Campigli Di Giammartino et al. find that RBBP6 is a component of a large multisubunit protein complex that mediates polyadenylation of mRNA precursors. Genome-wide analyses following RBBP6 knockdown revealed decreased transcript levels, especially of mRNAs with AU-rich 3′ UTRs such as c-Fos and c-Jun, and increased usage of distal poly(A) sites.
Keywords: polyadenylation, alternative polyadenylation, pre-mRNA 3′ end processing, AU-rich mRNA
Abstract
Polyadenylation of mRNA precursors is mediated by a large multisubunit protein complex. Here we show that RBBP6 (retinoblastoma-binding protein 6), identified initially as an Rb- and p53-binding protein, is a component of this complex and functions in 3′ processing in vitro and in vivo. RBBP6 associates with other core factors, and this interaction is mediated by an unusual ubiquitin-like domain, DWNN (“domain with no name”), that is required for 3′ processing activity. The DWNN is also expressed, via alternative RNA processing, as a small single-domain protein (isoform 3 [iso3]). Importantly, we show that iso3, known to be down-regulated in several cancers, competes with RBBP6 for binding to the core machinery, thereby inhibiting 3′ processing. Genome-wide analyses following RBBP6 knockdown revealed decreased transcript levels, especially of mRNAs with AU-rich 3′ untranslated regions (UTRs) such as c-Fos and c-Jun, and increased usage of distal poly(A) sites. Our results implicate RBBP6 and iso3 as novel regulators of 3′ processing, especially of RNAs with AU-rich 3′ UTRs.
The 3′ ends of nearly all polyadenylated RNAs are produced by a two-step reaction involving endonucleolytic cleavage of the transcript followed by synthesis of the poly(A) tail. This step in gene expression, which is necessary for mRNA stability, export, and translation (for review, see Moore and Proudfoot 2009), is mediated by a massive protein machinery (for review, see Proudfoot 2011; Xiang et al. 2014). The core 3′ processing machinery includes four multisubunit protein complexes: cleavage/polyadenylation specificity factor (CPSF), cleavage stimulatory factor (CstF), cleavage factor I (CFI), and CFII. Additional proteins, such as poly(A) polymerase (PAP), symplekin, poly(A)-binding protein II (PABPII), and RNA polymerase II (specifically the C-terminal domain [CTD] of its largest subunit), also play important roles. The site where polyadenylation occurs, the poly(A) site, is defined by multiple cis elements that contact several subunits of this machinery. Most transcripts contain more than one potential poly(A) site, and the selection of alternative sites is an important aspect of gene control (for review, see Di Giammartino et al. 2011; Elkon et al. 2013).
The core 3′ processing complex interacts with many additional factors. A large number of associated proteins were identified in a proteomic analysis of the human complex assembled on substrate RNA (Shi et al. 2009). Many of these (such as, for example, PARP-1) (Di Giammartino et al. 2013) are thought to connect 3′ processing to other nuclear events, while others (e.g., WDR33) were previously undiscovered components of the human core 3′ processing machinery. One protein that could conceivably fall in either category is RBBP6 (retinoblastoma-binding protein 6). RBBP6 is a large (∼250-kDa) multidomain protein that is similar in its N terminus to the yeast 3′ processing factor Mpe1, which is an integral subunit of the yeast CPF (cleavage and polyadenylation factor) complex and is required for 3′ end formation (Vo et al. 2001; Lee and Moore 2014). Mpe1 is required for cell viability, and absence of RBBP6 homologs leads to embryonic lethality in mice (Li et al. 2007), flies (Mather et al. 2005), and worms (Huang et al. 2013).
RBBP6 has a number of features that suggest important roles in linking 3′ end formation with other cellular processes. RBBP6 homologs all share three well-conserved domains at their N termini. The first is called the “domain with no name” or DWNN, which adopts a ubiquitin-like fold (Pugh et al. 2006). In addition to forming part of full-length RBBP6, this domain is also expressed in vertebrates as a small protein containing the DWNN and a short C-terminal tail (isoform 3 [iso3]) (Pugh et al. 2006), which has been shown to be down-regulated in several human cancers (Mbita et al. 2012). The second conserved domain is a CCHC zinc knuckle. This type of zinc finger is also found in a number of splicing factors and the 3′ processing factor CPSF30, where it functions in RNA binding (Barabino et al. 1997). The third domain is a RING finger, a domain found in E3-ubiquitin ligases. The RING domain of RBBP6 binds to YB-1, a multifunctional RNA-binding protein, and the transcriptional repressor ZBTB38. Both proteins were shown to be substrates of RBBP6 for ubiquitination, leading to their degradation by the proteasome (Chibi et al. 2008; Miotto et al. 2014). Mammalian RBBP6 also includes a long C-terminal extension containing several additional significant domains. One is an RS domain characteristic of SR proteins and other proteins involved in pre-mRNA splicing. Similar domains are also present in two other 3′ processing factors, CFI-68 and Fip1 (Boucher et al. 2001). RBBP6 was first identified as an interactor with the tumor suppressor protein Rb (Saijo et al. 1995; Sakai et al. 1995) and was subsequently shown to interact with another tumor suppressor, p53 (Simons et al. 1997). RBBP6 interferes with binding of p53 to DNA and also facilitates interaction between p53 and its negative regulator, Mdm2, leading to enhanced p53 ubiquitination and degradation. Moreover, disruption of RBBP6 in mice leads to early embryonic lethality, but a p53-null mutation partially rescues viability (Li et al. 2007). The RBBP6 C-terminal region contains domains responsible for interaction with both tumor suppressors.
Here we describe experiments that establish RBBP6 as a bona fide 3′ processing factor in vitro that functions in polyadenylation control in vivo. We show that nuclear extracts (NEs) prepared from HeLa cells following RBBP6 knockdown were defective in 3′ cleavage, but not poly(A) synthesis, and that activity could be rescued by adding a recombinant RBBP6 N-terminal derivative (RBBP6-N) containing only the DWNN, zinc knuckle, and RING domains. In vivo, RBBP6-N and endogenous RBBP6 coimmunoprecipitated with 3′ processing factors. The binding was particularly strong to CstF64 and is mediated by the DWNN. Consistent with this, RBBP6 iso3 outcompeted RBBP6-N for binding to CstF64 and inhibited cleavage when added to NEs or when overexpressed in cells. Genome-wide analyses following RBBP6 knockdown revealed a general down-regulation in transcript levels accompanied by increased usage of distal poly(A) sites. Interestingly, RNAs with AU-rich 3′ untranslated regions (UTRs), such as c-Fos and c-Jun, were especially enriched in the down-regulated transcripts, which we show resulted from defective 3′ processing coupled with degradation by the exosome.
Results
RBBP6 is a new 3′ processing factor
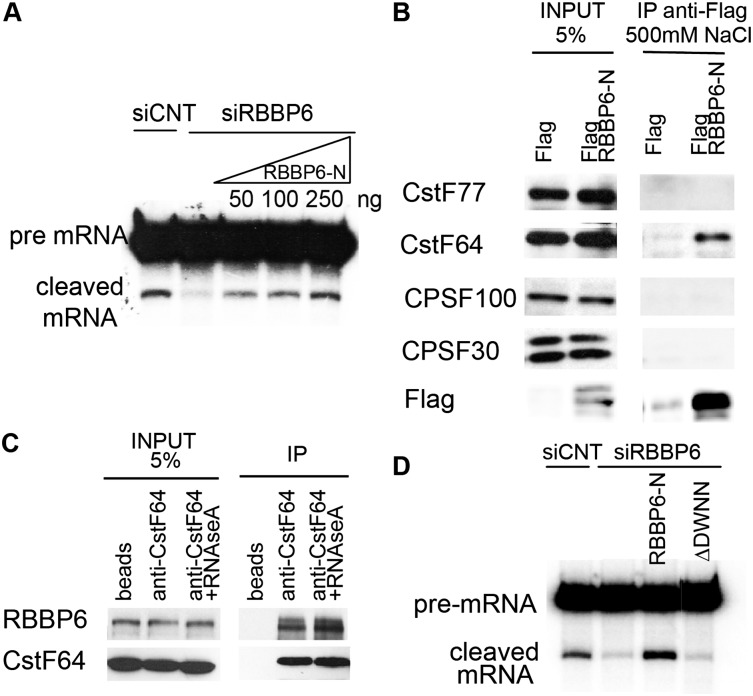
Our previous proteomic analysis showed that RBBP6 is physically associated with the active 3′ processing complex (Shi et al. 2009). To determine whether RBBP6 is in fact necessary for 3′ processing, we prepared NEs from HeLa cells treated for 72 h with siRNA against RBBP6 or a nontargeting siRNA (Fig. 1A is a Western blot showing knockdown efficiency) and used the NEs for in vitro 3′ processing assays. We took advantage of the fact that the two steps of pre-mRNA 3′ processing—cleavage and poly(A) synthesis, which are tightly coupled in vivo—can be analyzed separately in vitro (see the Materials and Methods). Figure 1B shows that upon RBBP6 knockdown, the NEs retained poly(A) synthesis activity when incubated with a pre-cleaved 32P-labeled simian virus 40 late (SVL) RNA (cf. lanes 1 and 2), while cleavage of a longer SVL RNA was largely inhibited (cf. lanes 3 and 4). Cleavage efficiencies observed in three separate experiments following RBBP6 knockdown were quantified and normalized to siCNT (Fig. 1C). RBBP6 depletion reduced cleavage activity by 80%, comparable with the extent of knockdown.
