HIV-1 Tat stimulates transcription elongation by recruiting the P-TEFb (CycT1:CDK9) CTD kinase to the HIV-1 promoter. Chen et al. show that Tat transactivation also requires the Ssu72 CTD Ser5P (S5P)-specific phosphatase. HIV-1 Tat interacts directly with Ssu72 and alters its cellular function to stimulate viral gene expression and facilitate the early S5P–S2P transition at the integrated HIV-1 promoter.
Keywords: HIV-1 Tat, Ssu72, CTD phosphatase, elongation
Abstract
HIV-1 Tat stimulates transcription elongation by recruiting the P-TEFb (positive transcription elongation factor-b) (CycT1:CDK9) C-terminal domain (CTD) kinase to the HIV-1 promoter. Here we show that Tat transactivation also requires the Ssu72 CTD Ser5P (S5P)-specific phosphatase, which mediates transcription termination and intragenic looping at eukaryotic genes. Importantly, HIV-1 Tat interacts directly with Ssu72 and strongly stimulates its CTD phosphatase activity. We found that Ssu72 is essential for Tat:P-TEFb-mediated phosphorylation of the S5P-CTD in vitro. Interestingly, Ssu72 also stimulates nascent HIV-1 transcription in a phosphatase-dependent manner in vivo. Chromatin immunoprecipitation (ChIP) experiments reveal that Ssu72, like P-TEFb and AFF4, is recruited by Tat to the integrated HIV-1 proviral promoter in TNF-α signaling 2D10 T cells and leaves the elongation complex prior to the termination site. ChIP-seq (ChIP combined with deep sequencing) and GRO-seq (genome-wide nuclear run-on [GRO] combined with deep sequencing) analysis further reveals that Ssu72 predominantly colocalizes with S5P–RNAPII (RNA polymerase II) at promoters in human embryonic stem cells, with a minor peak in the terminator region. A few genes, like NANOG, also have high Ssu72 at the terminator. Ssu72 is not required for transcription at most cellular genes but has a modest effect on cotranscriptional termination. We conclude that Tat alters the cellular function of Ssu72 to stimulate viral gene expression and facilitate the early S5P–S2P transition at the integrated HIV-1 promoter.
Functional studies of the HIV-1 transcriptional activator Tat have revealed new insights into the early steps of RNA polymerase II (RNAPII) elongation in mammalian cells. Expression of integrated HIV-1 provirus requires the host cell CycT1:CDK9/P-TEFb (positive transcription elongation factor-b) complex (Zhou et al. 2012; Kwak and Lis 2013). Limiting levels of CycT1 strongly restricts the expression of viral genes in latently infected resting memory CD4+ T cells and macrophages (Mbonye and Karn 2012; Ramakrishnan et al. 2012). P-TEFb phosphorylates multiple substrates at the core promoters, including Spt5/DSIF, NELF (negative elongation factor), and the H2BUb1 ubiquitin ligase UBE2A. P-TEFb is also an important RNAPII C-terminal domain (CTD) kinase that phosphorylates the heptad repeats during early elongation (Eick and Geyer 2013). The interaction of Tat with CycT1 directly stimulates P-TEFb kinase activity and targets the P-TEFb complex to HIV-1 TAR RNA. Additional interactions with the AFF4 subunit of the superelongation complex (SEC) (Luo et al. 2012) enhances binding to P-TEFb and TAR RNA and can further stimulate P-TEFb CTD kinase activity (He et al. 2010; Sobhian et al. 2010; Chou et al. 2013; Schulze-Gahmen et al. 2014). In addition, cellular levels of active P-TEFb are tightly regulated by the stress-sensitive HEXIM1:7SK snRNP complex (Zhou et al. 2012). Interestingly, the BRD4 bromodomain protein promotes P-TEFb occupancy and basal transcription from the HIV-1 core promoter but competes with Tat for P-TEFb and inhibits Tat transactivation (Mbonye and Karn 2012; Ramakrishnan et al. 2012; Zhou et al. 2012). Similarly, RNF20 is required for basal, but not Tat-activated, HIV-1 transcription (Brès et al. 2009), indicating that Tat bypasses the normal function of a subset of cellular transcription factors.
A key function of Tat is to counteract negative elongation factors at the basal promoter (Jadlowsky et al. 2014), including the RNA-binding NELF complex associated with RNAPII pausing (Kwak and Lis 2013). In addition to pausing, nascent HIV-1 transcripts are also subject to premature termination and processing by the Setx (Senataxin; Sen1 in Saccharomyces cerevisiae), Xrn2 5′-to-3′ exonuclease, and Microprocessor (Dgcr8/Dicer) complexes (Contreras et al. 2012; Wagschal et al. 2012). In yeast, the Sen1:Nrd1:Nab3 complex functions with the nuclear exosome to terminate snRNAs, cryptic unstable transcripts, and other noncoding RNAs (Buratowski 2009; Kuehner et al. 2011). This complex also regulates premature transcription termination at a subset of protein-coding genes (Grzechnik et al. 2014), including glucose-regulated genes, and negatively regulates Ras signaling (Darby et al. 2012). This attenuation process is RNA sequence-dependent and can be overcome by increased RNAPII elongation (Hazelbaker et al. 2013). At the HIV-1 promoter, stable short TAR-containing RNAs produced by Setx and the Microprocessor complex can accumulate and further down-regulate viral transcription, thereby reinforcing latency (Wagschal et al. 2012). Thus, the binding of pausing and premature termination factors to nascent viral transcripts, combined with highly limiting levels of P-TEFb, provide a strong barrier to viral gene expression in resting T cells. Further studies of the Tat transactivation mechanism are needed to develop this important paradigm for understanding the regulation of elongation by sequence-specific RNA-binding proteins.
Paused RNAPII complexes contain high levels of Ser5P (S5P)-CTD, which decline during the transition from initiation to elongation and again from elongation to termination, concomitant with increased S2P-CTD (Eick and Geyer 2013). Several phosphatases have been implicated to control S5P-CTD levels and coordinate these transitions at active genes. The Ssu72 CTD phosphatase selectively removes S5P and S7P without affecting S2P (Ganem et al. 2003; Krishnamurthy et al. 2004; Xiang et al. 2012; Zhang et al. 2012). In S. cerevisiae, Ssu72 predominantly occupies the 3′ end of genes, where it promotes termination in conjunction with either CPF/CPSF or Sen1:Nrd1:Nab3 complexes (Dichtl et al. 2002; He et al. 2003; Steinmetz and Brow 2003). At gene terminator regions, the Ssu72 CTD phosphatase activity can be stimulated by the symplekin (yeast Pta1) termination factor (Xiang et al. 2010). However, Ssu72 can also be detected at many core promoters, presumably through binding to TFIIB (St-Pierre et al. 2005). Both Ssu72 and TFIIB stabilize promoter–terminator gene loops (Singh and Hampsey 2007; Perkins et al. 2008). Intragenic gene looping appears to be an invariable downstream consequence of gene activation but is not required for transcription (Hampsey et al. 2011). Gene loops are associated with transcriptional memory, facilitated reinitiation, and association with nuclear pores (Tan-Wong et al. 2009; Hampsey et al. 2011) and can also confer directionality to bidirectional promoters (Tan-Wong et al. 2012). Although Ssu72 predominantly regulates S5P-CTD at the 3′ ends of genes, it may also act at the 5′ end of some genes and can stimulate transcription elongation in vitro (Dichtl et al. 2002, Reyes-Reyes and Hampsey 2007). Another S5P-CTD phosphatase, Rtr1/RPAP2, is implicated to regulate the S5P–S2P transition at gene promoters in yeast (Mosley et al. 2009), although the ability of Rtr1 to remove Y1P (Hsu et al. 2014) is also consistent with a role in termination. A third candidate, the small CTD phosphatase SCP-1, regulates S5P-CTD levels in the promoter region of neuronal genes (Eick and Geyer 2013). Unlike SCP1 and many CTD-binding proteins, the catalytic activity of Ssu72 requires the cis rather than the more prevalent trans configuration of the P-Ser5–Pro6 bond in the heptad repeats. Consequently, the Pin1 peptidyl-prolyl isomerase (Hanes 2014), which preferentially isomerizes this bond, is rate-limiting for Ssu72 activity in vivo (Werner-Allen et al. 2011).
