Abstract
White-spotting patterns in mammals can be caused by mutations in the gene KIT, whose protein is necessary for the normal migration and survival of melanocytes from the neural crest. The alpaca (Vicugna pacos) blue-eyed white (BEW) phenotype is characterized by 2 blue eyes and a solid white coat over the whole body. Breeders hypothesize that the BEW phenotype in alpacas is caused by the combination of the gene causing gray fleece and a white-spotting gene. We performed an association study using KIT flanking and intragenic markers with 40 unrelated alpacas, of which 17 were BEW. Two microsatellite alleles at KIT-related markers were significantly associated (P < 0.0001) with the BEW phenotype (bew1 and bew2). In a larger cohort of 171 related individuals, we identify an abundance of an allele (bew1) in gray animals and the occurrence of bew2 homozygotes that are solid white with pigmented eyes. Association tests accounting for population structure and familial relatedness are consistent with a proposed model where these alleles are in linkage disequilibrium with a mutation or mutations that contribute to the BEW phenotype and to individual differences in fleece color.
Keywords: association, coat color, KIT, pigmentation
Introduction
Alpacas (Vicugna pacos) were presumably domesticated and selected in large part for their highly valued fibre/fleece, which occurs in a wide range of colors and patterns. Mammalian pigmentation is controlled by the distribution of eumelanin (black/brown pigments) and phaeomelanin (red/yellow pigments) and appears to be determined by a small number of genes shared among different species (Jackson 1994; Barsh 1996; Newton et al. 2000). The genetics of alpaca coat-color inheritance is poorly understood, and most information on color inheritance was, until recently, based on breeder’s observations. However, breeders frequently report low predictability of fleece color outcomes in breeding programs, suggesting that coat color is inherited polygenically (Mcgregor 2006). Recent studies have begun to unravel the genetic complexity underlying fleece color determination in alpacas (Powell et al. 2008; Feeley and Munyard 2009; Cransberg and Munyard 2011; Feeley et al. 2011; Valbonesi et al. 2011). The coding region has been sequenced for MC1R, ASIP, TYRP1, MATP and TYR in a variety of alpaca fleece colors. Polymorphisms identified in MC1R are associated with the production of phaemelanin or eumelanin and mutations within ASIP are associated with black fibre (Feeley and Munyard 2009; Feeley et al. 2011).
As well as the type and amount of pigment, the development, migration, and survival of melanocytes also affects final pigmentation patterns. Genes known to cause spotting patterns through influencing melanocyte development include PAX3, SOX10, MITF, SLUG, EDN3, EDNRB, and KIT (Tachibana et al. 2003). Mutations in these genes result in a range of white-spotting phenotypes in a variety of mammals (e.g., Geissler et al. 1988; Hofstra et al. 1996; Destefano et al. 1998; Santschi et al. 1998; Southard-Smith et al. 1999; Yajima et al. 1999; Sánchez-Martín et al. 2002). Solid white alpacas are valued by breeders because their fleece can be more easily dyed to produce any desired color during processing and manufacturing. A range of white-spotting phenotypes are observed in alpacas including “white face,” “all white,” “blue-eyed white,” and a white spot anywhere on the body. Based on pedigree analysis, it has been proposed that the alpaca white-spotting allele is dominant to solid color (Merriwether and Merriwether 2003).
A further white-spotting pattern in alpacas occurs in gray alpacas. Gray alpacas typically have a white-spotting pattern of gray fleece separated by white fleece on the face, legs, and neck (sometimes referred to as “tuxedo”; Figure 1a). There is, however, variation in the extent of this pattern on the animal with some animals having less-extensive white patterning that doesn’t include the feet (Gregory DM of Chiverton Alpacas, personal communication). Therefore, we use the term “gray” here to refer to both a fleece color and a fleece pattern in alpacas. Based on herd book records, Paul (1999 , 2006) proposes that gray fleece in alpacas is the result of a dominant diluting allele operating on black and brown pigment, as well as minor white-spotting alleles producing the characteristic white face, neck, and feet of gray alpacas. The mutation causing gray has been hypothesized to be homozygous lethal as matings between gray alpacas lead to a 2:1 (66%) instead of 3:1 (75%) ratio of gray to nongray offspring. In other species, this ratio can indicate homozygote lethality (Pulos and Hutt 1969; Hintz and Van Vleck 1979; Russell 1979; Silvers 1979). It is not known whether this results from the diluting or spotting alleles associated with gray in alpacas. The observation of gray offspring from nongray parents suggests the presence of cryptic/hidden gray animals (Paul 1999, 2006). Evidence of embryonic lethality or sublethality of roan and white-spotting mutations in homozygotes has been reported in horses, rats, and mice (Gruneberg 1936; Hintz and Van Vleck 1979; Geissler et al. 1981; Niwa et al. 1991; Hosoda et al. 1994; Santschi et al. 1998).