Figure 1.
RBBP6 is an essential 3′ processing factor. (A) Western blot with the indicated antibodies of HeLa NEs following transfection with siRNA against RBBP6 (siRBBP6) or a nontargeting sequence (siCNT). (B) 3′ cleavage and polyadenylation assays were carried out using internally 32P-labeled SVL RNA substrate and HeLa NEs made after siRNA treatment. RNAs were purified, resolved by denaturing PAGE, and visualized by autoradiography. Positions of precursor and products are indicated. (C) Diagram representing the mean of three separate cleavage experiments following knockdown of RBBP6; standard error bars are indicated. (D) Schematic diagram of full-length RBBP6 iso1, RBBP6-N, ∆DWNN, DWNN only, and RBBP6 iso3. Gene maps of iso1 and iso3 are presented below each protein.
We next wished to determine whether the reduced 3′ processing activity of the NEs was a direct consequence of the absence of RBBP6. Because of the homology of the N-terminal region of RBBP6 with the yeast factor Mpe1 (Vo et al. 2001) and because all RBBP6 homologs include the three domains shown schematically in Figure 1D (RBBP6-N) but not the long C-terminal extension present only in vertebrates (Fig. 1D, top; Pugh et al. 2006), we suspected that the N-terminal part of the protein might be sufficient to support 3′ processing. Therefore, we expressed and purified the RBBP6-N derivative from Escherichia coli (Coomassie-stained gel shown in Supplemental Fig. 1A) and repeated the 3′ cleavage assay as in Figure 1B but adding increasing amounts of purified RBBP6-N, which fully restored cleavage activity of NEs made after RBBP6 knockdown (Fig. 2A).
Figure 2.
RBBP6-N interacts with CstF. (A) 3′ cleavage assay as in Figure 1B but adding increasing amounts of RBBP6-N that was previously purified from E. coli. (B) 293T cells were transfected with a Flag empty vector or Flag-RBBP6-N, and cell extracts in a buffer containing 500 mM NaCl were used to immunoprecipitate with anti-Flag followed by Western blot with the indicated antibodies. (C) HeLa NEs were used to immunoprecipitate endogenous CstF64 (with a polyclonal antibody) in the presence of 150 mM NaCl with or without prior incubation of the extract with 100 μg/mL RNase A for 15 min at 30°C. Western blots were carried out with anti-RBBP6 antibody and a monoclonal CstF64 antibody. (D) 3′ cleavage assay as in A: Two-hundred-fifty nanograms of wild-type or ∆DWNN RBBP6-N purified from E. coli was added to the NEs made after knockdown of RBBP6.
RBBP6-N interacts with CPSF/CstF
We next asked whether RBBP6 interacts in vivo with other 3′ processing factors. Since the N-terminal region was sufficient to restore 3′ cleavage after knockdown of the endogenous protein, we first used only that part of the protein to analyze its interactions. We cloned an RBBP6-N-expressing sequence into a Flag-tagged vector and transfected it into 293T cells. Supplemental Figure 1B shows that following immunoprecipitation with an anti-Flag antibody under mild conditions (150 mM NaCl), endogenous CPSF and CstF components coimmunoprecipitated with RBBP6-N. These interactions were specific to CPSF/CstF complexes because CFI25 and symplekin did not show a similar interaction with RBBP6. Moreover, when the same coimmunoprecipitation (co-IP) was performed under more stringent conditions (500 mM NaCl), only binding to CstF64 was observed (Fig. 2B). To provide evidence that the interaction between RBBP6 and CstF is physiologically relevant, we repeated the immunoprecipitation with endogenous proteins. Figure 2C shows that the anti-CstF64 antibody coimmunoprecipitated RBBP6, confirming the interaction (the reciprocal co-IP is shown in Supplemental Fig. 1C). Also, RNA does not mediate binding, since the interaction was resistant to RNase treatment (Fig. 2C).
We then asked whether RBBP6 is required for assembly of the 3′ processing complex. NE made after RBBP6 knockdown was briefly incubated with an in vitro transcribed SVL RNA under cleavage conditions to allow complex formation and then loaded onto a nondenaturing gel. No defects in 3′ complex formation were observed (Supplemental Fig. 1D), indicating that RBBP6 is not necessary for 3′ complex assembly.
Finally, as an additional way to characterize RBBP6 involvement in 3′ processing, we examined whether RBBP6 binds RNA. A gel shift assay with purified RBBP6-N and SVL RNA shows that this part of the protein could bind RNA in vitro (Supplemental Fig. 2A). Binding was resistant to the addition of up to 10-fold excess of a nonspecific competitor (tRNA), while it was reduced to 50% when equivalent amounts of cold and hot SVL were added to NEs and was eliminated completely by 10-fold excess of unlabeled competitor. As indicated by the gel shift assays, the interaction displayed a Kd of ∼60 nM (Supplemental Fig. 2B, first panel).
The DWNN is essential for 3′ processing activity
As mentioned above, RBBP6-N is comprised of three domains, of which the most N-terminal is the DWNN. This domain is particularly interesting because in addition to being present in full-length RBBP6, it is also expressed in vertebrates as a small protein, iso3 (see above; see the Discussion). Since the function of DWNN is unknown, we next investigated whether it contributes to RBBP6-N 3′ processing activity. To this end, we first expressed an RBBP6-N derivative that lacks the entire DWNN (ΔDWNN) and purified it from E. coli (Fig. 1D, schematic diagram; Supplemental Fig. 1A, Coomassie-stained gel of ΔDWNN). As shown in Figure 2D, ΔDWNN, in contrast to RBBP6-N, was unable to reconstitute cleavage activity of NEs prepared after RBBP6 knockdown.
The ΔDWNN protein could be defective in 3′ processing because it lost its ability to bind RNA or interact with CstF. Surprisingly, when a gel shift assay was done with purified ΔDWNN (Supplemental Fig. 2B, second panel), RNA-binding affinity was actually enhanced relative to RBBP6-N, indicating that the DWNN is in some way inhibitory to RNA binding. Consistent with this, a derivative consisting of the DWNN only or purified iso3 showed no RNA binding (Supplemental Fig. 2B, third and fourth panels). Addition of increasing amounts of the DWNN only (or iso3) did not affect the interaction between RNA and the ΔDWNN protein (Supplemental Fig. 2C), indicating that the inhibitory effect cannot be exerted in trans.
DWNN/iso3 binds CstF and inhibits cleavage in vitro
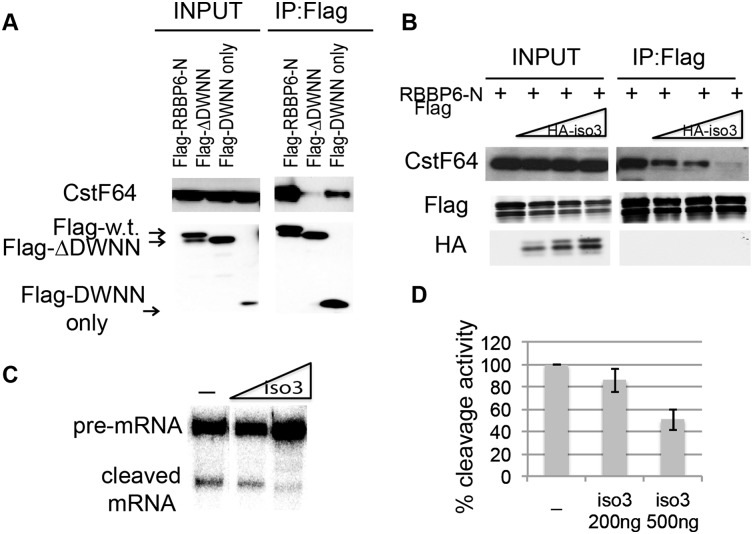
Another possible reason why ΔDWNN was unable to reconstitute cleavage activity of NEs after RBBP6 knockdown could be that it lost its ability to interact with CstF. To test this possibility, we transfected 293T cells with Flag-tagged RBPP6-N or Flag-ΔDWNN vectors, prepared extracts, and performed immunoprecipitation with an anti-Flag antibody as in Figure 2B. Figure 3A shows that while, as expected, RBBP6-N bound to endogenous CstF, ΔDWNN did not. The Flag-DWNN-only protein, on the other hand, was sufficient to bind CstF. These results show that the DWNN is necessary and sufficient for the interaction between RBBP6 and CstF.