Here we show that HIV-1 Tat, like the human symplekin termination factor, interacts directly with the C terminus of Ssu72 and strongly stimulates its CTD phosphatase activity in vitro. In turn, Ssu72 potently synergizes with Tat in a phosphatase-dependent manner to activate transcription from an integrated HIV-1:Luc promoter in HeLa cells. Moreover, we show that Ssu72 is recruited by Tat, together with P-TEFb and AFF4, to the 5′ end of the integrated HIV-1 proviral promoter in the 2D10 T-cell line, a model for HIV-1 latency (Kim et al. 2011). Knockdown of Ssu72 impairs Tat transactivation in HeLa and 2D10 T cells and affects S5P–RNAPII levels at the HIV-1 promoter in vivo. In cell-free kinase assays, the removal of S5P-CTD by Ssu72 is essential for Tat:P-TEFb-mediated S2P-CTD. ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) and GRO-seq (genome-wide nuclear run-on [GRO] combined with deep sequencing) analysis of Ssu72 in human embryonic stem cells reveals that Ssu72 predominantly localizes with S5P–RNAPII at promoters, with much lower levels detected in gene terminator regions, although a subset of active genes, including NANOG, contains high levels of Ssu72 at both promoter and terminator regions. Unlike P-TEFb, Ssu72 is dispensable for transcription at many cellular genes. Taken together, these findings suggest that Tat uses Ssu72 as an anti-termination factor and stimulates the catalytic activity of both Ssu72 and P-TEFb to coordinate the loss of S5P-CTD with the acquisition of S2P-CTD during early elongation. Because Ssu72 is not commonly required for cellular gene transcription, it could be a useful target to selectively disrupt viral gene expression in infected cells.
Results
HIV-1 Tat interacts directly with the Ssu72 RNAPII CTD phosphatase
To identify novel regulators of HIV-1 transcription, we carried out a proteomics analysis of Tat complexes using the multidimensional protein identification technology (MudPIT) approach. To start, the full-length HIV-1 HA-Tat (amino acids 1–101) protein was affinity-purified from a Tet-OFF/Tat-ON HeLa cell line (Supplemental Fig. S1A). Mass spectrometry analysis of this fraction (Fig. 1A) revealed the presence of several proteins known to be required for Tat activity, including CycT1, AFF4, and SirT1, and a CDK9 T-loop phosphatase, PPM1G (McNamara et al. 2013). Consistent with previous reports (Sobhian et al. 2010), the Tat fractions also contained inhibitory 7SK snRNP subunits MECPE and LARP7 and not HEXIM1. Interestingly, we noted that these fractions were also enriched for the Ssu72 CTD phosphatase, which has not been previously reported to affect Tat transactivation. Binding to native Ssu72 was confirmed by Western blot of HA-Tat immunoprecipitates isolated from Tat-expressing HeLa cells (Fig. 1B).
Figure 1.
HIV-1 Tat interacts directly with the human Ssu72 CTD phosphatase. (A) MudPIT analysis of affinity-purified HA-Tat101-associated protein complexes from a stable Tet-OFF/Tat-ON HeLa cell line. (B) Immunoblot analysis of anti-HA immunoprecipitates from HA-Tat HeLa stable cells with the indicated antibodies. (C) Immunoblot analysis of anti-Tat serum immunoprecipitates from 2D10 Jurkat cells before and after TNF-α treatment. (D) Recombinant S-tagged Ssu72 and S-tagged CycT1 (amino acids 1–303) were incubated with recombinant GST-Tat101. The recovered pull-down products (PD) were detected by immunoblot analysis with anti-GST antibody (to detect GST and GST-Tat proteins) and HRP-conjugated S-protein (to detect Ssu72 and Cyclin T1). (E) The pull-down assays were performed as in D for S-tagged Ssu72, with indicated GST-Tat fragments. The amounts of GST and GST-Tat proteins were monitored by Ponceau staining (PS). The regions of Tat binding to Ssu72 are shown in the schematic diagram at the bottom of the panel. (F) Recombinant full-length and truncated GST-Ssu72 were incubated with recombinant Flag-Tat101 (GST was cleaved after purification). The recovered pull-down products were detected by immunoblot with anti-Flag (to detect Flag-Tat101) and anti-GST (to detect GST and GST-Ssu72) antibodies. Arrowheads indicate GST-Ssu72 fusion proteins. (G) HA-Tat HeLa stable cells were transfected with Flag-tagged full-length or coiled-coil domain truncated (∆CC) Ssu72. The immunoprecipitation experiments were performed as in B. The region of Ssu72 binding to Tat is shown in the schematic diagram at the bottom of the panel. (H) Recombinant GST-CycT1 (amino acids 1–303) was incubated with recombinant S-tagged Ssu72 in the absence or presence of recombinant Flag-Tat101. The pull-down products were detected by immunoblot with the indicated antibodies.
To address whether Ssu72 is also present in native HIV-1 Tat complexes assembled in infected T cells, we analyzed extracts from 2D10 T (Jurkat) cells, which contain a single proviral insertion of HIV-1NL4-3 and express Tat (amino acids 1–86) under the control of the proviral long terminal repeat (LTR) promoter (Kim et al. 2011). Tat expression is induced rapidly in response to T-cell receptor signaling in these cells, dependent on activation of ERK kinases and CDK9 phosphorylation at S175 (Mbonye et al. 2013), and can also be induced strongly, but with much slower kinetics, in response to TNF-α signaling. The 2D10 HIV-1NL4-3 transgene encodes the fluorescent d2EGFP protein in place of the viral Nef gene, which can be used as a reporter for transcription activation. Activation of the latently infected 2D10 cells by TNF-α treatment induced HIV-1 mRNA with delayed (4–6 h) kinetics, as measured by quantitative RT–PCR (qRT–PCR) (Supplemental Fig. S1B), and strongly increased d2EGFP expression within 5–6 h, as measured by flow cytometry (Supplemental Fig. S1C). The native HIV-1 Tat protein expressed from the endogenous LTR promoter was readily detected by immunoblot following TNF-α treatment (Fig. 1C). Importantly, endogenous Ssu72 coimmunoprecipitated with Tat and P-TEFb in extracts from TNF-α-treated 2D10 cells (Fig. 1C). In contrast, Tat did not associate with SCP1, a distinct RNAPII CTD S5P-specific phosphatase.
To assess whether Ssu72 associates with Tat or P-TEFb, wild-type and mutant GST-Tat proteins were purified and analyzed for binding to recombinant S-tagged human Ssu72 in pull-down experiments. As shown in Figure 1D, purified S-Ssu72 bound strongly to GST-Tat (amino acids 1–86) but not to GST-Tat (amino acids 1–48), whereas recombinant S-CycT1 (amino acids 1–333) recognized both GST-Tat proteins, and no binding to GST was observed. Additional mapping experiments revealed weak but specific binding of Ssu72 to GST-Tat (amino acids 1–54) and stronger binding to longer Tat fragments that include the Arg-rich motif (Fig. 1E). However, removal of the Tat N-terminal Pro-rich motif (GST-Tat [amino acids 38–72]) also destroyed binding to Ssu72 (Fig. 1E), indicating that the interaction requires both the N-terminal domain and CTD of Tat.
Human Ssu72 contains an N-terminal phosphatase domain and a C-terminal coiled-coil domain, which binds to symplekin. In vitro pull-down experiments showed that recombinant Flag-Tat binds full-length GST-Ssu72 as well as mutants that lack the phosphatase domain but did not recognize a mutant lacking the C terminus (Fig. 1F). Further mapping revealed that in-frame deletion of the Ssu72 coiled-coil domain (Ssu72ΔCC) destroyed binding to Ssu72 in vivo (Fig. 1G). Interestingly, S-CycT1 (amino acids 1–333) was unable to bind to Ssu72 directly but was able to do so in the presence of Tat (Fig. 1H, cf. lanes 4 and 6). Sucrose gradient density fractionation of 2D10 cell extracts before and after TNF-α signaling further revealed that a fraction of virus-expressed Tat cosediments with native Ssu72 (Supplemental Fig. S1D). At present, it is unclear whether Ssu72 can be incorporated into the Tat:P-TEFb complex or forms a distinct complex with Tat in infected cells.
