Abstract
The four mammalian Pellinos (Pellinos 1, 2, 3a, and 3b) are E3 ubiquitin ligases that are emerging as critical mediators for a variety of immune signaling pathways, including those activated by Toll-like receptors, the T-cell receptor, and NOD2. It is becoming increasingly clear that each Pellino has a distinct role in facilitating immune receptor signaling. However, the underlying mechanisms by which these highly homologous proteins act selectively in these signaling pathways are not clear. In this study, we investigate whether Pellino substrate recognition contributes to the divergent functions of Pellinos. Substrate recognition of each Pellino is mediated by its noncanonical forkhead-associated (FHA) domain, a well-characterized phosphothreonine-binding module. Pellino FHA domains share very high sequence identity, so a molecular basis for differences in substrate recognition is not immediately apparent. To explore Pellino substrate specificity, we first identify a high-affinity Pellino2 FHA domain-binding motif in the Pellino substrate, interleukin-1 receptor-associated kinase 1 (IRAK1). Analysis of binding of the different Pellinos to a panel of phosphothreonine-containing peptides derived from the IRAK1-binding motif reveals that each Pellino has a distinct phosphothreonine peptide binding preference. We observe a similar binding specificity in the interaction of Pellinos with a number of known Pellino substrates. These results argue that the nonredundant roles that Pellinos play in immune signaling are in part due to their divergent substrate specificities. This new insight into Pellino substrate recognition could be exploited for pharmacological advantage in treating inflammatory diseases that have been linked to the aberrant regulation of Pellinos.
Pellinos make up one of several families of E3 ubiquitin ligases that have emerging roles in regulating diverse pathways in innate and adaptive immune receptor signaling.1−4 There are four mammalian Pellinos: Pellino1, Pellino2, and two splice variants of Pellino3, Pellino3a and Pellino3b. Sequence analysis and in vitro ubiquitination assays indicate that all mammalian Pellinos are active RING E3 ubiquitin ligases.5−7 All Pellinos have been implicated in polyubiquitination events that occur immediately following the stimulation of Toll-like receptors (TLRs) and the interleukin-1 (IL-1) receptor (IL-1R).8−10 Pellino2 plays a role in IL-1-dependent K63-linked polyubiquitination of IL-1R-associated kinase 1 (IRAK1),9 which is required for the subsequent activation of nuclear factor κB (NF-κB).11,12 Pellino1 and -3 are dispensable for IL-1R-mediated NF-κB activation. Rather, these Pellinos are critical regulators of TLR3- and/or TLR4-dependent signaling pathways, although some debate about the precise role of Pellinos in these pathways remains.8,13,14 Upon TLR3/4 stimulation, Pellino1 has been implicated in the ubiquitination of receptor-interacting serine/threonine protein kinase 1 (RIP1) and the subsequent activation of NF-κB.8 However, it has also been reported that the E3 ubiquitin ligase activity of Pellino1 is not required for RIP1 ubiquitination or NF-κB activation following TLR3 stimulation.13 This second study implicates Pellino1 E3 ubiquitin ligase activity in the upregulation of type I interferon expression. Pellino3 has also been implicated in the regulation of the TLR3-mediated induction of type I interferon expression. However, Pellino3 negatively regulates this pathway by modulating the ubiquitination of the tumor necrosis factor receptor-associated factor 6 (TRAF6).14 Other recent studies have demonstrated that Pellinos are also critical mediators of immune signaling pathways that are independent of Toll-like and IL-1 receptors. For example, Pellino3 regulates tumor necrosis factor α (TNFα)-mediated apoptosis and is critical for mediating signaling following the stimulation of the another innate immune receptor, nucleotide-binding oligomerization domain-containing protein 2 (NOD2).15,16 Pellino1 plays a role in maintaining proper T-cell receptor (TCR) activation and therefore is also involved in adaptive immunity.17
Clearly, Pellinos have nonredundant and contrasting roles in regulating immune receptor signaling. To mediate these various functions, Pellinos interact with a diverse set of substrates, and these interactions must be regulated. It is unlikely that this occurs simply by the availability of a particular Pellino during a specific immune response. Most cells of the immune system have comparable expression levels of two or more Pellinos, with the exception of B and T cells that express predominantly Pellino1.1 Pellinos must be phosphorylated to become active E3 ubiquitin ligases,5,18 and for Pellino1, this activating phosphorylation is mediated by different kinases in a cell and/or pathway specific manner, suggesting that phosphorylation can dictate functional specificity.19 It also seems reasonable to suggest that there is functional selectivity at the level of Pellino substrate recognition and that this contributes to ensuring the fidelity of each Pellino in immune signaling pathways. Given the extremely high degree of sequence similarity among Pellinos (>70% identical sequences among the human Pellinos), the molecular basis for such specific substrate recognition is not immediately apparent.
We have previously shown that the human Pellino2 substrate recognition domain comprises a noncanonical example of a forkhead-associated (FHA) domain.20 The high level of sequence identity among Pellinos strongly suggests that all Pellinos will contain such an FHA domain. FHA domains are protein interaction modules that bind to phosphorylated threonine (pT) in the context of a specific recognition sequence.21−25 In this study, we investigate the binding of all four mammalian Pellinos to phosphorylated targets. We identify a high-affinity Pellino2-binding region in IRAK1 using GST-tagged Pellino pull-downs of mutated and truncated IRAK1 overexpressed in human embryonic kidney 293T (HEK293T) cells. We show that all Pellinos bind to a peptide derived from this region of IRAK1, but with different affinities. Drawing on what is known about the determinants of FHA domain binding specificities, we generate a panel of phosphorylated peptides and show that each Pellino has a distinct pT peptide binding specificity. GST pull-downs of other Pellino substrates expressed in HEK293T cells corroborate our finding that Pellinos have different specificities. The observed specificity correlates with the known physiological roles of each Pellino and, to some extent, with our observed pT peptide binding specificities. These results argue that the nonredundant roles of Pellinos in immune receptor signaling are in part due to their different substrate specificities.
Experimental Procedures
Expression Vectors
All proteins are human homologues except for mouse Pellino1, which differs by only one amino acid from the human homologue (S146 is asparagine in human). The cDNA for RIP1 (Open Biosystems) was inserted into pcDNA4/HisMaxC (Invitrogen). IRAK1 and TRAF6 in pcDNA4/HisMaxC were generous gifts from L. Jensen (Temple University, Philadelphia, PA). Expression plasmids for GST-Pellinos and His6-Pellinos were previously described.20 The appropriate coding regions for truncated IRAK1 and Pellinos were inserted into pcDNA4/HisMaxC and into a pET21-derived HTUA vector (generously provided by G. Van Duyne, University of Pennsylvania, Philadelphia, PA), respectively. The QuikChange site-directed mutagenesis strategy (Stratagene) was used for creating alterations in pcDNA4/HisMaxC-IRAK1-197 and in GST-Pellinos.
Protein Purification
GST- and His6-Pellinos were expressed in Escherichia coli BL21(DE3) pLys cells and purified essentially as described previously,20 with the following modifications. Glutathione agarose beads were incubated with GST-Pellinos, washed three times in 50 mM sodium phosphate and 300 mM NaCl (pH 7.4), and resuspended in 20 mM sodium phosphate and 150 mM NaCl (pH 7.4) (PBS). To remove the His6 tag, His6-Pellinos with TEV protease26 (∼100 μg for every 5 mg of His6-Pellino) were dialyzed against 10 mM HEPES, 300 mM NaCl, and 5 mM β-mercaptoethanol (βME) (pH 7.0). Dialysis was performed overnight at 4 °C for Pellino1 and -2 and at 22 °C for Pellino3a and -3b. Digested, untagged Pellinos were purified by cation exchange chromatography (Source S, GE Healthcare) at pH 6 (Pellino1, -2, and -3b) or pH 5.5 (Pellino3a), followed by size exclusion chromatography (Superose 12, GE Healthcare) in 10 mM HEPES, 150 mM NaCl, and 5 mM βME (pH 7.0).