Figure 3.
The DWNN of RBBP6 is required for cleavage activity, while iso3 inhibits 3′ processing. (A) Co-IP experiment as in Figure 2B but using either wild-type (w.t.), ΔDWNN, or DWNN-only constructs of RBBP6-N. (B) Co-IP as in Figure 2B but with increasing amounts of HA-tagged RBBP6iso3. (C) 3′ cleavage assay with increasing amounts of His-RBBP6 iso3 purified from E. coli. (D) Diagram representing the mean of three separate experiments as in Figure 1C, with standard error bars.
In light of the above, an intriguing possibility is that RBBP6 and iso3 might compete for binding to CstF. To test this, we transfected fixed amounts of Flag-RBBP6-N with increasing amounts of HA-iso3 and performed an immunoprecipitation with anti-Flag antibody as above. As can be seen in Figure 3B, iso3 was indeed able to outcompete RBBP6-N for binding to CstF. We then examined whether adding increasing amounts of His-tagged iso3 to the NE would inhibit 3′ cleavage (a Coomassie-stained gel of His-iso3 is shown in Supplemental Fig. 1A). Indeed, purified iso3 inhibited cleavage of SVL pre-mRNA when added to the NE (Fig. 3C; quantitation of three independent experiments is shown in Fig. 3D). Inhibition was also observed using NE prepared from HeLa cells overexpressing HA-iso3 (Supplemental Fig. 2D). These results support a competition model in which the relative levels of the two RBBP6 isoforms affects the efficiency of 3′ processing (Discussion; see below).
RBBP6 modulates alternative polyadenylation (APA)
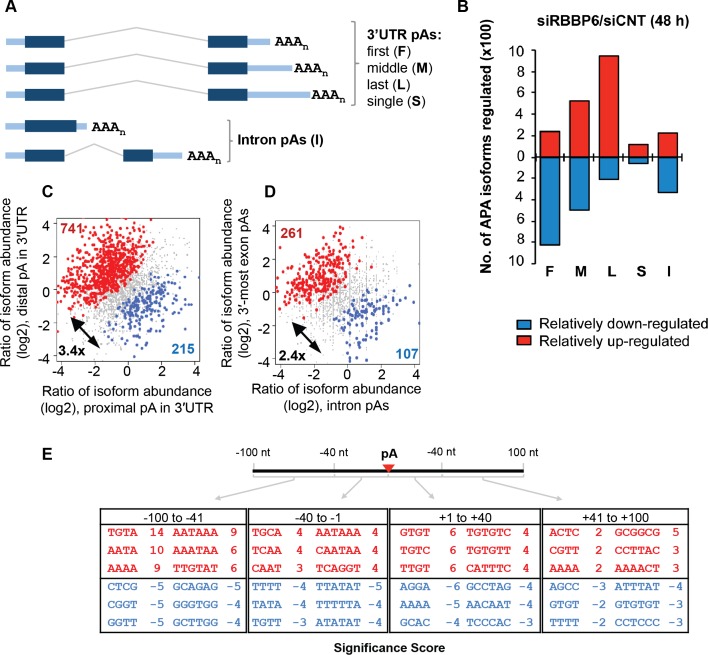
We next wished to investigate the functions of RBBP6 in vivo, including the possibility that the protein plays a role in APA. Most human genes encode transcripts with more than one potential poly(A) site, and APA is a widespread mechanism that generates mRNA isoforms with alternative 3′ ends (for review, see Di Giammartino et al. 2011; Elkon et al. 2013; Tian and Manley 2013). A change in the levels or activity of core polyadenylation factors has been shown to affect APA globally (Gruber et al. 2012; Yao et al. 2012). Given the above results implicating RBBP6 in 3′ processing, we wondered whether lowering RBBP6 levels in cells would affect APA. To this end, we used 3′ region extraction and deep sequencing (3′ READS) (Hoque et al. 2013) to detect APA changes following RBBP6 knockdown in MCF-7 cells. (We used MCF-7 cells because, as mentioned above, RBBP6 can interact with p53 and we wanted to carry out the knockdown in the background of a functional p53, which is not the case in HeLa cells.) Supplemental Figure 3A shows a Western blot following knockdown of RBBP6 for 48 or 72 h. The total number of poly(A) site-supporting (PASS) reads is shown in Supplemental Figure 3B, and the relative abundance of a specific polyadenylated isoform was defined as the fraction of PASS reads corresponding to that isoform over all PASS reads derived from the relevant gene (see the Materials and Methods for more details). We examined APA events based on the poly(A) site types depicted in Figure 4A and found a general lengthening of 3′ UTRs after siRNA treatment for 48 h (Fig. 4B). Among the up-regulated isoforms after RBBP6 knockdown, 48% used the last poly(A) site of the gene (L), and only 10% used the first (F), while among isoforms that decreased in abundance, 45% used the first poly(A) site, and only 10% used the last. This trend could also be seen when the two most abundant 3′ UTR APA isoforms of each gene were compared (Fig. 4C): Greater than threefold more genes had the distal site isoform increased compared with those with the proximal site isoform elevated (741 vs. 215). In addition, intronic APA isoforms were decreased (Fig. 4D): About 2.5-fold more genes displayed increased levels of isoforms using 3′-most exon poly(A) sites as compared with genes having elevated amounts of isoforms using upstream (intronic) poly(A) sites (261 vs. 107) (Fig. 4D). These results together indicate a global shift to distal poly(A) sites in RBBP6 knockdown cells, regardless of the poly(A) site location. siRNA treatment for 72 h gave similar results (Supplemental Fig. 3C–E).
Figure 4.
RBBP6 levels modulate APA. (A) Schematic of different poly(A) site types analyzed in this study. (F) First poly(A) site in the 3′ UTR; (M) middle poly(A) site in the 3′ UTR; (L) last poly(A) site in the 3′ UTR; (S) single poly(A) site in the 3′ UTR; (I) intronic poly(A) site; (pA) poly(A) site. (B) Summary of up-regulated or down-regulated APA events. RNA samples were processed with the 3′ READS method following knockdown of RBBP6 in MCF-7 cells for 48 h. Poly(A) sites are divided into five types, as in A. Up-regulated and down-regulated APA isoforms are shown in red and blue boxes, respectively. (C) Analysis of 3′ UTR APA. Significant events are colored with red [distal poly(A) site isoform relatively up-regulated] or blue [proximal poly(A) site isoform relatively up-regulated]. Only the two most abundant isoforms from each gene based on 3′ READS read number were analyzed. (D) Analysis of intronic APA events. Significant events are colored with red [3′-most exon poly(A) site isoforms relatively up-regulated] or blue [intronic region poly(A) site isoforms relatively up-regulated]. (E) Significant cis elements associated with poly(A) sites of regulated isoforms. Tetramers and hexamers in four surrounding regions were examined; i.e., −100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt. Numbers are −log10(P-value). The positive sign indicates up-regulation (in red), and the negative sign indicates down-regulation (in blue).
We then examined cis elements surrounding the poly(A) sites whose isoforms were altered in siRBBP6-treated cells (Fig. 4E). Canonical cis elements, including upstream UGUA elements, AAUAAA hexamers, and downstream U/G-rich elements (see Tian and Manley 2013), were significantly associated with poly(A) sites of up-regulated isoforms (Fig. 4E, in red), indicating that strong poly(A) sites were used preferentially in RBBP6 knockdown cells. Several other sequence elements, whose roles in poly(A) site usage are not clear, were associated with poly(A) sites of down-regulated isoforms (Fig. 4E, in blue). Taken together, our genomic analysis of APA suggests that RBBP6 plays a role in global APA, facilitating proximal (and weak) poly(A) site usage, consistent with its positive function in 3′ end processing.
