Fanconi anemia (FA) is an autosomal recessive genetic disorder caused by defects in FA genes responsible for processing DNA interstrand cross-links (ICLs). FA-associated nuclease (FAN1) is recruited to lesions by a monoubiquitinated FANCI–FANCD2 (ID) complex and participates in ICL repair. Here, Gwon et al. determined the crystal structure of Pseudomonas aeruginosa FAN1 (PaFAN1) lacking the UBZ (ubiquitin-binding zinc) domain in complex with 5′ flap DNA. The PaFAN1 structure provides insights into how FAN1 integrates with the FA complex to participate in ICL repair.
Keywords: Fanconi anemia nuclease 1, interstrand cross-link repair, crystal structure
Abstract
Fanconi anemia (FA) is an autosomal recessive genetic disorder caused by defects in any of 15 FA genes responsible for processing DNA interstrand cross-links (ICLs). The ultimate outcome of the FA pathway is resolution of cross-links, which requires structure-selective nucleases. FA-associated nuclease 1 (FAN1) is believed to be recruited to lesions by a monoubiquitinated FANCI–FANCD2 (ID) complex and participates in ICL repair. Here, we determined the crystal structure of Pseudomonas aeruginosa FAN1 (PaFAN1) lacking the UBZ (ubiquitin-binding zinc) domain in complex with 5′ flap DNA. All four domains of the right-hand-shaped PaFAN1 are involved in DNA recognition, with each domain playing a specific role in bending DNA at the nick. The six-helix bundle that binds the junction connects to the catalytic viral replication and repair (VRR) nuclease (VRR nuc) domain, enabling FAN1 to incise the scissile phosphate a few bases distant from the junction. The six-helix bundle also inhibits the cleavage of intact Holliday junctions. PaFAN1 shares several conserved features with other flap structure-selective nucleases despite structural differences. A clamping motion of the domains around the wedge helix, which acts as a pivot, facilitates nucleolytic cleavage. The PaFAN1 structure provides insights into how archaeal Holliday junction resolvases evolved to incise 5′ flap substrates and how FAN1 integrates with the FA complex to participate in ICL repair.
DNA interstrand cross-linking agents, such as cisplatin (cis-diammineplatinum [II] dichloride) and mitomycin C (MMC), covalently link the two DNA strands in a double-strand helix, preventing strand separation and blocking the progression of replication forks (RFs), thus promoting chromosomal instability (Auerbach and Wolman 1976). To maintain genomic stability, interstrand cross-links (ICLs) must be repaired during or after replication. Several models for the repair of ICLs have been proposed (McCabe et al. 2009; Moldovan and D’Andrea 2009). One recent model is the fork traverse model, in which the translocase allows replication to continue without blockage by the ICL (Huang et al. 2013). Single-molecule analysis suggests that this model may account for the majority of ICL repair. Another well-established model is the fork convergence model based on the replication of a cross-linked plasmid in Xenopus egg extracts (Räschle et al. 2008; Knipscheer et al. 2012). In this model, fork collision activates repair processes required for resumption of replication: The two leading strands progressing from opposite directions converge at the ICL, stalling some distance from the lesion. Next, one of the two leading strands directly approaches the cross-link, and duel incisions are formed on either side of the ICL to unhook the cross-link from one DNA strand. At this point, the DNA structure is expected to contain two flaps or one flap and one splayed arm on either side of the cross-link (Räschle et al. 2008; Knipscheer et al. 2009). Subsequently, translesion DNA synthesis bypasses the lesion, and the unhooked ICL is removed. Finally, homologous recombination (HR) completes the repair (Long et al. 2011; Kim and D’Andrea 2012).
Formation of ICLs activates the Fanconi anemia (FA) pathway to resolve damage in higher eukaryotes (Thompson and Hinz 2009; Kee and D’Andrea 2010). FA is a genetic disorder caused by mutations in any of 15 genes (Wang 2007). FA patients suffer hypersensitivity to ICL agents and exhibit congenital defects, bone marrow failures, and cancer susceptibility. An important event in FA-associated ICL repair is the initial recognition of stalled RFs by the FANCM–FAAP24 complex (Gari et al. 2008). The FANCM–FAAP24 complex activates the ataxia telangiectasia and Rad3-related (ATR) signaling pathway and recruits the FA core complex, which is composed of multiple subunits (A, B, C, E, F, G, L, and M) (Garcia-Higuera et al. 2001; Pichierri and Rosselli 2004; Ciccia et al. 2007; Ling et al. 2007; Smogorzewska et al. 2007; Collis et al. 2008; Schwab et al. 2010). Another important event is the monoubiquitination of FANCD2 by FANCL, the E3 ligase subunit of the FA core (Moldovan and D’Andrea 2009; Smogorzewska et al. 2007; Knipscheer et al. 2009; Thompson and Hinz 2009). Monoubiquitinated FANCD2 then acts as a landing pad for recruiting ubiquitin-binding zinc (UBZ) domain-containing proteins, such as FA-associated nuclease 1 (FAN1) and SLX4, to the lesion via its monoubiquitin group (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010).
FAN1 is a structure-specific nuclease implicated in ICL repair. Although recent studies suggest that Slx4-bound XPF–ERCC1 plays an important role in unhooking the ICL in the FA pathway, FAN1 is likely to be involved in other steps in ICL repair (Kim et al. 2011, 2013; Hodskinson et al. 2014; Klein-Douwel et al. 2014). It has been proposed that this nuclease might participate in incision of the lagging strand template flanking the ICL, removal of the fully unhooked ICL, or removal of a 5′ flap that arises by strand invasion during the HR-dependent step (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). However, it is unclear at which point FAN1 is involved. In vitro, FAN1 displays both endonuclease and exonuclease activities toward various substrates with a strong preference for 5′ flap DNA (Kratz et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010).
FAN1 involvement in the FA pathway is indicated by the observation that FAN1 depletion in human cells induces sensitivity to ICL-inducing agents, chromosome instability of MMC-treated cells, and defects in HR—observations consistent with the phenotypes observed for cells expressing a ubiquitination-deficient FANCD2 mutant (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Germline mutations in human FAN1 cause karyomegalic interstitial nephritis, a kidney disease with renal tabular degeneration, suggesting that FAN1 may be particularly important for maintaining kidney function (Zhou et al. 2012). Depletion of FAN1 in zebrafish resulted in activation of a DNA damage response, apoptosis, and kidney cysts (Zhou et al. 2012). Furthermore, the FAN1 locus has been linked to schizophrenia and autism (Ionita-Laza et al. 2014).
FAN1 is a novel structure-selective nuclease consisting of four characterized domains: UBZ; SAF-A/B, Acinus, and PIAS (SAP); tetratricopeptide repeat (TPR); and viral replication and repair (VRR) nuclease (VRR nuc) (Kinch et al. 2005; Iyer et al. 2006). The protein is present in most eukaryotes and some bacterial species. However, monocellular eukaryote and bacterial FAN1 homologs do not possess the UBZ domain, and their precise role remains to be determined (MacKay et al. 2010), although the fission yeast homolog is phenotypically linked to cross-link repair (Fontebasso et al. 2013). At present, FAN1 homologs are the only known eukaryotic proteins that possess the VRR nuc domain. Although the UBZ domain is critical for localization of FAN1 to the site of damage via FANCD2, other parts of FAN1 also significantly contribute to its localization (Liu et al. 2010; Smogorzewska et al. 2010). Despite its well-characterized biochemical functions, it is unclear how FAN1 recognizes and cleaves its substrates. Since FAN1 may be involved in unhooking ICLs, trimming the unhooked ICL, and/or removal of 5′ flaps during HR repair, understanding the structure of FAN1 in complex with 5′ flap DNA should provide a better insight into how FAN1 is involved in the ICL repair pathway and possibly other DNA repair pathways.
Here we describe the crystal structure of Pseudomonas aeruginosa FAN1 (PaFAN1) bound to 5′ flap DNA in the absence (3.2 Å resolution) and presence (4 Å resolution) of metal ions. Based on these structures, we provide a basis for the mechanism by which FAN1 recognizes and resolves 5′ flap DNA, whose structure resembles that of lagging strand templates near a cross-link and HR intermediate during DNA repair. Biochemical and structural studies of PaFAN1 show that UBZ-deficient FAN1 efficiently binds and processes DNA. Four domains (an uncharacterized N-terminal domain [NTD] plus the SAP, TPR, and VRR nuc domains) are organized such that each domain plays a specific role in prenick duplex, post-nick duplex, and 5′ flap ssDNA recognition and bending DNA at the nick. The unique architecture by which the catalytic subdomain is connected to the six-helix bundle and the TPR domain explains why FAN1 incises the scissile phosphate 5 nucleotides (nt) downstream from the junction and cannot resolve intact Holliday junctions (HJs). A clamping motion of the two C-terminal domains toward the two NTDs may facilitate DNA incision. Interestingly, a UBZ-deficient human FAN1 did not incise the 5′ flap or nicked HJs in the presence of the FANCI–FANCD2 (ID) complex, suggesting that the two proteins cannot simultaneously bind. The structure provides insights into the evolutionary relationship between archaeal HJ resolvases and the bacterial and eukaryotic FAN1 homologs that incise 5′ flap DNA and how FAN1 integrates with the FA complex during ICL repair.