GST Pull-Down Assays
The culturing of HEK293T cells and the GST pull-down assays of Xpress-tagged IRAK1, IRAK1 truncation variants, TRAF6, and RIP1 were performed as described previously.20 For dephosphorylation of IRAK1–197, HEK293T cells overexpressing Xpress-IRAK1–197 were lysed in 50 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 5 mM βME, and 0.1% IGEPAL (octylphenoxypolyethoxyethanol, Sigma) supplemented with protease inhibitor cocktail (Sigma) and incubated for 1 h at 37 °C with or without calf intestinal alkaline phosphatase [CIP, 10 units/100 μL of lysate (New England Biolabs)]. CIP treatment was stopped with 5 mM EDTA and 1 mM sodium vanadate or PhosSTOP (Roche). Western blots were probed with α-Xpress (Invitrogen) or α-phosphothreonine (Invitrogen or Cell Signaling) antibodies, and detection used horseradish peroxidase conjugated to α-mouse and α-rabbit (GE Healthcare) antibodies, or IRDye680RD-α-rabbit and IRDye800CW-α-mouse (Li-COR Biosciences) antibodies.
Fluorescence Polarization Peptide Binding Assays
C-Terminally amidated peptides were purchased from Bio Basic Inc., AnaSpec, or the Keck Biotechnology Resource Laboratory (Yale University, New Haven, CT). These peptides were N-terminally labeled with fluorescein 5-isothiocyanate (FITC) or 5,6-carboxyfluorescein (5,6-FAM) by the manufacturer, or in house by using amine-reactive FITC (Invitrogen) in 0.1 M sodium bicarbonate (pH 9.0). Binding reaction mixtures (100 μL) contained 50 nM fluorescein-labeled peptide and increasing concentrations of Pellinos (0–190 μM) in PBS. Fluorescence polarization for each sample was measured at 25 °C using a Beacon 2000 system (Panvera), use of which was generously provided by G. Van Duyne. Data were fit to a simple binding isotherm using Prism (GraphPad Software). Polarization values were normalized to the value without added Pellino. For binding experiments with low-affinity peptides, the highest protein concentrations did not reach saturation. In curve fitting for these data, Bmax was constrained to the Bmax values obtained from the fit to data for the same Pellino binding to the wild-type IRAK1 pT141 peptide. All binding experiments were performed at least three times.
Results
The Pellino FHA Domain pT Peptide-Binding Pocket Is Essential for Interaction with IRAK1
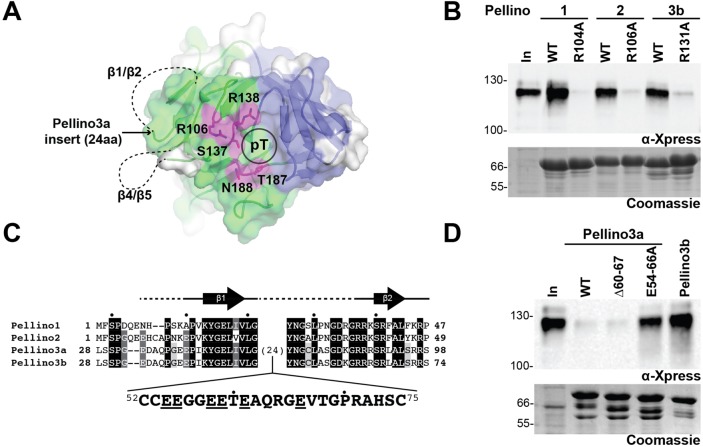
The binding pocket for the phosphothreonine moiety of a phosphorylated FHA substrate23 is highly conserved in Pellino2.20 We previously showed that substitution of alanine at highly conserved R106 in the predicted pT-binding pocket of the Pellino2 FHA domain abolished the interaction of Pellino2 with phosphorylated IRAK1 (pIRAK1) in a GST pull-down assay (Figure 1A).20 This qualitative binding assay takes advantage of the fact that IRAK1 is highly phosphorylated when transiently overexpressed in HEK293 cells. GST-Pellino can pull down this phosphorylated IRAK1 from HEK293T cell lysates. Using this assay, we now show that substitution of alanine at the analogous, highly conserved arginine in the pT-binding pocket in Pellino1 (R104A) and Pellino3b (R131A) also ablates binding of pIRAK1 to GST-Pellino (Figure 1B). These data confirm that the pT-binding pocket is critical for interaction of Pellino1, -2, and -3b with pIRAK1.
Figure 1.
The Pellino FHA domain phosphothreonine (pT)-binding pocket is essential for interaction with phosphorylated IRAK1 (pIRAK1). (A) Cartoon representation of the Pellino2 FHA domain structure (PDB entry 3EGA) looking onto the face of the pT-binding pocket (circled). The canonical FHA domain and the FHA wing are colored green and blue, respectively. Amino acids that are strongly implicated in binding to the phosphorylated peptides and proteins are shown as sticks and colored magenta. Highlighted in white on the molecular surface are amino acids that are not conserved among the four Pellinos (see also Figure S1 of the Supporting Information). Dashed lines represent two disordered loops located near the pT-binding pocket: amino acids 28–37 (β1/β2) and 120–129 (β4/β5). The 24-amino acid insertion in Pellino3a would be located at the start of the disordered β1/β2 loop. (B) Substitution of alanine at a key arginine in the FHA domain pT-binding pocket of Pellino1 (R104A), Pellino2 (R106A), and Pellino3b (R131A) ablates pIRAK1 interaction in a GST pull-down assay. The indicated wild-type (WT) or mutated GST fusions of Pellinos were incubated with HEK293T lysates that contained overexpressed, phosphorylated Xpress-tagged IRAK1. Following washing, bound proteins were eluted by denaturation, analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and visualized by Western blotting with an α-Xpress antibody to detect total IRAK1 and Coomassie blue staining to detect the GST fusion proteins. “In” indicates 4% of total lysate used for each pull-down assay. Positions of molecular mass markers are shown. (C) Sequence alignment of the β1/β2 region of the four Pellinos used in this study (mouse Pellino1 and human Pellino2, -3a, and -3b). The sequence for the 24-amino acid glutamate-rich insert unique to Pellino3a is shown. This Pellino3a insert is highly conserved across species. Structural information for the Pellino2 FHA domain (PDB entry 3EGA) is shown above the sequences. Arrows indicate the location of β-strands 1 and 2, and the dashed lines indicate regions of disorder in the crystal structure. Dots above the sequences indicate every 10th amino acids in Pellino3a, starting at amino acid 30. (D) GST pull-down assays exactly as described for panel B show that Pellino3a and a Pellino3a variant with a truncation in the glutamate-rich insert (Δ60–67) do not interact with pIRAK1. Substitution of alanine for every glutamate in the Pellino3a insertion (E54–66A) confers pIRAK1 binding to Pellino3a.
We also previously showed that Pellino3a does not associate with pIRAK1 in this GST pull-down assay.20 Pellino3a differs from Pellino3b only in the presence of a glutamate-rich 24-amino acid insertion (Figure 1C). This insertion is highly conserved among mammals, suggesting that it is important for the substrate specificity and/or function of Pellino3a. This insertion is predicted to lie adjacent to the FHA domain pT peptide-binding site (Figure 1A) and may occlude the binding site or compete for substrate binding as a phospho-mimetic motif. A Pellino3a variant lacking the seven central amino acids of the inset (Pellino3a Δ60–67) is unable to bind pIRAK1 (Figure 1D), whereas a Pellino3a variant with alanine substituted for all six glutamates in the insertion (Pellino3a E54–66A) gains the ability to interact with pIRAK1 almost to the extent observed for Pellino3b (Figure 1D). These observations argue against simple occlusion of the pT peptide-binding site. Rather, we suggest that the glutamates in the Pellino3a insertion mimic phosphorylated residues, bind to the pT peptide-binding pocket, and block pIRAK1 binding. These experiments affirm that the Pellino pT peptide-binding pocket is critical for mediating interaction with pIRAK1 and suggest that local sequence differences between Pellinos may be responsible for the different substrate specificities implied by functional studies.