Ssu72 stimulates HIV-1 Tat transactivation in a CTD phosphatase-dependent manner
Although Ssu72 can regulate transcription elongation at some yeast genes, it is unclear whether it does so in mammalian cells. Given its association with Tat, we next tested whether ectopic Ssu72 affects the activity of the integrated HIV-1:Luc reporter gene in HeLa cells in the presence or absence of Tat (Fig. 2). These experiments were carried out with and without the Pin1 peptidyl-prolyl isomerase, which is critical for Ssu72 catalytic activity. Interestingly, we found that Ssu72 and Pin1 synergistically induce basal HIV-1 core promoter activity in HIV-1:Luc cells (Fig. 2A). Interestingly, Ssu72 was a relatively weak activator in the absence of Pin1, suggesting that the response may depend on Pin1-mediated isomerization of the CTD. Ssu72 and Pin1 also activated an unintegrated HIV-1:Luc reporter gene in cotransfection experiments in HeLa cells (Supplemental Fig. S2A). We did not observe activation of the p21 promoter or TOP-FLASH reporter genes, indicating that Ssu72 transactivation is promoter-specific. These data indicate that both Ssu72 and Pin1 are limiting for HIV-1 core promoter activity in vivo.
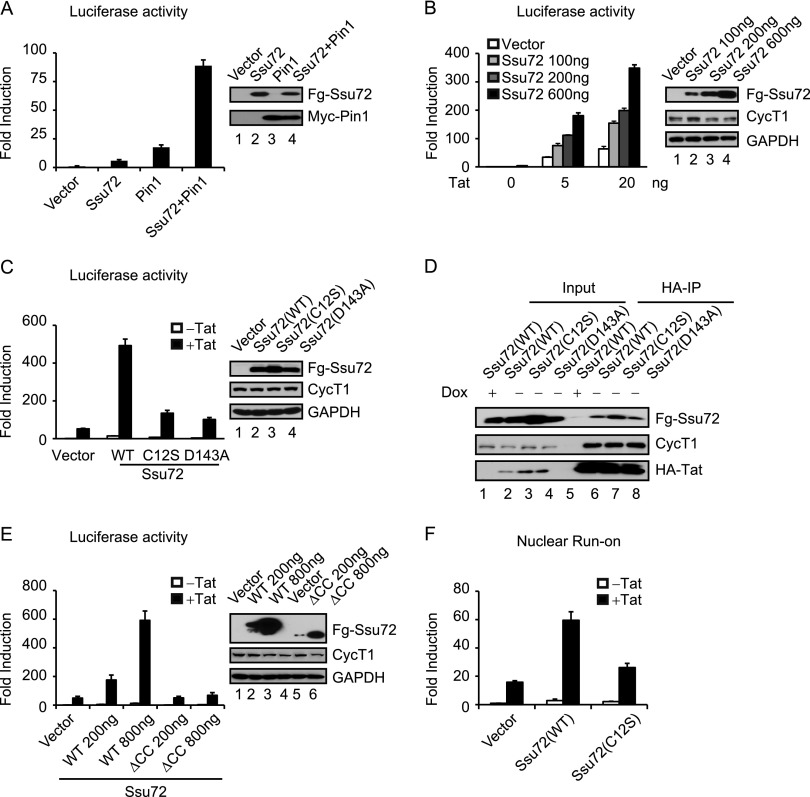
Figure 2.
Ssu72 synergizes with Tat to activate HIV-1 transcription. (A) Luciferase activities were measured in extracts of HIV-1:Luc HeLa cells transfected with the indicated constructs. The protein expression was monitored by immunoblot analysis. (B) Luciferase activities were measured in extracts transfected with the indicated amounts of Ssu72 expression constructs in the presence or absence of Tat. The protein lysates were analyzed by immunoblot with the indicated antibodies. (C) Luciferase assays were performed as in B to compare the activity of wild type or catalytically inactive mutants (C12S and D143A) of Ssu72. (D) Immunoblot analysis of anti-HA immunoprecipitates from HA-Tat HeLa stable cells transfected with Flag-tagged Ssu72 (wild type, C12S, and D143A). (E) Luciferase assays were performed as in B to analyze the activity of full-length Ssu72 and the Ssu72∆CC mutant. (F) Wild-type Ssu72 or Ssu72 C12S expression constructs were transfected in the HIV-1:Luc HeLa cells in the absence or presence of Tat. Nuclear run-on analysis was performed as described in the Materials and Methods. Error bars represent the standard deviation obtained from three independent experiments.
We next tested whether Ssu72 also stimulates Tat transactivation of the integrated HIV-1:Luc reporter in HeLa cells. Importantly, Ssu72 functioned synergistically with Tat in a dose-dependent manner to activate HIV-1 expression (Fig. 2B). The synergy between Tat and Ssu72 did not require coexpression with Pin1. Presumably, in this case, the Tat-stimulated Ssu72 relies on the endogenous Pin1 PPIase. Consistent with this scenario, knockdown of Pin1 reduced Tat transactivation in vivo (Supplemental Fig. S2B), although we cannot rule out the possibility that Tat might have cryptic PPIase activity. Ssu72 stimulation of Tat was TAR-dependent in vivo (Supplemental Fig. S2C). Further analysis revealed that point mutations in the Ssu72 catalytic site (C12S) or D loop (D143A) effectively eliminated Ssu72 stimulatory activity (Fig. 2C), indicating that transcriptional activation requires Ssu72 phosphatase activity. Western blot analysis confirmed expression of the mutant proteins (Fig. 2C), and coimmunoprecipitation experiments showed that they retained the ability to bind Tat (Fig. 2D). Moreover, a mutant Ssu72 protein lacking the C-terminal coiled-coil domain (ΔCC), which destroys binding to Tat, was unable to stimulate Tat transactivation in vivo (Fig. 2E). To confirm that the effect of Ssu72 is transcriptional, we carried out nuclear run-on experiments in the presence of biotin-UTP and recovered newly formed transcripts on strepavidin beads. As shown in Figure 2F, Ssu72 strongly stimulated nascent mRNA synthesis in the presence of Tat, whereas the catalytically inactive C12S mutant had no effect. The effect of Tat in the nuclear run-on assay was dose-dependent and had no effect on the endogenous p21 gene used as a control (Supplemental Fig. S2D). We conclude that Ssu72 binds to Tat and activates viral transcription in a phosphatase-dependent manner.
Tat stimulates Ssu72 RNAPII CTD phosphatase activity in vitro
Previous studies have shown that the binding of symplekin to the C terminus of Ssu72 strongly stimulates its CTD phosphatase activity (Xiang et al. 2010). Because Tat also interacts with this region, we addressed whether it might also affect Ssu72 catalytic activity. For these studies, in vitro phosphorylation experiments were carried out using a TFIIH:CDK7-phosphorylated GST-CTD (human) substrate containing 52 heptad repeats (Fig. 3A). Extensive S5P was confirmed by immunoblot with the H14 antibody, and we also detected high levels of S7P using the 4E12 antibody. Incubation of the hyperphosphorylated S5P-, S7P-CTD with purified wild-type Ssu72 resulted in a dose-dependent loss of S5P and S7P in vitro (Fig. 3A, lane 5). Consequently, the number of unphosphorylated CTD repeats increased in these reactions, as detected with the 8WG16 antibody, and the overall migration of the CTD increased. As expected, CTD phosphorylation was unaffected by the Ssu72 C12S mutant (Fig. 3A, lane 6). Interestingly, the loss of S5P and S7P was accelerated in reactions containing Tat (Fig. 3A, cf. lanes 4,5 and lanes 9,10). A detailed kinetic analysis (Fig. 3B) confirmed that Tat stimulates Ssu72-mediated removal of S5P and S7P, most notably at the 60-min and 90-min time points, and results in a more extensively dephosphorylated CTD, as indicated by immunoblot with the 8WG16 antibody. Moreover, the stimulation of Ssu72 activity by Tat was dose-dependent (Fig. 3C), and the highest level of Tat used in this study had no effect on S5P-CTD levels in the absence of Ssu72. To assess whether Tat stimulation of Ssu72 requires the Arg-rich motif, we identified several point mutations, including R49A and R53A, that reduce binding of Tat to Ssu72 in vitro (Fig. 3D). Further analysis revealed that these mutants were unable to stimulate Ssu72 CTD phosphatase activity in vitro (Fig. 3E). We conclude that Tat binds to the C terminus of Ssu72 and strongly stimulates its CTD phosphatase activity.