Results
The PaFAN1 homolog exhibits a strong preference for 5′ flap and nicked DNA
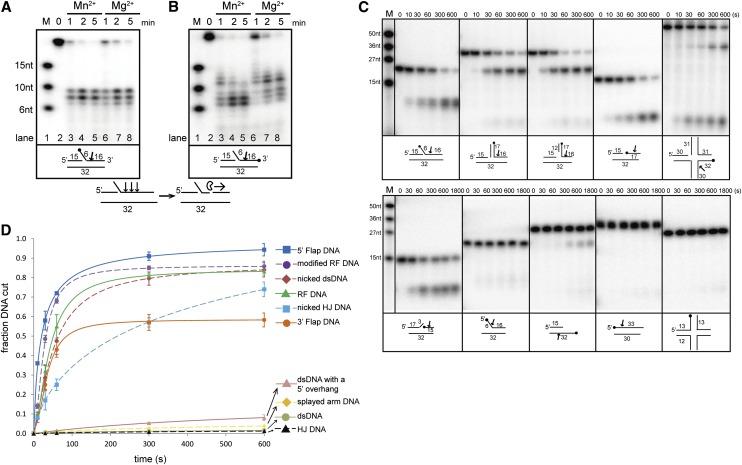
PaFAN1 (UniProt ID Q9I2N0) shares 24% sequence identity (36% similarity) with human FAN1 (Supplemental Fig. 1; Kratz et al. 2010). Although the protein lacks a UBZ domain, it exhibits all of the key enzymatic features of FAN1, including endonuclease and 5′–3′ exonuclease activities in the presence of Mg2+ or Mn2+ ions (Fig. 1A,B; Supplemental Fig. 2A–C). In addition, PaFAN1 efficiently cleaves the nicked duplex DNA and the 5′ nicked strand of the 3′ flap substrate (Fig. 1C,D). The enzyme incises the third to fifth nucleotide downstream from the junction between the prenick and post-nick duplexes and successively cleaves the DNA downstream from this point. PaFAN1 also exhibited moderate nuclease activities toward an RF and the nicked HJ (Fig. 1C,D). However, excess amounts of the nuclease or long reaction times were required to incise splayed-arm DNA, the complementary strand of the flap strand in 5′ flap DNA, or duplex DNA with a 5′ overhang.
Figure 1.
Biochemical characterization and overall structure of PaFAN1 bound to DNA. Endonuclease (A) and exonuclease (B) activities of PaFAN1 in the presence of Mn2+ and Mg2+ ions. The labeled site is marked with a filled circle. Although PaFAN1 exhibited nuclease activity in the presence of Mg2+ or Mn2+, nucleolytic activity was more efficient in the presence of Mn2+, especially when FAN1 mutant proteins were used (see Fig. 4A; Supplemental Fig. 6). PaFAN1 incises the third, fourth, or fifth phosphate group (arrow) downstream from the junction between the ssDNA and dsDNA and cleaves further in the 3′ direction. (C) Nuclease activities of PaFAN1 toward various substrates in the presence of Mn2+ ions. (Top) 5′ flap (lanes 2–7), RF (lanes 8–13), modified RF (lanes 14–19), nicked dsDNA (lanes 20–25), and nicked HJ (lanes 26–31). (Bottom) 3′ flap (lanes 2–7), splayed arm (lanes 8–13), dsDNA with a 5′ overhang (lanes 14–19), dsDNA (lanes 20–25), and intact HJ (lanes 26–31). See Supplemental Figure 8 for detailed substrate structures. (D) The cleavage products shown in C have been quantified. The fraction of DNA cut is the ratio of the relevant cleavage product to total DNA (cleaved plus uncleaved DNA). Data were plotted as a function of time using Origin software. From these data, we calculated observed rates of cleavage of 5′ flap DNA (>0.046 sec−1; blue square and line), modified RF (>0.043 sec−1; purple circle and dash), nicked dsDNA (>0.042 sec−1; brown diamond and dash), RF (>0.040 sec−1; green triangle and line), 3′ flap DNA (>0.016 sec−1; orange circle and line), nicked HJ (>0.014 sec−1; cyan square and dash), dsDNA with a 5′ overhang (>0.0015 sec−1; pink triangle and line), splayed-arm DNA (>0.0014 sec−1; yellow diamond and dash), dsDNA (>0.0002 sec−1; gray circle and line), and intact HJ (>0.0002 sec−1; black triangle and dash).
Overall structure
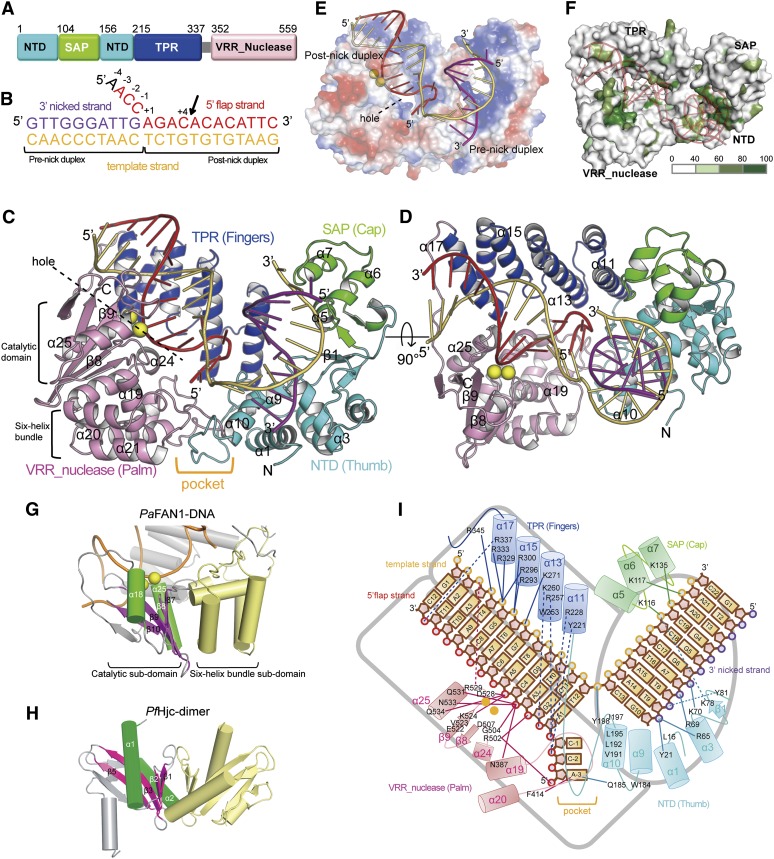
The 3.2 Å crystal structure of PaFAN1 bound to a 5′ flap 22-base DNA molecule resembles a curved right hand (57 × 17 × 53 Å) in which the NTD (residues 1–103 and 156–214) forms the thumb and the TPR (215–337) and VRR (352–559) domains represent fingers and a palm, respectively (Fig. 2A–D; crystallographic statistics in Supplemental Table 1). The SAP domain (104–155) caps the thumb of the hand. A conserved and basic groove (30 Å wide and 20 Å deep) runs between the TPR and VRR nuc domains and extends toward the NTD, where another shallow groove continues to the SAP domain in a nearly perpendicular direction (Fig. 2E,F). The bound DNA is shown in Figure 2, B, C, and E, with the “3′ nicked strand” in purple (residues G1 to G10), the “5′ flap strand” in red (A-3 to C12; “−3” means 3 nt toward the 5′ end from the junction), and the “template strand” in orange (G1 to C22). The last nucleotide of the 5′ flap is disordered.
Figure 2.