Identification of an IRAK1-Derived Pellino-Binding Domain
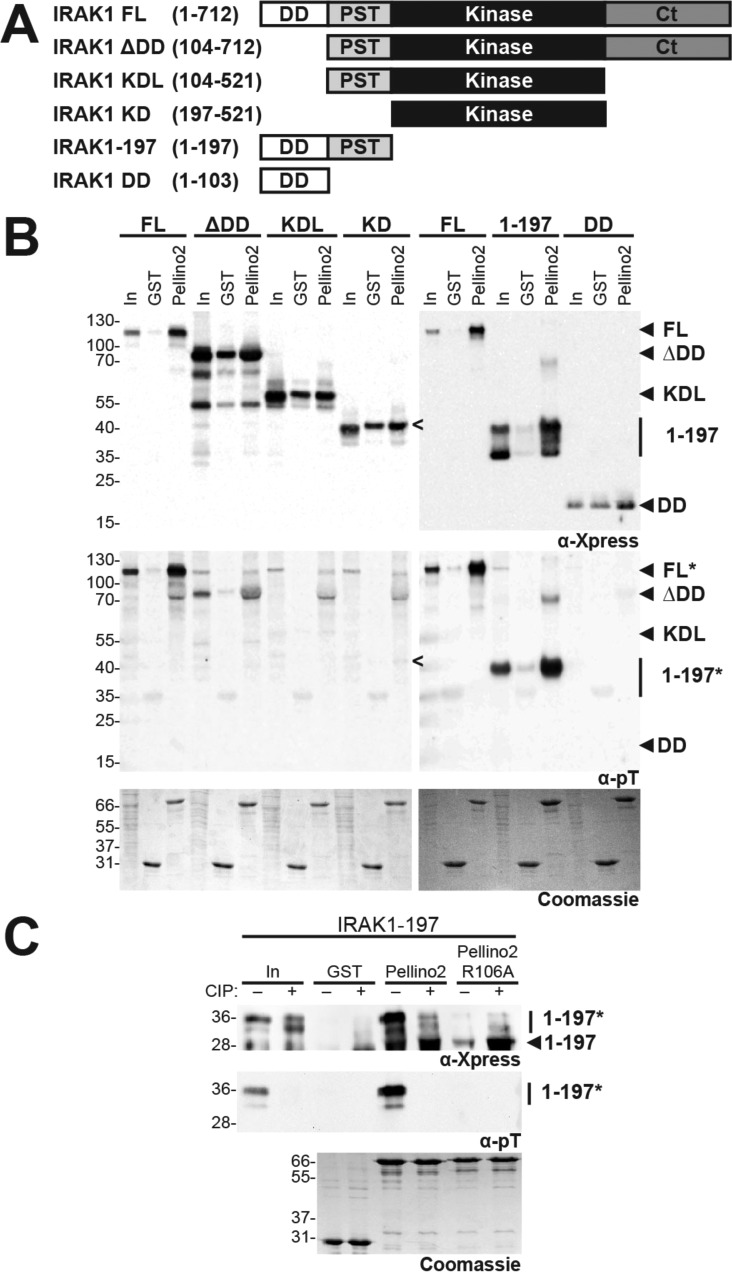
We next sought to identify a specific Pellino-binding domain in IRAK1. We chose to use Pellino2 for this analysis as there is strong evidence that IRAK1 is a physiologically relevant substrate for Pellino2.9 We performed an IRAK1 truncation analysis to identify regions of IRAK1 that can interact with Pellino2. IRAK1 truncation variants (Figure 2A) were transiently overexpressed in HEK293T cells and qualitatively tested for their ability to associate with GST-Pellino2 (Figure 2B). None of the IRAK1 truncation variants that contain the kinase domain (ΔDD, KDL, and KD) interacts with GST-Pellino2 above the relatively high background levels observed with GST alone (Figure 2B). Consistent with this, these kinase domain-containing IRAK1 fragments are not robustly phosphorylated, as assessed by α-pT immunoblots (Figure 2B, middle panel).
Figure 2.
Pellino2 interacts with IRAK1–197 in a phosphorylation-dependent manner. (A) Schematic representations of IRAK1 and truncation variants indicating the amino acids of full length (FL) IRAK1 that are included in each variant generated. Abbreviations: DD, death domain; PST, proline-, serine-, and threonine-rich domain; KD, kinase domain; Ct, C-terminal domain. (B) GST-Pellino2 pull-down analysis of the indicated IRAK1 variants expressed in HEK293T cells, essentially as described in the legend of Figure 1B. For each IRAK1 variant, three samples were analyzed: 4% of the HEK293T cell lysate (In) and samples from GST alone and GST-Pellino2 pull-down assays. Total Xpress-tagged protein was detected with an α-Xpress antibody (top), the extent of phosphorylation detected with the α-phosphothreonine (α-pT) antibody (middle), and equal loading of GST and GST-Pellino2 verified with Coomassie staining (bottom). Only IRAK1 FL and IRAK1–197 show robust interaction with Pellino2. IRAK1–197 migrates as several discrete bands, the range of which is indicated by a vertical line. The faster-migrating species are not observed on the α-pT blot and presumed to represent unphosphorylated IRAK1–197. The expected position of each IRAK1 variant is shown, as are positions of molecular weight markers. For the sake of clarity, the position of IRAK1 KD is indicated with a less than sign on the left blots, because this species runs at the same position as phosphorylated IRAK1–197. An asterisk after the protein name indicates that this species is detected on the α-pT blot. (C) Pellino2–IRAK1–197 interaction is ablated when IRAK1–197 is treated with CIP or when the Pellino2 FHA domain pT peptide-binding pocket contains an alanine substitution at a key arginine (Pellino2 R106A). GST pull-down assays were performed as described for panel B. For the sake of clarity, the α-Xpress blot (top panel) was cropped below the 28 kDa molecular mass marker to cut off a strong nonspecific band seen in the CIP-treated samples. The uncropped blot is shown in Figure S2 of the Supporting Information.
By contrast, we observe robust, phosphorylation-dependent interaction of GST-Pellino2 with a fragment containing only the first 197 amino acids of IRAK1 (IRAK1–197). This fragment contains a death domain (DD) followed by a proline-, serine-, and threonine-rich (PST) domain. Substitution of alanine at several serine and threonine sites in the PST domain impairs IRAK1 ubiquitination and function.27 Further, multiple serines and threonines in the PST domain of IRAK1 have been shown to be phosphorylated in cells.28 At least three species of IRAK1–197 are observed in the HEK293T lysates (In) with the α-Xpress antibody. Only the slowest-migrating form of IRAK1–197 contains significant phosphothreonine (Figure 2B, middle panel) and is highly enriched in the GST-Pellino2 pull-down fraction (Figure 2B, top panel). Enzymatic dephosphorylation of IRAK1–197 results in undetectable levels of pT in this sample and abolishes interaction with GST-Pellino2 (Figure 2C). In addition, the Pellino2 pT-binding pocket variant R106A does not interact with IRAK1–197 (Figure 2C). IRAK1–197 lacks the IRAK1 kinase domain and must be phosphorylated by kinases endogenous to HEK293T cells. We presume that the same endogenous kinases that can phosphorylate the kinase-dead IRAK1 variant (IRAK1 K239A) in IRAK1-null HEK293 cells (I1a cells)29 also phosphorylate IRAK1–197. A shorter truncation variant that lacks the PST domain and comprises only the IRAK1 DD is not phosphorylated in HEK293T cells and shows no binding to GST-Pellino2 (Figure 2B). This suggests that the Pellino2-binding region may lie within the PST domain or that this domain is required for appropriate phosphorylation of IRAK1–197.
