Abstract
To explore the structural basis for odorant specificity in odorant receptors of the human malaria vector mosquito, Anopheles gambiae, odorant-binding subunits (Agam\Ors) expressed in Xenopus oocytes in combination with Agam\Orco (coreceptor subunit) were assayed by 2-electrode voltage clamp against 25 structurally related odorants. Agam\Or13 and Agam\Or15 display 82% amino acid identity and had similar, but somewhat distinct odorant response profiles. The ratio of acetophenone to 4-methylphenol responses was used in a mutation-based analysis of Agam\Or15, interchanging 37 disparate residues between Agam\Or15 and Agam\Or13. Eleven mutations caused significant changes in odorant responsiveness. Mutation of alanine 195 resulted in the largest shift in response ratio from Agam\Or15 toward Agam\Or13. Concentration–response analysis for a series of mutations of residue 195 revealed a large effect on acetophenone sensitivity, with EC50 values varying by >1800-fold and correlating with residue side chain length. Similar results were obtained for propiophenone and benzaldehyde. But, for other odorants, such as 4-methylphenol, 4-methylbenzaldehyde, and 4-methylpropiophenone, the effect of mutation was much smaller (EC50 values varied by ≤16-fold). These results show that alanine 195, putatively located at the second extracellular loop/fourth transmembrane domain interface, plays a critical role in determining the odorant response specificity of Agam\Or15.
Key words: electrophysiology, insect, olfaction, receptor, structure, Xenopus oocytes
Introduction
Animals navigate myriad chemosensory cues, which mediate their behavior. This is particularly true for olfaction, where thousands of odors must be routinely recognized and discriminated. The ability to perceive and analyze odorants is especially important for insects, because olfaction facilitates their most basic survival functions: feeding, mating behavior, and oviposition (Carey and Carlson 2011). Insects are vectors for many human diseases and mosquitoes are the most potent of these vectors, transmitting the causative agents of malaria, dengue fever, and yellow fever, that together place nearly half of the world’s population at risk of infection. Of these, human malaria is by far the most deadly and is carried by a variety of anophelines, most notably Anopheles gambiae, which transmits Plasmodium pathogen during obligate blood feeding by reproductive females and accounts for the majority of malarial transmission in sub-Saharan Africa (Takken and Knols 1999). More than 200 million cases of malaria occur each year, resulting in more than 600000 deaths (World Malaria Report 2013). In light of the direct impact on blood-feeding selection and other critical behaviors in A. gambiae and other vectors, an increased understanding of the intricacies of the insect olfactory system is a high priority in the design of novel strategies for the control of insect vector borne diseases.
Insects rely on a large array of odorant receptors (ORs) to detect the multitude of odorant compounds (Vosshall et al. 1999). Insect ORs are odorant gated, nonselective cation channels, composed of multiple subunits in an unknown stoichiometry (Sato et al. 2008; Wicher et al. 2008). Each subunit has a cytoplasmic amino-terminus and 7 transmembrane domains (TMDs) (Benton et al. 2006; Lundin et al. 2007; Smart et al. 2008). Each insect OR complex contains one of a large family of highly variable odorant-binding (or “tuning”) subunits that confer odorant specificity and at least one copy of Orco, the highly conserved OR coreceptor subunit (Larsson et al. 2004; Nakagawa et al. 2005; Hallem and Carlson 2006). Orco and odorant-binding subunits interact directly (Neuhaus et al. 2005; Benton et al. 2006) and in the absence of Orco, functional receptor complexes do not form (Larsson et al. 2004; Nakagawa et al. 2005; Hallem and Carlson 2006; Wanner et al. 2007; Sato et al. 2008; Nichols and Luetje 2010). ORs composed of different odorant-binding subunits and Orco subunits display altered ion permeability (Pask et al. 2011) and sensitivity to channel block (Nichols et al. 2011), suggesting that the ion channel is intrinsic to the OR complex and that odorant-binding subunits and Orco each make contributions to the structure of the ion pore. These receptors have also been proposed to initiate or be modified by second messenger cascades (Wicher et al. 2008; Nakagawa and Vosshall 2009). Insect ORs share little or no genetic similarity to any known receptors or channels of tetrapods (Benton et al. 2006), making them ideal targets for insect-specific control strategies.
Rational design of compounds targeting insect ORs can benefit from a detailed structural map of the OR-odorant-binding site. However, while much work has focused on identifying odorant ligands for specific receptors (Hallem and Carlson 2006; Wanner et al. 2007; Pelletier et al. 2010; Wang et al. 2010; Wanner et al. 2010; Mitchell et al. 2012), only limited progress has been made in exploring the structural basis for receptor function (Nichols and Luetje 2010; Pellegrino et al. 2011; Leary et al. 2012; Kumar et al. 2013; Xu and Leal 2013). Here, we report a structural study of Agam\Or15, a tuning OR from the malaria vector, A. gambiae, which uses a combination of mutational and functional analyses to identify several amino acid residues that serve as determinants of odorant sensitivity. For one of these residues (alanine 195), predicted to be located at the interface between extracellular loop (ECL) 2 and TMD 4, we conduct a detailed exploration of the effects of mutation on odorant specificity.