Knockdown of RBBP6 decreases expression of mRNAs with AU-rich 3′ UTRs
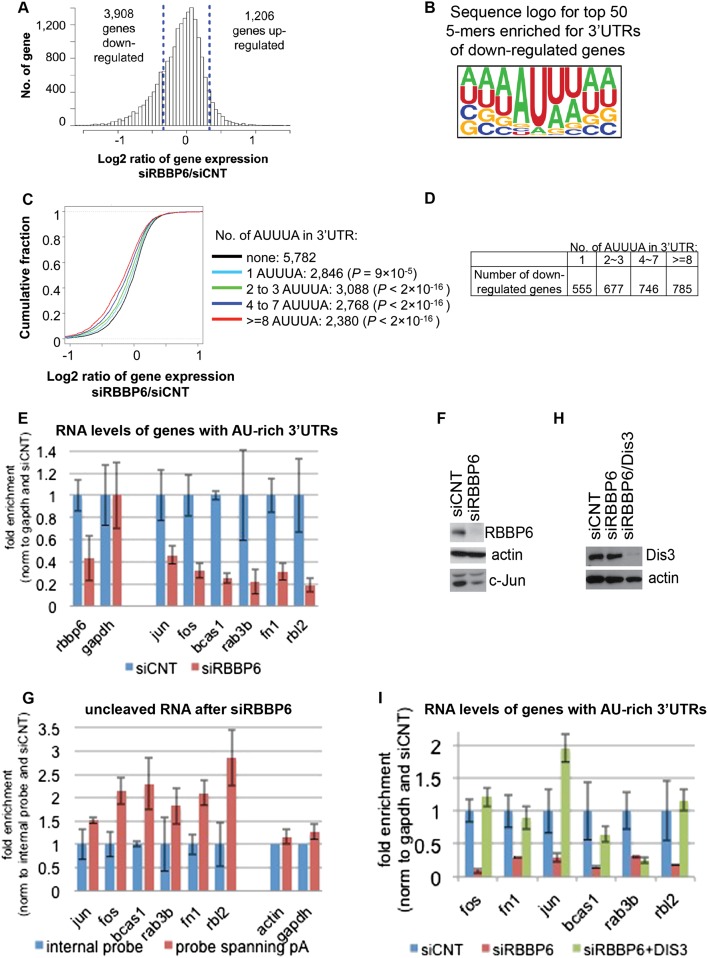
We next asked whether RBBP6 knockdown, in addition to altering APA, might also affect transcript abundance. To this end, we used Affymetrix genome-wide exon arrays, which can be more sensitive than deep sequencing in analyzing genes expressed at low levels (Guo et al. 2013). We found that after 72 h, RBBP6 knockdown caused decreased expression of 3908 genes compared with 1206 genes that displayed increased expression (Fig. 5A). A similar bias in numbers of up-regulated versus down-regulated genes was also observed with the 3′ READS data (Supplemental Fig. 4A). Interestingly, the disease and biological function term “cancer” was found to be most significantly associated with down-regulated genes by ingenuity pathway analysis (IPA) (Supplemental Fig. 4B), suggesting that RBBP6 plays a role in expression of cancer-related genes.
Figure 5.
RBBP6 knockdown leads to down-regulation of ARE-containing transcripts. (A) Histogram for gene expression changes based on microarray data of MCF-7 cells after siRNA to RBBP6 (siRBBP6) or nontargeting sequence (siCNT). Dotted lines indicate one standard deviation value of gene expression changes. (B) Sequence logo of top 50 pentamers associated with the 3′ UTRs of down-regulated genes. Pentamer sequences are shown in Supplemental Figure 4C. (C) Cumulative fraction analysis of genes with different numbers of AREs in the 3′ UTR. The Kolmogorov-Smirnov (KS) test P-value for difference in data distribution between genes with AREs and those without is indicated. (D) The table shows the number of down-regulated genes based on AUUUA repeats in the 3′ UTRs. (E) RNA was extracted from MCF-7 after siRBBP6 or siCNT, and RT-qPCR was used to calculate the relative amount of the indicated transcripts as normalized to siCNT and gapdh. (F) Western blot with the indicated antibodies after knockdown of RBBP6 in MCF-7 cells. (G) RNA was extracted from MCF-7 after siRBBP6 or siCNT, and RT-qPCR was used to calculate the relative amount of the indicated uncleaved transcripts using primers spanning the last poly(A) site of each gene and normalizing to siCNT and an internal probe for each gene. (H) Western blot with the indicated antibodies after knockdown of RBBP6 or RBBP6 and Dis3 in MCF-7 cells. (I) RNA was extracted from MCF-7 after siCNT,siRBBP6, or siRBBP6 together with siDis3. RT-qPCR was used to calculate the relative amount of the indicated transcripts as normalized to siCNT and gapdh. For all RT-qPCR analyses, results from three independent experiments are shown, represented as mean and standard error.
We next examined the 3′ UTRs of affected transcripts using the microarray data. Based on analysis of pentamers, we found that, following RBBP6 knockdown, pentamers rich in A and U residues were highly enriched in 3′ UTRs of down-regulated genes as compared with 3′ UTRs of unaffected genes (Fig. 5B). AU-rich elements (AREs) are found in 3′ UTRs of many mRNAs and constitute one of the most common determinants of RNA stability (for review, see Gingerich et al. 2004). AREs are defined as sequences with frequent A and U residues, the best characterized of which have a core sequence of AUUUA within a U-rich context. AUUUA, in fact, was one of the significant pentamers identified to be enriched in the 3′ UTRs of down-regulated genes (Supplemental Fig. 4C, highlighted in red), with a P-value of 10−119 (Fisher’s exact test). We then compared genes with different numbers of AUUUA motifs. As shown in Figure 5C, genes with more AUUUA motifs in their 3′ UTRs tended to be more down-regulated, further indicating that genes with AREs are down-regulated in siRBBP6 cells (the table in Fig. 5D shows the numbers of down-regulated genes based on number of AUUUA repeats in the 3′ UTR).
Given the abundance of AREs in the 3′ UTRs of down-regulated genes, we wished to determine whether miRNA-binding sites were also overrepresented in such genes. To this end, we determined whether potential miRNA-binding sites were enriched in 3′ UTRs of down-regulated genes versus genes that showed no change in expression following RBBP6 knockdown. Supplemental Figure 4D shows that in the top 50 hits of all possible 6-mers, several potential miRNA-binding sites, such as target seeds for mir-374, mir944, and mir1277, were enriched. However, since these are exclusively AU-rich motifs (ATTATA, ATAATT, and TATATT, respectively), we conclude that miRNA-binding sites, unlike AREs, were not overrepresented in the RBBP6-sensitive transcripts.
We next wished to validate the microarray data. We first analyzed by RT-qPCR the levels of a number of mRNAs with AU-rich 3′ UTRs that appeared to be down-regulated by RBBP6 knockdown in the above microarray analysis. Figure 5E shows that the levels of six AU-rich transcripts analyzed—c-Jun, c-Fos, Bcas1, Rab3b, Rbl2, and Fn1, all of which encode proteins implicated in cancer—were indeed all decreased following RBBP6 knockdown. c-Jun protein levels were also reduced by RBBP6 knockdown (Fig. 5F). Essentially identical results were obtained with a second RBBP6 siRNA (Supplemental Fig. 5A).
One explanation for the down-regulation of the ARE containing mRNAs was that these naturally unstable transcripts were further destabilized in the absence of RBBP6. To test this possibility, we measured the half-life of c-Jun and c-Fos mRNAs after RBBP6 knockdown by actinomycin D chase (Supplemental Fig. 5B). No significant differences between siCNT- and siRBBP6-treated cells were observed. Similar results were obtained using a tet-off β-globin mRNA-based assay in which the c-Fos ARE was introduced in the globin 3′ UTR, or the whole c-Jun 3′ UTR was inserted in place of the original 3′ UTR. No change in stability was observed for either transcript following RBBP6 knockdown and addition of tet (Supplemental Fig. 5C).