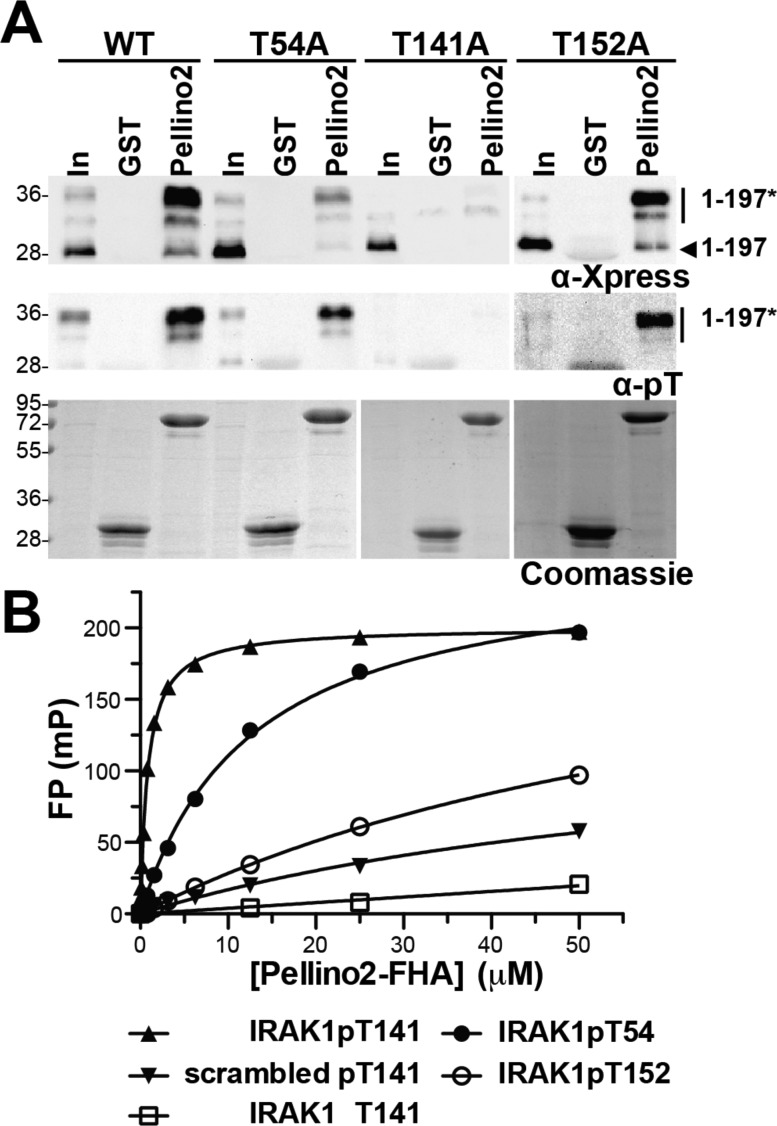
Next, we sought to identify which threonines in IRAK1–197 are important for interaction with Pellino2. There are nine threonines in IRAK1–197: five are in the presumed unstructured PST domain and four in the DD. On the basis of a structure-based sequence alignment with the IRAK2 DD (PDB entry 3MOP),30 only three of the four threonines in the IRAK1 death domain are predicted to be surface-exposed. We individually mutated to alanine each of the eight presumed surface-exposed threonines in IRAK1–197, expressed each mutated IRAK1–197 in HEK293T cells, and tested the mutants for interaction with GST-Pellino2 (Figure 3A and Figure S3A of the Supporting Information). Six of these IRAK1–197 variants behave like the wild type. However, alanine substitution at T54 or T141 significantly disrupts GST-Pellino2 interaction (Figure 3A). In both cases, the IRAK1–197 variants show significantly reduced levels of phosphorylation (Figure 3A, middle panel). While the phosphorylated fraction of IRAK1–197 T54A still associates with Pellino2, no phosphorylated IRAK1–197 T141A is pulled down with GST-Pellino2. One explanation for these results is that T54 in the DD is important for the phosphorylation of IRAK1–197, whereas T141 in the PST domain represents a phosphorylation site that is directly recognized by Pellino2. T141 is one of several sites in the PST domain that have been confirmed to be phosphorylated in full length IRAK1 from cells by mass spectrometry.28 Substitution of alanine at T141 in the context of full length IRAK1 does not abolish binding of Pellino2, suggesting that there are additional phosphorylated sites that can interact with GST-Pellino in the hyperphosphorylated, overexpressed IRAK1 from HEK293T cells (Figure S3B of the Supporting Information).
Figure 3.
Pellino2 specifically interacts with a peptide motif around pT141 of IRAK1–197. (A) GST-Pellino2 was used to pull down Xpress-tagged IRAK1–197 variants with alanine substituted at eight different threonines (see also Figure S3A of the Supporting Information), exactly as described in the legend of Figure 2B. T54A and T141A show reduced interaction with Pellino2. T152A and five other substitution mutants look like wild-type IRAK1–197 (Figure S3A of the Supporting Information). The T54A and T141A variants of IRAK1–197 also show reduced levels of threonine phosphorylation. (B) Binding of the indicated fluorescein-labeled peptides to increasing concentrations of the Pellino2 FHA domain (amino acids 15–275 of Pellino2). Binding is detected by a change in the fluorescence polarization (FP) of the fluorescein. A representative experiment is shown for each peptide. The curves indicate the fit to a simple binding isotherm for the data sets shown. Mean KD values from at least three independent experiments are listed in Table 1.
Pellino2 Specifically Binds to an IRAK1pT141-Derived Phosphopeptide
Synthetic phosphorylated peptides have been widely used to study the binding specificity of FHA domains,23,24,31−33 so we next asked whether Pellino2 can interact with a peptide comprising amino acids 137–145 of IRAK1 with a phosphothreonine at position 141 (IRAK1pT141). We tested binding of the more stable Pellino2 truncation variant (Pellino2 FHA domain, residues 15–275) to fluorescently labeled phosphopeptides in a quantitative fluorescence polarization assay. IRAK1pT141 binds to the Pellino2 FHA domain with a KD of 0.82 μM (Figure 3B and Table 1), which is comparable to the affinities that have been reported for other FHA domains binding to peptides containing their preferred phosphopeptide recognition motif.23,24,31−33 The affinity for a scrambled version of pT141 is more than 100-fold weaker, and no binding is detected for the nonphosphorylated version of this peptide. These data demonstrate that high-affinity binding to Pellino2 relies not only on a phosphorylated threonine but also on the local sequence context around this moiety. In agreement with our IRAK1–197 mutational analysis, phosphothreonine-containing peptides derived from the sequences around IRAK1 T54 and T152 (IRAK1pT54 and -pT152, respectively) show affinities for the Pellino2 FHA domain markedly weaker than that measured for IRAK1pT141 (Figure 3B and Table 1). These results further support the argument that the local sequence context around the pT is important for optimal Pellino2 binding. In addition, these peptide binding studies confirm our identification of the region around T141 as a high-affinity Pellino2-binding site in IRAK1.
Table 1. Binding of IRAK1-Derived Peptides to the Pellino2 FHA Domaina.