Figure 1.
Photograph of an alpaca with (a) the typical gray fleece phenotype characterized by white face, chest, and feet (left) standing next to an all white alpaca (right) and (b) the BEW phenotype, which is characterized by a solid white coat, blue irides, and often deafness.
White alpacas are sometimes afflicted with the blue-eyed white (BEW) phenotype (Figure 1b), which is characterized by a solid white coat, blue irides, and often deafness (Gauly et al. 2005). The BEW trait is common in alpacas, although its prevalence is hard to measure because the International Alpaca Registry does not include eye-color information. Although the deafness does not pose a welfare concern for an alpaca in a herd situation, many breeders regard the BEW phenotype as a defect or undesirable trait. The inheritance pattern of the BEW phenotype has not been proven, but breeders have postulated that it results from an interaction (of unknown type) between the gray gene and the white-spotting gene because BEW animals typically have many white and/or gray relatives in their pedigree (Merriwether and Merriwether 2003; Paul 2006). However, no studies have been done on a molecular level to understand the possible identity of these genes.
The KIT gene, which encodes the c-kit receptor tyrosine kinase, has been implicated in pigment variations, including white-spotting phenotypes, in a range of species (Chabot et al. 1988; Geissler et al. 1988; Fleischman et al. 1991). In humans, a heterozygous deletion of KIT leads to a pigmentation defect with random patches of unpigmented skin and hair called piebaldism, a condition similar to the white-spotting phenotype in mice (Fleischman et al. 1991). In pigs, the dominant white coat-color phenotype is caused by 2 mutations within the KIT gene, which lead to an absence of melanocytes in the hair follicles and hair bulbs (Johansson Moller et al. 1996; Marklund et al. 1998). Four depigmentation phenotypes (tabiano, roan, sabino-1, dominant white) have been mapped to a region of the equine Chromosome 3 harbouring the KIT locus (Marklund et al. 1999; Mau et al. 2004; Brooks and Bailey 2005). Furthermore, the cattle KIT locus is considered the main candidate gene for the spotting (S) locus (Reinsch et al. 1999; Fontanesi et al. 2010). In domestic cats (Felis catus), markers close to KIT are associated with white spotting (Cooper et al. 2006). Investigations into differential gene expression between colors of alpaca fibre found KIT expression was lowest in white animals and highest in bay animals (intermediate expression in black animals; Munyard 2011). Therefore, the alpaca KIT gene is a major candidate for white-spotting patterns.
Because of the importance of the all (solid) white phenotype and the BEW phenotype in the alpaca industry, our goal was to 1) identify genetic markers associated with these phenotypes and 2) to characterize the occurrence of these markers in known pedigrees and the general population. Here, we report the first evidence that the BEW phenotype is controlled by mutation/s at or nearby the KIT locus and provide diagnostic opportunities and practical guidelines for breeders.
Materials and Methods
Animals
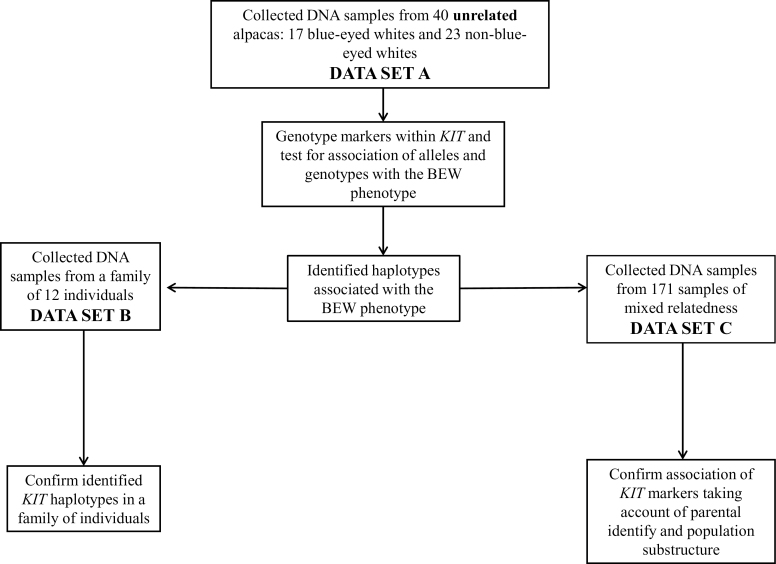
Three data sets were used to investigate the genetic basis of the BEW phenotype (Figure 2). First, a sample of 40 unrelated individuals (17 BEWs; 23 non-BEWs: 2 black, 4 brown, 4 fawn, 6 gray, 4 white, and 3 multicolored animals; Table 1) were studied to examine association of KIT markers with the BEW phenotype (data set A). These individuals were determined to be unrelated according to 3 generations of pedigree data. Haplotypes were constructed manually from inspection of family relationships within a pedigree of 12 individuals (data set B) to confirm the associated haplotypes identified in data set A. To examine the phenotypic effects of individual haplotypes, a larger sample of 171 individuals (29 BEWs, 66 black, 23 white, 13 gray, 21 brown, 8 fawn, and 11 multicolored) of mixed relatedness was studied (data set C). Blood samples were collected by collaborating veterinarians. Pedigree information was accessed through the IAR (International Alpaca Registry) database on the Australian Alpaca Association website (http://www.alpaca.asn.au/). Blood was stored in PaxGeneTM tubes (PreAnalytiX), and genomic DNA was extracted using a commercially available kit according to the manufacturer’s instructions (AxyPrep blood genomic DNA miniprep kit, Axygen Biosciences, USA). Some DNA samples were also extracted from saliva swabs (Oragene; DNAGenotek, Kanata, Ontario, Canada) according to manufacturer instructions. Eye and coat color were reported by sample collectors, and photos of animals were obtained where possible.