Another explanation for the enrichment of ARE-containing mRNAs in the down-regulated genes could be that these RNAs are short-lived, and therefore reductions in their levels would occur more rapidly. An extension of this idea is that all short-lived mRNAs might appear as RBBP6 targets. To address this question, we identified a number of short-lived transcripts that do not contain AREs in their 3′ UTRs (Yang et al. 2003). Many of these mRNAs (e.g., DUSO6, half-life 25 min; HNF3, half-life 32 min; and SHP, half-life 35 min) were not down-regulated following RBBP6 knockdown, indicating that not all unstable mRNAs are affected by RBBP6 knockdown. In addition, among our top 20 down-regulated genes, we identified four transcripts (TFPI, TCN1, GTF2H5, and ABCA12) with half-lives of >20 h (Schueler et al. 2014), indicating that some very stable mRNAs can also be down-regulated by RBBP6 knockdown. We then used a recently published data set containing global-scale information on mRNA half-lives (Schueler et al. 2014) and compared this with gene expression changes based on our microarray data following RBBP6 knockdown. The graph in Supplemental Figure 5D confirms that there is no correlation between transcript half-life and gene expression changes after knockdown (Pearson correlation coefficient of −0.048 and −0.144 for MCF7 and HEK293, respectively).
RBBP6 knockdown impairs 3′ end processing of ARE-containing transcripts
We next examined whether 3′ end formation of the AU-rich transcripts was impaired after RBBP6 knockdown. For this, we used RT-qPCR with primers spanning the poly(A) site of AU-rich mRNAs to detect possible 3′ cleavage defects in siRBBP6-treated compared with siCNT-treated cells [if more than one poly(A) site was present, the most distal one was selected]. Results were normalized to an internal region in each gene so that any effect that RBBP6 knockdown might have on total transcript levels would not influence results. Figure 5G shows that the AU-rich mRNAs were indeed less efficiently cleaved after RBBP6 knockdown. No significant differences were observed for actin and GAPDH mRNAs, indicative of specificity in the response to RBBP6 knockdown. To confirm that this was not the result of a change in stability in the uncleaved RNA following RBBP6 knockdown, we measured the half-lives of endogenous uncleaved c-Jun and c-Fos pre-mRNAs. Supplemental Figure 6A shows that there was no change in stability of either transcript after RBBP6 knockdown.
Defects in 3′ processing in yeast are known to be coupled to exosome-mediated degradation of the unprocessed RNAs (Hilleren et al. 2001; Milligan et al. 2005; Kazerouninia et al. 2010). We therefore suspected that the observed decreased accumulation of ARE-containing transcripts could be a consequence of exosomal degradation of the uncleaved transcripts. To test this idea, we carried out a double knockdown of RBBP6 and a catalytic subunit of the exosome, Dis3. Figure 5H shows a Western blot indicating knockdown efficiency, and Figure 5I presents the RT-qPCR analysis, normalized to siCNT. While RBBP6 knockdown alone again decreased the abundance of AU-rich RNAs (Fig. 5I, cf. blue and red bars), double knockdown of RBBP6 and Dis3 restored AU-rich RNA accumulation, with the exception of Rab3b, to levels detected in siCNT-treated cells (Fig. 5I, cf. blue and green bars). The exosome has two catalytic subunits, Dis3 and Exosc10 (also known as Rrp6), both of which have overlapping and specific roles in degrading distinct classes of substrates (Gudipati et al. 2012). Consistent with this, we obtained similar results for c-Fos, c-Jun, and Fn1 transcripts following RBBP6/Exocs10 double knockdown but not for Bcas, Rab3b, and Rbl2 (Supplemental Fig. 6B).
Increased RBBP6 iso3 expression down-regulates ARE-rich mRNA expression and 3′ processing
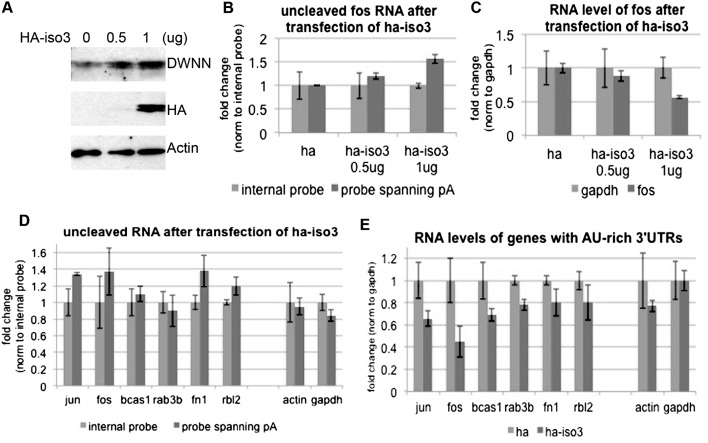
Our in vitro data provided evidence that RBBP6 iso3 competes with RBBP6 and thereby reduces 3′ cleavage efficiency. If iso3 functions similarly in vivo, then iso3 overexpression should repress ARE-containing mRNA accumulation by inhibiting 3′ cleavage, similar to the effects brought about by RBBP6 knockdown. We examined this by transfecting MCF-7 cells with HA vector alone or increasing amounts of a plasmid encoding HA-iso3 and then measuring 3′ cleavage of endogenous c-Fos RNA, analogous to what was done in Figure 5E. Figure 6A shows a Western blot with anti-HA antibody to visualize the transfected HA-iso3 and anti-DWNN antibody to detect endogenous and transfected iso3 protein levels. The amount of unprocessed c-Fos transcript indeed accumulated in a dose-dependent manner relative to the increasing amount of iso3 (shown by RT-qPCR in Fig. 6B). As in the case of RBBP6 knockdown, this was accompanied by a dose-dependent decrease in total c-Fos RNA accumulation, as normalized to gapdh (Fig. 6C). Supplemental Figure 6, C and D, shows that processing of gapdh and actin transcripts was not significantly affected by HA-iso3 overexpression. We then confirmed by RT-qPCR that AU-rich transcripts and in particular c-Jun, c-Fos, Fn1, and Rbl2, but not actin and gapdh, were cleaved less efficiently following expression of HA-iso3 (Fig. 6D; Supplemental Fig. 6E, t-test values). Note that inhibition was less than achieved by siRNA, which might reflect lower transfection efficiency of plasmids compared with siRNAs and/or incomplete competition of transfected iso3 with endogenous iso1. Also, as expected from the defects in 3′ cleavage and consistent with the effects of knockdown analyzed above, accumulation of all of the AU-rich mRNAs was repressed by HA-iso3 expression, while actin and gapdh mRNAs were affected minimally or not at all (Fig. 6E).
Figure 6.
RBBP6 isoform3 inhibits cleavage of AU-rich mRNAs and reduces their expression level. (A) MCF7 cells were transfected for 48 h with HA or increasing amounts of HA-iso3, and cell lysates were analyzed by Western blots with the indicated antibodies. (B) RNA was extracted from MCF-7 cells after transfection with an empty HA vector or increasing amounts of HA-tagged RBBP6 isoform3 (HA iso3). RT-qPCR was used to calculate the relative amount of uncleaved fos transcripts using primers spanning the last poly(A) site normalizing to an internal probe. (C) RNA was extracted from MCF-7 cells after transfection with an empty HA vector or increasing munts of HA-iso3. RT-qPCR was used to calculate the relative amount of fos as normalized to gapdh and transfection with HA vector. (D) RNA was extracted from MCF-7 cells after transfection with an empty HA vector or of HA-tagged RBBP6 iso3. RT-qPCR was used to calculate the relative amount of the indicated uncleaved transcripts using primers spanning the last poly(A) site of each gene. Values were normalized to an internal probe of each gene. (E) RNA was extracted from MCF-7 cells after transfection with an empty HA vector or HA-iso3. RT-qPCR was used to calculate the relative amount of the indicated transcripts as normalized to gapdh and transfection with HA vector. For all RT-qPCR analyses, results from three independent experiments are shown, represented as mean and standard error.
Discussion
In this study, we used biochemical and global analyses to characterize the function of RBBP6. Our results showed that RBBP6 stimulates 3′ cleavage in vitro and interacts with CstF and CPSF. Binding is mediated by an unusual ubiquitin-like domain, the DWNN, and is especially strong to CstF. RBBP6 does not appear to be a core CstF subunit (e.g., Takagaki et al. 1990) but, similarly to another 3′ processing factor, symplekin, may serve to help link CstF and CPSF, as was, in fact, suggested for yeast Mpe1 (Vo et al. 2001). In addition, we described a novel mechanism to modulate 3′ processing, in which a truncated RBBP6 isoform produced by alternative RNA processing (iso3) competes with the functional protein to control cleavage efficiency. Finally, we showed that RBBP6 levels can modulate APA and affect accumulation of specific target transcripts. Below we discuss how these and other properties of RBBP6 contribute to 3′ processing and regulation of gene expression.