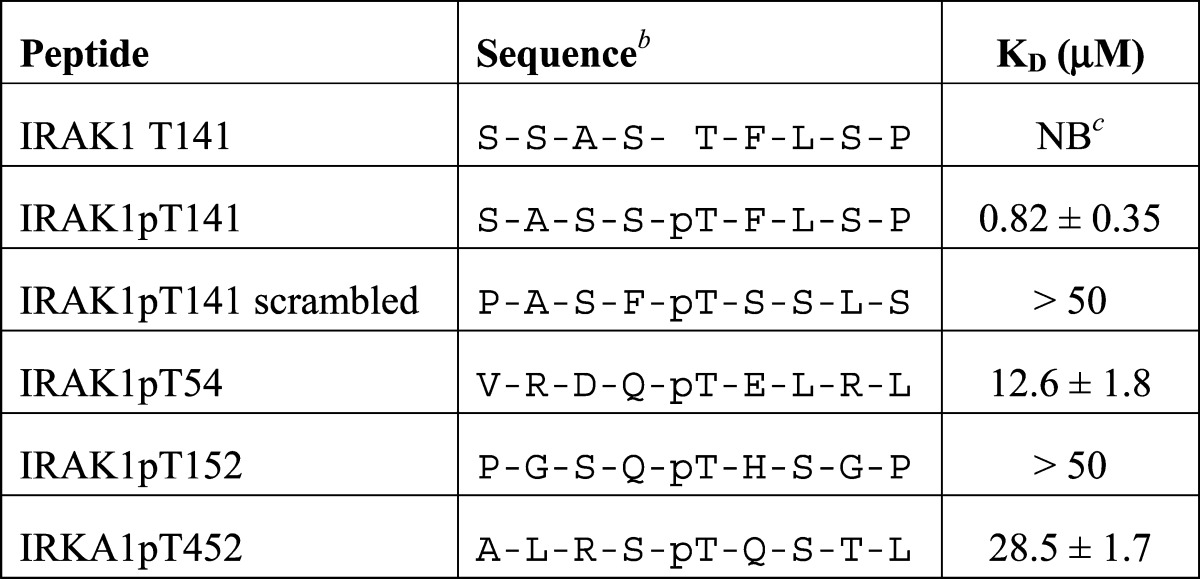
KD values for binding of the Pellino2 FHA domain to the indicated peptides were determined from at least three independent fluorescence polarization assays (Figure 3B). Baseline-corrected data were fit to a simple binding equation.
All peptides include an N-terminal tyrosine to allow for spectroscopic quantification.
No binding detected.
Pellinos Have Different Phosphothreonine Peptide Binding Specificities
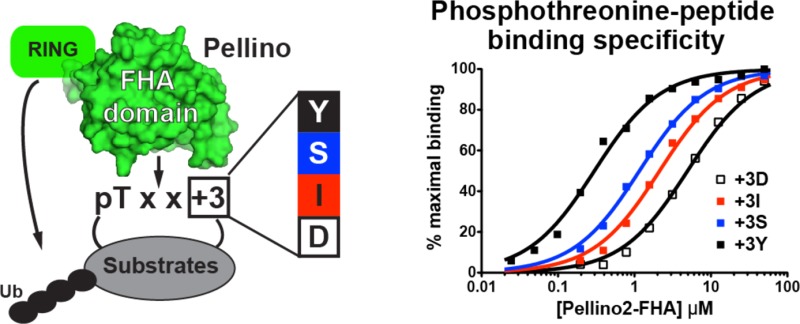
To further assess the peptide binding specificity of Pellinos, we turned to the rich information available about the binding specificity of FHA domains.25 The major determinant of pT peptide recognition for many FHA domains is the amino acid located three positions C-terminal to the phosphorylated threonine (+3 position).23 Peptide library screens performed on a diverse selection of FHA domains have identified four FHA pT recognition motifs with different amino acids at position +3: pTxxD, pTxxI/L, pTxxY/M, and pTxxS/A.23 The motif around T141 in IRAK1 falls in the last of these four groups with a serine at position +3. Henceforth, the peptide based on this IRAK1 sequence is termed pT141+3S. To determine whether pTxxS is the optimal Pellino2 pT recognition motif, we analyzed binding to the Pellino2 FHA domain of peptides with aspartate, isoleucine, and tyrosine in place of serine at position +3 [pT141+3D, pT141+3I, and pT141+3Y, respectively (Table 2)]. The Pellino2 FHA domain binds most tightly to pT141+3Y with a KD 4.6-fold stronger than that for pT141+3S. Aspartic acid at position +3 is the most unfavorable binding motif with a KD 5-fold weaker than that of pT141 + 3S, whereas isoleucine at position +3 shows a KD 2.6-fold weaker than that of pT141+3S. These data indicate that the Pellino2 FHA domain prefers the pTxxY-binding motif 4.6-, 12-, and 23-fold over the pTxxS, pTxxI, and pTxxD motifs, respectively (Figure 4A).
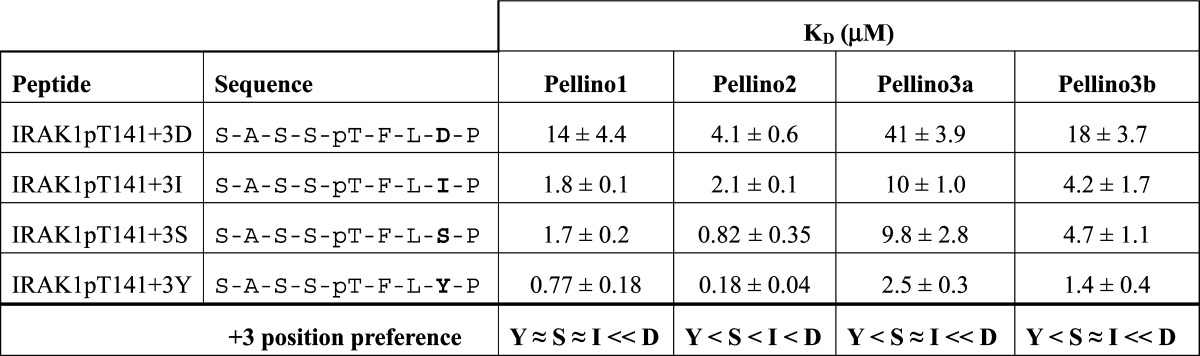
Table 2. Binding of IRAK1pT141+3 Position Variants to Pellino FHA Domainsa.
KD values for the binding of peptides to the FHA domains of Pellino1, -2, -3a, and -3b were determined as described in the footnotes of Table 1.
Figure 4.
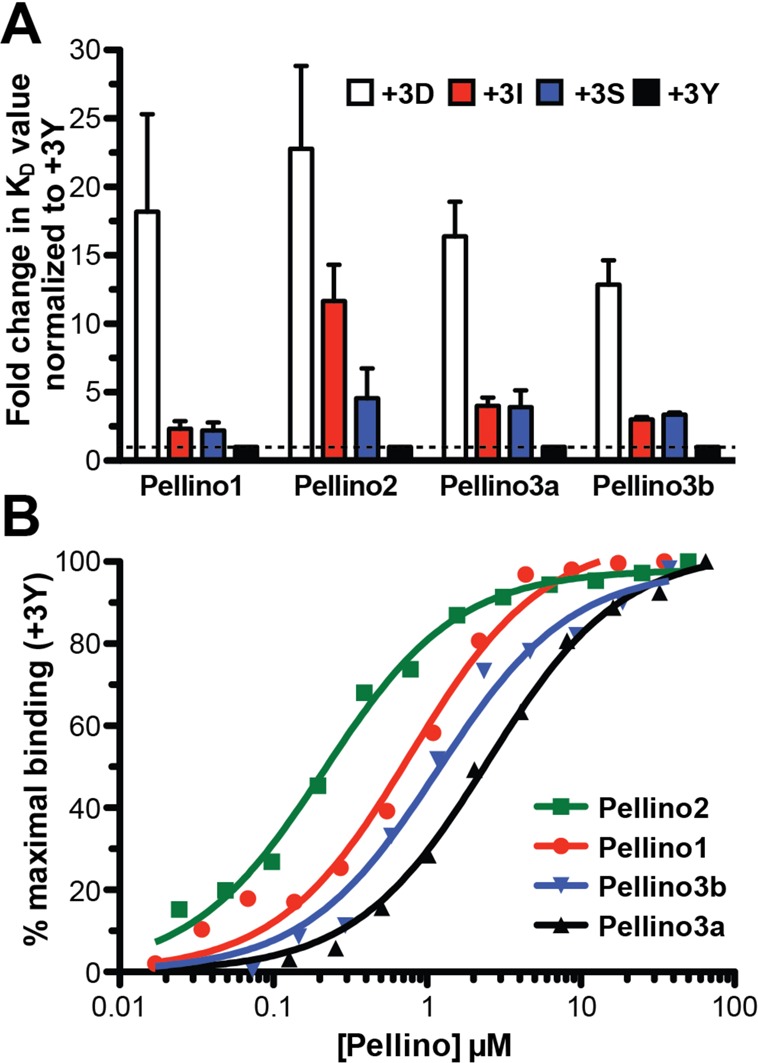
Pellinos have different phosphothreonine peptide binding specificities. (A) The KD for the binding of each Pellino FHA domain to IRAKpT141 (+3S) and to peptides with D, I, or Y substituted at position +3 were determined and are presented as the fold change in the KD relative to the binding to the pT141+3Y peptide (+3Y). Each Pellino binds the +3Y peptide with the highest affinity (Table 2), which is normalized to 1 as indicated by the dashed line. The fold changes in KD values for binding to the pT141+3D (+3D, white), pT141+3I (+3I, red), and pT141+3S (+3S, blue) are shown. (B) Fluorescence polarization binding assay for each Pellino with the pT141+3Y peptide, exactly as described in the legend of Figure 3B. A representative experiment is shown for the FHA domain of Pellino1 (red), Pellino2 (green), Pellino3a (black), and Pellino3b (blue). Curves indicate the fit to a simple binding isotherm for each data set shown. Mean KD values for at least three independent experiments are listed in Table 2.