Overall structure of the PaFAN1–DNA complex. (A) Schematic diagram of the domain organization of PaFAN1. (B) The 4-nt 5′ flap DNA used in crystallization. In the text, each nucleotide is numbered from the 5′ end of the 3′ nicked or template strand. For the 5′ flap strand, each nucleotide is numbered from the junction. The substrate is labeled prenick and post-nick duplex based on the 3′ nick position. The three strands are labeled as 3′ nicked strand (purple), 5′ flap strand (red), and template strand (orange). The cleavage site is marked with an arrow, and the terminal nucleotide (A-4; black) in the 5′ flap strand is disordered in the structure. (C) A ribbon representation of PaFAN1 showing the NTD (cyan), the SAP domain (green), the TPR domain (blue), and the VRR nuc domain (pink). The 3.2 Å PaFAN1–DNA structure is shown in C–I and Figure 3, A–D, and metal ions are modeled from the superimposed 4.0 Å metal-bound structure. The 5′ flap DNA is shown in purple for the 3′ nicked strand, red for the 5′ flap strand, and orange for the template strand. Active site metal ions are shown as yellow spheres. The pocket for 5′ flap binding is marked with an orange label. See Supplemental Movie 1. (D) Alternative view of the complex shown in C. (E) Corresponding view of the complex shown in C, presented as an electrostatic color-coded surface representation. (F) FAN1 is colored according to the degree of conservation in the FAN1 orthologs of 13 species shown in Supplemental Figure 1. More conserved residues are colored deep green. The view is the same as that in D. (G) Overall structure of the VRR nuc domain of PaFAN1. The six-helix bundle subdomain (yellow) is connected to the catalytic subdomain. Helices α18 and α25 of PaFAN1 are shown in green, and the four strands (β7–β10) are shown in magenta. The helices in gray are from the TPR domain. (H) Dimer structure of the archaeal HJ resolvase (PfHjc). Helices and strands of PfHjc corresponding to PaFAN1 are also shown in green and magenta. (I) Molecular details of the PaFAN1–DNA interaction. Hydrogen bonds and ion pairs between FAN1 and the DNA molecule are indicated by arrows. Mn2+ ions are shown as orange spheres.
The DNA threads its way within a central groove between the TPR and VRR domains and is sharply bent (by ∼70°) toward the shallow groove from the NTD to the SAP domain, thereby making contact with all domains in PaFAN1 (Fig. 2E,F). The bound DNA is bent at the junction between the prenick and post-nick duplexes by the protrusion of the helix α10 wedge of the NTD.
At the interface of the NTD, TPR domain, and VRR domain, the loop α10–α11, two helices (α11 and α13), and α19–α20 form a pocket in which the 5′ flap ssDNA is accommodated (Supplemental Fig. 3A–D). The TPR and VRR nuc domains are loosely associated with each other, forming an elliptical hole in which the ssDNA can protrude near the pocket (Fig. 2C,E).
Specific features of individual domains
The NTD and SAP domain form an elongated α/β structure whose axis is nearly parallel to that of the prenick duplex (Fig. 2C). The NTD consists of four helices (α1–α4) packed by two helices (α9 and α10) and three strands (β1, β2, and β5). The SAP domain is composed of a short two-stranded sheet (β3 and β4) and four helices (α5–α8) (Fig. 2C; Supplemental Fig. 4A).
The TPR domain consists of four inner layer helices (α11, α13, α15, and α17) packed by three outer layer helices (α12, α14, and α16) in a two-layered wall (Supplemental Fig. 4B). In the inner layer, the latter two helices (α15 and α17) are tilted by >40° relative to the first two helices (α11 and α13), forming a concave that accommodates the post-nick duplex.
The VRR nuc domain can be divided into a catalytic and a flap-binding subdomain. The catalytic domain forms the α/β fold with its central five-stranded sheet flanked by a helix and a bundle of helices on each side (Fig. 2G; Supplemental Fig. 4C). This domain is similar to that of type II restriction enzymes (Kosinski et al. 2005) and single-domain VRR nuc proteins from phage or bacteria (Pennell et al. 2014) and is most similar to the HJ cleavage enzyme of Pyrococcus furiosus (PfHjc [1GEF]) (Fig 2H; Nishino et al. 2001). The catalytic subdomain of PaFAN1 VRR nuc and PfHjc share low sequence identity (11.5%) and 3.0 Å root mean square deviation for 69 Cα atoms (Fig. 2G,H). The two α helices (α18 and α25) and four β strands (β7–β10) of PaFAN1 correspond well to the equivalent parts of PfHjc (Supplemental Fig. 4D–F). In the single-domain VRR nuc or PfHjc, the central four- or five-stranded sheet is surrounded by helices on each side and interacts with the hydrophobic β sheet of the opposing molecule to form a dimer. Such a dimeric arrangement of the single-domain VRR nuc or PfHjc allows these proteins to cleave intact HJs (Nishino et al. 2001; McGregor et al. 2005; Pennell et al. 2014). In contrast, the VRR nuc domain of FAN1 contains a six-helix bundle inserted between the second (β7) and third (β8) strand of the β sheet, at the equivalent position of an opposing molecule in the dimeric HJ resolvase (PfHjc or Bacillus subtilis RecU [1ZP7]). This six-helix bundle is important, since the α19–α20 and α21–α22 loops participate in the formation of a pocket for 5′ flap binding (Fig. 2C,D; Supplemental Fig. 3A,C). Thus, the VRR nuc domain of FAN1 has a dual role: 5′ flap recognition by the six-helix bundle and active site formation by the central a/β catalytic subdomain. This unique feature of the VRR nuc domain in PaFAN1 endows it with strong 5′ flap nuclease activity but prevents it from resolving intact HJs.
A striking observation is that all four domains are extensively involved in binding to DNA and fulfil unique roles in the recognition of various parts of the sharply bent DNA (Fig. 2C,D,I): (1) The NTD blocks the path of the prenick and post-nick duplexes via the α10 helix and an α9–α10 loop and recognizes the 3′ end of the 3′ nicked strand. (2) The SAP and TPR domains augment the overall protein–DNA interaction by interacting with the 3′ and 5′ ends of the template strand, respectively. (3) Regions of the NTD, TPR domain, and VRR (six-helix bundle) domain form a pocket that houses the 5′ flap. (4) The two-metal active site is positioned 5 nt downstream from the six-helix bundle of the VRR nuc domain. Interactions between the four domains and the 5′ flap DNA are almost completely achieved by phosphate oxygen atoms, which explains the structure-specific nuclease activity of PaFAN1.
Prenick DNA recognition by the NTD and SAP domain
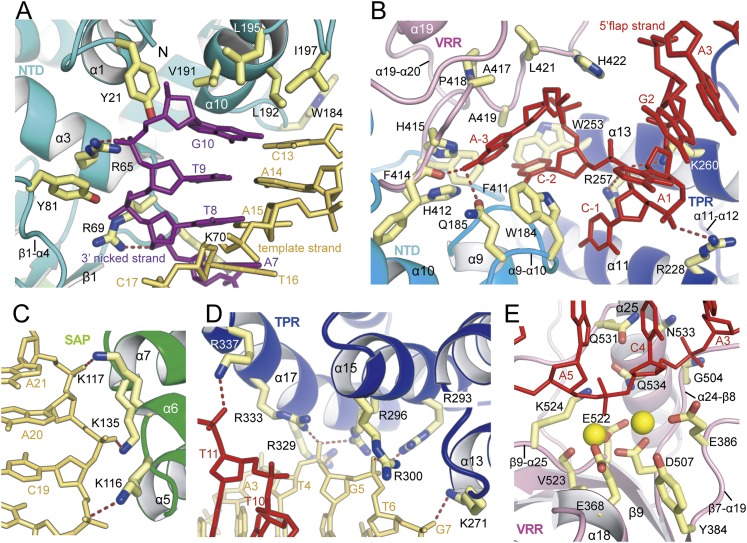
The 3′ nicked strand and its template strand form bipartite interactions with basic residues from the NTD and the SAP domain, respectively (Figs. 2C, 3A; Supplemental Fig. 4A). Within the prenick segment, the bound DNA is continuously stacked from residues C22 to C13, with distinct breaks at C13 and T12 of the template strand (Fig. 2C,D; Supplemental Fig. 5A). This break is achieved by protrusion of the conserved and fully exposed hydrophobic side chains (V191, L195, L192, and I197) of the α10 helix (Fig. 3A). These residues point toward the base planes of the G10:C13 pair at the 3′ nicked end, block the path of a prenick DNA, and sharply kink the template strand. In addition, Trp184 (α9–α10) is packed against the C-1 and C-2 of the 5′ flap to reinforce bending of the substrate DNA (Fig. 3A,B).
Figure 3.
Close-up view of the interactions between the FAN1 and DNA substrate. (A) Close-up view of the prenick segment binding to the NTD. Four hydrophobic residues at the α10 wedge face the terminal bases G10:C13, and tyrosine residues and basic residues make contact with the phosphate oxygen atoms of the 3′ nicked and template strands. (B) Recognition of the 5′ flap part by the pocket composed of TPR domain (purple), VRR nuc domain (pink), and NTD (cyan). Recognition of the 5′ flap by FAN1 is augmented via binding of the phosphate of C-1 by the α13 helix (R257 and K260) and of the phosphate of A1 by the α11 helix (R228) See also Figure 2I and Supplemental Figure 3. (C) Three bases (K116, K135, and K117) from the SAP domain recognize the phosphate oxygen atoms adjacent the 5′ end of the template strand. (D) Recognition of the post-nick segment by the four inner layer helices from the TPR domain. Basic residues from the α15 and α17 helices bind the phosphate groups near the 3′ end of the template strand, whereas the α11 and α13 helices participate in binding to the 5′ flap region. K271 (α13) binds the phosphate oxygen atoms of G7. R293, R296, and R300 (α15) interact with the phosphate oxygen atoms of T6, and R296, R329, and R333 (α17) bind the phosphate oxygen atoms of G5 of the template strand. (E) The incised phosphodiesters bound to the active site at the VRR nuc domain. The 4.0 Å metal-bound PaFAN1–DNA structure is shown. Two metal ions are shown as yellow spheres. The A5 is 5 nt downstream from the junction between the prenick and post-nick DNA. For an overall scheme of the FAN1–DNA interaction, see Figure 2I.