Figure 2.
Flow chart diagram describing the use of 3 data sets to investigate the genetic inheritance of the BEW phenotype.
Table 1.
Details of coat-color phenotype, eye-color phenotype, sex, and KIT marker genotypes of unrelated alpacas used in the current study. Dam and sire fleece color are also provided. Fleece color is denoted for animals where pedigree records of fleece color were available. Individuals with assumed recombination events are bolded (recombinant for bew1 haplotype)
| Sex | Fleece color | Eye color | Dam fleece color | Sire fleece color | 49774 genotype | KIT10 genotype | KIT3 genotype |
|---|---|---|---|---|---|---|---|
| Female | White | Blue | White | White | 236/241 | 266/278 | -/- |
| Female | White | Blue | White | White | 234/236 | 266/275 | 255/255 |
| Female | White | Blue | - | - | 236/241 | 266/275 | 268/270 |
| Male | White | Blue | White | White | 241/241 | 272/278 | 239/270 |
| Female | White | Blue | White | White | 236/241 | 266/278 | 268/270 |
| Male | White | Blue | White | Silver gray | 236/241 | 266/278 | 268/270 |
| Female | White | Blue | White | White | 236/241 | 266/278 | 268/270 |
| Female | White | Blue | Brown/white | White | 234/236 | 266/275 | 253/253 |
| Male | White | Bluea | White | Fawn | 236/241 | 266/278 | -/- |
| Female | White | Blue | Fawn | White | 236/241 | 266/272 | 239/268 |
| Female | White | Blue | - | - | 236/241 | 266/278 | 268/270 |
| Female | White | Blue | Fawn | Rose gray | 236/241 | 266/278 | 268/270 |
| Female | White | Blue | Silver gray | White | 236/241 | 266/278 | 268/270 |
| Female | White | Blue | Black | White | 236/241 | 266/278 | 268/270 |
| - | White | Blue | - | - | 236/241 | -/- | -/- |
| Male | White | Blue | Fawn | White | 236/241 | 266/272 | -/- |
| Male | White | Blue | - | - | 236/241 | 266/278 | 268/270 |
| Female | Black | Pigmented | Black | Fawn | 233/236 | 275/278 | 253/253 |
| Female | Black | Pigmented | Black | Black | 247/247 | 239/239 | 257/257 |
| Male | Brown | Pigmented | Fawn | Fawn | 234/243 | 266/275 | 239/253 |
| Male | Brown | Pigmented | - | - | 241/247 | 239/278 | 257/270 |
| Female | Brown | pigmented | Brown | Brown | 234/241 | 275/284 | 253/253 |
| Female | Brown | Pigmented | Brown | Rose gray | 236/241 | 220/220 | 268/270 |
| Female | Fawn | Pigmented | Brown | Brown | 234/241 | 278/281 | 255/270 |
| Female | Fawn | Pigmented | White | Brown | 234/243 | 239/275 | 253/257 |
| Female | Fawn | Blue | White | White | 236/236 | 266/272 | 268/270 |
| Male | Fawn | Pigmented | Brown | Fawn | 234/247 | 239/275 | 253/257 |
| Female | Rose gray | Pigmented | White | Fawn | 234/236 | 266/275 | 251/268 |
| Female | Silver gray | Pigmented | Silver gray | Brown/silver gray | 234/243 | 266/275 | 251/268 |
| Male | Silver gray | Pigmentedb | - | - | 234/236 | 266/278 | 255/268 |
| Female | Rose gray | Pigmented | Rose gray | Rose gray | 234/236 | 266/275 | 251/268 |
| Female | Silver gray | Pigmented | - | - | 236/247 | 239/266 | -/- |
| Female | Silver gray | Pigmentedb | Silver gray | Silver gray | 236/243 | 239/266 | 257/268 |
| Female | Black/white | Pigmented | Silver gray | White | 236/241 | 263/278 | 255/270 |
| Female | Brown/white | Pigmented | Brown | Rose gray | 236/247 | 239/266 | 257/268 |
| Female | Black/white | Blue | White | Black | 234/236 | 266/278 | -/- |
| Female | White | Pigmented | White | Fawn | 234/247 | 239/275 | 251/257 |
| Female | White | Pigmented | - | - | 234/241 | 275/278 | 253/270 |
| Female | White | Pigmented | - | - | 236/243 | 272/278 | 268/270 |
| Female | White | Pigmented | White | White | 236/241 | 275/278 | 270/272 |
-/- denotes where genotypes could not be obtained due to poor amplification.