Materials and methods
Materials
Xenopus laevis frogs were purchased from Nasco. The care and use of frogs in this study were approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. Odorants and other chemicals were from Sigma-Aldrich. Agam\Or15, Agam\Or13, and Agam\Orco were cloned and inserted in expression vectors (pSP64T-Oligo for Agam\Or15 and Agam\Or13; pT7TS for Agam\Orco) as previously described (Wang et al. 2010). Mutations were introduced using QuikChange Lightning kits (Stratagene). Each mutant construct was verified by sequencing.
Expression of ORs in Xenopus oocytes
Oocytes were surgically removed from mature X. laevis frogs and follicle cells removed by treatment with collagenase B (Roche Applied Science) for 2h at room temperature. Capped cRNA encoding each OR subunit was generated using SP6 (Agam\Or15 and 13) or T7 (Agam\Orco) mMessage mMachine kits (Ambion). Oocytes were injected with 25ng of cRNA encoding each subunit and incubated at 18 °C in Barth’s saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.6, and 150 µg/mL ceftazidime) for 2–6 days prior to electrophysiological recording.
Electrophysiology and data capture
Odorant-induced currents were recorded under 2-electrode voltage clamp using an automated electrophysiology system (OpusXpress 6000A; Molecular Devices). Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). Odorant stock solutions (usually 1M) were prepared in dimethyl sulfoxide. Odorants were diluted from stock into ND96 on the day of experimentation. Odorants were applied, unless otherwise noted, for 20 s at 1.65mL/min, with extensive washing in ND96 (4.6mL/min) between applications. Micropipettes were filled with 3M KCl and had resistances of 0.2–2.0 MΩ. The holding potential was −70 mV. Current responses, filtered (4-pole, Bessel, low pass) at 20 Hz (−3 db) and sampled at 100 Hz, were captured and stored using the OpusXpress 1.1 software (Molecular Devices).
Data analysis
Concentration–response analysis was done by using PRISM 5 (GraphPad). Concentration–response curves (CRCs) were fit according to the equation: I = I max/(1 + (EC50/X)n), where I represents the current response at a given concentration of odorant, X; I max is the maximal response; EC50 is the concentration of odorant yielding a half-maximal response; and n is the apparent Hill coefficient. In Figure 2, we were interested in comparing EC50 and maximal response values for 2 odorants (Ace and 4-mp) at each receptor. For this reason, all odorant responses for a given receptor were normalized to the response of the same oocyte to a concentration of Ace that was near the EC50 for that receptor (5 µM Ace for Agam\Or13, 30 µM Ace for Agam\Or15). Normalized data were then combined and fit using the above equation to establish EC50 and maximal response (efficacy) values. In Figure 5, we were only interested in comparing the EC50 values. For this reason, the data set from each individual cell was fit to the above equation. Each data set was then normalized to that cell’s fit maximum. The normalized data for all cells expressing a given receptor were then combined and fit using the above equation to establish an EC50. Statistical significance was assessed by 2-tailed, unpaired t-test, or one-way ANOVA followed by a Dunnett’s or Bonferroni’s posttest, as appropriate.
Figure 2.
Agam\Or13 and Agam\Or15 differ in their responsiveness to 4-methylphenol and acetophenone. (A) Concentration–response relationships for activation of Agam\Or13 + Agam\Orco by acetophenone (Ace) and 4-methylphenol (4-mp). Current responses to each concentration of odorant (n = 5) were normalized to the response of the same oocyte to 5 µM Ace. EC50 values for Ace (6.7±1.8 µM) and 4-mp (45±19 µM) were significantly different (P < 0.01, t-test). Maximal response values for Ace (3.4±0.2) and 4-mp (4.9±0.5) were not significantly different (P = 0.077, t-test). (B) Concentration–response relationships for activation of Agam\Or15 + Agam\Orco by acetophenone (Ace) and 4-methylphenol (4-mp). Current responses to each concentration of odorant (n = 4) were normalized to the response of the same oocyte to 30 µM Ace. EC50 values for Ace (18±9 µM) and 4-mp (71±12 µM) were significantly different (P < 0.01, t-test). Maximal response values for Ace (1.9±0.2) and 4-mp (0.64±0.03) were significantly different (P < 0.05, t-test). (C) An oocyte expressing Agam\Or13 + Agam\Orco was challenged with challenged 20 s applications of 1mM 4-mp and 1mM Ace. (D) An oocyte expressing Agam\Or15 + Agam\Orco was challenged 20 s applications of 1mM 4-mp and 1mM Ace.