The α1, α3, and α10 helices as well as the N terminus, α3–β1, and β1–α4 loops form a shallow groove for the 3′ end of the 3′ nicked strand that cements the FAN1–prenick duplex complex (Figs. 2C, 3A). The three basic residues (R65, R69, and K70) and conserved tyrosine residues (Y21 and Y81) in this groove form a track for the four phosphate oxygen (G10, T9, T8, and A7) atoms adjacent to the 3′ nick (Fig. 3A). The significance of this domain is illustrated by functional experiments in fission yeast, in which expression of a truncated FAN1∆N193 phenocopied a FAN1-deficient strain with respect to its response to a cross-linking agent (Fontebasso et al. 2013). The NTD possesses a fully exposed binding site for the 3′ nicked end, which is in contrast to the deep binding pocket for the 3′ flap in flap endonuclease 1 (FEN1, also known as 3Q8K) (Tsutakawa et al. 2011) or for the 5′ nicked end in Mus81–Eme1 (MUS81, also known as 4P0S and 4P0R) (Gwon et al. 2014). Therefore, the 3′ nicked end can be further extended, which explains why FAN1 incises the 5′ nicked strand of the 3′ flap DNA (Fig. 1D).
The SAP domain further stabilizes binding of the prenick segment by FAN1. The helix–hairpin–helix (α5–α6) and helix α7 of the SAP domain recognize three consecutive phosphate groups (C19, A20, and A21) at the 3′ end of the template strand via the basic residues K116, K135, and K117 (Fig. 3C). Support of prenick segment binding by the SAP domain is crucial, as multiple mutations in this domain in fission yeast or truncation of the SAP domain in Caenorhabditis elegans FAN1 resulted in hypersensitivity to a cross-linking agent (Kratz et al. 2010; MacKay et al. 2010; Fontebasso et al. 2013).
The TPR domain recognizes post-nick DNA
The four parallel inner helices of the TPR domain are arranged in the order of α11, α13, α15, and α17 and form a nearly flat structure. Basic residues are positioned at each of the parallel inner layer helices and run along the concave surface to interact with the phosphate backbone of the post-nick duplex (Fig. 3B,D; Supplemental Figs. 1, 4B). TPR–DNA binding in FAN1 is markedly different from the binding of RNA with 5′ phosphates to human interferon-induced proteins with TPRs, where the short RNA is embedded in a deep cleft formed by the TPR motifs (Abbas et al. 2013); this is the only reported example of an interaction between the TPR fold and nucleic acids. The root mean square deviation of the TPR domains between PaFAN and IFIT is 2.0 Å for 100 Cα atoms.
The two inner helices (α11 and α13) contribute to recognition of the 5′ flap, whereas the three helices (α13, α15, and α17) bind the phosphates adjacent to the 5′ end of the template strand to reinforce DNA binding by FAN1 (Figs. 2I, 3D). The interactions between the template strand of the duplex DNA and FAN1 are largely fulfilled by the TPR domain and may contribute to the positioning of the 5′ flap strand, thereby helping to position the scissile bond at the active site.
5′ flap recognition by the pocket
The 5′ flap ssDNA and the junction between the prenick and post-nick segments are accommodated by the pocket (8 Å deep, 8 Å long, and 14 Å wide) formed by the NTD (α9–α10 and α10–α11), TPR domain (α11, α11–α12, and α13), and VRR nuc domain (α19–α20) (Supplemental Fig. 3A–D). In the pocket, the last base (A1) of the post-nick duplex is displaced out of its duplex interaction and, together with the 5′ flap ssDNA, forms a U-shaped structure (Fig. 3B). W184 is at the path of the post-nick duplex and interrogates the bases of C-1, C-2, and A-3, altering the direction of the 5′ flap, and moves the bases of C-2 and A-3 into the pocket. An elliptical hole (formed by α13, α15, α16–α17, α19–α20, and α23) with a size sufficient to allow only the passage of ssDNA is located close to the phosphate of C-1. However, the flap DNA end does not protrude into this hole and instead turns in the opposite direction, which is wide open.
The active site is located 5 nt downstream from the junction
The second binding of the 5′ flap strand in the VRR nuc domain occurs at the tip between the β8 and β9 strands, which is located around the fourth (C4) and fifth (A5) phosphates downstream from the junction between the prenick and post-nick duplexes (Figs. 2B, 3E). To examine whether metal ions bind around this region, we incubated the PaFAN1–5′ flap DNA complex crystal in a buffer containing 50 mM MnCl2 for 1 h. Increased soaking time significantly decreased the diffraction quality, presumably because it induced conformational changes in the PaFAN1–DNA complex. An electron density map with the phases calculated after molecular replacement using the metal-free PaFAN1–DNA complex as a search model showed pronounced positive density near the fifth phosphate downstream from the junction (Supplemental Fig. 5B).
The active site pocket formed by the β7–α19, α24–β8, and β9–α25 loops; the α18 helix; and the β9 strand contains highly conserved catalytic carboxylates in a PDLXnEVKX3D motif (Supplemental Fig. 1). The fourth phosphate from the junction interacts with the side chains of N387, Q531, and N533 and the main chain of G504. The β9–α25 loop (R528) is inserted into the major groove and interacts with the phosphate of A3 of the template strand, further augmenting the binding of the post-nick segment by FAN1 (Figs. 2I, 3E). Despite the limited resolution, the metal-bound PaFAN1–DNA structure clearly revealed the active site: The fifth phosphate is bound by two Mn2+ ions and K524. Metal 1 is coordinated with D507, E522, and V523 (main chain oxygen) and the phosphate oxygen, while metal 2 is liganded by E386 and D507 and the phosphate oxygen and lies close to Y384. E368 is >3 Å distant from metal 1. The two metal ions are separated by 3.8 Å. Mutational analysis near this region showed that D507A (equivalent to human FAN1 D960A, E522A [E975A], and K524A [K977A] mutants that lack the nuclease activities of human FAN1 in vitro and D651A [equivalent to PaFAN1 D507], E666Q [E522], or K668A [K524]) endowed hypersensitivity to a fission yeast strain harboring any of these mutants in vivo (Kratz et al. 2010; MacKay et al. 2010; Fontebasso et al. 2013). Mutation of the residue equivalent to D507 results in karyomegalic interstitial nephritis, demonstrating the significance of FAN1 nuclease activity in preventing kidney failure (Zhou et al. 2012). The present structure suggests that cleavage would release a fragment containing the region 4 nt downstream from the junction, consistent with an earlier observation (MacKay et al. 2010).
In vitro and in vivo functional analysis of the key motifs in each domain
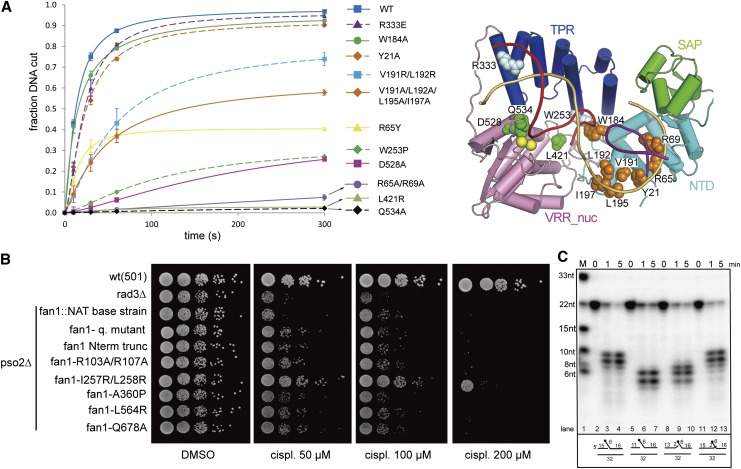
To verify systematically the importance of residues in the individual domains of PaFAN1 for DNA cleavage, we generated a series of PaFAN1 mutants in each domain and performed 5′ flap DNA cleavage assays (Fig. 4A; Supplemental Fig 6A–D). In the NTD, we tested Y21A, R65Y, R65A/R69A, W184A, and two wedge (V191R/L192R and V191A/L192A/L195A/I197A) mutants (Fig. 3A). The R65A/R69A mutant was very inefficient for incision of a 5′ flap substrate in the presence of Mn2+ (Fig. 4A). Thus, 3′ nicked end recognition and its correct positioning by the NTD are important for PaFAN1 nuclease activity. Interestingly, the two wedge mutants retained substantial activity, although this was notably diminished. This observation indicates that the wedge has a clear impact on nuclease activity. However, the space between the wedging residues and the terminal base pair of the prenick might not be critical. The W184A mutant did not alter nuclease activity, indicating that stacking of the 5′ flap by W184 is not critical for the function of FAN1.