asome pigment bleed through; bwith blue speckles
Microsatellite Marker and Allele Detection
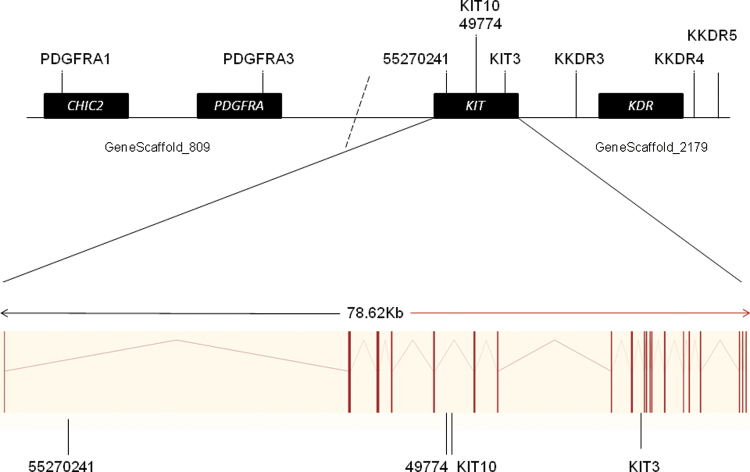
Tandem repeats within the GeneScaffold_2179 of the V. pacos genome sequence, vicPac1, (http://www.ensembl.org/), which contains the KIT gene, were identified using an online repeat finder tool (Benson 1999). The vicPac1 genome sequence is a 2.51X assembly with sequencing reads aligned to the human genome. Comparative analysis of the human, bovine, dog, and mouse genomes (http://www.ensembl.org/) places the PDGFRA gene downstream of KIT and KDR upstream of KIT in these species. Repeats within and flanking PDGFRA and KDR were obtained from GeneScaffold_809 and GeneScaffold_2179 of the V. pacos genome sequence, respectively, to examine the possibility of either gene contributing to the BEW trait (Figure 3). Marker order in a de novo assembled alpaca genome sequence was identical to order in the low coverage, vicPac1 genome (Johnson WE, unpublished data). Primers were designed using Primer3 (Rozen and Skaletsky 2000). The 5‵ end of the forward primer from each pair was modified with a M13(-21) universal sequence tag (5‵-TGTAAAACGACGGCCAGT-3‵) to enable the incorporation of the universal fluorescent-labeled M13(-21) primer (Schuelke 2000). The polymerase chain reaction (PCR) products were then analyzed on a 3730XL capillary analyser (Applied Biosystems) by Macrogen, Korea, and allele sizes were scored using GeneMapper 4.0 software (Applied Biosystems). We genotyped 9 KIT-associated microsatellite markers of which 4 were within KIT and 5 were in the adjacent regions (see Supplementary Material online).
Figure 3.
The positions of microsatellite markers used in this study, relative to gene positions. Markers were derived from 2 scaffolds from the Vicugna pacos Ensembl genome (GeneScaffold_809 and GeneScaffold_2179). The dotted line represents the hypothetical join between these scaffolds based on conservation of this gene order in human, bovine, murine, and canine genomes (http://www.ensembl.org/). The lower part of the figure shows the 29 exons of the alpaca KIT gene as vertical bars separated by intronic sequences.
PCR Conditions
Amplification of microsatellite loci was carried out in a 12 µL volume consisting of approximately 50–100ng of DNA template, 0.0885 µL 10 µM forward primer, 0.35 µL 10 µM reverse primer, 0.35 µL 10 µM fluorescent-labeled M13(-21) primer (FAM, VIC, NED, or PET), 0.2mM of each dNTP, 2.5mM MgCl2, 5 × MgCl2-free buffer, and 1 U⁄µL GoTaq Flexi Taq DNA polymerase (Promega). The cycling protocol was 5min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at the optimized annealing temperature, 40 s at 72°C. This was followed by 8 cycles of 30 s at 94°C, 45 s at 53°C and 45 s at 72°C. The final extension step was at 72°C for 10min.