We next asked whether the observed Pellino2 peptide binding preference is also seen for the other Pellinos. We measured binding to the same panel of fluorescently labeled peptides using Pellino1, -3a, and -3b truncations that contain only the FHA domain [amino acids 2–287, 40–305, and 2–304, respectively (Table 2 and Figure 4)]. All Pellinos bind to the original IRAK1-derived pT141+3S, which suggests that phosphorylation of T141 plays a role in association of IRAK1 with all GST-Pellinos. Pellino3a shows the weakest binding to pT141+3S (KD of 9.8 μM), which is consistent with the lack of detectable binding between GST-Pellino3a and pIRAK1 (Figure 1D).
All Pellinos bind most tightly to the pT141+3Y peptide and substantially disfavor binding to the pT141+3D motif. Differences in the extent to which each Pellino favors the pTxxY motif over the pTxxI and pTxxS motifs arise. As described above, Pellino2 clearly discriminated among all three, with KD values increasing in the following order: pTxxY < pTxxS < pTxxI. By contrast, Pellino1 shows minimal discrimination in binding between the peptides with tyrosine, serine, and isoleucine at position +3 (KD values of 0.77, 1.7, and 1.8 μM, respectively). Pellino3a and -3b, which differ only by the 24-amino acid insertion (Figure 1C), have very similar +3 motif preferences, although the affinity in each case is ∼2-fold weaker for Pellino3a than for Pellino3b. Like Pellino1, the Pellino3 isoforms do not discriminate between serine and isoleucine at position +3. Unlike Pellino1, the Pellino3 isoforms do show a clear preference for tyrosine at this location (see Table 2 for a summary of the +3 position binding preferences). The observed binding preferences revealed from this panel of pT peptides suggest that each Pellino will have a distinct set of preferred substrates.
Pellinos Have Distinct Specificities for RIP1 and TRAF6
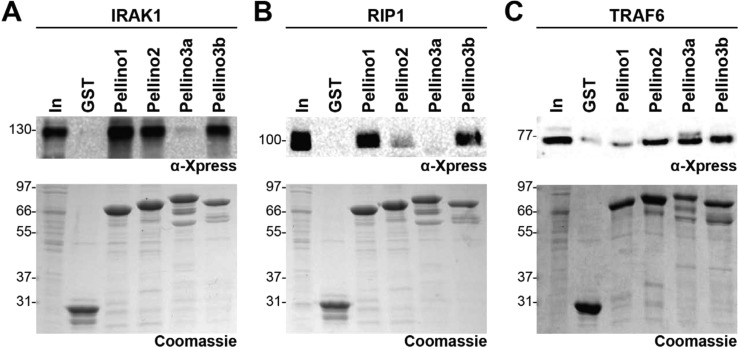
Knockout studies in mice indicate that Pellino1 and Pellino3 have nonredundant functions in TLR signaling. These divergent functions have been attributed to differential polyubiquitination of the substrates RIP1 and TRAF6 by Pellino1 and -3, respectively.8,14 On the basis of our observation that each Pellino has a different peptide binding profile, we postulated that Pellino1 selectively targets RIP1 over TRAF6, whereas Pellino3 is selective for TRAF6. We tested this hypothesis by examining the interaction of GST-Pellinos with these proteins transiently expressed in HEK293T cells. We find that Pellino1 interacts robustly with RIP1 and IRAK1, but not with TRAF6, consistent with our prediction (Figure 5). Pellino3a, which fails to interact with either IRAK1 or RIP1 in this GST pull-down assay, is able to interact with TRAF6. Similar amounts of TRAF6 are pulled down with Pellino2, -3a, and -3b (Figure 5C). Finally, we observe that Pellino2 shows weak interaction with RIP1. The different substrate interaction preferences we observe in this assay can be attributed solely to differences in the binding specificity of each Pellino. Because substrate binding is a prerequisite for substrate ubiquitination, this binding selectivity of Pellinos is likely responsible, at least in part, for their different cellular functions.
Figure 5.
Pellinos have different specificities for IRAK1, RIP1, and TRAF6. GST-Pellino pull-down results for (A) Xpress-tagged IRAK1, (B) Xpress-tagged RIP1, and (C) Xpress-tagged TRAF6 from lysates of HEK293T cells overexpressing each Xpress-tagged protein. Samples were processed as described in the legend of Figure 1. IRAK1 is pulled down to approximately the same extent by Pellino1, -2, and -3b and not at all by Pellino3a. RIP1 interacts robustly with only Pellino1 and -3b. TRAF6 interacts with Pellino2, -3a, and -3b but not with Pellino1.
Pellino3 was recently shown to interact with RIP1 to regulate TNFα-induced apoptosis.16 Although RIP1 does not appear to be a Pellino3 substrate in this context, both Pellino3 isoforms associate with RIP1.16 This result is in contrast with our pull-down assays that show interaction of RIP1 with Pellino3a and not with Pellino3b (Figure 5B). It is of note that Pellino3a binds pT peptides only ∼2-fold more weakly than Pellino3b (Table 2 and Figure 3B). The discrepancy in Pellino3a–RIP1 interaction may therefore be due to differences in pull-down assay conditions.
Distribution of FHA-Binding Motifs in Pellino Substrates
We next asked whether it is possible to rationalize the Pellino specificity differences observed in our simple GST pull-down assay based only on our peptide binding results and the distribution of pT+3 motifs in Pellino substrates (Table 3 and Table S1 of the Supporting Information), with no consideration of the in vivo phosphorylation state of these motifs. Pellino1 binds robustly to IRAK1 and RIP1 but not to TRAF6. This correlates with a requirement for pTxxY- or pTxxS-binding motifs, which are found in IRAK1 and RIP1, but not in TRAF6. These motifs are also found in cRel, another known Pellino1 substrate.17 Pellino2 interacts more robustly with IRAK1 than with RIP1 and TRAF6. This may reflect the strong preference of Pellino2 for pTxxY motifs (Figure 4B), which are more frequent in IRAK1 than in the other two substrates. It is hard to rationalize the binding of Pellino3a to TRAF6 and not to IRAK1 and RIP1 based on these very simple considerations. We note that TRAF6 and RIP2 (a recently discovered Pellino3 substrate15) both contain pTxxI motifs that are not found in the other surveyed substrates. However, the peptide binding experiments do not suggest that Pellino3a shows any significant preference for this motif.