Figure 4.
Endonuclease and exonuclease activities of wild-type and mutant PaFAN1. (A) The 5′ flap DNA nuclease activities of various PaFAN1 mutants were examined at different times (10–300 sec) and quantified. (Blue square and line) Wild type (WT); (purple triangle and dash) R333E; (green circle and line) W184A; (orange diamond and dash) Y21A; (cyan square and dash) V191R/L192R; (orange diamond and line) V191R/L192R/L195A/I197A; (yellow triangle and line) R65Y; (green diamond and dash) W253P; (magenta square and line) D528A; (purple circle and line) R65A/R69A; (gray triangle and line) L421R; (black diamond and dash) Q534A. Assay conditions were the same as those in Figure 1C. The 5′ flap DNA was incubated with the indicated FAN1 protein in the presence of Mn2+ ions. Cleavage products were resolved by denaturing polyacrylamide gel electrophoresis and visualized by phosphoimaging. A schematic diagram of the positions of the mutated residues is shown in the right panel. NTD residues are shown as orange spheres, TPR residues are pale cyan spheres, and VRR nuc residues are green spheres. (B) Sensitivity of point mutations in the conserved residues of S. pombe Fan1 to cisplatin (cispl). A pso2-deleted background was used to compare the effect of the mutations on the hypersensitive double-mutant fan1-deleted pso2-deleted strain. Logarithmically growing cultures were serially diluted and spotted onto a YEA-DMSO control plate and YEA plates containing the cross-linking agent cisplatin at the concentrations indicated. A checkpoint-deficient rad3-deleted strain was used as a hypersensitive control to test the efficacy of the cisplatin used. (C) To identify the reference point of the incision, PaFAN1 was incubated with 5′ flap substrates with various gap sizes. Detailed substrate structures are shown in Supplemental Figure 8.
The W253P mutant, which was expected to perturb the α13 helix of the TRP domain, showed very weak nuclease activity, demonstrating the importance of the TRP domain for the function of FAN1 (Figs. 3D, 4A; Supplemental Fig. 6A,B). Next, we examined L421 at the VRR nuc domain, which forms a pocket for the base of the 5′ flap (Fig. 3B). The L421R mutation significantly decreased nuclease activity on the 5′ flap substrate, demonstrating the importance of the pocket (Fig. 4A). Q534 is near the second metal-binding site and is a possible ligand for a nucleophilic water molecule. The Q534A mutation had the most severe effect among all of the mutants tested.
To characterize the in vivo significance of residues whose mutations reduced FAN1 nuclease activity, we generated single and multipoint mutations in the NTD, TPR domain, and VRR nuc domain of Schizosaccharomyces pombe Fan1. The corresponding fan1Sp mutants were introduced into a pso2-deleted “base strain” using recombinase-mediated cassette exchange (Watson et al. 2008; Fontebasso et al. 2013), and the cisplatin sensitivity of the strain harboring the fan1 mutant was examined. We used five different mutants that exhibited reduced nuclease activities (Figs. 2I, 4A): R103A/R107A (NTD mutant: R65A/R69A of PaFAN1), I257R/L258R (wedge mutant: V191R/L192R), A360P (TPR mutant: W253P), L564R (VRR nuc mutant: L421R), and Q678A (VRR nuc mutant: Q534A). Four of the mutants (NTD, TPR, and both VRR nuc mutants), but not the I257R/L258R wedge mutant, displayed a marked hypersensitivity to doses of cisplatin as low as 50 µM (Fig. 4B). The strains harboring these four fan1 mutants were as sensitive as those harboring an N-terminal truncation mutant (Δ1–193) or the fan1 quadruple mutant (R160E/R164E/K171E/R173E) to a low dose of cisplatin. At a high dose of cisplatin (>100 µM), the four mutants phenocopied the pso2-d fan1∷NAT (null) double-mutant strain. However, the strain harboring the wedge mutant (I257R/L258R) was resistant to cisplatin, which is consistent with the moderately reduced nuclease activity seen for the equivalent PaFAN1 protein, suggesting that the amino acid identity of the wedging residues is not critical for Fan1 nuclease activity. Presumably, the prenick and post-nick duplexes of the substrate are separated regardless of the amino acid type in the wedge helix. Collectively, the in vitro and in vivo results demonstrate the importance of Arg65, Arg69 (NTD), Trp253 (TPR), Leu421, and Glu534 (VRR nuc) for ICL repair.
Determinants for the incision site
To identify the reference point from which FAN1 cleaves the scissile bond of dsDNA, we examined the incision site of the 5′ flap DNA with various gap sizes (Fig. 4C). PaFAN1 cleaved 8, 9, or 10 nt downstream from the 5′ end of the flap (or 2, 3, or 4 nt from the junction between the prenick duplex and ssDNA of the 5′ flap DNA) (Fig. 4C, lanes 2–4). However, the protein incised 4, 5, or 6 nt downstream from the 5′ end of the flap when a 4-nt gap was introduced between the prenick and post-nick duplexes (Fig. 4C, lanes 5–7). We then introduced a 2-nt 3′ flap (Fig. 4C, lanes 11–13) or a 2-nt gap to the 2-nt 3′ flap DNA (Fig. 4C, lanes 8–10). Extension of the 3′ nicked strand did not alter the cleavage site by FAN1 (Fig. 4C, lanes 11–13 vs. lanes 2–4). When the prenick strand had 13 bases and two additional unpaired bases (Fig. 4C, lanes 8–10), FAN1 incised 6, 7, or 8 nt from the end of the 5′ flap (or 0, 1, or 2 nt from the junction between the prenick duplex and ssDNA), suggesting that the junction between ssDNA and the prenick duplex is a reference point.
Conformational changes in PaFAN1
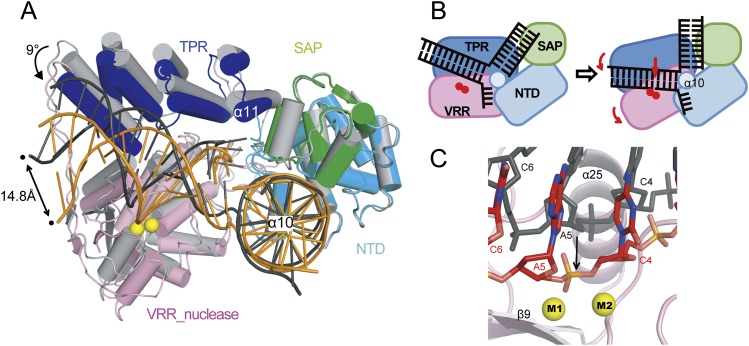
Binding of Mn2+ ions induced significant conformational changes in both DNA and PaFAN1. When the N-terminal region (NTD and SAP domain) of the metal-free and metal-bound structures were superimposed, the overall Cα of the TPR or VRR nuc domains moved by 9° and as much as 7.5 Å (TPR) and 8.0 Å (VRR) toward the NTD (Poornam et al. 2009). The α10 helix serves as a pivot for rotating the C-terminal domains (TPR and VRR nuc) toward the NTDs (and SAP) in the presence of Mn2+ ions (Fig. 5A,B). Accordingly, the post-nick duplex also shifts closer to the prenick duplex, and this movement notably distorts the DNA: The majority of base planes in the prenick and post-nick duplexes are disoriented, and base pairs are partly perturbed (Supplemental Fig. 7A–D). Furthermore, the overall shape of the 5′ flap structure becomes more compressed (Supplemental Fig. 7E,F). Importantly, shifting of the post-nick duplex places the fifth phosphate closer to the metal-binding site by 4.5 Å and facilitates its incision (Fig. 5C).
Figure 5.
Metal-induced conformational changes in FAN1. (A) Alignment of PaFAN1–DNA in complexes in the absence (FAN1 in light gray; DNA in black) and presence (FAN1 in green, blue, and pink; DNA in orange) of Mn2+ ions after superposition of their NTDs. Arrows indicate the magnitude of the conformational changes on the transition from the metal-free to the metal-bound complex. For transition from the metal-free to the metal-bound FNA1–DNA complex, see Supplemental Movie 2. (B) Schematic diagram for the clamping motion mechanism of PaFAN1 for substrate cleavage. (C) Close-up view of the phosphate atoms undergoing a shift toward the metal ions. Phosphates in the absence (light gray) and presence (orange) of metal ions are shown.