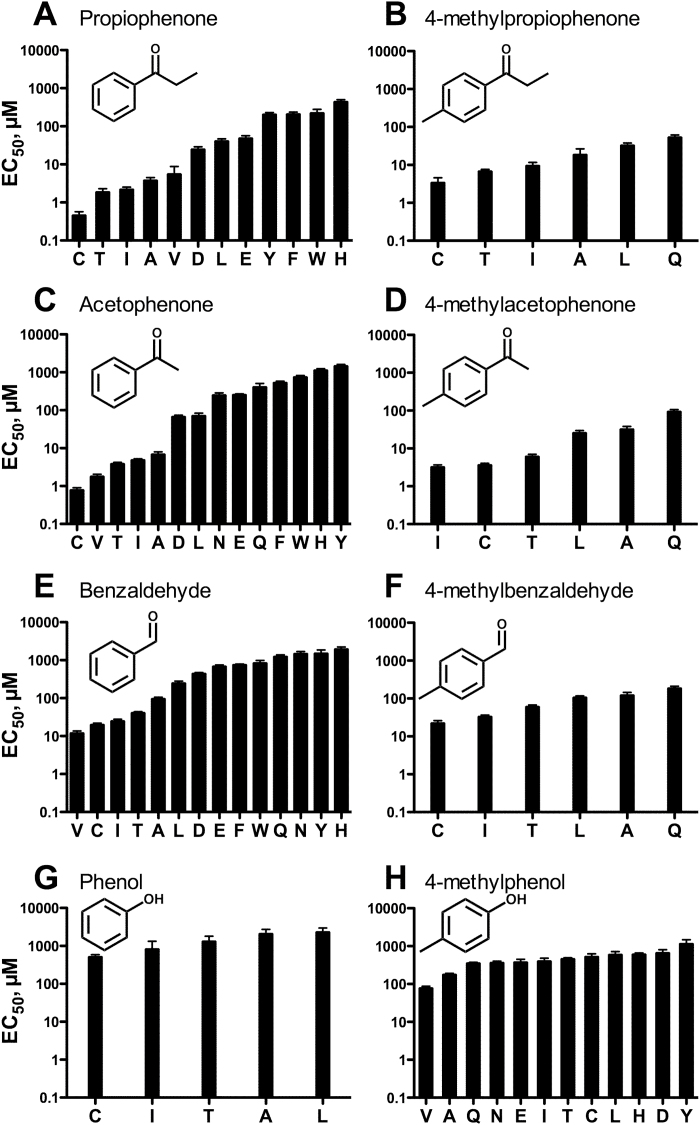
Figure 5.
The effect of mutation at position 195 of Agam\Or15 on odorant sensitivity is not consistent among odorants. CRCs were constructed (see Methods) for 8 odorants at a series of Agam\Or15 receptors with various residues at position 195. EC50 values (mean ± SEM) for 1 odorant at a series of mutants are presented in each panel. (A) propiophenone, (B) 4-methylpropiophenone, (C) acetophenone, (D) 4-methylacetophenone, (E) benzaldehyde, (F) 4-methylbenzaldehyde, (G) phenol, and (H) 4-methylphenol. Numerical values and statistical analyses are provided in Supplementary Table 2.
Results
Determinants of receptor function can be identified by conducting a mutagenesis-based screen that interchanges disparate residues of homologous, yet functionally distinct receptor subunits (Luetje et al. 1998). In this context, a mutation-induced shift in a functional characteristic of one subunit (odorant specificity, in this case) toward that of its homologue can be taken as an indication that the mutated position is involved, in some way, with the functional property under study. We examined 2 related odorant-binding subunits, Agam\Or13 and Agam\Or15, which exhibit 82% amino acid identity (317 of 387 residues are identical) and have similar, but somewhat distinct odorant response profiles (Wang et al. 2010). Of the 70 disparate residues between these 2 subunits, we chose to examine 37 residues that fall within algorithmically predicted TM and EC domains. While the general features of OR TM topology have been established for several OR subunits from 3 different species (Benton et al. 2006; Lundin et al. 2007; Smart et al. 2008; Jordan et al. 2009; Tsitoura et al. 2010), the topology of the vast majority of insect OR subunits can only be contemplated using computational topology algorithms. For this reason, the topological location of these residues should be considered approximate.