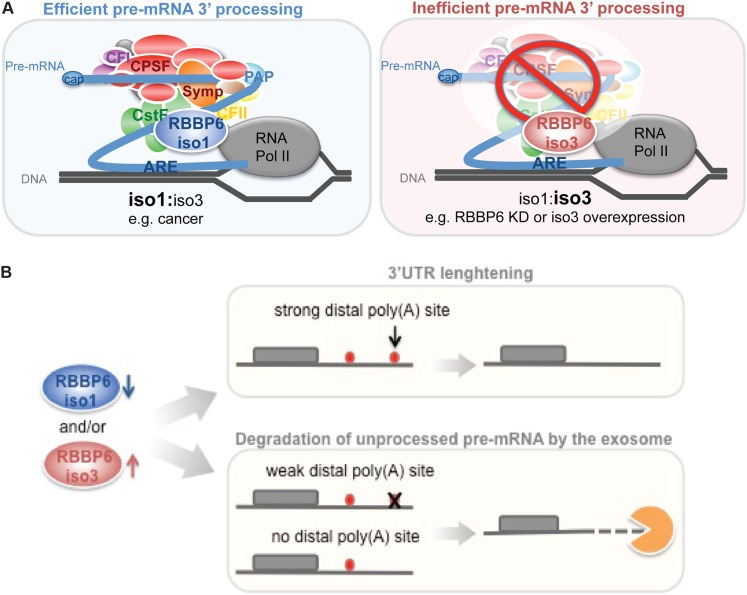
Studies of RBBP6 to date have focused mostly on its RB- and p53-binding domains; this likely reflects how the protein was initially discovered (Sakai et al. 1995; Simons et al. 1997). However, these domains are found exclusively in mammalian homologs of the protein, while the first three domains are present in all eukaryotes. Our data show that these N-terminal domains are necessary and sufficient for RBBP6 function in cleavage, supporting the idea that 3′ processing is the primary role of RBBP6. In fact, the complex domain composition of RBBP6 suggests that the protein plays roles in multiple cellular events, perhaps functioning to integrate such pathways with pre-mRNA processing. Indeed, RBBP6 has been reported to have roles in cell proliferation (Gao et al. 2002) and differentiation (Witte and Scott 1997; Ji et al. 2009), and RBBP6 levels are increased in a number of tumors (Yoshitake et al. 2004; Motadi et al. 2011; Chen et al. 2013). In contrast to the behavior of full-length RBBP6, also known as iso1, iso3 is down-regulated in several tumors (Mbita et al. 2012). Our results suggest a novel mechanism for how differences in the ratio between these two isoforms could help control cell proliferation. In this model (Fig. 7A), alterations in the levels of the two isoforms modulates 3′ processing efficiency and thereby APA by affecting the competition between iso1 and iso3. In cancer cells, there is less of the inhibitory iso3 and more functional iso1, which together result in enhanced 3′ processing activity. Such increased activity is expected to favor the use of proximal poly(A) sites, leading to shorter 3′ UTRs, a known property of cancer cells that contributes to activation of certain oncogenes (Mayr and Bartel 2009). Our data further suggest that this includes proteins such as c-Fos and c-Jun, as mRNAs encoding these proteins are among the strongest RBBP6 targets. It is possible that interactions between iso1 and p53 or Rb also influence RBBP6 activity in 3′ processing, although future experiments are required to address this.
Figure 7.
Model for RBBP6-mediated regulation of 3′ processing. (A) RBBP6 iso1 competes with iso3 in binding to CstF64. When iso1 is up-regulated and/or iso3 is down-regulated, such as in cancer cells, iso1 can bind to CstF64, and pre-mRNA 3′ processing functions properly; when the opposite is true (for example, after knockdown of iso1 or overexpression of iso3), 3′ cleavage is inhibited by binding of iso3 to CstF64, resulting in down-regulation of gene expression, especially of ARE-containing transcripts. (B) RBBP6 affects APA and gene expression. In genes with two or more APA sites, when RBBP6 levels are low and/or iso3 levels are high, the more distant and typically strongest poly(A) site will be used, resulting in 3′ UTR lengthening. However, if there is only a single poly(A) site or if the distal site is not strong enough, then the pre-mRNA will not be cleaved, and failure to process the RNA will lead to its degradation by the exosome.
We showed that RBBP6 isoforms iso1 and iso3 have opposing functions in 3′ processing, which provides a new mechanism of regulating mRNA processing. Interestingly, iso1 and iso3 expression, in addition to being inversely correlated in several cancers, is also inversely regulated during differentiation of C2C12 cells such that iso3 is up-regulated, and iso1 down-regulated (Ji and Tian 2009). How this alternative processing is regulated is an important question. The poly(A) site used in iso3 production is found in intron 3 (Fig. 1D), and its usage might thus be determined by a competition between splicing and polyadenylation, which could be modulated in several ways (e.g., Kaida et al. 2010; Luo et al. 2013). Alternatively, this could be regulated by changes in intron 3 splicing efficiency, consistent with well-documented changes in splicing that occur during differentiation and disease (for review, see David and Manley 2010; Singh and Cooper 2012). Finally, an attractive idea is the possibility of autoregulation of the two isoforms. Thus, in cells with less iso3, as in cancer cells, 3′ processing will be more efficient, resulting in increased usage of the proximal poly(A) site and enhanced iso3 production and vice versa with low iso1 levels.
Our results showing global changes in APA following RBBP6 knockdown indicate that total levels of the protein as well as differences in the iso1/iso3 ratio can modulate APA. Other studies have shown that decreased expression of 3′ processing factors often correlates with global 3′ UTR lengthening. For example, most core polyadenylation factors were found to be down-regulated in differentiated embryonic tissues as compared with induced pluripotent stem cells, and this correlated with global 3′ UTR lengthening (Ji et al. 2009). The same was observed during differentiation of C2C12 myoblasts into myotubes (Ji and Tian 2009). Our data strengthen the idea that changes in abundance of a single 3′ processing factor can modulate APA, similar to what has been shown for CstF64 (Yao et al. 2012), CFI68 (Gruber et al. 2012), and CFI25 (Masamha et al. 2014).
Despite binding of RBBP6 to RNA, we were unable to obtain evidence for RNA-binding specificity by either using a variety of RNAs for gel shift assays or performing SELEX-seq (systematic evolution of ligands by exponential enrichment combined with deep sequencing) with RBBP6-N and an in vitro synthesized library (DC Di Giammartino, W Li, B Tian, and J Manley, unpubl.). However, this is not unprecedented, as there are many examples of proteins involved in RNA processing that interact with RNA but display limited or no specificity, including other components of the 3′ complex, such as FIP1, CPSF100, and CPSF30 (Xiang et al. 2014). Consistent with our data, a recent study, published while this report was under revision, found that the zinc and RING domains of Mpe1 are required for RNA binding and that the DWNN is important for interaction with CPF but not for RNA binding (Lee and Moore 2014). The same study also showed that Mpe1 is required for the previously established ubiquitination of PAP (Mizrahi and Moore 2000) and for polyadenylation activity. However, the significance of this with respect to mammalian systems is unclear, as there is currently no evidence that mammalian PAP is ubiquitinated, and, in any event, our results show that RBBP6 depletion inhibits cleavage but not polyadenylation.
Reduced levels of RBBP6 in cells results in decreased transcript levels of ∼4000 genes. To our knowledge, other measurements of global transcript levels following knockdown of a 3′ processing factor in mammalian cells have not been reported. However, previous studies in yeast showed that generation of aberrant transcripts by mutating either PAP (Milligan et al. 2005) or the splicing factor PRP2 (Bousquet-Antonelli et al. 2000) caused exosome-mediated degradation of the unprocessed RNAs and reduced levels of mRNAs in these cells. Our findings that reducing the efficiency of 3′ cleavage in human cells by decreasing the level of RBBP6 also leads to degradation of unprocessed transcripts by the exosome extends this mechanism to mammalian cells.