The ID complex inhibits incision of the 5′ flap by UBZ-deficient FAN1
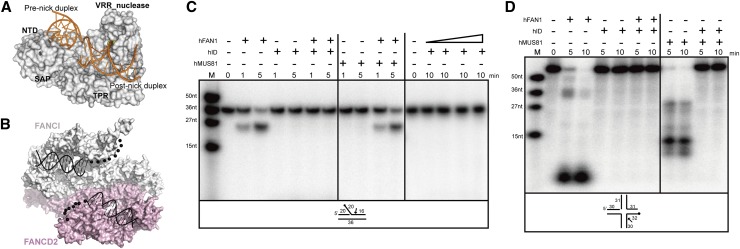
Prior to the binding of FAN1, the ID complex recognizes ICL damage (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Crystal structures of the ID complex and PaFAN1 show that these proteins extensively interact with the flap or splayed-arm DNA (Fig 6A,B; Joo et al. 2011). Thus, it is possible that both proteins cannot simultaneously bind to the lesion. Alternatively, DNA binding by the ID complex may expose some part of the DNA, rendering it accessible to FAN1. To test this possibility, we preincubated 5′ flap DNA in the presence of the ID complex or MUS81 (as a control) and added human FAN1 lacking the UBZ domain (Fig. 6C). The human FAN1 efficiently incised the 5′ flap DNA in both the absence and presence of MUS81. However, the ID complex protected 5′ flap DNA from FAN1, and no FAN1-mediated cleavage was observed, even in the presence of an excess amount of FAN1 (Fig. 6C, lanes 7,8,14–17). We also examined a nicked HJ for protection by the ID complex; human FAN1 did not incise a nicked HJ in the presence of the ID complex (Fig. 6D). These data suggest that UBZ-deficient FAN1 cannot bind DNA in the presence of the ID complex and may help explain why the UBZ domain is required to anchor FAN1 close to the lesion via a monoubiquitinated ID complex (see the Discussion).
Figure 6.
Human FAN1 lacking a UBZ domain cannot incise the 5′ flap or nicked HJ in the presence of the ID complex. (A) Surface representation of PaFAN1 bound to DNA. (B) Surface representation of the model of ID bound to DNA. The model was based on data from Joo et al. (2011) (Protein Data Band [PDB], 3S4W and 3S4Z). (C) Nuclease activities of human FAN1 lacking a UBZ domain (lanes 1–12, 25 nM; lanes 13–17, 250, 500, 2500, 10,000 nM) on 5′ flap DNA (5 nM) (Supplemental Fig. 8N) were examined in the presence of 25 nM human ID complex and quantified. The assay was also performed in the presence of 25 nM hMus81–Eme1 complex instead of the ID complex. Assay buffer condition was the same as that used in Figure 1C. (D) Nuclease activities of human FAN1 lacking a UBZ domain (25 nM) on nicked HJ DNA (5 nM) were examined in the presence of 25 nM human ID complex.
Discussion
Multiple structure-selective nucleases are involved in various steps of ICL repair by the FA pathway (Kim and D’Andrea 2012). FAN1 may be involved in unhooking of the ICL, removal of the unhooked ICL, or incision of the 5′ flap intermediate during HR, although the exact step or steps remain to be clarified (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Here we report the structural and functional features of a bacterial FAN1 homolog bound to 5′ flap DNA. Despite the limited sequence identity to mammalian FAN1 and absence of a UBZ domain, the PaFAN1 homolog (with four conserved domains) exhibited a strong preference toward 5′ flap and nicked DNA and demonstrated in vitro properties similar to those of human FAN1. However, unlike human FAN1, PaFAN1 exhibited weak nuclease activity toward splayed-arm DNA, suggesting that some differences exist between these proteins (Fig. 1C,D).
FAN1 shares several common features with other structure-selective nucleases
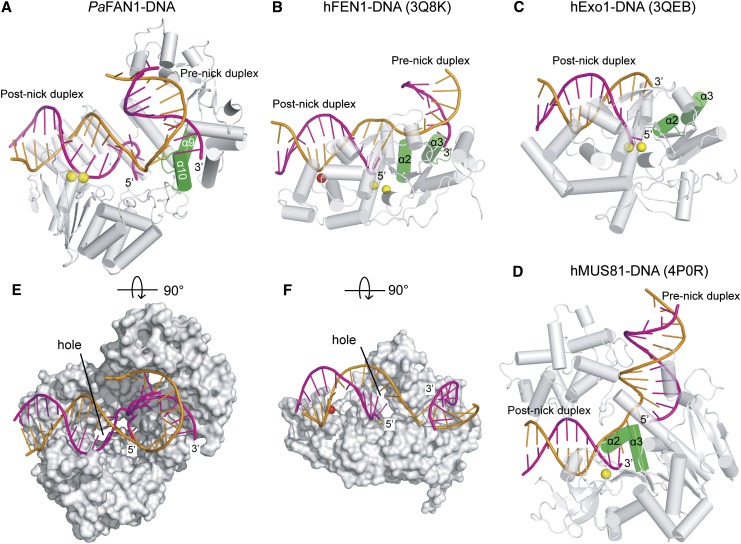
Structural studies on flap structure-selective nucleases such as FEN1/ExoI and MUS81, which preferably incise 5′ and 3′ flap DNA substrates, respectively, reveal that these nucleases share several common structural features for substrate recognition and cleavage (Orans et al. 2011; Tsutakawa et al. 2011, 2014; Gwon et al. 2014). FAN1 also shares conserved features with these nucleases (Fig. 7A–D): First, PaFAN1 contains two discontinuous dsDNA-binding sites for 5′ flap DNA, which are separated by a helical DNA turn; one binding site is formed at the NTD and SAP domain for the prenick duplex, and the other binding site is formed by the C-terminal domains (TPR and VRR nuc) for the post-nick DNA duplex. None of these regions share noticeable similarity with the corresponding part of FEN1 (3Q8K), ExoI (3QEB), or MUS81 (4P0S and 4P0R). However, the separate duplex DNA-binding activity of PaFAN1 facilitates positioning of the flap ssDNA in the pocket and presumably plays a role similar to that of other structure-selective nucleases.
Figure 7.
Structural comparison of four flap structure-selective nucleases. Four structure-selective nucleases were compared: PaFAN1 (A), hFEN1-DNA (3Q8K) (B), hExo1-DNA (3QEB) (C), and hMUS81-DNA (4P0S and 4P0R) (D). Post-nick DNA is in the same orientation. The wedge helix is shown in green. (E) Surface representation of PaFAN1 bound to 5′ flap DNA. A hole is marked. The figure is an orthogonal view of A. (F) Surface representation of hFEN1 with 5′ flap DNA. The figure is an orthogonal view of B.
Second, the hydrophobic helix is located in the direct path of the substrate DNA, resulting in abrupt contortion at a nick, which may be a hallmark of the flap structure-selective nuclease family (Fig. 7A). When each post-nick segment from FEN1 and PaFAN1 is superimposed, the prenick segment of PaFAN1 is located at a different position relative to the prenick segment of FEN1. Interestingly, multiple mutations of the hydrophobic residues in the helical wedge did not significantly reduce PaFAN1 nuclease activity, and the cells harboring such mutations did not exhibit hypersensitivity (Fig. 4A,B). Thus, it is possible that the wedge helix functions in a sequence-independent manner. Another important feature is the recognition of the template strand by the four domains, which is dominant in the FAN1–DNA interaction: A number of phosphate groups from the template strand make contact to the TPR and SAP domains of PaFAN1 (Fig. 2I). In addition, several phosphate groups from the 5′ flap strand and 3′ nicked strand interact with the VRR nuc domain and NTD, respectively. These extensive interactions with the template, 5′ flap, and 3′ nicked strand impose FAN1 specificity for the dsDNA.
FEN1 and ExoI contain a hole between the helical arch and the active site in which the 5′ flap can protrude (Orans et al. 2011; Tsutakawa et al. 2011). PaFAN1 also contains a hole that is only suitable for the passage of ssDNA at the interface of the TPR and VRR nuc domains. However, in our crystal structure, flap ssDNA did not pass through this hole. Instead, the 5′ flap ssDNA of PaFAN1 makes a U-turn in the opposite direction of the hole. This feature is consistent with biochemical data showing that PaFAN1 can incise the RF or nicked HJ. Although PaFAN1 may undergo a substantial conformational change resulting in enlargement of the hole and the passage of dsDNA, nicked HJ would still encounter difficulty in passing through the hole. In contrast, the opposite side of the hole, where the duplex or bulky segments could be located, is wide open.
FEN1/ExoI and MUS81 have a binding pocket for 1 nt at the 3′ flap and 5′ flap, respectively. Binding of 1 nt at the 3′ flap (or 5′ flaps for MUS81) to this pocket is believed to initiate the disorder-to-order structural transition that is critical for substrate recognition and incision. It is unclear whether FAN1 undergoes a similar structural transition. However, bases at the end of the 3′ nicked strand are paired and bind to the fully exposed binding site of PaFAN1. Thus, the 3′ nicked end can be further extended.
In FEN1 and Exo1, the two bases near the junction must be unpaired to move the scissile bond within catalytic distance from the metal. This base unpairing presumably occurs because ssDNA is required to bend and move to the active site of FEN1 or Exo1. Unlike FEN1, junction recognition in PaFAN1 is achieved by the pocket, which is located quite far (a few nucleotides distant) from the active site. Thus, the dsDNA at the active site of PaFAN1 may not need to bend and maintain base-pairing. Nevertheless, we note that the structure of the metal-bound PaFAN1–DNA complex was determined after soaking the crystals in a metal solution. Thus, we cannot exclude the possibility that the crystal packing force may restrict the movement of the protein and DNA in the crystal. In fact, the bases of C4 and C5 at the active site are slightly disoriented and partly perturbed by metal ions.