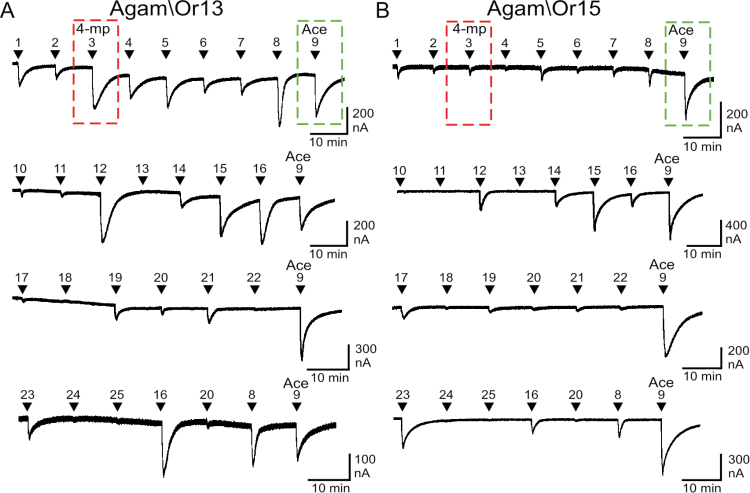
To comprehensively examine the odor space around each OR in order to highlight a difference in odorant specificity that could serve as a functional probe in our mutation screen, we expressed Agam\Or13 and Agam\Or15 in Xenopus oocytes (each in combination with Agam\Orco, which will not be subsequently mentioned) and screened with a panel of 25 odorants that included several compounds previously shown to activate these ORs (Wang et al. 2010), as well as a series of structurally related compounds (Supplementary Table 1). It is important to recognize that differences in current response amplitude upon application of an agonist to individual Xenopus oocytes is not an appropriate metric with which to assess differences in receptor function. This is because receptor expression levels can vary greatly among wild-type and mutant receptors, among individual oocytes and among oocyte batches (Luetje et al. 2013). For this reason, we sought to identify a ratio of odorant responses (which would not depend on receptor expression levels) with which to functionally distinguish Agam\Or13 and Agam\Or15. As seen in Figure 1, the set of odorants that could activate each of these 2 ORs was similar. However, the relative amplitude of the response to acetophenone (#9, Ace), with respect to the responses to several other odorants, was quite different between the 2 receptors. For Agam\Or13, the 4-methylphenol (#3, 4-mp) response was nearly identical in amplitude to the response to Ace. In contrast, the Ace response of Agam\Or15 was much larger than the 4-mp response. The ratio of the responses to these 2 odorants could provide a measure of odorant responsiveness that is independent of receptor expression level, thus avoiding the problems outlined above.
Figure 1.
Screening of Agam\Or13 and Agam\Or15 reveals differences in relative odorant sensitivity. Oocytes expressing Agam\Or13 + Agam\Orco (A) or Agam\Or15 + Agam\Orco (B) were challenged with 20 s applications of various odorants, each applied at 100 µM. Odorants: 1) 2-methylphenol, 2) 3-methylphenol, 3) 4-methylphenol (4-mp), 4) 4-ethylphenol, 5) methyl benzoate, 6) indole, 7) 3-methylindole, 8) benzaldehyde, 9) acetophenone (Ace), 10) phenylacetaldehyde, 11) 3-phenyl-1-propanol, 12) salicylaldehyde, 13) trifernal, 14) thioanisole, 15) 4-methylacetophenone, 16) 4-methylbenzaldehyde, 17) 2-acetylpyridine, 18) 2-acetylpyrazine, 19) anisyl acetate, 20) anisyl alcohol, 21) 4-butylbenzaldehyde, 22) p-cymene, 23) anisole, 24) benzoic acid, and 25) phenylacetic acid. Each current recording is from a separate oocyte and Ace was included at the end of each recording for comparison. Current responses to 4-mp and Ace applications in the top traces are indicated by dashed boxes.
Concentration–response curves for Ace and 4-mp activation of Agam\Or13 and Agam\Or15 receptors were generated to better understand the differences in odorant responsiveness between these receptors (Figure 2A,B). For each receptor, current responses to various concentrations of Ace and 4-mp were normalized to a fixed concentration of Ace applied to the same oocyte. Thus, both the potency (EC50) and maximal response (efficacy) of these 2 odorants could be compared. The EC50 values for Ace activation of Agam\Or13 (7±2 µM) and Agam\Or15 (18±9 µM) were similar, as were the EC50 values for 4-mp activation (45±19 µM and 71±12 µM, respectively). However, the relative efficacy values were dramatically different, with 4-mp displaying efficacy similar to that of Ace at Agam\Or13, but significantly lower efficacy than that of Ace at Agam\Or15. From this analysis, we decided to use a concentration of 1mM in our analysis. While this concentration is relatively high, it yields a response ratio that would be most useful in distinguishing Agam\Or13 and Agam\Or15: the Ace/4-mp response ratio for Agam\Or13 was 0.43±0.05, while the Ace/4-mp response ratio for Agam\Or15 ratio was 6.5±0.5 (Figure 2C,D). Agam\Or15 generally yielded larger odorant response amplitudes than did Agam\Or13 (in Figure 2, compare panels C and D). For this reason, we chose Agam\Or15 as the “platform” upon which we sequentially mutated amino acid residues to match the corresponding residues of Agam\Or13. The 37 positions that we examined are shown in Figure 3.
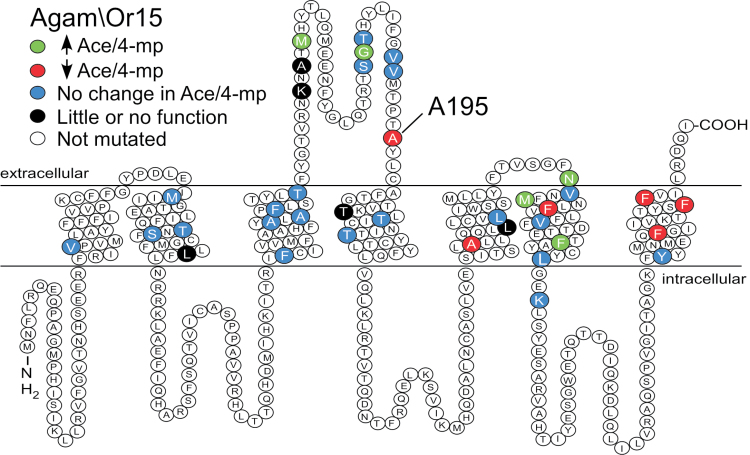
Figure 3.