Our results, taken together, suggest how RBBP6 can both modulate APA and control transcript levels (Fig. 7B). When levels of active RBBP6 are low, which can be achieved by either down-regulation of total RBBP6 levels or increasing the abundance of the inhibitory iso3, pre-mRNA 3′ processing efficiency of RBBP6-sensitive transcripts is reduced, resulting in increased usage of more distal and conserved poly(A) sites (causing the observed overall 3′ UTR lengthening). However, if there is no downstream poly(A) site or if the distal site is itself weak, then the pre-mRNA will not be efficiently processed, leading to degradation of the aberrant transcript by the exosome and decreased mRNA accumulation. Indeed, our early studies on IgM H-chain pre-mRNA processing indicated that both mechanisms, a switch in APA and a change in transcript abundance due to inefficient processing, can in fact occur with the same gene when levels of a specific 3′ processing factor, CstF64, vary (Takagaki et al. 1996; Takagaki and Manley 1998).
In conclusion, we showed that RBBP6 is a functionally important component of the human 3′ processing machinery. Our studies also defined a new mechanism, competition between negative- and positive-acting RBBP6 isoforms, for regulating 3′ processing efficiency and thus gene expression. Finally, we showed that alterations in RBBP6 levels in cells affect not only APA but also the abundance of specific transcripts—notably, ARE-containing mRNAs. These and other features of RBBP6 point to an important role for the protein in regulation of gene expression during normal cell growth and disease.
Materials and methods
Cell culture and transfections
HeLa, 293T, and MCF-7 cells were cultured in DMEM with 10% fetal bovine serum. siRNA (50 nM) against RBBP6 (CGAAAGAAGAAUAUACUGA) or nontargeting control was transfected with Lipofectamine RNAimax (Invitrogen), and NEs were made 72 h post-transfection (as described in Kleiman and Manley 2001). RNA was extracted 48 or 72 h post-transfection as indicated. If double knockdown was carried out, we first transfected siRNA against 20 nM Exosc10 (CAUUAAGGAUCGAAGUAAA) or 20 nM Dis3 (AGGUAGAGUUGUAGGAAUA) for 24 h and then transfected siRNA against RBBP6 and waited an additional 48 h.
Lipofectamine 2000 (Invitrogen) was used for transient transfection of Flag- or HA-tagged RBBP6 constructs into 293T cells; cells were collected 24 h post-transfection for immunoprecipitation. Lipofectamine LTX (Invitrogen) was used for transient transfection of HA-iso3 in MCF-7 cells; cells were collected 48 h post-transfection for Westerns.
Plasmids and protein purification from bacteria
RBBP6-N and its truncations were amplified from HeLa cDNA and cloned in p3XFlag-CMV14 using HindIII and XbaI for mammalian expression or in pRSETC using XhoI and KpnI for expression from bacteria. RBBP6 iso3 was amplified from HeLa cDNA and cloned into pCMV-HA for mammalian expression using BglII and NotI or into pRSETC using XhoI and KpnI. For protein expression in E. coli, the plasmids were transformed into BL21 bacteria, and nickel-NTA-agarose beads (Qiagen) were used for protein purification followed by a second purification with Dynabeads (Invitrogen) to obtain more pure and concentrated protein.
In vitro 3′ processing assays
32P-labeled SVL full-length or precleaved RNA substrates were prepared as described previously (Ryner et al. 1989). For 3′ cleavage assays, reaction mixtures consisted of 40% NE, 0.5 ng of labeled RNA, 0.25 U of RNasin (Promega), 1 mM 3′dATP (Trilink), 2.5% polyvinyl alcohol (PVA), 20 mM creatine phosphate (Sigma), 8 mM Tris (pH 7.9), 10% glycerol, 25 mM ammonium sulfate, 0.2 mM DTT, and 0.2 mM PMSF. Polyadenylation assays contained the same reagents, with the omission of 3′dATP and addition of 1 mM MgCl2 and 1 mM ATP. Reaction mixtures were incubated for up to 90 min at 30°C followed by proteinase K treatment, phenol/chloroform extraction, ethanol precipitation, and separation on 6% urea-acrylamide gels.
Immunoprecipitation and Western blot
Following transfection, cells were collected in two packed cell volumes (PCVs) of lysis buffer (10 mM Tris at pH 7.4, 150 mM NaCl or 500 mM where noted, 0.5% NP40, 0.25% sodium deoxycholate, 0.5 mM EDTA, inhibitors) and lysed using a syringe. The lysate was centrifuged at maximum speed for 15 min at 4°C, and 2 vol of lysis buffer was then added to the supernatant. Preclearing of the supernatant was carried out for 30 min at 4°C with IgG agarose beads followed by immunoprecipitation with anti-Flag M2 agarose for 3 h at 4°C. Where noted, 100 μg/mL RNase A was added for 15 min at 30°C prior to adding the antibody. Beads were then washed three times in wash buffer (10 mM Tris at pH 7.4, 300 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 1 mM EDTA, inhibitors), and immunoprecipitated proteins were eluted from beads using 100 μg/mL 3xFlag peptide. Proteins were resolved on SDS-PAGE, and Westerns were carried out using the indicated antibodies: anti-Flag antibody (Sigma), anti-actin (Sigma), anti-Rbbp6 (Santa Cruz Biotechnology), anti-Dis3 and anti-Exosc10 (Novus), and anti c-Jun (Santa Cruz Biotechnology). Anti-CPSF100 and anti-CstF64 (Takagaki et al. 1990) were made in our laboratory, and all other antibodies for 3′ processing factors were from Bethyl Laboratories. Anti-DWNN is a monoclonal antibody from the laboratory of Dr. David Pugh (University of the Western Cape, South Africa). As secondary antibodies, we used HRP-conjugated anti-mouse or anti-rabbit (Sigma). The signal was detected using the ECL Western blotting system from GE Healthcare.
Gel shift assays
Gel shift assays were carried out by incubating the indicated amounts of recombinant proteins with 32P-labeled SVL RNA in a buffer containing 2 μg/mL heparin, 20 mM Tris (pH 7.9), 0.2 mM EDTA, 20% glycerol, and 250 mM NaCl. After 10 min of incubation at 30°C, samples were loaded onto a 5% nondenaturing polyacrylamide gel. The gel was then dried and analyzed by PhosphorImager (Molecular Dynamics Storm 860).
RT–PCR
Real-time PCR was performed in 96-well plates with Maxima SYBR Green (Roche) using StepOnePlus (Applied Biosystems). RNA extraction was carried out with Trizol (Invitrogen) followed by DNase I treatment (Fermentas). cDNA was produced with Maxima reverse transcriptase (Fermentas) following the manufacturer’s protocol. In order to quantify the RNA expression level, cDNA was amplified using primers for the indicated genes (sequences available on request), and qPCR data were analyzed by ΔΔCT method, normalizing to the gapdh gene and siCNT. For calculating the relative cleavage efficiency after knockdown or double knockdown, cDNA was amplified using primers that span the poly(A) site of each of the indicated genes [the last poly(A) site was used if more than one were present], and data were analyzed by normalizing to the value of internal primers for each gene and siCNT.
Affymetrix microarray
Fifty nanomolar RBBP6 siRNA or control siRNA was transfected in MCF7 with RNAimax (Invitrogen) for 72 h. RNA was purified using the RNeasy kit (Qiagen) followed by on-column DNase treatment (Qiagen).
The GeneChip wild-type terminal labeling and control kit combined with the Ambion wild-type expression kit were then used, and RNA was hybridized to GeneChip Exon 1.0 ST arrays (Affymetrix) according to standard protocols.
3′ READS
Total RNA was processed by the 3′ READS method as described by Hoque et al. (2013). The reverse sequencing protocol was used, which generates reads corresponding to the antisense strand of transcript. Data analysis was carried out as previously described (Hoque et al. 2013). Briefly, we first removed 5′ adapter sequences from reads, and reads with lengths <15 nucleotides (nt) after this step were discarded. We then mapped reads against the hg19 genome sequence using bowtie 2 (version 2.1.0) (Langmead and Salzberg 2012) with the following setting: “−local −5 4.” We used only reads with mapping quality score (MAPQ) ≥ 10 and required mismatches to be ≤5% of the read. Reads with two or more unaligned Ts at the 5′ end were called PASS reads, which were used to identify poly(A) sites. Poly(A) sites located within 24 nt of each other were clustered, but each cluster did not span >48 nt. Poly(A) sites mapped to the genome were further assigned to genes using gene models defined by RefSeq, Ensembl, and University of California at Santa Cruz Known Gene databases. The 3′ ends of the gene models were extended by 4 kb to include downstream poly(A) sites, but the extension did not go beyond the transcription start site of the downstream gene. To reduce false poly(A) sites, we further required that the number of PASS reads for a poly(A) site was ≥5% of all PASS reads for the gene and also detected in at least two samples.