Table 3. Frequencies of FHA Domain Peptide-Binding Motifs in Pellino Substratesa.
| pTxxD | pTxxI/L | pTxxS/A | pTxxY/M | |
|---|---|---|---|---|
| IRAK1 | – | –/3 | 1/5 | 2/– |
| RIP1 | – | –/4 | 5/– | 1/2 |
| TRAF6 | 4 | 2/2 | –/– | –/2 |
| RIP2 | – | 2/3 | 4/– | 2/1 |
| cRel | 3 | –/5 | 6/– | 1/1 |
For motifs with two possible amino acids at position +3, the number of motifs with the amino acids analyzed in this study (I, S, and Y) is listed first, followed by the number of occurrences of the motif with the other amino acid (L, A, and M). See also Table S1 of the Supporting Information.
Discussion
We find that the Pellino2 FHA domain binds to pT141+3Y with a KD of 180 nM, which is comparable to the tightest known FHA domain–pT peptide interaction, the interaction of the Rv0020c FHA domain with a highly optimized library screen-derived peptide (KD of ∼100 nM).24 The interaction of Pellino2 with the pT141+3Y peptide has an affinity 3–14-fold higher than those for the other three Pellinos (Figure 4B). If this high-affinity binding is an intrinsic property of Pellino2 compared to the other members of this family of E3 ligases, this may explain the difficulty in identifying the true physiologically relevant Pellino2 substrates on the basis of experiments using overexpression or incomplete knockdown of this protein. Alternatively, high-affinity binding may be a property of all Pellino FHA domains, but we have not identified the optimal binding motif for Pellino1, -3a, or -3b in this study.
On the basis of our peptide binding analysis, all four Pellinos show the weakest binding to the IRAK1pT141-derived peptide variant containing the pTxxD motif, in each case, at least 13 times weaker than for the pT141+3Y peptide (Table 2 and Figure 4A). This trend against aspartate (a phospho-mimetic residue) at position +3 could reflect negative selection of binding sites with a phosphorylated residue in this position. Phosphorylation of the serine of a pTxxS motif would be expected to reduce the level of Pellino binding and could represent a regulatory mechanism of Pellino substrate recognition. There is evidence to suggest that this form of regulation may occur in the PST domain of IRAK1. IRAK1 is known to become phosphorylated at a number of serines and threonines in the PST domain, including T141 and S141, based on mass spectrometry analyses of IRAK1 from TLR7-stimulated cells.28 In that study, peptides were identified with either one or both of these two amino acids phosphorylated. Our pT peptide binding analysis suggests that phosphorylation of only T141 would promote Pellino binding, while simultaneous phosphorylation of T141 and S144 would disfavor Pellino binding. Two other IRAK1-binding partners have been reported to bind this same region of the IRAK1 PST, the phosphorylation specific prolyl isomerase Pin1 and the substrate recognition domain of SCF-β-TrCp E3 ubiquitin ligase (β-TrCp).28,34 The interaction of these two proteins requires phosphorylation of serines N- or C-terminal to T141, in contrast to Pellinos, binding of which we propose is disfavored by additional local phosphorylation. Thus, the IRAK1 phosphorylation state may dictate which post-translational modifiers can associate with IRAK1 and, hence, influence the regulation of downstream TLR signaling events.
Pellino phosphorylation has previously been shown to enhance the catalytic activity of Pellinos in vitro(5,18) (Figure S1 of the Supporting Information). It is unclear whether phosphorylation affects Pellino substrate specificity, as has been observed for E3 ubiquitin ligase Nedd4-2, in which phosphorylation of Nedd4-2 inhibits substrate binding as part of a regulatory mechanism for maintaining proper epithelial Na+ transport.35 We speculate that phosphorylation on surface loops of Pellinos could inhibit substrate binding in a manner that mirrors the autoinhibition of Pellino3a by the Pellino3a specific insertion (Figure 1C,D). The β4/β5 loop (Figure 1A and Figure S1 of the Supporting Information) represents a possible site for such regulation. This loop contains several serines and threonines, is predicted to lie close to the phosphothreonine-binding site, and is relatively poorly conserved among Pellinos. Interestingly, S125 and T127 in this loop were identified as sites of phosphorylation of Pellino1 by IRAK1 and IRAK4,18 although in this case Pellino1 phosphorylation was implicated in activation of the E3 ligase activity. More studies are required to evaluate the effect of Pellino phosphorylation on substrate binding.
There are clear links between several Pellinos and the pathogenesis of specific inflammatory diseases. Pellino1 is a critical mediator of microglia activation and contributes to the onset of the mouse model for multiple sclerosis, autoimmune encephalomyelitis (EAE).36 Pellino3 deficiency enhances the pathogenesis of experimental murine models of colitis, and this correlates with the abnormally low Pellino3 expression levels that are consistently seen in colon samples from Crohn’s disease patients.15 Our study demonstrating specificity in the binding of Pellinos to phosphorylated peptides and to intact substrates serves as encouragement that agents can be developed to specifically target an individual Pellino for pharmacologic intervention with minimal off-target effects.
Acknowledgments
We thank Mark Lemmon, Michael May, and members of the Ferguson and Lemmon laboratories for insightful discussion and critical reading of the manuscript; Chun-Chi Lin for technical assistance; and Nisha Ninan, Kathryn Sarachan, and the Van Duyne laboratory for assistance with the fluorescence polarization peptide binding assays.
Glossary
Abbreviations
- 5,6-FAM
5,6-carboxyfluorescein
- CIP
calf intestinal alkaline phosphatase
- DD
death domain
- FHA
forkhead-associated
- FITC
fluorescein 5-isothiocyanate
- HEK
human embryonic kidney
- IGEPAL
octylphenoxypolyethoxyethanol
- IL-1
interleukin-1
- IL-1R
interleukin-1 receptor
- IRAK1
interleukin-1 receptor-associated kinase 1
- NF-κB
nuclear factor κB
- NOD2
nucleotide-binding oligomerization domain-containing protein 2
- PDB
Protein Data Bank
- PST
proline-, serine-, and threonine-rich domain
- pT
phosphorylated threonine
- RIP1 and -2
receptor-interacting serine/threonine protein kinases 1 and 2, respectively
- TCR
T-cell receptor
- TLR
Toll-like receptor
- TNFα
tumor necrosis factor α
- TRAF6
tumor necrosis factor receptor-associated factor 6
- βME
β-mercaptoethanol.