Location of mutated residues within a predicted secondary structure of Agam\Or15. Positions that differ between Agam\Or15 and Agam\Or13 in the predicted TM and EC regions are highlighted: black indicates mutants that were either nonfunctional (L81W, T206I) or did not meet our minimum signal amplitude criteria of 100 nA (K158D, A160V, L270V); blue indicates mutations that did not significant alter the Ace/4-mp response ratio from that of WT Agam\Or15; green indicates mutations that shifted the response ratio to values significantly higher than that of WT Agam\Or15; red indicates mutations that significantly shifted the response ratio from that of WT Agam\Or15 toward that of WT Agam\Or13. The image was constructed by hand, based on TMD locations estimated using TMRPres2D (Spyropoulos et al. 2004).
Each Agam\Or15 substitution variant was assayed for the ratio of Ace and 4-mp responses at 1mM (as in Figure 2C,D) and the response ratio was compared to that of Agam\Or15 and Agam\Or13 (Figure 4). Based on a lack of any odorant response, it is likely that 2 mutants (L81W and T206I) did not form functional receptors. Three mutants (K158D, A160V, and L270V) yielded only very small odorant responses that did not allow detailed analysis (data not shown). Of the remaining 32 mutants, the majority (21 mutants) displayed odorant response ratios that were not significantly different from Agam\Or15 (indicated by blue in Figures 3 and 4), while 11 mutants had significantly different odorant response ratios. Five of these mutants (M162V, G181T, N289D, M293I, and F307I) had odorant response ratios that were significantly higher than that of WT Agam\Or15 (indicated by green in Figures 3 and 4), while 6 mutants (A195I, A264V, F296L, F369L, F379I, and F380Y) displayed odorant response ratios that were shifted toward that of Agam\Or13 (indicated by red in Figures 3 and 4), suggesting that these positions may play a role in odorant responsiveness. Because mutations that shift the response ratio from that of Agam\Or15 toward that of Agam\Or13 are easier to interpret, we chose to focus among the latter 6 mutants for further study. In particular, the A195I mutation yielded the greatest shift toward Agam\Or13, with an odorant response ratio of 2.7±0.1, leading us to examine this position in more detail. When the reverse substitution was made in Agam\Or13 (I195A), the resulting mutant displayed an odorant response ratio (2.5±0.1) that was significantly increased from that of WT Agam\Or13 (Figure 4).
Figure 4.
Odorant response ratios of Agam\Or15 mutants. The Ace/4-mp odorant response ratios for 32 Agam\Or15 mutants and the Agam\Or13, I195A mutant, as well as WT Agam\Or15 and WT Agam\Or13, are presented as mean ± SEM (n = 3–16). The odorant response ratio was determined by dividing the 1mM Ace peak amplitude by the 1mM 4-mp peak amplitude for each individual oocyte. For the mutants of Agam\Or15, differences from WT Agam\Or15 were assessed by 1-way ANOVA and Dunnett’s posttest (*P < 0.05, **P < 0.01, ***P < 0.001). For the mutant of Agam\Or13, difference from WT Agam\Or13 was assessed using an unpaired t-test (††† P < 0.001). The values for the A195I mutant of Agam\Or15 and the I195A mutant of Agam\Or13 are highlighted with hashed bars. Bars representing data for the Agam\Or15 mutants are colored to correspond to Figure 3: blue = no change in Ace/4-mp ratio; green = increase in Ace/4-mp ratio; red = decrease in Ace/4-mp ratio.
