 1997. Cloning and disruption of the ornithine decarboxylase gene of Ustilago maydis: evidence for a role of polyamines in its dimorphic transition. Microbiology 143:2237-2245. [DOI] [PubMed] [Google Scholar]
- 17.Hamasaki-Katagiri, N., C. W. Tabor, and H. Tabor. 1997. Spermidine biosynthesis in Saccharomyces cerevisiae: polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase). Gene 187:35-43. [DOI] [PubMed] [Google Scholar]
- 18.Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung. 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:265-272. [DOI] [PubMed] [Google Scholar]
- 20.Ito, H., H. Hibasami, K. Shimura, J. Nagai, and H. Hidaka. 1982. Antitumor effect of dicyclohexylammonium sulfate, a potent inhibitor of spermidine synthase against P388 leukemia. Cancer Lett. 15:229-235. [DOI] [PubMed] [Google Scholar]
- 21.Kaouass, M., M. Audette, D. Ramotar, S. Verma, D. De Montigny, I. Gamache, K. Torossian, and R. Poulin. 1997. The STK2 gene, which encodes a putative Ser/Thr protein kinase, is required for high-affinity spermidine transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:2994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khomutov, R. M., T. Hyvonen, E. Karvonen, L. Kauppinen, T. Paalanen, L. Paulin, T. Eloranta, R. L. Pajula, L. C. Andersson, and H. Poso. 1985. 1-Aminooxy-3-aminopropane, a new and potent inhibitor of polyamine biosynthesis that inhibits ornithine decarboxylase, adenosylmethionine decarboxylase and spermidine synthase. Biochem. Biophys. Res. Commun. 130:596-602. [DOI] [PubMed] [Google Scholar]
- 23.Kingsbury, J. M., Z. Yang, T. M. Ganous, G. M. Cox, and J. H. McCusker. 2004. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology 150:1547-1558. [DOI] [PubMed]
- 24.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, Pa.
- 25.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landry, J., and R. Sternglanz. 2003. Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem. Biophys. Res. Commun. 303:771-776. [DOI] [PubMed] [Google Scholar]
- 28.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, S., A. Da'dara, K. Lüersen, C. Wrenger, R. Das Gupta, R. Madhubala, and R. D. Walter. 2000. In the human malaria parasite Plasmodium falciparum, polyamines are synthesized by a bifunctional ornithine decarboxylase, S-adenosylmethionine decarboxylase. J. Biol. Chem. 275:8097-8102. [DOI] [PubMed] [Google Scholar]
- 32.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegg, A. E., K. C. Tang, and J. K. Coward. 1982. Effects of S-adenosyl-1,8-diamino-3-thiooctane on polyamine metabolism. Biochemistry 21:5082-5089. [DOI] [PubMed] [Google Scholar]
- 35.Perfect, J. R., D. L. Toffaletti, and T. H. Rude. 1993. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaller, M. A., J. Riley, and T. Gerarden. 1990. Polyamine depletion and growth inhibition of Cryptococcus neoformans by α-difluoromethylornithine and cyclohexylamine. Mycopathologia 112:27-32. [DOI] [PubMed] [Google Scholar]
- 37.Ramos, F., E. Dubois, and A. Pierard. 1988. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur. J. Biochem. 171:171-176. [DOI] [PubMed] [Google Scholar]
- 38.Roberts, S. C., Y. Jiang, A. Jardim, N. S. Carter, O. Heby, and B. Ullman. 2001. Genetic analysis of spermidine synthase from Leishmania donovani. Mol. Biochem. Parasitol. 115:217-226. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schmeding, K. A., S. C. Jong, and R. Hugh. 1981. Sexual compatibility between serotypes of Filobasidiella neoformans (Cryptococcus neoformans). Curr. Microbiol. 5:133-138. [Google Scholar]
- 41.Shantz, L. M., and A. E. Pegg. 1999. Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int. J. Biochem. Cell Biol. 31:107-122. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Suvarna, K., A. Bartiss, and B. Wong. 2000. Mannitol-1-phosphate dehydrogenase from Cryptococcus neoformans is a zinc-containing long-chain alcohol/polyol dehydrogenase. Microbiology 146:2705-2713. [DOI] [PubMed] [Google Scholar]
- 44.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, T., and T. J. Thomas. 2001. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 58:244-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres-Guererro, H., and J. C. Edman. 1994. Melanin-deficient mutants of Cryptococcus neoformans. J. Med. Vet. Mycol. 32:303-313. [PubMed] [Google Scholar]
- 48.Urrestarazu, L. A., C. W. Borell, and J. K. Bhattacharjee. 1985. General and specific controls of lysine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 9:341-344. [DOI] [PubMed] [Google Scholar]
- 49.Varma, A., and K. J. Kwon-Chung. 1999. Characterization of the glyceraldehyde-3-phosphate dehydrogenase gene [correction of glyceraldehyde-3-phosphate gene] and the use of its promoter for heterologous expression in Cryptococcus neoformans, a human pathogen. Gene 232:155-163. [DOI] [PubMed] [Google Scholar]
- 50.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 51.Wallace, H. M., A. V. Fraser, and A. Hughes. 2003. A perspective of polyamine metabolism. Biochem. J. 376:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, W. H., P. L. Gunyuzlu, and J. H. Toyn. 2001. Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of β-alanine production from spermine. J. Biol. Chem. 276:10794-10800. [DOI] [PubMed] [Google Scholar]
- 53.Williamson, P. R. 1997. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front. Biosci. 2:e99-e107. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z., R. C. Pascon, A. Alspaugh, G. M. Cox, and J. H. McCusker. 2002. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology 148:2617-2625. [DOI] [PubMed] [Google Scholar]
- 55.Zabriskie, T. M., and M. D. Jackson. 2000. Lysine biosynthesis and metabolism in fungi. Nat. Prod. Rep. 17:85-97. [DOI] [PubMed] [Google Scholar]
- 56.Zaret, K. S., and F. Sherman. 1985. α-Aminoadipate as a primary nitrogen source for Saccharomyces cerevisiae mutants. J. Bacteriol. 162:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.