Figure 3.
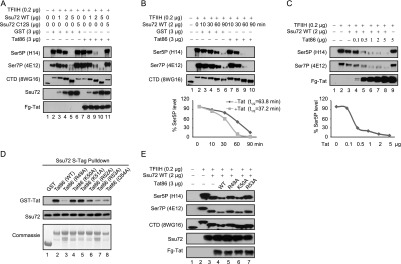
Tat stimulates Ssu72 CTD phosphatase activity in vitro. (A) TFIIH-phosphorylated GST-CTD was incubated with the indicated amounts of recombinant human GST-Ssu72 or catalytically inactive GST-Ssu72 (C12S) in the presence of GST or GST-Tat86. The phosphatase assay products were detected with the indicated antibodies by immunoblot analysis. (B) Kinetics of Tat-stimulated Ssu72 phosphatase activity were monitored for the indicated times. The intensity of the S5P bands was quantitated and plotted. The half-time for reduction of S5P levels in the absence or presence of GST-Tat86 was calculated as described in the Supplemental Material. (C) The phosphatase assays were performed as in A except that the indicated amounts of GST-Tat86 were included. (D) In vitro purified recombinant S-tagged Ssu72 was incubated with GST, wild-type GST-Tat86, or mutant GST-Tat86 proteins as indicated. The recovered pull-down products were subjected to immunoblot analysis with anti-GST (to detect GST or GST-Tat86) or anti-Ssu72 antibodies. The amounts of GST and GST-Tat86 proteins were monitored by Commassie staining. (E) The phosphatase assays were performed as in A for GST-Ssu72 except that the wild-type or mutant GST-Tat86 proteins were used, as indicated.
Ssu72 is required for Tat:P-TEFb to access the S5P-CTD in vitro
Paused RNAPII complexes, which form at the HIV-1 promoter prior to Tat:P-TEFb transactivation, are highly phosphorylated at the S5 position due to the action of the TFIIH:CDK7. Thus, the substrate for Tat:P-TEFb complexes at the proviral promoter is not the unphosphorylated CTD but an extensively S5P–RNAPII. However, it was recently reported that CTD peptides containing S5P can potently inhibit phosphorylation by P-TEFb in vitro (Czudnochowski et al. 2012), indicating that a S5P-specific CTD phosphatase may be required for P-TEFb to access the CTD. Consequently, we next addressed whether Ssu72 can act coordinately with Tat:P-TEFb in cell-free kinase assays using purified recombinant enzyme complexes (Fig. 4). As expected, Tat strongly stimulated P-TEFb-mediated phosphorylation of the unphosphorylated GST-CTD substrate, as assessed using the 3E10 (S2P-specific) and H14 (S5P-specific) antibodies (Fig. 4A). In contrast, Tat did not significantly enhance dual S2-, S5-CTD phosphorylation in these reactions, as detected with the H5 antibody (Eick and Geyer 2013). Previous studies have shown that Tat stimulates P-TEFb catalytic activity without changing the CTD substrate specificity (Czudnochowski et al. 2012). Consistent with those conclusions, increased levels of P-TEFb resulted in extensive CTD phosphorylation even in the absence of Tat (data not shown).
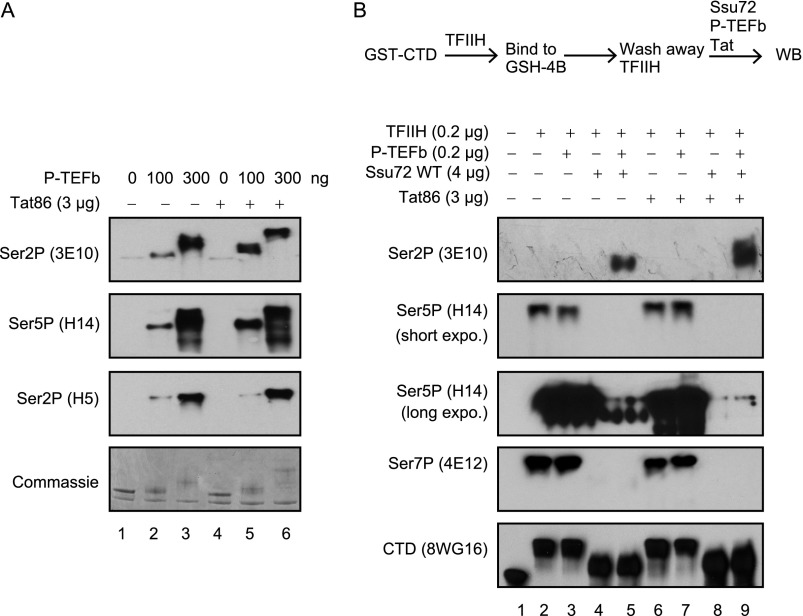
Figure 4.
Ssu72 is essential for Tat:P-TEFb to phosphorylate the S5P-CTD in vitro. (A) The GST-CTD (52 repeats) substrate was incubated with indicated amounts of P-TEFb in the absence or presence of Tat (GST-Tat86). CTD phosphorylation was monitored by immunoblot with the indicated antibodies. (B) In vitro phosphorylation of the TFIIH:CDK7-phosphorylated GST-CTD substrate by recombinant Tat:P-TEFb complexes was monitored in the presence or absence of recombinant Ssu72, as indicated. The experimental procedure is outlined above the panel, and CTD phosphorylation was monitored by immunoblot with the indicated antibodies. Note that each reaction contained identical levels of the S5P-CTD substrate.
We next addressed whether the CTD phosphatase activity of Ssu72 is necessary for P-TEFb to phosphorylate the S5P-GST-CTD substrate in vitro (Fig. 4B). In these experiments, P-TEFb and Tat were incubated alone or in the presence of Ssu72 with a GST-CTD substrate that had been previously phosphorylated by TFIIH:CDK7. The TFIIH:CDK7-phosphorylated CTD was extensively phosphorylated at S5 and S7 but not S2 (Fig. 4B, cf. lanes 1 and 2). However, P-TEFb was unable to phosphorylate this CTD substrate at the S2 position (Fig. 4B, lane 3), indicating that it cannot function on this substrate. Addition of Ssu72 to these reactions substantially reduced S5P and S7P levels (Fig. 4B, lane 4) and, when further incubated with P-TEFb, allowed robust S2P to occur (Fig. 4B, lane 5). Because Ssu72 removes S5P and S7P without affecting S2P, the CTD in these reactions is predominantly phosphorylated at the S2 position. Most importantly, addition of Tat to these reactions further stimulated the removal of residual S5P by Ssu72 (H14 antibody, long exposure in Fig. 4B, cf. lanes 4 and 8) and also increased P-TEFb-induced S2P levels (Fig. 4B, cf. lanes 4,5 with 8,9). These data indicate that Ssu72 can cooperate with Tat:P-TEFb to create a CTD that is highly enriched in S2P. Although the S5P–S2P switch is extensive in these reactions, the effects on total CTD phosphorylation are likely to be more nuanced in vivo, where Tat may reset the overall S2P, S5P, and S7P pattern as required to attract CTD-binding proteins to the RNAPII elongation complex.
Ssu72 affects transcription and S5P–RNAPII levels at the HIV-1 promoter in vivo
We also examined the effect of depleting native Ssu72 on Tat transactivation and RNAPII phosphorylation in vivo using the HeLa HIV-1:Luc reporter cell line. Knockdown of endogenous Ssu72 with two different siRNAs reduced Tat transactivation in these cells to an extent comparable with that observed by depletion of the superelongation subunit AFF1 (Fig. 5A). In contrast, Tat transactivation was unaffected by knockdown of two unrelated CTD phosphatases, RPAP2 or SCP1 (Fig. 5B), indicating that these factors do not function at the HIV-1 promoter.