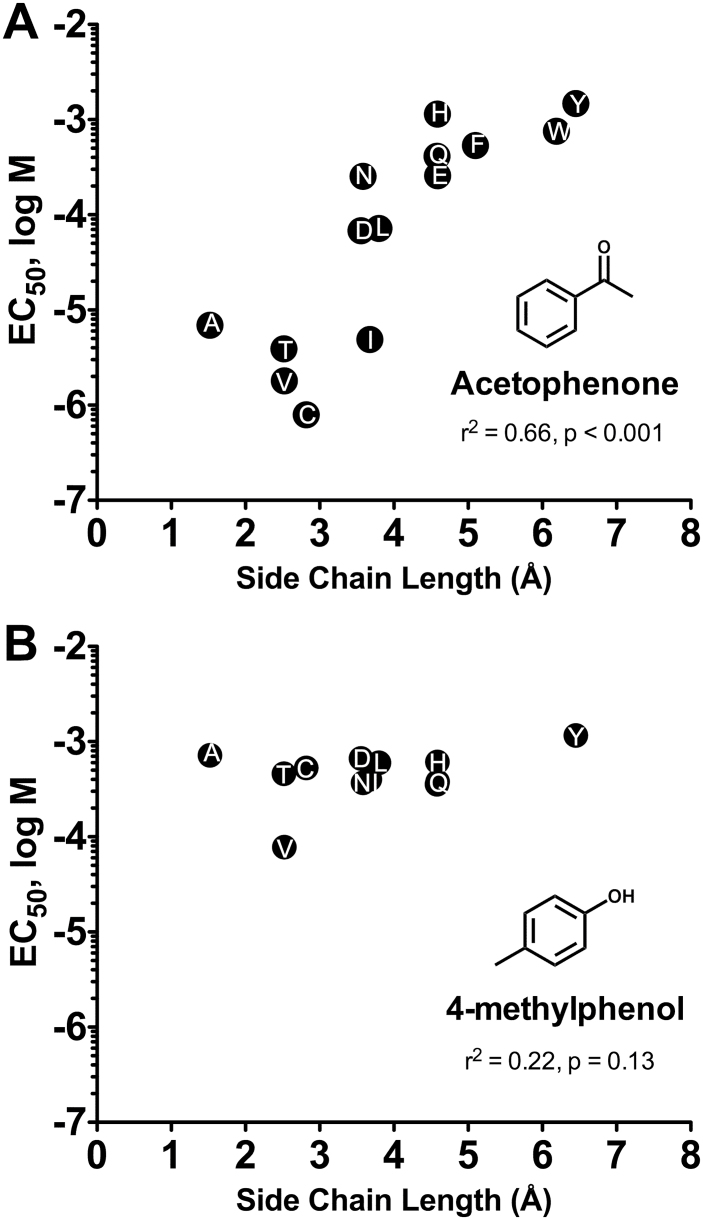
The results presented in Figure 4 suggest that position 195 (and several other positions) is involved in determining the responsiveness of these receptors to odorants. To gain more insight, we generated 14 additional mutations at this site. We constructed CRCs for Ace activation of 12 of these mutants (A195G and A195K were nonfunctional), as well as wild-type Agam\Or15 and the Agam\Or15 A195I mutant (Figure 5C, Supplementary Table 2). When we compared the EC50 for Ace activation of WT Agam\Or15 and various substitution mutants, we found substantial variation. For example, the EC50 for Ace activation of the A195Y mutant (1472±142 µM) was 1863-fold greater than the EC50 for Ace activation of the A195C mutant (0.79±0.11 µM, P < 0.001). Overall, Ace sensitivity correlated well with amino acid side chain length at this position (Figure 6A), although there was no obvious correlation with other side chain properties, such as polarity, hydrophobicity, or hydrogen bonding potential.
Figure 6.
Correlation between side chain length at position 195 of Agam\Or15 and acetophenone sensitivity. (A) EC50 values for acetophenone activation of WT Agam\Or15 and mutant subunits with various substitutions at position 195, taken from Supplementary Table 2, are plotted against amino acid side chain length (Kasahara et al. 2011). A significant correlation was observed. (B) EC50 values for 4-methylphenol activation of WT Agam\Or15 and mutant subunits with various substitutions at position 195, taken from Supplementary Table 2, are plotted against amino acid side chain length. No significant correlation was observed.
In contrast to what we observed for Ace, there was little variation among the WT and mutant receptors in sensitivity to 4-mp (Figure 5H, Supplementary Table 2). The difference in EC50 for 4-mp activation of A195Y (1160±310 µM) and A195C (528±100 µM) was only 2-fold (P < 0.05), while the greatest difference in EC50, between A195Y and A195V (78±9 µM), was only 15-fold (P < 0.001). There was no significant correlation between EC50 for 4-methylphenol activation and side chain length (Figure 6B). We conducted similar analyses for 6 compounds that are structurally related to Ace and 4-mp. Mutations at position 195 also had a large effect on sensitivity to propiophenone and benzaldehyde (Figure 5A,E), with EC50 values again correlating with side chain length (r 2 = 0.44, P < 0.05 and r 2 = 0.42, P < 0.05, respectively). In contrast, the effect of mutation on sensitivity to 4-methylpropiophenone, 4-methylacetophenone, 4-methylbenzaldehyde, and phenol was much smaller, with no significant correlation between EC50 and side chain length. Overall, minor alterations to the ligand structure had large effects on the extent to which mutation could alter the CRC. For example, addition of a methyl to acetophenone, resulting in 4-methylacetophenone, severely reduced the effect of amino acid substitution, with the EC50 values for activation of A195Q and A195C differing by 521-fold for Ace, but only 26-fold for 4-methylacetophenone. A similar effect can be seen when the 4-methyl group is added to propiophenone and benzaldehyde. The differential effect of mutation on the CRCs for different odorants is also illustrated by the variation in odorant rank orders of potency for the various mutant receptors (Supplementary Table 3).