One important biochemical feature of FAN1 is its broad substrate specificity. In addition to 5′ flap and nicked DNA, PaFAN1 can cleave RFs and nicked HJs and weakly cleave splayed-arm DNA and dsDNA with a 5′ overhang. Although all four domains are essential for binding and incising DNA substrates, the TPR and VRR nuc domains are the minimal domains required to recognize the dsDNA segment (post-nick duplex); the TPR domain recognizes the template stand, and the VRR nuc domain binds to the cleavage site. Thus, we predict that the C-terminal domains (TPR and VRR nuc) provide a frame for dsDNA binding (Fig. 2C,D). Nevertheless, the C-terminal domains may not be sufficient for efficient substrate incision (Fig. 1C,D; Kratz et al. 2010; MacKay et al. 2010). Furthermore, in fission yeast, a truncation of the NTD and SAP domain, which are responsible for recognition of the prenick duplex, exhibited sensitivity toward an ICL agent that was as severe as that of a FAN1-null mutant, demonstrating the importance of DNA recognition by these domains in resolving ICLs (Fontebasso et al. 2013).
PaFAN1 may incise the scissile bond via a clamping mechanism
Interestingly, all four domains of PaFAN1 are involved in the recognition of the DNA substrate, which suggests a possible transition of the flexible linked domains that wrap around the DNA. Despite the limited resolution of our structure and crystal packing barrier, comparative analysis of the metal-free and metal-bound PaFAN1–DNA complex suggested that FAN1 may incise the scissile bond via a clamp-like motion whereby C-terminal domains (VRR and TRP) undergo rigid body rotation relative to the NTDs (TPR), with the helix wedge as a pivot (Fig. 5B). Although not directly analogous, Mus81–Eme1 also recognizes the 3′ flap substrate through four domains (nuclease, nuclease-like, and two helix–hairpin–helix [HhH2] domains) and undergoes a gross rigid body movement of the HhH2 domain to place the scissile bond into the active site (Chang et al. 2008; Gwon et al. 2014). Such rotation has been proposed to provide Mus81–Eme1 with exonuclease activity. Mre11–Rad50 is another multidomain nuclease, which has endonuclease and exonuclease activities (Lim et al. 2011; Shibata et al. 2014). The Mre11 dimer undergoes domain rotation to facilitate the substrate cleavage (Williams et al. 2008; Sung et al. 2014). Thus, it is possible that FAN1 also exhibits processive exonuclease activity via the repeated up and down motions of the four domains. However, it is unclear whether DNA binding induces the conformational change and, if so, how a significant structural transition occurs upon DNA binding, as a DNA-free PaFAN1 structure is not currently available. Further studies will be necessary to verify this model using single-molecule FRET analyses and/or crystal structure analysis of PaFAN1 in the absence of DNA.
FAN1 and the ID complex may not bind simultaneously at an equivalent site
The structure of PaFAN1 bound to 5′ flap DNA reveals that the four domains of FAN1 lacking a UBZ domain are sufficient and essential for the recognition of flap DNA. This is consistent with previous studies demonstrating that removal of other domains notably diminishes its localization to the lesion (Liu et al. 2010; Smogorzewska et al. 2010). During ICL repair, FAN1 is expected to be anchored to the lesion through the monoubiquitinated ID complex (Räschle et al. 2008; Knipscheer et al. 2009). The crystal structure and modeling studies of the ID complex bound to splayed-arm DNA reveal that the ID complex forms a pseudo twofold symmetric arrangement, resulting in extensive binding to the lesion (Fig. 6A,B; Joo et al. 2011). In this case, it would be impossible for FAN1 to bind the equivalent ICL site in the presence of the ID complex. The results of the competition analysis in the present study demonstrate that FAN1 (lacking the UBZ domain) cannot incise the 5′ flap or nicked HJ DNA in the presence of the nonubiquitinated ID complex (Fig. 6C,D). Thus, while the ID complex acts as a landing pad for FAN1 localization to the lesion, some additional mechanism is needed for FAN1 to be able to incise the substrate in the presence of the ID complex.
Klein-Douwel et al. (2014) showed that the ID complex also binds to dsDNA and exhibits a spreading effect, and it is thus possible that the ID complex binds some distance away from the lesion junction. This may allow recruitment of FAN1 and direct competition with the lesion-associated ID complex to allow FAN1 access to the lesion. Alternatively, once the localized ID complex is monoubiquitinated, it recruits UBZ-containing FAN1 through ubiquitin-mediated interactions, and the ID complex undergoes conformational changes and/or relocalization that allows FAN1 access to the lesion so that it can participate in ICL repair.
Archaeal HJ resolvases may have adapted to integrate into the FA complex
Structures of phage and bacterial single-domain VRR nucs or archaeal and bacterial HJ resolvases show that these proteins form a dimer that is essential for intact HJ cleavage (Nishino et al. 2001; McGregor et al. 2005; Pennell et al. 2014). In PaFAN1, the six-helix bundle subdomain is fused to the catalytic subdomain and is located at the opposing monomer of the single-domain VRR nuc or archaeal Hjc. The whole VRR nuc domain of PaFAN1 is further packed against the TPR domain. As a result, only one incision can be made by this nuclease. Thus, the significance of the six-helix bundle subdomain in PaFAN1 is that it not only plays a critical role as a reference point for 5′ flap cleavage but also limits the substrate specificity of PaFAN1 to 5′ flap or nicked DNA.
We speculate that a few domains have been added to the single-domain VRR nuc proteins and bacterial HJ resolvases to form the bacterial or eukaryotic FAN1 homologs during evolution. Considering their broad substrate specificity, it is possible that bacterial and eukaryotic FAN1 homolog functions in various repair processes. It is notable that no proteins in the core FA pathway have been found in bacteria or unicellular eukaryotes. At a later stage in evolution, the UBZ domain may have been added to unicellular eukaryotic FAN1 homologs. The core complex-dependent ubiquitination of the ID complex may have evolved in parallel to both promote the local concentration of structure-selective nucleases and allow a precise handover between the ID complex and these nucleases at specific structures. The extensive interaction between UBZ-lacking PaFAN1 and DNA, the recent finding that FAN1 can repair damaged DNA in fission yeast (Fontebasso et al. 2013), and the FA-independent but FAN1-dependent repair of DNA damage observed in chicken cells (Yoshikiyo et al. 2010) all support the possibility that FAN1 participates in other DNA repair pathways in addition to the FA pathway. The structural and functional properties of PaFAN1 presented here provide insights into how FAN1 recognizes and incises 5′ flap DNA and how this nuclease might have been incorporated into the FA pathway.
Materials and methods
Protein expression and purification
The full-length gene encoding PaFAN1 (residues 1–559; UniProt number Q9I2N0) was amplified by PCR and inserted into pET28a. Escherichia coli Rosetta (DE3) containing the plasmid was grown in LB medium. His-tagged protein was purified by Ni2+-NTA affinity chromatography. Fractions containing FAN1 were further purified using anion exchange chromatography and dialyzed against a buffer containing 20 mM Bis-Tris–propane–HCl (BTP-HCl) (pH 7.0), 0.2 M NaCl, and 5 mM DTT. Proteins were concentrated to 12 mg/mL by ultrafiltration and stored at −80°C. A selenomethionine (Se-Met)-substituted PaFAN1 protein was produced in E. coli B834(DE3) in M9 minimal medium. The protein was purified by the same method used for the purification of native protein. The human FAN1 (residues 330–1017) was amplified by PCR, inserted into pET28a, and purified by the same protocol used for the purification of PaFAN1. The full-length human ID complex and human Mus81 (246–551)–Eme1 (178–570) complex were prepared as previously described (Yuan et al. 2009; Gwon et al. 2014).
Crystallization and data collection
Crystals of the PaFAN1–DNA complex were grown by a hanging drop vapor diffusion method at 18°C. Crystals containing a 22-base-pair (bp) DNA molecule with a 4-nt 5′ flap were grown in crystallization buffer containing 20% (w/v) polyethylene glycol (PEG) 8000, 0.1 M HEPES (pH 7.0), and 5 mM DTT. For Mn2+-bound FAN1–DNA crystals, crystals were soaked in crystallization buffer containing 50 mM MnCl2 for 1 h. Diffraction data were collected at −170°C using crystals flash-frozen in crystallization buffer containing 30% (v/v) glycerol. Diffraction data were collected from FAN1–DNA and FAN1–DNA–Mn2+ crystals at 0.9795 Å and 0.9792 Å, respectively, on Beamline 5C at the Pohang Accelerator Light Source and Beamline 19ID at the Advanced Photon Source. Diffraction data were integrated and scaled using the HKL2000 package (Supplemental Table 1; Otwinowski and Minor 1997).