Figure 5.
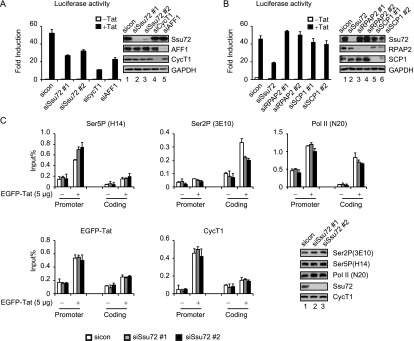
Ssu72 promotes Tat transactivation and regulates S5P-CTD levels at the HIV-1 promoter in vivo. (A) Luciferase activities were measured in extracts of HIV-1:Luc HeLa cells transfected with different siRNAs against Ssu72, Cyclin T1, or AFF1 in the presence or absence of Tat101. The efficiency of siRNA-mediated knockdown was monitored by immunoblot. (B) HIV-1:Luc HeLa cells were transfected with siRNAs specific for Ssu72, RPAP2, or SCP1 in the presence or absence of Tat, as indicated. Luciferase assays were performed as in A. (C) HIV-1:Luc HeLa cells were first transfected with control or Ssu72 siRNAs for 24 h and then were transfected with 5 µg of EGFP or EGFP-Tat101 for another 24 h. The cell extracts were subjected to ChIP analysis with the indicated antibodies. Error bars represent the standard deviation obtained from three independent experiments. Protein lysates were monitored by immunoblot with the indicated antibodies.
In S. cerevisiae, Ssu72 localizes predominantly at the 3′ end of genes, where it removes residual S5P-CTD in connection with increased S2P and loading of the CPF/CPSF termination complex and contacts factors such as TFIIB at the promoter to initiate gene looping (Hampsey et al. 2011). To investigate whether Ssu72 affects RNAPII phosphorylation at the HIV-1 promoter, we analyzed S5P–RNAPII levels by ChIP in HeLa HIV-1:Luc cells. Interestingly, knockdown of Ssu72 increased S5P–RNAPII at the HIV-1 promoter and decreased S2P at the 3′ end of the proviral coding region (Fig. 5C). In contrast, reduced levels of Ssu72 had no effect on the occupancy of Tat, P-TEFb, or total RNAPII. Moreover, Ssu72 knockdown had no effect on global S5P–RNAPII levels, indicating that it is not a global CTD S5P-specific phosphatase in these cells. These studies suggest that Ssu72 controls the RNAPII S5P–S2P transition at the integrated proviral promoter.
Tat recruits Ssu72 together with P-TEFb to the integrated HIV-1 proviral promoter in 2D10 T cells
To explore the role of Ssu72 within the context of activation of latent viral transcription in T cells, we used the 2D10 system (Kim et al. 2011), which provides a useful model to study the induction of HIV-1 transcription in response to TNF-α or TCR signaling. In this system, Tat is expressed from the endogenous HIV-1 LTR, with different kinetics depending on the signaling pathway used. Tat expression was detected 6 h after exposure to TNF-α (Fig. 6A; Supplemental Fig. S3A) and was accompanied by strong induction of the d2EGFP reporter gene (Supplemental Fig. S1C). In contrast, TNF-α treatment caused a rapid loss of IκB and nuclear translocation of NFκB within 30 min (Supplemental Fig. S3B) and induced downstream target genes such as IκBα and A20 (data not shown). Immunoblot analysis confirmed Ssu72 expression in 2D10 cells, and knockdown of Ssu72 with either of two different siRNAs significantly decreased Tat expression from the integrated provirus in these cells (Fig. 6B). Analysis of proviral mRNA levels by qRT–PCR (Supplemental Fig. S3C) and expression of the d2EGFP reporter gene (Supplemental Fig. S3D) revealed that Ssu72 is required for optimal HIV-1 transcription in these cells.
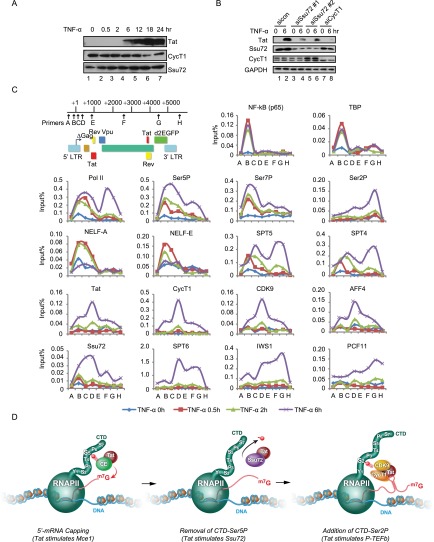
Figure 6.
Tat recruits Ssu72 to the integrated HIV-1 proviral promoter in 2D10 T cells. (A) Immunoblot analysis of Tat and cellular protein expression in TNF-α treated 2D10 cells for the indicated times. (B) Immunoblot analysis of protein expression of 2D10 cells transfected with Ssu72 or Cyclin T1 siRNAs with the indicated antibodies. (C) ChIP analysis of the recruitment of virus-encoded Tat and host cell factors to the integrated provirus in TNF-α-induced 2D10 cells. Schematic representation of the genomic organization of the lentiviral vector and the relative locations of the primers used for ChIP are shown at the top of the panel. (D) Model depicting three different steps regulated by Tat to facilitate the transition from promoter clearance to early elongation at the HIV-1 promoter. See the Discussion for details.
Changes at the integrated provirus in response to TNF-α signaling were examined by ChIP analysis in these cells (Fig. 6C). Previous studies have shown that the HIV-1 proviral promoter is generally devoid of active chromatin modifications in the absence of signaling, and basal transcription is very low (Mbonye and Karn 2012). As shown in Figure 6C, NFκB and TBP were rapidly recruited to the HIV-1 promoter in response to TNF-α signaling (0.5 h, red line). Interestingly, the NELF complex subunits (NELF-A and NELF-E) were also recruited at this time, which suggests that NFκB initially recruits NELF and induces RNAPII pausing at the HIV-1 LTR, which may contribute to the delay of viral transcription compared with that of TNF-α-induced cellular genes. In support of this scenario, the levels of S5P–RNAPII and S7P–RNAPII at the HIV-1 promoter, which were low prior to TNF-α stimulation, also increased strongly at the 5′ end of the transgene at the early time points (0.5 h and 2 h). These findings show that TNF-α signaling leads to the assembly of a paused RNAPII complex at the proviral promoter, which is only overcome at later time points (6 h) when Tat is expressed.
Consistent with the time course of Tat expression by immunoblot, our ChIP experiments revealed that Tat was not present at the HIV-1 promoter at 2 h but could be readily detected after 6 h of treatment with TNF-α (Fig. 6C, purple line). Importantly, Tat occupancy correlated with the loss of the NELF-A and NELF-E subunits. Moreover, the DSIF complex subunits Spt4 and Spt5, which were recruited together with NELF to the HIV-1 5′ LTR but not the coding region at early time points (0.5 h and 2 h), moved into the coding region upon Tat binding at the 6-h time point. Interestingly, Tat occupancy peaked downstream from the viral promoter and declined prior to reaching the 3′ end of the provirus, indicating that Tat does not travel with DSIF and the RNAPII elongation complex to the transcription termination site (TTS). Occupancy of CycT1 and CDK9 (P-TEFb) increased dramatically at the 6-h time point and followed a profile identical to that seen with Tat. Moreover, the SEC subunit AFF4 also showed the same pattern. Levels of S5P–RNAPII and S7P–RNAPII at the promoter also increased at the early 6-h time point, indicating that, in addition to increasing S2 phosphorylation, Tat may also increase P-TEFb phosphorylation at some S5 and S7 residues, as required to recruit specific CTD-binding proteins to the elongation complex.