Marker-Phenotype Association Analyses
SamplePower (IBM SPSS Version 3.0) was applied to estimate the power of the case–control association tests. The criterion for significance (alpha) was set at 0.050, and the tests were 2-tailed. Marker diversity was analyzed for data set A using GenAlex version 6.1 (Peakall and Smouse 2006). A case–control analysis based on χ2 and Fisher’s exact tests for genotypes were conducted within GraphPad Prism using 40 unrelated animals (data set A). Both test types were performed, as chi-squared tests approximate a P value, whereas Fisher’s tests provide an exact P value. Chi-squared tests give an easy-to-interpret chi-squared value to indicate the strength of association. To account for multiple testing, we used the Bonferroni correction and considered significant only those markers for which raw P < 0.05/7 = 7.14 × 10–3.
The genotypes of a further 60 genome-wide microsatellite markers were used to measure and adjust for possible unmatched population structure in cases and controls of data set C. These microsatellite markers were from previously published studies of Camelidae species (Lang 1996; Penedo 1998; Penedo et al. 1999; Sarno et al. 2000; Mariasegaram et al. 2002; Bustamante et al. 2003; Evdotchenko 2003) or were identified in the Ensembl vicPac1 genome sequence (see Supplementary Material online). STRAT software was used to conduct association tests while controlling for population stratification (as assessed by microsatellite markers), which can cause spurious associations (Pritchard et al. 2000). STRAT works with the output of the STRUCTURE program (Pritchard and Rosenberg 1999; Pritchard et al. 2000; Falush et al. 2003). We used the default settings assuming correlated allele frequencies and the admixture model for one population. We ran STRUCTURE for 50 000 burn-in steps followed by 20 000 replications. We used 10 000 permutations to calculate P values within STRAT. Another possible cause of spurious association is family structure. Therefore, tests for association were also performed using the SAS surveyreg procedure (version 9.1, SAS Institute). This procedure was used to fit a linear regression model, which controlled for parent identity. Tests for association of alleles with gray fleece and white fleece were conducted using data set C.
Results
The Blue-eyed White Phenotype Is Associated with Two KIT Alleles
About 7 of the 9 markers tested were polymorphic and used in subsequent analyses. The number of alleles at each marker ranged between 5 and 9 (see Supplementary Material online). Shannon’s Information Index (Sherwin et al. 2006) was between 1.800 (KIT3) and 1.296 (KKDR4) and observed heterozygosity ranged from 0.676 (PDGFRA1) to 0.949 (KIT10). The study had a power of greater than 99.9% to detect significant differences in genotype frequencies between cases and controls at 49774. The genotypes of markers within KIT (49774, KIT10 and KIT3) showed a significant association with the BEW phenotype (Table 2). The microsatellite 49774 showed the strongest association with the BEW phenotype (χ2 = 21.56, bonferroni corrected P = <0.0007) for the genotypes, whereas tests for association of individual alleles with the BEW phenotype were significant but showed weaker association than tests for association with genotypes (χ2 = 6.93 and χ2 = 13.19). The other markers within KIT, KIT10, and KIT3 showed the same pattern of high association with genotypes. The genotypes 236/241, 266/278, and 268/270 at markers 49774, KIT10, and KIT3, respectively, were significantly associated with the BEW phenotype. This suggested that 2 mutations contribute to the BEW phenotype. The markers flanking KIT did not show a significant association (bonferroni corrected P > 0.065) for genotypes with the BEW phenotype. The significance of association of marker 49774 with the BEW phenotype was tested in a larger cohort of 171 individuals (data set C) and significance remained after correcting for population substructure (χ2 = 77.385, P ≤ 1.0 × 10–4) and parental identity (P = < 0.0001; Table 3). Two individuals with colored fleece and blue eyes were included in this study and did not have the BEW-associated KIT genotype.
Table 2.
Results of chi-square tests, as well as their error probabilities (P), for associations of intragenic KIT marker genotypes (49774, KIT10, KIT3) and markers flanking KIT (KKDR4, KKDR5, PDGFRA1, PDGFRA3) with the BEW phenotype. All tests were 2-sided with an α value of 0.05 and used 40 unrelated individuals
| Trait | Marker | χ2 | Chi-square P value | Fisher’s exact test P value |
|---|---|---|---|---|
| BEW | PDGFRA1 | 6.728 | 0.0095 (0.0665) | 0.0252 (0.1764) |
| BEW | PDGFRA3 | 0.02724 | 0.8689 (>1.00) | 1.000 (>1.00) |
| BEW | 49774 | 21.56 | <0.0001 (<0.0007) | <0.0001 (<0.0007) |
| BEW | KIT10 | 12.82 | 0.0003 (0.021) | 0.0009 (0.0063) |
| BEW | KIT3 | 10.61 | 0.0011 (0.0077) | 0.0024 (0.0168) |
| BEW | KKDR4 | 2.332 | 0.1267 (0.8869) | 0.1642 (>1.00) |
| BEW | KKDR5 | 2.692 | 0.1009 (0.7063) | 0.1872 (>1.00) |
Bonferroni corrected P values are provided in brackets () alongside raw P values.