Discussion
Our results demonstrate that amino acid position 195 plays a critical role in determining Agam\Or15 odorant sensitivity and specificity. That said, where might position 195 be located with respect to the important functional components of the receptor, such as the odorant-binding site? Alteration of a CRC by a site-specific substitution should not be taken as direct evidence that the mutated position is located at the agonist-binding site (Colquhoun 1998). This is because the liganded gating equilibrium constant (E) for a receptor, which underlies the CRC, is a product of the unliganded gating equilibrium constant (E0) and the ratio of the equilibrium ligand dissociation constants for the inactive and activated forms of the receptor (λ) (Jadey et al. 2011). Mutations almost anywhere in a receptor structure can alter E0 (Jadey et al. 2011) and thus the CRC. For example, a point mutation (L9’S) within the ion pore forming second TMD of nicotinic acetylcholine receptor (nAChR) subunits can dramatically alter the acetylcholine CRC for these receptors (Labarca et al. 1995; 2001), despite this position being >50Å away from the agonist-binding sites in this receptor class (Hibbs and Gouaux 2011). Mutations that alter E through a change in E0 are independent of agonist (Jadey et al. 2011), making the effect on CRCs similar for different agonists. Indeed, the L9’S mutation in the α4 subunit of a neuronal nAChR has a similar effect on the CRCs for several agonists (Labarca et al. 2001). Positions at or near to the ligand-binding site of nAChRs have been shown to be able to alter λ (Jadey et al. 2011). Mutation at such positions should have differential effects on the CRCs for different agonists. Thus, if position 195 of Agam\Or15 were distal to the odorant-binding site, the effect of mutations at this site might be expected to be similar for multiple odorant agonists. The results we present in Figure 5 clearly show that the effect of mutation at position 195 is not similar for the various odorants. For some ligands, such as Ace, the effect of mutation is profound, while for other ligands, such as 4-mp, the effect of mutation is modest.
Can we take these results to mean that position 195 is at or near the odorant-binding site of Agam\Or15? While this is a strong possibility, some caution is warranted. It is unclear to what extent results obtained with nAChRs can be extrapolated to other receptors. Also, some receptors display a phenomenon known as “stimulus trafficking”, which is thought to be the result of the receptor assuming different active conformations (activating different signal transduction pathways) in response to different agonists (Molero and Miller 1991; Spengler et al. 1993; Gudermann et al. 1996; Kenakin 1997). Thus, mutations at position 195 could be differentially affecting such receptor behavior, which might then be reflected in a differential effect on CRCs.
If position 195 were physically proximate to the odorant-binding site of Agam\Or15, what role might it play? The lack of correlation with side chain properties (other than size) suggests that this residue would not be interacting directly with odorant ligands. However, the side chain could be facing away from the odorant-binding cavity, perhaps interacting with other parts of the OR complex. An alteration of the size of the side chain at this position might then shift the structure of the binding site through a repositioning of the peptide backbone. The location of this position near the interface between ECL 2 and TMD 4 is consistent with the proposed location of the binding site amongst the EC halves of the TMDs (Guo and Kim 2010). The interface between ECL 2 and TMD 3 has also been implicated in determining odorant sensitivity. The binding of a competitive antagonist can protect a residue in this region of a Drosophila melanogaster odorant-binding subunit from covalent modification (Nichols and Luetje 2010) and a residue in the same region has been shown to be a determinant of pheromone specificity in an odorant-binding subunit from the European Corn Borer, Ostrinia nubilalis (Leary et al. 2012). ECL 2 itself has been proposed to serve as a lid for a binding site formed by the TMDs (Xu and Leal 2013). Residues within TMD 2 and TMD 5 have also been implicated as determinants of ligand sensitivity (Pellegrino et al. 2011; Kumar et al. 2013). Thus, it is becoming possible to envision a binding site for odorants located among the TMDs of these OR subunits.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was supported by grants from the National Institutes of Health [RO1 DC011091 to C.W.L. and RO1 AI056402 to L.J.Z.] and the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative [VCTR121 to L.J.Z.]. D.T.H. was supported, in part, by T32 NS007044.
Supplementary Material
Acknowledgments
We thank A. Castro and B. Sherman for oocyte preparation.