Of note, Ssu72 levels were low at the promoter at the early time points (0.5 h and 2 h) but increased dramatically in the presence of Tat and spread into the proviral coding region (Fig. 6C). Ssu72 occupancy mirrored that of the Tat:P-TEFb complex, which peaked in the middle of the coding region and declined before reaching the 3′ end of the provirus. In contrast, the Spt6 elongation factor, which is critical for Tat transactivation, was only detected with Tat at the 6-h time point but traveled with RNAPII to the 3′ end of the provirus, similar to the pattern observed for Spt4 and Spt5. Interestingly, Iws1, a partner of Spt6 in S2P–RNAPII complexes (Yoh et al. 2007), was also only detected at 6 h but peaked strongly in the middle and at the 3′ end of the transgene, consistent with the high levels of S2P–RNAPII at this location. Last, the Pcf11 termination factor, which binds S2P-CTD, was detected only at 6 h and predominantly at the 3′ end of the provirus. Knockdown of Tat in these cells greatly reduced CycT1/P-TEFb levels at the HIV-1 LTR and reduced RNAPII phosphorylation without affecting binding of NFκB or TBP, confirming the essential role for Tat in the recruitment and activation of P-TEFb (Supplemental Fig. S3E). Moreover, analysis of the control transactivation-deficient 2B2D cell line, in which the integrated provirus encodes the inactive C22G-Tat mutant, revealed that TNF-α induces binding of NFκB and S5P–RNAPII to the proviral promoter but only marginal expression of the Tat C22G protein, and S2P–RNAPII levels did not increase in the coding region (Supplemental Fig. S3F).
Taken together, these data show that Tat transactivation is accompanied by the release of the NELF pausing complex and assembly of productive RNAPII elongation complexes that carry the Spt4:Spt5 and Spt6:Iws1 complexes to the 3′ end of the gene. The Tat:P-TEFb and AFF4/SEC complexes colocalize with RNAPII through a portion of the coding region but are displaced prior to the peak of S2P-CTD at the 3′ end of the coding region. Ssu72 levels are also strongly increased in the presence of Tat and then decline, together with Tat:P-TEFb and AFF4/SEC, in the middle of the coding region. Tat modestly increased S5P–RNAPII and S7P–RNAPII, consistent with the recruitment and activation of P-TEFb, which is balanced by the increased levels of Ssu72 and correlates with increased elongation and S2P–RNAPII throughout the coding and termination region. A model depicting the effects of Tat on P-TEFb, Ssu72, and mRNA capping during the early stages of elongation is shown in Figure 6D (see the Discussion for details).
Human Ssu72 colocalizes with S5P–RNAPII but is dispensable for transcription at many human embryonic stem cell (hESC) genes
In budding yeast, Ssu72 is part of the CPF/CPSF termination complexes and shows highest occupancy at the 3′ end of genes (Hampsey et al. 2011), with a minor peak at active promoters, which correlates with gene looping. Due to potential redundancy with other RNAPII CTD S5P-specific phosphatases (including, especially, RPAP2), it is unclear whether Ssu72 is a necessary elongation factor for mammalian genes. Our initial experiments indicated that this may not be the case because Ssu72 was dispensable for mRNA accumulation at the c-Fos and the Wnt-induced AXIN2 genes. Thus, Ssu72 may not be generally required for P-TEFb-regulated transcription.
To better define Ssu72 occupancy at cellular genes, ChIP-seq analysis was carried out in hESCs under self-renewal growth conditions (Fig. 7). A total of 16,326 Ssu72 peaks were mapped genome-wide in these cells. In contrast to the pattern observed in yeast but similar to the pattern observed at the HIV-1 promoter, human Ssu72 was preferentially recruited to host cell promoters and 5′ untranslated regions (UTRs) (Fig. 7A; Supplemental Fig. S4A). This is consistent with previous reports that human Ssu72, unlike the yeast Ssu72, is not an integral component of transcription termination complexes (Xiang et al. 2010). As shown in Figure 7A, significant levels of Ssu72 were also associated with noncoding RNA genes. Ssu72 occupancy peaked most strongly at gene transcription start sites (TSSs), declined sharply throughout the coding region, and peaked again, but more modestly, in the region beyond the 3′ TTS (Fig. 7B). In general, Ssu72 aligned with promoter-proximal S5P–RNAPII genome-wide, although some variation in the pattern of Ssu72 binding was observed at individual genes. For example, the POU5F1 gene showed the typical Ssu72 distribution at the promoter, whereas the NANOG gene contained similar levels of Ssu72 and S5P–RNAPII at the promoter and terminator regions (Fig. 7C). Overall, 5950 genes were found to contain Ssu72 near the TSS, whereas 702 genes contained Ssu72 both at the TSS and downstream from the TTS (Supplemental Fig. S4B; Supplemental Table S1).
Figure 7.
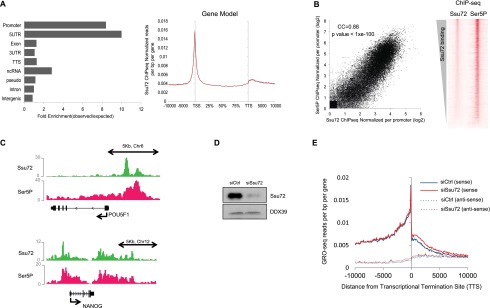
ChIP-seq and GRO-seq analysis of Ssu72 in hESCs. (A, left) Genomic distribution of Ssu72 ChIP-seq peaks. (Right) Metagene representation of the Ssu72 ChIP-seq profile. (B) The graph at the left shows a statistically significant (CC = 0.88; P-value < 1 × 10−100) correlation between the S5P–RNAPII and Ssu72 ChIP-seq peaks around gene TSSs. The right panel shows a heat map analysis of the correlation of Ssu72 and S5P–RNAPII occupancy. (C) Ssu72 and S5P–RNAPII binding at the POU5F1 and NANOG genes, as captured from the Integrative Genomics Viewer genome browser. Images show visualization of WIG files. Gene diagrams are shown at the bottom, with scale bars above. (D) Immunoblot from H1 hESCs transfected with control (Ctrl) or Ssu72 siRNAs for 48 h. DDX39 antibody was used as a loading control. (E) GRO-seq read count covering 10 kb from the gene TTS in siCtrl and siSsu72 hESCs, as indicated. The read counts from both the sense and antisense strands are plotted, as indicated in the legend.
The potential influence of Ssu72 on hESC gene transcription was evaluated by siRNA knockdown and GRO-seq analysis (Fig. 7D,E). Depletion of Ssu72 did not affect nascent transcription at most genes, indicating that it is not a general elongation factor in hESCs. Consistent with a possible role in cotranscriptional termination, a modest readthrough of transcription was observed at the 3′ TTSs of many genes, which was also apparent globally (Fig. 7E; Supplemental Fig. S4C). Taken together, these findings indicate that Ssu72 colocalizes with S5P–RNAPII in hESCs but, unlike P-TEFb, is not generally required for transcription.
Discussion
The Tat transactivation mechanism
Transcription elongation is invariably coupled with phosphorylation changes in the RNAPII CTD that reflect the interplay of specific CTD kinases and phosphatases. We show here that the HIV-1 Tat transactivator binds to Ssu72 and strongly stimulates its CTD phosphatase activity in vitro. Ssu72 also acts synergistically with Tat to stimulate HIV-1 transcription in a manner that requires the Ssu72 CTD phosphatase activity and the C-terminal motif that binds Tat. These results are supported by the finding that knockdown of Ssu72 in HIV-1:Luc HeLa cells or in latently infected 2D10 cells reduces Tat expression and increases S5P–RNAPII levels at the integrated proviral promoter. In cell-free kinase assays, Tat stimulates the catalytic activity of both Ssu72 and P-TEFb on CTD substrates. Removal of S5P-CTD by Ssu72 was essential to enable Tat:P-TEFb to phosphorylate this substrate, consistent with the observation that S5P blocks P-TEFb phosphorylation of CTD peptides in vitro (Czudnochowski et al. 2012). Thus, Tat activates Ssu72 to overcome transcriptional attenuation and pausing of the S5P–RNAPII complex at the integrated proviral promoter. Through stimulation of Ssu72 activity, Tat might also simultaneously reinforce Ssu72-dependent intragenic looping of the provirus (Perkins et al. 2008). Tat-dependent recruitment and activation of the P-TEFb kinase can then promote elongation and accelerate the early S5P–S2P transition and loading of CTD-binding factors necessary for cotranscriptional splicing and viral gene expression.