Table 3.
Patterns of allelic variation of microsatellite 49774 in various color classes of alpaca fleece in a cohort of 171 related individuals
| Genotype | Blue-eyed white | Black | Brown | Fawn | Gray | Multi | White with dark eyes |
|---|---|---|---|---|---|---|---|
| 234/236 | 2 | 5 | 5 | 1 | 7 | 3 | 6 |
| 234/234 | 0 | 16 | 2 | 0 | 1 | 0 | 2 |
| 241/247 | 0 | 1 | 2 | 2 | 0 | 0 | 0 |
| 234/241 | 0 | 6 | 2 | 2 | 1 | 7 | 4 |
| 247/247 | 0 | 5 | 2 | 0 | 0 | 0 | 0 |
| 234/247 | 0 | 24 | 6 | 0 | 0 | 0 | 1 |
| 236/247 | 1 | 2 | 1 | 1 | 4 | 1 | 0 |
| 234/249 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 236/241 | 22 | 0 | 0 | 1 | 0 | 0 | 1 |
| 234/243 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 241/241 | 2 | 0 | 0 | 0 | 0 | 0 | 8 |
| 236/243 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 243/243 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 232/234 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 236/236 | 2 | 0 | 1 | 1 | 0 | 0 | 1 |
| Total | 29 | 66 | 21 | 8 | 13 | 11 | 23 |
Haplotype Analysis
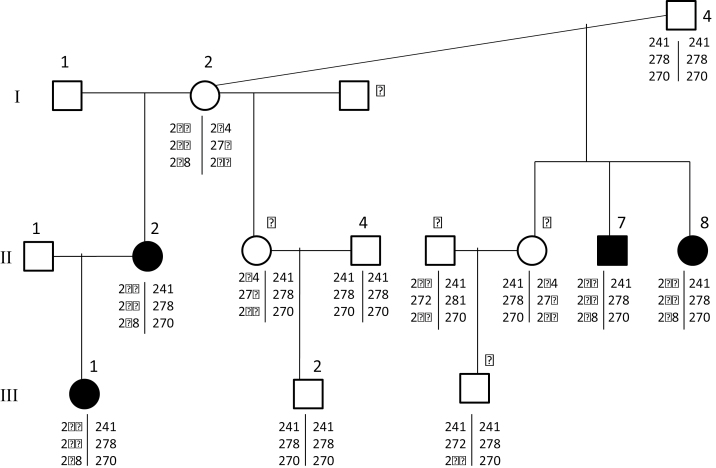
A family of 12 individuals was genotyped for the KIT markers associated with the BEW phenotype, and no Mendelian errors were identified in microsatellite genotypes (Figure 4). The association of 2 haplotypes with the BEW phenotype was also evident in the pedigree data. Indeed it can be seen that all affected individuals in the pedigree possessed the same KIT haplotypes as affected individuals in the unrelated animals in data set A. Examination of microsatellite genotypes within a pedigree allowed the identification of 2 haplotypes bew1 (236 at 49774, 266 at KIT10, 268 at KIT3) and bew2 (241 at 49774, 278 at KIT10, 270 at KIT10). The haplotypes bew1 and bew2 were present in affected (BEW) offspring, whereas unaffected parents carried either haplotype in homozygous or heterozygous form. The bew1 haplotype of I-2 was inherited by all her offspring with blue eyes, and none of her offspring with dark eyes. All offspring of I-2 also inherited the bew2 allele from their sire; however, only the offspring that also inherited bew1 from their dam are BEW. There was complete cosegregation of the bew1/bew2 genotype and the BEW phenotype within this pedigree.
Figure 4.
Mendelian inheritance of alpaca KIT alleles within a family segregating the BEW phenotype. Individuals shaded black have the BEW phenotype, whereas unshaded individuals are animals with white fleece and pigmented irides. For each individual, the genotypes are given at markers 49774, KIT10, and KIT3 in this order.
Once the BEW-associated haplotypes had been confirmed in a family, individuals within data set A were examined for recombination events that might suggest a smaller critical region. As only 3 markers showed significant association with the BEW phenotype, a conservative approach was taken when identifying recombination events, and only individuals who carried at least 2 consecutive alleles of haplotype bew1 or bew2 were considered. If one pertains to the assumption that the mutation causing the gray fleece phenotype is homozygous lethal and that the bew1 haplotype is in linkage disequilibrium with this mutation, then 2 individuals in Table 1 (bolded) suggest that this mutation is 3′ of 49774.