Structure determination and refinement
The structure of the PaFAN1–DNA complex was determined by single wavelength anomalous diffraction (SAD) data using Se-Met in which 12 selenium sites in the asymmetric unit were found using the PHENIX program (Adams et al. 2010). After density modification (solvent flattening and twofold noncrystallographic symmetry averaging), an electron density map generated at a resolution of 3.5 Å using the PHENIX program clearly revealed positions for most of the main chain for FAN1 and the backbone of the DNA molecule. An initial model was built on the 3.5 Å experimental electron density map. Successive rounds of model building using COOT and refinement using PHENIX were performed to build most of the model (Emsley and Cowtan 2004). When the Rfree value of the model reached ∼31%, we refined the model against native data to a 3.2 Å resolution. Further refinements included interactive cycles of bulk solvent corrections, overall B-value refinement, positional and individual B-value refinement, TLS refinement, and manual building. Initial phases for the metal-bound PaFAN1–5′ flap DNA complex were obtained by molecular replacement using the structure of 3.2 Å native PaFAN1–DNA as a search model at a resolution of 4.0 Å with Phaser software (Adams et al. 2010). The calculated electron density clearly revealed the Mn2+ ion sites. Interactive cycles of rigid body refinement, overall B-value refinement, positional and group B-value refinement, TLS refinement, and manual building improved the model. Prior to refinement, 5% of the reflections were randomly omitted to monitor the Rfree value. Details of refinement statistics are listed in Supplemental Table 1.
Nuclease activity assay
A cleavage reaction solution containing 10 nM various 32P-labeled DNA substrates and 50 nM PaFAN1 in reaction buffer (25 mM Tris-HCl at pH 7.4, 5 mM β-mercaptoethanol, 50 mM NaCl, 100 μg/mL bovine serum albumin, 5% glycerol) containing 5 mM MnCl2 or MgCl2 was incubated for various times (10–600 [or 1800] sec) at 37°C. The reaction was stopped by adding the same volume of 2× reaction stop buffer (95% formamide, 18 mM EDTA, 0.025% sodium dodecyl sulfate, 0.01% bromophenol blue) followed by 5 min of boiling at 100°C. The products were resolved on 15% denaturing polyacrylamide gels containing 7 M urea in 1× TBE (Tris-Borate-EDTA) for 120 min at 13 Vcm−1. Cleaved substrates were quantified by phosphorimage analysis using MultiGauge version 3.0 (Fujifilm). Error bars in the figures represent the standard deviation from the mean. See Supplemental Figure 8 for detailed structures of substrates.
For the competition assay, 25 nM human ID complex was mixed with 5 nM 5′ flap or nicked HJ DNA in a 5:1 ratio and preincubated for 30 min. Excess human FAN1 lacking the UBZ domain (25 nM∼10 uM) was added to the reaction solution and incubated for various times. The product was resolved as described above.
In vivo survival assays
The S. pombe fan1 mutants were created by site-directed mutagenesis using a Stratagene QuikChange kit with pAW8-fan1+ as the PCR template. Cre-lox recombinase-mediated cassette exchange was used to integrate the mutant fan1 genes into the S. pombe genome at the endogenous fan1 locus (Watson et al. 2008) using base strain YFS228 (fan1∷loxP-ura4+-loxM3, pso2∷kanMX6). Correct integration of the fan1 mutant alleles was confirmed by sequencing. Mutant fan1 S. pombe strains created are listed in Supplemental Table 2.
For survival analysis, strains were inoculated in 100 mL of YE medium and grown at 30°C to a density of 1 × 107 cells per milliliter. Four serial 1/10 dilutions were prepared from each culture, and 10 μL were spotted onto YEA plates supplemented with increasing doses of the DNA cross-linking agent cisplatin (Sigma). As the cisplatin stock was dissolved in DMSO, cells were also grown on a YEA control plate supplemented with DMSO (YEA-DMSO). Plates were incubated for 3 d at 30°C.
Accession numbers
Coordinates and structure factors have been deposited to the Research Collaboratory for Structural Bioinformatics (RCSB): 4R8A for the metal-free PaFAN1–DNA complex and 4R89 for the metal-bound PaFAN1–DNA complex.
Supplementary Material
Acknowledgments
We thank Dr. John Rouse, Dr. Christophe Lachaud, Dr. Seung-Jae Lee, Dr. Sang Uk Kim, Dr. Dong Yeup Lee, and Dr. Philip Jeffrey for helpful comments. We also thank anonymous reviewers for insightful suggestions. This work was supported by grants from the National R&D Program for Cancer Control, Ministry for Health and Welfare (1020280), National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST, nos. 2012004028, 2012-054226, and 20120008833), a rising star program (Pohang University of Science and Technology [POSTECH]), the BK21 program (Ministry of Education), National Institutes of Health (HL105631 to Y.Z.) and the European Research Council (268788-SMI-DDR).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.248492.114.
References
- Abbas YM, Pichlmair A, Górna MW, Superti-Furga G, Nagar B. 2013. Structural basis for viral 5′-PPP–RNA recognition by human IFIT proteins. Nature 494: 60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD, Wolman SR. 1976. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature 261: 494–496 [DOI] [PubMed] [Google Scholar]
- Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y. 2008. Crystal structure of the Mus81–Eme1 complex. Genes Dev 22: 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani EH, Joenje H, McDonald N, de Winter JP, et al. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell 25: 331–343 [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsí Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. 2008. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fontebasso Y, Etheridge TJ, Oliver AW, Murray JM, Carr AM. 2013. The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast. DNA Repair (Amst) 12: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262 [DOI] [PubMed] [Google Scholar]
- Gari K, Décaillet C, Stasiak AZ, Stasiak A, Constantinou A. 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Gwon GH, Jo A, Baek K, Jin KS, Fu Y, Lee JB, Kim Y, Cho Y. 2014. Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates. EMBO J 33: 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. 2014. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF–ERCC1 in DNA crosslink repair. Mol Cell 54: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. 2013. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell 52: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Xu B, Makarov V, Buxbaum JD, Roos JL, Gogos JA, Karayiorgou M. 2014. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc Natl Acad Sci 111: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Babu MM, Aravind L. 2006. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle 5: 775–782 [DOI] [PubMed] [Google Scholar]
- Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP. 2011. Structure of the FANCI–FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 333: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD. 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD. 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26: 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. 2013. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 121: 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch LN, Ginalski K, Rychlewski L, Grishin NV. 2005. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res 33: 3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Douwel D, Boonen RA, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. 2014. XPF–ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54: 460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Schärer OD, Walter JC. 2012. Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol Biol 920: 221–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Feder M, Bujnicki JM. 2005. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J. 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. 2011. Crystal structure of the Mre11–Rad50–ATPγS complex: understanding the interplay between Mre11 and Rad50. Genes Dev 25: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. 2007. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J 26: 2104–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. 2010. FAN1 acts with FANCI–FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Long DT, Räschle M, Joukov V, Walter JC. 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Déclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KM, Olson SB, Moses RE. 2009. DNA interstrand crosslink repair in mammalian cells. J Cell Physiol 220: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor N, Ayora S, Sedelnikova S, Carrasco B, Alonso JC, Thaw P, Rafferty J. 2005. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure 13: 1341–1351 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. 2009. How the fanconi anemia pathway guards the genome. Annu Rev Genet 43: 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K. 2001. Dissection of the regional roles of the archaeal Holliday junction resolvase Hjc by structural and mutational analyses. J Biol Chem 276: 35735–35740 [DOI] [PubMed] [Google Scholar]
- Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. 2011. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145: 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pennell S, Déclais AC, Li J, Haire LF, Berg W, Saldanha JW, Taylor IA, Rouse J, Lilley DM, Smerdon SJ. 2014. FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain. Cell Reports 8: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F. 2004. Fanconi anemia proteins and the s phase checkpoint. Cell Cycle 3: 698–700 [PubMed] [Google Scholar]
- Poornam GP, Matsumoto A, Ishida H, Hayward S. 2009. A method for the analysis of domain movements in large biomolecular complexes. Proteins 76: 201–212 [DOI] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. 2010. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 29: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiácovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Li F, Park YB, Kim JS, Kim AK, Song OK, Kim J, Che J, Lee SE, Cho Y. 2014. DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA. EMBO J doi: 10.15252/embj.201488299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. 2009. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res 668: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. 2011. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. 2014. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 19: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8: 735–748 [DOI] [PubMed] [Google Scholar]
- Watson AT, Garcia V, Bone N, Carr AM, Armstrong J. 2008. Gene tagging and gene replacement using recombinase-mediated cassette exchange in Schizosaccharomyces pombe. Gene 407: 63–74 [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J. 2010. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci 107: 21553–21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, El Hokayem J, Zhou W, Zhang Y. 2009. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J Biol Chem 284: 24443–24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. 2012. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 44: 910–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.