These conclusions are supported by ChIP analysis of the proviral genome in 2D10 cells, which shows that the virus-encoded Tat protein recruits Ssu72, P-TEFb, and AFF4/SEC to the HIV-1 promoter concomitant with the release of NELF and movement of RNAPII elongation complexes containing Spt4, Spt5, and Spt6 into the proviral coding region. Interestingly, Tat, Ssu72, P-TEFb, and AFF4/SEC did not travel with RNAPII to the 3′ end of the provirus, consistent with previous studies showing that P-TEFb does not colocalize with the peak of S2P–RNAPII at the 3′ end of cellular genes (Bartkowiak et al. 2010). In yeast, Ssu72 is commonly associated with the S2P–S5P transition at gene terminator regions, whereas Rtr1 (human RPAP2) controls promoter-proximal S5P levels for many active genes. However, Ssu72 is also required for transcription at a subset of yeast genes (Dichtl et al. 2002, Reyes-Reyes and Hampsey 2007). Our nuclear run-on experiments establish a transcriptional role for catalytically active Ssu72 in Tat transactivation, indicating that it can also function as a gene-specific coactivator in mammalian cells.
These new findings, combined with earlier studies, indicate that Tat controls multiple steps at the HIV-1 core promoter immediately following promoter clearance (Fig. 6D). First, Tat binds to the Mce1 capping enzyme on the S5P–RNAPII and stimulates guanylation of nascent HIV-1 transcripts (Chiu et al. 2001, 2002). Biochemical reconstitution of this reaction indicates that Tat binds viral mRNAs cotranscriptionally and can stimulate capping in early elongation complexes prior to the synthesis of the TAR RNA stem–loop structure. Following successful mRNA capping, our findings here indicate that Tat recruits and stimulates Ssu72 to remove the TFIIH:CDK7-mediated S5P-CTD. This will release factors bound to the S5P-CTD, including the capping enzymes, and potentially also the Setx premature termination complex. Thus, stimulation of Ssu72 phosphatase activity by Tat may allow it to counteract premature termination and processing of nascent transcripts by the human Setx complex (Contreras et al. 2012; Wagschal et al. 2012), in essence functioning as a gene-specific anti-termination factor. Tat then stimulates P-TEFb to phosphorylate the CTD at the S2 position to effect the early S5P–S2P transition and stimulates Spt5 and other factors at the promoter to promote elongation.
Because P-TEFb also phosphorylates the CTD at the S5 and S7 positions, Tat may balance the activities of Ssu72 and P-TEFb to create the CTD phosphorylation pattern needed to attract specific CTD-binding proteins to the elongation complex. As expected, we found that Tat:P-TEFb promotes the release of NELF and enables Spt5/DSIF to travel with the elongation complex. High-affinity binding of Tat:P-TEFb to TAR RNA may further enhance CTD phosphorylation and elongation complex assembly. Although the model outlined in Figure 6D suggests that Tat acts in successive steps to promote elongation, these reactions may be carried out in concert in vivo to ensure that short attenuated transcripts are not capped and cannot compete with full-length transcripts for export and translation. Our mapping experiments indicate that Ssu72 does not interfere with binding of Tat to P-TEFb, and it will be important to learn whether Ssu72 can function within the Tat:P-TEFb:TAR RNA complex. Alternatively, multiple Tat proteins may function at the proviral promoter in distinct complexes that contain P-TEFb, Ssu72, or the capping enzymes.
The observation that Tat is induced with very different kinetics in T cells in response to TNF-α (4–6 h) or TCRα (0.5 h) signaling in 2D10 cells strongly suggests that multiple mechanisms exist to overcome pausing and premature termination. In yeast, attenuation at the IMD2 gene, which is most pronounced under limiting nucleotide levels, is relieved through switching to an alternative downstream TSS that bypasses the terminator complex (Jenks et al. 2008). At the S. cerevisiae FKS2 cell wall gene, premature termination is overcome by the osmotic stress-induced MAP kinase Mpk1, which binds Paf1c and blocks recruitment of the Sen1:Nrd1:Nab3 termination complex (Kim and Levin 2011). The rapid induction of HIV-1 transcription and Tat expression also depends on activation of MAPK/ERK kinases and correlates with CDK9-S175 phosphorylation (Mbonye et al. 2013). Our data show that HIV-1 Tat induction in TNF-α signaling cells, which assemble high levels of NELF and paused S5P–RNAPII at the proviral promoter but do not activate MAPK/ERK kinases, proceeds through the recruitment and activation of Ssu72. Transcription elongation is highly efficient in TCRα signaling cells, and paused S5P–RNAPII complexes do not accumulate to high levels at the promoter. Consequently, it will be important to learn whether Tat can use different mechanisms to overcome pausing and premature termination, depending on the cellular environment of the integrated provirus.
Human Ssu72 colocalizes with RNAPII CTD-S5P genome-wide
To further characterize human Ssu72, we carried out ChIP-seq and GRO-seq studies in hESCs under self-renewal conditions. These studies revealed that the genome-wide occupancy pattern for human Ssu72 is opposite to that seen in yeast, with the highest peaks detected at gene promoters, and a much smaller peak in the terminator region. The Ssu72-binding pattern closely matches that of S5P–RNAPII in hESCs, which is predominantly found at promoter-proximal and 5′ UTRs. These findings are consistent with reports that mammalian Ssu72, unlike yeast Ssu72, is not an integral subunit of human cleavage/polyadenylation complexes. Heat map analysis indicates that Ssu72 levels correlate with active genes, and GRO-seq analysis revealed little effect on transcription at most hESC genes. Interestingly, we did detect a modest cotranscriptional readthrough at gene terminator regions genome-wide in Ssu72-depleted cells, which may reflect the ability of the symplekin terminator protein to bind and stimulate Ssu72 catalytic activity (Xiang et al. 2010). Ssu72 was also dispensable for transcription at several Wnt and Activin-induced genes that we tested in hESCs (data not shown). The predominant role of Ssu72 at hESC promoters may be related to gene looping, which has been suggested to facilitate transcription reinitiation, directionality, and epigenetic memory (Grzechnik et al. 2014). Interestingly, some active genes, such as NANOG, contained high levels of Ssu72 at both the 5′ and 3′ ends of the gene, whereas Ssu72 was restricted to the promoter region at other active genes, such as POU5F1. Although it is not clear whether 3′ enrichment of Ssu72 correlates with promoter–terminator gene looping, these findings indicate that the pattern of Ssu72 occupancy and, presumably, also its function may differ among active genes.
It will be important to learn whether human Ssu72 affects transcription at other genes (for example, the subset of cellular genes that require Pin1) and whether other activators bind and stimulate Ssu72 activity. It also remains to be seen whether Tat regulates Ssu72 catalytic activity through allosteric effects on the phosphatase domain, as seen with human symplekin (Xiang et al. 2010). Given the strong genetic interactions between Ssu72 and TFIIB, it is interesting that TFIIB was recently reported to be dispensable for transcription at many genes (Gelev et al. 2014). Like Ssu72, TFIIB is required for gene looping (Singh and Hampsey 2007; Perkins et al. 2008) and therefore may have other roles outside of transcription. Because TFIIB is required for the transcription and replication of many viruses and is also needed for transcription at cellular genes required to support virus infection (Gelev et al. 2014), it might also serve as a useful target for selective inhibitors of virus replication. Additional studies of the role of premature termination in HIV-1 latency and the mechanisms used by Tat to counteract this process could suggest new strategies to reactivate viral gene expression for more effective therapeutic intervention.
Materials and methods
Additional details are provided in the Supplemental Material.
Affinity purification of HA-Tat-associated protein complexes and MudPIT analysis
The affinity purification was performed as previously described (Choi et al. 2013). MudPIT protein identification methods and analysis are provided in the Supplemental Material.
siRNA transfection in 2D10 Jurkat cells
Specific siRNAs (Ambion) were transfected using the Stemfect RNA transfection kit (Stemgent). The final concentration of each siRNA was 50 nM, and cells were collected 48 h after transfection. Specific siRNA sequences are listed in the Supplemental Material.