The Distribution of KIT Alleles in Alpacas of Different Fleece Colors
To explore the phenotypic significance of the individual alleles associating with the BEW phenotype, we extended our study to examine a larger cohort of individuals of mixed relatedness (data set C). The bew1 haplotype was present in animals of all color classes but in high abundance in gray animals. The test for association with gray fleece had a power of 90.1% to detect significance using the frequency distribution of the 236 allele at 49774. There was evidence for association of 49774 with gray fleece color even when taking into account possible population structure using STRAT software (χ2 = 11.771, P = 2.0 × 10–3). The level of significance for marker 49774 (with gray fleece) was reduced for regression analysis after accounting for shared parentage (P = 0.0872). There were 13 gray individuals in the 171 samples studied. Eleven of 13 (84.61%) gray individuals (including rose and silver grays) possessed the bew1 allele at marker 49774, 10 at KIT10 and 11 at KIT3 (Table 3). No bew1 homozygotes were identified in the 13 gray individuals. Although 5 of 171 (3.21%) individuals were homozygous for the 236 allele at 49774 (Table 3), no individuals of any color had the bew1 haplotype at all 3 markers. This haplotype appears to be frequent in the alpaca population with 13 of 23 (56.52%) non-BEW unrelated individuals possessing the bew1 allele at marker 49774.
The 241 allele at marker 49774 was present in animals of all fleece colors. Nine individuals (8 solid white and 1 BEW) were homozygous at all 3 markers for the bew2 haplotype. The test for association with white fleece had a power of greater than 99.9% to detect significance using the frequency distribution of the 241/241 genotype at 49774. Evidence for association was detected for 49774 with white fleece after correcting for population structure (χ2 = 33.844, P ≤ 0.01 × 10–3) and shared parents (P = 0.0002). Eight of 23 (34.78%) white animals with pigmented eyes were homozygous for the bew2 allele (Table 3). This allele appears to be common in the alpaca population with 7 of 23 (30.43%) non-BEW unrelated animals carrying the allele.
Discussion
Mammalian coat and skin color appear to be determined by a similar set of genes across different species (Jackson 1994; Barsh 1996; Newton et al. 2000). Most notably, these include melanocortin-receptor 1 (MC1R), Agouti signaling protein (ASIP), tyrosinase (TYR), microphthalmia-associated transcription factor (MITF), membrane-associated transporter protein (MATP), mast cell growth factor (MGF), α-melanocyte stimulating hormone (α-MSH), melanophilin (MLPH), tyrosinase-related protein 1 (TYRP1), oculocutaneous albinism type 2 (OCA2), and c-kit (KIT). The KIT gene, which encodes the c-kit receptor tyrosine kinase, has been implicated in pigment variations (primarily white-spotting phenotypes) in a range of species. The swine dominant white phenotype is caused by 2 mutations in the KIT gene: one is a gene duplication, and the other is a splice-site mutation in intron 17 leading to skipping of exon 17 of one of the copies (Marklund et al. 1998). Characterization of the equine KIT locus has found multiple independent mutations that are presumed to be responsible for dominant white-spotting (Haase et al. 2007) Furthermore, cattle breeds carrying different putative alleles at the spotting locus show genetic heterogeneity within the KIT gene (Fontanesi et al. 2010). KIT has been implicated in white-spotting patterns in many species but also in roan coat color in horses and pigs (Marklund et al. 1999; Cho et al. 2011).
In alpacas, the only molecular genetic studies of coat color have been to screen candidate genes for coat color dilution, absence of dark fibre, black fleece, and lightness of fibre (Feeley and Munyard 2009; Cransberg and Munyard 2011; Feeley et al. 2011; Guridi et al. 2011). KIT is one of the major mammalian coat color loci and affects pigmentation through influencing the migration of melanocytes from the neural crest during embryogenesis (Chabot et al. 1988; Geissler et al. 1988), and is therefore a reasonable candidate gene for white-spotting patterns in alpacas, as well as the rarer BEW phenotype.
Genetic case–control studies such as this must be conducted mindful of the possibility of false positive results (Lohmueller et al. 2003; Price et al. 2010). To decrease the likelihood of spurious results, we first tested for association of microsatellite markers with the BEW phenotype using a set of individuals known to be unrelated for at least 3 generations (data set A), and a strong association was identified between KIT genotype and the BEW phenotype. We then confirmed the presence of haplotypes identified in data set A, in a family (data set B), and indeed the same two haplotypes bew1 and bew2 were present in affected (BEW) individuals. We also tested a larger cohort containing some related individuals for association of KIT markers with the BEW phenotype (data set C). To address the possibility that our results were caused by spurious associations due to unknown population structure, we genotyped 171 cases and controls with 60 genome-wide microsatellite markers. We also used a second association test that controlled for one generation of pedigree relatedness. We were, however, only able to access pedigree information for animals postimportation to Australia, and it is therefore possible that related animals were inadvertently included but not controlled for. This larger data set also showed strong association of the BEW phenotype with KIT marker genotypes. The 3 data sets (A, B, and C) including a group of unrelated individuals, a pedigree, and a larger cohort of samples all supported the involvement of 2 KIT alleles with the BEW phenotype.