References
- Benton R, Sachse S, Michnick SW, Vosshall LB. 2006. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4(2):e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Carlson JR. 2011. Insect olfaction from model systems to disease control. Proc Natl Acad Sci USA. 108(32):12987–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. 1998. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol. 125(5):924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudermann T, Kalkbrenner F, Schultz G. 1996. Diversity and selectivity of receptor-G protein interaction. Annu Rev Pharmacol Toxicol. 36:429–459 [DOI] [PubMed] [Google Scholar]
- Guo S, Kim J. 2010. Dissecting the molecular mechanism of Drosophila odorant receptors through activity modeling and comparative analysis. Proteins. 78(2):381–399 [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. 2006. Coding of odors by a receptor repertoire. Cell. 125(1):143–160 [DOI] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E. 2011. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 474(7349):54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadey SV, Purohit P, Bruhova I, Gregg TM, Auerbach A. 2011. Design and control of acetylcholine receptor conformational change. Proc Natl Acad Sci USA. 108(11):4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MD, Anderson A, Begum D, Carraher C, Authier A, Marshall SD, Kiely A, Gatehouse LN, Greenwood DR, Christie DL, et al. 2009. Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem Senses. 34(5):383–394 [DOI] [PubMed] [Google Scholar]
- Kasahara T, Shimogawara K, Kasahara M. 2011. Crucial effects of amino acid side chain length in transmembrane segment 5 on substrate affinity in yeast glucose transporter Hxt7. Biochemistry. 50(40):8674–8681 [DOI] [PubMed] [Google Scholar]
- Kenakin T. 1997. Pharmacologic analysis of drug-receptor interaction. 3rd ed. Philadelphia (PA): Lippincott Raven. p. 491 [Google Scholar]
- Kumar BN, Taylor RW, Pask GM, Zwiebel LJ, Newcomb RD, Christie DL. 2013. A conserved aspartic acid is important for agonist (VUAA1) and odorant/tuning receptor-dependent activation of the insect odorant co-receptor (Orco). PLoS One. 8(7):e70218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C, Nowak MW, Zhang H, Tang L, Deshpande P, Lester HA. 1995. Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature. 376(6540):514–516 [DOI] [PubMed] [Google Scholar]
- Labarca C, Schwarz J, Deshpande P, Schwarz S, Nowak MW, Fonck C, Nashmi R, Kofuji P, Dang H, Shi W, et al. 2001. Point mutant mice with hypersensitive alpha 4 nicotinic receptors show dopaminergic deficits and increased anxiety. Proc Natl Acad Sci USA. 98(5):2786–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 43(5):703–714 [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Jr, Macallister IE, Kavanaugh MP, Wanner KW. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 109(35):14081–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetje CW, Maddox FN, Harvey SC. 1998. Glycosylation within the cysteine loop and six residues near conserved Cys192/Cys193 are determinants of neuronal bungarotoxin sensitivity on the neuronal nicotinic receptor alpha3 subunit. Mol Pharmacol. 53(6):1112–1119 [PubMed] [Google Scholar]
- Luetje CW, Nichols AS, Castro A, Sherman BL. 2013. Functional assay of mammalian and insect olfactory receptors using Xenopus oocytes. Methods Mol Biol. 1003:187–202 [DOI] [PubMed] [Google Scholar]
- Lundin C, Käll L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, Heijne Gv, Nilsson I. 2007. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 581(29):5601–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RF, Hughes DT, Luetje CW, Millar JG, Soriano-Agatón F, Hanks LM, Robertson HM. 2012. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae . Insect Biochem Mol Biol. 42(7):499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero X, Miller LJ. 1991. The gall bladder cholecystokinin receptor exists in two guanine nucleotide-binding protein-regulated affinity states. Mol Pharmacol. 39(2):150–156 [PubMed] [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K. 2005. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 307(5715):1638–1642 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Vosshall LB. 2009. Controversy and consensus: noncanonical signaling mechanisms in the insect olfactory system. Curr Opin Neurobiol. 19(3):284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Störtkuhl K, Hatt H. 2005. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster . Nat Neurosci. 8(1):15–17 [DOI] [PubMed] [Google Scholar]
- Nichols AS, Chen S, Luetje CW. 2011. Subunit contributions to insect olfactory receptor function: channel block and odorant recognition. Chem Senses. 36(9):781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols AS, Luetje CW. 2010. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J Biol Chem. 285(16):11854–11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask GM, Jones PL, Rützler M, Rinker DC, Zwiebel LJ. 2011. Heteromeric anopheline odorant receptors exhibit distinct channel properties. PLoS One. 6(12):e28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB. 2011. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature. 478(7370):511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Hughes DT, Luetje CW, Leal WS. 2010. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One. 5(4):e10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. 2008. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 452(7190):1002–1006 [DOI] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. 2008. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 38(8):770–780 [DOI] [PubMed] [Google Scholar]
- Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg PH, Journot L. 1993. Differential signal transduction by five splice variants of the PACAP receptor. Nature. 365(6442):170–175 [DOI] [PubMed] [Google Scholar]
- Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. 2004. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics. 20(17):3258–3260 [DOI] [PubMed] [Google Scholar]
- Takken W, Knols BG. 1999. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 44:131–157 [DOI] [PubMed] [Google Scholar]
- Tsitoura P, Andronopoulou E, Tsikou D, Agalou A, Papakonstantinou MP, Kotzia GA, Labropoulou V, Swevers L, Georgoussi Z, Iatrou K. 2010. Expression and membrane topology of Anopheles gambiae odorant receptors in lepidopteran insect cells. PLoS One. 5(11):e15428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 96(5):725–736 [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. 2010. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae . Proc Natl Acad Sci USA. 107(9):4418–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, Linn CE, Robertson HM, Luetje CW. 2010. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis . PLoS One. 5(1):e8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner KW, Nichols AS, Walden KK, Brockmann A, Luetje CW, Robertson HM. 2007. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc Natl Acad Sci USA. 104(36):14383–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. 2008. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 452(7190):1007–1011 [DOI] [PubMed] [Google Scholar]
- World Malaria Report. 2013. Geneva (Switzerland): World Health Organization Press [Google Scholar]
- Xu P, Leal WS. 2013. Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem Biophys Res Commun. 435(3):477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.