RNAPII CTD kinase assays
Purified recombinant GST-CTD (400 ng) was incubated with 200 ng of recombinant TFIIH (Invitrogen) or P-TEFb (Millipore) in kinase reaction buffer (50 mM Tris-HCl at pH 7.5, 100 mM KCl, 10 mM MgCl2) supplemented with 5 mM DTT and 500 µM ATP for 1 h at 30°C. Reactions were stopped by adding SDS-loading buffer, and the products were resolved on an 8% SDS-PAGE gel and detected by Western blot.
CTD phosphatase assays
TFIIH-phosphorylated GST-CTD was passed through an Illustra Micro-Spin G-25 column (GE Healthcare) to remove unincorporated ATP. Purified recombinant Ssu72 was then mixed with the hyperphosphorylated CTD in phosphatase reaction buffer (50 mM Tris-HCl at pH 6.5, 20 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.055% Tween 20, 2% glycerol, 1 mM DTT) and incubated for 2 h at 30°C. The reaction was stopped with SDS-loading buffer, and the products were resolved on an 8% SDS-PAGE gel and analyzed by Western blot.
ChIP-seq
Approximately 1 × 106 H1 hESCs growing on mTeSR1 medium were used for each ChIP. The ChIP protocol was described previously (Choi et al. 2013). Briefly, cells were first fixed with 2 mM DSG cross-linker (Sigma, 80424) for 45 min and washed twice with 1× PBS, and then 1% FA was added for 15 min. Approximately 0.5 mg of protein was used for each immunoprecipitation. Five micrograms of Ssu72 (Genetex, GTX116436) or 3 µg of S5P-RNAPII (Millipore, 05-623) antibody was added, and the samples were incubated overnight at 4°C. DNA purification was performed using a Qiaquick PCR purification kit (Qiagen, 28106). Before sequencing, ChIP-specific signals were tested by qPCR. DNA library preparation and high-throughput sequencing (HiSeq 2500) were carried out at the Next-Generation Sequencing Core (Salk Institute).
GRO-seq
hESCs were transfected with a scramble or Ssu72-specific siRNAs (Ambion). Forty-eight hours later, cells were collected to proceed with the nuclear run-on experiment. Nucleus isolation, run-on, RNA fragmentation, and enzymatic TAP and PNK treatments were performed according to Core et al. (2008). The BrU-RNA library was prepared using the NEBNext multiplex small RNA library prep set for Illumina (New England Biolabs, E7300S/L). The bar-coded, retrotranscribed, and amplified DNA was sequenced using an Illumina HiSeq 2500 system.
Data accessibility
The Ssu72 hESC ChIP-seq and GRO-seq data have been deposited in the NCBI Gene Expression Omnibus public repository under the accession number GSE61393.
Supplementary Material
Acknowledgments
We thank Chris Benner (Razavi Newman Integrative Genomics and Bioinformatics Core, The Salk Institute), Sven Heinz, and Manching Ku (Next-Generation Sequencing Core, The Salk Institute) for critical bioinformatics analysis and support for the ChIP-seq and GRO-seq studies, and W. Travis Beggren in the Salk Stem Cell Core Facility for assistance with hESC cultures. Y. C. gratefully acknowledges post-doctoral fellowship support from the Blasker-Rose-Miah Fund and the Margaret T. Morris Foundation. Proteomics analysis was carried out by Patricia G. Tu and J.J.M. and was supported by grants from the National Center for Research Resources (5P41RR011823) and the National Institutes of Health (NIH) (GM103533) to J.R.Y. III. This study was funded by NIH grants to J.K. (DP1-DA028869 and U19-AI096113) and K.A.J. (AI044615).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.250449.114.
References
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoshida T, Pickle L, Jones KA. 2009. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell 36: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 531–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Estarás C, Moresco JJ, Yates JR III, Jones KA. 2013. α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev 27: 2473–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Coronel E, Ho CK, Schwer B, Shuman S, Rana TM. 2001. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem 276: 12959–12966 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell 10: 585–597 [DOI] [PubMed] [Google Scholar]
- Chou S, Upton H, Bao K, Schulze-Gahmen U, Samelson AJ, He N, Nowak A, Lu H, Krogan NJ, Zhou Q, et al. 2013. HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc Natl Acad Sci 110: E123–E131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Benkirane M, Kiernan R. 2012. Premature termination of transcription: the beginning of the end. Transcription 4: 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Bösken CA, Geyer M. 2012. Serine-7 but not serine-5 can prime RNA polymerase II CTD for P-TEFb recognition. Nat Commun 3: 824. [DOI] [PubMed] [Google Scholar]
- Darby MM, Serebreni L, Pan X, Boeke JD, Corden JL. 2012. The Saccharomyces cerevisiae Nrd1-Nab3 transcription termination pathway acts in opposition to Ras signaling and mediates response to nutrient depletion. Mol Cell Biol 32: 1762–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell 10: 1139–1150 [DOI] [PubMed] [Google Scholar]
- Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113: 8456–8490 [DOI] [PubMed] [Google Scholar]
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J 22: 1588–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelev V, Zabolotny JM, Lange M, Hiromura M, Yoo SW, Orlando JS, Kushnir A, Horikoshi N, Paquet E, Bachvarov D, et al. 2014. A new paradigm for transcription factor TFIIB functionality. Sci Rep 4: 3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Tan-Wong SM, Proudfoot NJ. 2014. Terminate and make a loop: regulation of transcriptional directionality. Trends Biochem Sci 39: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. 2011. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul 51: 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD. 2014. The ESS1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle. Biochim Biophys Acta 1839: 316–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. 2013. Kinetic competition between RNA polymerase II and Sen1-dependent transcription termination. Mol Cell 49: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17: 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PL, Yang F, Smith-Kinnaman W, Yang W, Song JE, Mosley AL, Varani G. 2014. Rtr1 is a dual-specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J Mol Biol 426: 2970–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadlowsky JK, Wong JY, Graham AC, Dobrowolski C, Devor RL, Adams MD, Fujinaga K, Karn J. 2014. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol 34: 1911–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MH, O’Rourke TW, Reines D. 2008. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol Cell Biol 28: 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. 2011. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell 144: 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol 410: 896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C. 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Lis JT. 2013. Control of transcriptional elongation. Annu Rev Genet 47: 483–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547 [DOI] [PubMed] [Google Scholar]
- Mbonye U, Karn J. 2012. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virol 454-455: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, Cooper M, Chance MR, Karn J. 2013. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog 9: e1003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RP, McCann JL, Gudipaty SA, D’Orso I. 2013. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Reports 5: 1256–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 34: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. 2008. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell 29: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Chiang K, Liu H, Budhiraja S, Donahue H, Rice AP. 2012. Making a short story long: regulation of P-TEFb and HIV-1 transcriptional elongation in CD4+ T lymphocytes and macrophages. Biology 1: 94–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes M, Hampsey M. 2007. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol Cell Biol 27: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Gahmen U, Lu H, Zhou Q, Alber T. 2014. AFF4 binding to Tat–P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV-1 promoter. eLife 3: e02375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Hampsey M. 2007. A transcription-independent role for TFIIB in gene looping. Mol Cell 27: 806–816 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Liu X, Kha LC, Zhu X, Ryan O, Jiang Z, Zacksenhaus E. 2005. Conserved and specific functions of mammalian Ssu72. Nucleic Acids Res 33: 464–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23: 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 23: 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, et al. 2012. Microprocessor, Setx, Xrn2, and Rrp6 cooperate to induce premature termination of transcription by RNAPII. Cell 150: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. 2011. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 286: 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Behav MM, Manley JL, Tong L. 2010. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Manley JL, Tong L. 2012. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev 26: 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 21: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. 2012. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 287: 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ssu72 hESC ChIP-seq and GRO-seq data have been deposited in the NCBI Gene Expression Omnibus public repository under the accession number GSE61393.