Gene expression analysis
Raw data from the Affymetrix GeneChip Human Exon 1.0 ST array were normalized by the RMA method in the Affymetrix Power Tools (APT) program, and probe sets with detection above background (DABG) P-value < 0.05 in at least one sample group were used for further analysis. To eliminate the potential effect of APA in 3′ UTRs on expression analysis of a gene, we used only probe sets mapped to the coding sequence (CDS) to represent the expression level of a gene. We used t-test P-value < 0.05 or log2(ratio) > 1× standard deviation of log2(ratio) of all genes to select significantly regulated genes.
For 3′ READS data, all PASS reads for a gene were summed to represent the expression level of the gene. The read number of a gene was normalized to the total number of PASS reads from the sample. The resultant value, reads per million (RPM), was used to represent the expression level of a gene. To examine gene regulation, we used the set of genes showing no expression change in the microarray data (<1% difference in probe set intensity) as a reference and compared other genes with the reference set using the Fisher’s exact test. P-value < 0.01 and fold change > 1.3 were used to select significantly regulated genes.
Cis element analysis
The 3′ UTR sequence of genes were based on the last poly(A) site identified using our 3′ READS libraries or RefSeq-annotated 3′ end if no reads were available. To identify cis elements associated with regulated genes, we enumerated the number of occurrence of each 5-mer in 3′ UTRs of genes that were up-regulated, down-regulated, or not significantly changed based on the microarray analysis. We then used the Fisher’s exact test to examine the significance of association of each 5-mer with each 3′ UTR group.
APA analysis for 3′ READS
For 3′ READS data, the relative abundance of a poly(A) site isoform was defined as the fraction of PASS reads supporting the poly(A) site over all PASS reads supporting the gene. To analyze APA changes, we compared poly(A) site isoform abundance between the RBBP6 knockdown sample and the siRNA control sample. For analysis of APA events in the 3′-most exons, we compared the top two poly(A) site isoforms with highest abundance. For analysis of APA events in upstream regions, we compared the summed isoform abundance for all poly(A) sites in upstream regions with that for all poly(A) sites in the 3′-most exon. Difference in abundance of >5% and P-value < 0.05 (Fisher’s exact test) were used as the cutoff to select significantly regulated APA events.
Supplementary Material
Acknowledgments
We thank Dr. Orit Rozenblat-Rosen for sharing antibodies to 3′ processing factors, and Dr. David Pough for the anti-DWNN antibody. We also thank our laboratory members for helpful discussions. Tian’s laboratory is supported by National Institutes of Health (NIH) GM084089. Manley’s laboratory is supported by NIH GM28983.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245787.114.
References
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716 [DOI] [PubMed] [Google Scholar]
- Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. 2001. A genome-wide survey of RS domain proteins. RNA 7: 1693–1701 [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Chen J, Tang H, Wu Z, Zhou C, Jiang T, Xue Y, Huang G, Yan D, Peng Z. 2013. Overexpression of RBBP6, alone or combined with mutant TP53, is predictive of poor prognosis in colon cancer. PLoS ONE 8: e66524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibi M, Meyer M, Skepu A, Rees DJ, Moolman-Smook JC, Pugh DJ. 2008. RBBP6 interacts with multifunctional protein YB-1 through its RING finger domain, leading to ubiquitination and proteosomal degradation of YB-1. J Mol Biol 384: 908–916 [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24: 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, Manley JL. 2013. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell 49: 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506 [DOI] [PubMed] [Google Scholar]
- Gao S, Witte MM, Scott RE. 2002. P2P-R protein localizes to the nucleolus of interphase cells and the periphery of chromosomes in mitotic cells which show maximum P2P-R immunoreactivity. J Cell Physiol 191: 145–154 [DOI] [PubMed] [Google Scholar]
- Gingerich TJ, Feige JJ, LaMarre J. 2004. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim Health Res Rev 5: 49–63 [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2012. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol 9: 1405–1412 [DOI] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Sheng Q, Li J, Samuels D, Shyr Y. 2013. Large scale comparision of gene expression levels by microarrays and RNAseq using TCGA data. PLoS ONE 8: e71462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Ma X, Zhao Y, Miao L. 2013. The C. elegans homolog of RBBP6 (RBPL-1) regulates fertility through controlling cell proliferation in the germline and nutrient synthesis in the intestine. PLoS ONE 8: e58736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasin M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerouninia A, Ngo B, Martinson HG. 2010. Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA 16: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. 2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SD, Moore CL. 2014. Efficient mRNA polyadenylation requires a ubiquitin-like domain, a zinc knuckle, and a RING finger domain, all contained in the Mpe1 protein. Mol Cell Bio doi: 10.1128/MCB.00077-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Deng B, Xing G, Teng Y, Tian C, Cheng X, Yin X, Yang J, Gao X, Zhu Y, et al. 2007. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci 104: 7951–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Ji Z, Pan Z, You B, Hoque M, Li W, Gunderson SI, Tian B. 2013. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet 9: e1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to gliobastoma tumour suppression. Nature 510: 412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather A, Rakgotho M, Ntwasa M. 2005. SNAMA, a novel protein with a DWNN domain and a RING finger-like motif: a possible role in apoptosis. Biochim Biophys Acta 1727: 169–176 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbita Z, Meyer M, Skepu A, Hosie M, Rees J, Dlamini Z. 2012. De-regulation of the RBBP6 isoform 3/DWNN in human cancers. Mol Cell Biochem 362: 249–262 [DOI] [PubMed] [Google Scholar]
- Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. 2005. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol 25: 9996–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Chibi M, Xie P, Koundrioukoff S, Moolman-Smook H, Pugh D, Debatisse M, He F, Zhang L, Defossez PA. 2014. The RBBP6/ZBTB38?MCM10 axis regulates the DNA replication and common fragile site stability. Cell Reports 7: 575–587 [DOI] [PubMed] [Google Scholar]
- Mizrahi N, Moore C. 2000. Posttranslational phosphorylation and ubiquitination of the Saccharomyces cerevisiae poly(A) polymerase at the S/G2 stage of the cell cycle. Mol Cell Biol 20: 2794–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Motadi LR, Bhoola KD, Dlamini Z. 2011. Expression and function of retinoblastoma binding protein 6 (RBBP6) in human lung cancer. Immunobiology 216: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh DJ, Ab E, Faro A, Lutya PT, Hoffmann E, Rees JG. 2006. DWNN, a novel ubiquitin-like domain, implicates RBBP6 in mRNA processing and ubiquitin-like pathways. BMC Struct Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Takagaki Y, Manley JL. 1989. Sequences downstream of AAUAAA signals affect pre-mRNA cleavage and polyadenylation in vitro both directly and indirectly. Mol Cell Biol 9: 1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M, Sakai Y, Kishino T, Nikawa N, Matsuura Y, Morino K, Tamai K, Taya Y. 1995. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics 27: 511–519 [DOI] [PubMed] [Google Scholar]
- Sakai Y, Saijo M, Coelho K, Kishino T, Nikawa N, Taya Y. 1995. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics 30: 98–101 [DOI] [PubMed] [Google Scholar]
- Schueler M, Munschauer M, Gregersen LH, Fnzel A, Loewer A, Chen W, Landthaler M, Dieterih C. 2014. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol 15: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J., Manley J.L. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V. 1997. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene 14: 145–155 [DOI] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. 2012. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18: 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38: 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo LT, Minet M, Schmitter JM, Lacroute F, Wyers F. 2001. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol 21: 8346–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte MM, Scott RE. 1997. The proliferation potential protein-related (P2P-R) gene with domains encoding heterogeneous nuclear ribonucleoprotein association and Rb1 binding shows repressed expression during terminal differentiation. Proc Natl Acad Sci 94: 1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Tong L, Manley JL. 2014. Delineating the structural blue print of the pre-mRNA 3′-end processing machinery. Mol Cell Biol 34: 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64–RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci 109: 18773–18778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake Y, Nakatsura T, Monji M, Senju S, Matsuyoshi H, Tsukamoto H, Hosaka S, Komori H, Fukuma D, Ikuta, Y, et al. 2004. Proliferation potential-related protein, an ideal esophageal cancer antigen for immunotherapy, identified using complementary DNA microarray analysis. Clin Cancer Res 10: 6437–6448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.