Supporting Information Available
Sequence alignment of the mammalian Pellinos used in this study (Figure S1), uncropped versions of the Western blots in Figure 2 (Figure S2), additional GST-Pellino2 pull-down assays of IRAK1–197 and full length IRAK1 mutants (Figure S3), and FHA domain-binding motifs found in Pellino substrates (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by American Heart Association Grant 09GRNT2120055. Y.-S.H. was supported in part by National Institutes of Health Training Grant T32-GM008275 and American Heart Association Grant 11PRE7420022.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jin W.; Chang M.; Sun S.-C. (2012) Peli: A family of signal-responsive E3 ubiquitin ligases mediating TLR signaling and T-cell tolerance. Cell. Mol. Immunol. 9, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynagh P. N. (2009) The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol. 30, 33–42. [DOI] [PubMed] [Google Scholar]
- Schauvliege R.; Janssens S.; Beyaert R. (2007) Pellino Proteins: Novel Players in TLR and IL-1R Signalling. J. Cell. Mol. Med. 11, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynagh P. N. (2014) The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Rev. Immunol. 14, 122–131. [DOI] [PubMed] [Google Scholar]
- Ordureau A.; Smith H.; Windheim M.; Peggie M.; Carrick E.; Morrice N.; Cohen P. (2008) The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J. 409, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. P.; Hanly J. A.; Moynagh P. N. (2007) Kinase-active Interleukin-1 Receptor-associated Kinases Promote Polyubiquitination and Degradation of the Pellino Family Direct Evidence for Pellino Proteins Being Ubiquitin-Protein Isopeptide Ligases. J. Biol. Chem. 282, 29729–29737. [DOI] [PubMed] [Google Scholar]
- Schauvliege R.; Janssens S.; Beyaert R. (2006) Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 580, 4697–4702. [DOI] [PubMed] [Google Scholar]
- Chang M.; Jin W.; Sun S.-C. (2009) Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 10, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W.; Yu M.; Zhou H.; Cui W.; Wang J.; DiCorleto P.; Fox P.; Xiao H.; Li X. (2012) Pellino 2 Is Critical for Toll-like Receptor/Interleukin-1 Receptor (TLR/IL-1R)-mediated Post-transcriptional Control. J. Biol. Chem. 287, 25686–25695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H.; Qian W.; Staschke K.; Qian Y.; Cui G.; Deng L.; Ehsani M.; Wang X.; Qian Y. W.; Chen Z. J.; Gilmour R.; Jiang Z.; Li X. (2008) Pellino 3b Negatively Regulates Interleukin-1-induced TAK1-dependent NF κB Activation. J. Biol. Chem. 283, 14654–14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze D. B.; Wu C. J.; Thomas J. A.; Landstrom A.; Ashwell J. D. (2008) Lys63-Linked Polyubiquitination of IRAK-1 Is Required for Interleukin-1 Receptor- and Toll-Like Receptor-Mediated NF-κB Activation. Mol. Cell. Biol. 28, 3538–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-J.; Conze D. B.; Li T.; Srinivasula S. M.; Ashwell J. D. (2006) Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 8, 398–406. [DOI] [PubMed] [Google Scholar]
- Enesa K.; Ordureau A.; Smith H.; Barford D.; Cheung P. C. F.; Patterson-Kane J.; Arthur J. S. C.; Cohen P. (2012) Pellino1 Is Required for Interferon Production by Viral Double-stranded RNA. J. Biol. Chem. 287, 34825–34835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siednienko J.; Jackson R.; Mellett M.; Delagic N.; Yang S.; Wang B.; Tang L. S.; Callanan J. J.; Mahon B. P.; Moynagh P. N. (2012) Pellino3 targets the IRF7 pathway and facilitates autoregulation of TLR3- and viral-induced expression of type I interferons. Nat. Immunol. 13, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Yang S.; Wang B.; Humphries F.; Jackson R.; Healy M. E.; Bergin R.; Aviello G.; Hall B.; McNamara D.; Darby T.; Quinlan A.; Shanahan F.; Melgar S.; Fallon P. G.; Moynagh P. N. (2013) Pellino3 ubiquitinates RIP2 and mediates Nod2-induced signaling and protective effects in colitis. Nat. Immunol. 14, 927–936. [DOI] [PubMed] [Google Scholar]
- Yang S.; Wang B.; Tang L. S.; Siednienko J.; Callanan J. J.; Moynagh P. N. (2013) Pellino3 targets RIP1 and regulates the pro-apoptotic effects of TNF-α. Nat. Commun. 4, 2583. [DOI] [PubMed] [Google Scholar]
- Chang M.; Jin W.; Chang J.-H.; Xiao Y.; Brittain G. C.; Yu J.; Zhou X.; Wang Y.-H.; Cheng X.; Li P.; Rabinovich B. A.; Hwu P.; Sun S.-C. (2011) The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 12, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.; Peggie M.; Campbell D. G.; Vandermoere F.; Carrick E.; Cohen P. (2009) Identification of the phosphorylation sites on the E3 ubiquitin ligase Pellino that are critical for activation by IRAK1 and IRAK4. Proc. Natl. Acad. Sci. U.S.A. 106, 4584–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh E. T.; Arthur J. S.; Cheung P. C.; Akira S.; Toth R.; Cohen P. (2012) Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system. Biochem. J. 441, 339–346. [DOI] [PubMed] [Google Scholar]
- Lin C.-C.; Huoh Y.-S.; Schmitz K. R.; Jensen L. E.; Ferguson K. M. (2008) Pellino proteins contain a cryptic FHA domain that mediates interaction with phosphorylated IRAK1. Structure 16, 1806–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D.; Henckel J.; Fersht A. R.; Jackson S. P. (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4, 387–394. [DOI] [PubMed] [Google Scholar]
- Durocher D.; Jackson S. P. (2002) The FHA domain. FEBS Lett. 513, 58–66. [DOI] [PubMed] [Google Scholar]
- Durocher D.; Taylor I. A.; Sarbassova D.; Haire L. F.; Westcott S. L.; Jackson S. P.; Smerdon S. J.; Yaffe M. B. (2000) The molecular basis of FHA domain: Phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6, 1169–1182. [DOI] [PubMed] [Google Scholar]
- Pennell S.; Westcott S.; Ortiz-Lombardía M.; Patel D.; Li J.; Nott T. J.; Mohammed D.; Buxton R. S.; Yaffe M. B.; Verma C.; Smerdon S. J. (2010) Structural and Functional Analysis of Phosphothreonine-Dependent FHA Domain Interactions. Structure 18, 1587–1595. [DOI] [PubMed] [Google Scholar]
- Mahajan A.; Yuan C.; Lee H.; Chen E. S. W.; Wu P.-Y.; Tsai M.-D. (2008) Structure and function of the phosphothreonine-specific FHA domain. Sci. Signaling 1, re12.and references cited therein. [DOI] [PubMed] [Google Scholar]
- Tropea J. E., Cherry S., and Waugh D. S. (2009) Expression and purification of soluble His6-tagged TEV protease (Doyle S. A., Ed.) pp 297–307, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Yao J.; Kim T. W.; Qin J.; Jiang Z.; Qian Y.; Xiao H.; Lu Y.; Qian W.; Gulen M. F.; Sizemore N.; DiDonato J.; Sato S.; Akira S.; Su B.; Li X. (2007) Interleukin-1 (IL-1)-induced TAK1-dependent versus MEKK3-dependent NFκB activation pathways bifurcate at IL-1 receptor-associated kinase modification. J. Biol. Chem. 282, 6075–6089. [DOI] [PubMed] [Google Scholar]
- Tun-Kyi A.; Finn G.; Greenwood A.; Nowak M.; Lee T. H.; Asara J. M.; Tsokos G. C.; Fitzgerald K.; Israel E.; Li X.; Exley M.; Nicholson L. K.; Lu K. P. (2011) Essential role for the prolyl isomerase Pin1 in Toll-like receptor signaling and type I interferon-mediated immunity. Nat. Immunol. 12, 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Commane M.; Burns C.; Vithalani K.; Cao Z.; Stark G. R. (1999) Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol. Cell. Biol. 19, 4643–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-C.; Lo Y.-C.; Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein N. K.; Williams R. S.; Rakovszky M. L.; Cui D.; Green R.; Karimi-Busheri F.; Mani R. S.; Galicia S.; Koch C. A.; Cass C. E. (2005) The Molecular Architecture of the Mammalian DNA Repair Enzyme, Polynucleotide Kinase. Mol. Cell 17, 657–670. [DOI] [PubMed] [Google Scholar]
- Alderwick L. J.; Molle V.; Kremer L.; Cozzone A. J.; Dafforn T. R.; Besra G. S.; Fütterer K. (2006) Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M. S. Y.; Grant R.; Manke I.; Minn K.; Yu X.; Yaffe M. B.; Chen J. (2007) RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell 131, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W.; Xiao N.; Xiao H.; Zhou H.; Yu M.; Gu J.; Li X. (2012) β-TrCP-mediated IRAK1 degradation releases TAK1-TRAF6 from the membrane to the cytosol for TAK1-dependent NF-κB activation. Mol. Cell. Biol. 32, 3990–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger R. S.; Lebowitz J.; Li H.; Alzamora R.; Wang H.; Johnson J. P.; Hallows K. R. (2009) Functional regulation of the epithelial Na+ channel by IκB kinase-β occurs via phosphorylation of the ubiquitin ligase Nedd4-2. J. Biol. Chem. 284, 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Jin J.; Chang M.; Chang J.-H.; Hu H.; Zhou X.; Brittain G. C.; Stansberg C.; Torkildsen Ø.; Wang X.; Brink R.; Cheng X.; Sun S.-C. (2013) Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 19, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.