The analysis presented here demonstrates a strong association but not unequivocal relationship between the BEW phenotype and KIT genotype. We therefore postulate that these microsatellites are in linkage disequilibrium with the causative mutation, which may be in KIT or a nearby promoter or enhancer region. The finding of no significant association of markers in the genes neighboring KIT (PDGFRA and KDR) supports the conclusion that the mutations associated with the bew1 and bew2 haplotypes lie close to the KIT gene. It is also possible that the BEW phenotype is caused by a single mutation that has multiple KIT haplotypes, that is, that recombination may have placed the causative mutation on a second haplotype. However, under these circumstances we would expect to see a class of BEW animals that are homozygous for the bew1 allele, as well as a class of BEW animals that are bew2 homozygotes. However, of the 5 animals that have the 236/236 genotype at 49774, only 2 are BEW and only 2 of 10 animals with the genotype 241/241 are BEW. The finding of strong association of KIT genotype with the BEW phenotype at 3 different markers within KIT suggested the presence of 2 haplotypes that contribute to the BEW phenotype. This is in agreement with breeder’s suggestion that 2 alleles are required for the expression of the BEW trait. The findings in this study suggest that 2 haplotypes representing 2 mutations contribute to the BEW phenotype. We propose that the BEW phenotype is the result of the cumulative hypopigmentation effects of 2 KIT mutations.
The involvement of a common allele with the gray and BEW phenotype has been proposed previously (Paul 1999; Merriwether and Merriwether 2003; Paul 2006) although this study is the first to use molecular genetics to investigate this. Upon examination of a larger cohort of individuals, it was evident that an association exists between the bew1 haplotype and the gray fleece phenotype. Most gray individuals (11 of 13) carried 1 copy of the bew1 haplotype. The association of the bew1 haplotype with gray fleece was not as strong as the association of bew1/bew2 with the BEW phenotype. This is not surprising as the bew1 haplotype is present in many nongray individuals. This finding is in agreement with suggestions by Paul (1999, 2006) that there are hidden gray animals that carry a gray allele but do not have a base fleece coat color which allows its visualization, that is, white-spotting patterns maybe hard to see in animals whose base coat color is light such as fawn or white. The findings in this study are in agreement with findings by Paul (1999, 2006) regarding the dominance of the gray phenotype and the involvement of a white-spotting gene, such as KIT with the gray phenotype. Based on its function in other species, KIT is not a likely candidate for pigment dilution; however, the bew1 allele at KIT may control the patterning of white fleece on a gray alpaca. No individuals were found to be homozygous for the bew1 haplotype. Five individuals had the bew1 allele at 49774 but not the bew1 alleles at KIT10 or KIT3. It is likely that these individuals carry a recombination event between marker 49774 and the bew1 mutation or genotyping error. Therefore, it is probable that these 5 exceptions do not carry the mutation associated with the bew1 haplotype. The genotypic distributions of marker genotypes at KIT add further support to the hypothesis of Paul (1999 , 2006) that a mutation associated with gray fleece is homozygous lethal. It is not known whether this allele is homozygous lethal only in a gray genetic background; however, our data suggests that the bew1 mutation is homozygous lethal in most, if not all genetic backgrounds, as no individuals with the full bew1 haplotype were identified in the 171 individuals studied. This is not uncommon in other species where mutations in the KIT locus often produce lethal or sublethal phenotypes in homozygous condition (Russell 1979; Silvers 1979; Nocka 1990).
The phenotypic effect of the mutation associated with the bew2 haplotype is less clear. There was a tendency for bew2/bew2 animals to have white fleece; however, none of these animals had blue eyes. The data in Table 3 demonstrate that the bew2 allele is present in a wide variety of color classes. The bew2/bew2 genotype did not correlate with all individuals with white fleece, suggesting that white fleece has multiple genetic origins. This may also, in part, be a function of errors in the alpaca pedigree database records where owners are misassigning individuals with subtle white-spotting patterns as solid white. The phenotypic effect of the bew2 haplotype and associated mutation requires further investigation such as functional studies of melanocyte distribution in bew2/bew2 animals.
There is considerable debate among breeders regarding the defectiveness of the BEW phenotype. Many believe the BEW phenotype to be a genetic flaw requiring purging from the national stock. Others believe that the deafness of a BEW alpaca does not lead to any animal welfare issues as the animals quickly compensate with their other senses and by following the cues of other animals in the herd. Given that the haplotypes bew1 and bew2 are relatively common in the alpaca population, alpaca breeders who wish to avoid mating BEWs may inadvertently do so by mating individuals carrying bew1 and bew2. This study is the first to report on the genetic inheritance of this trait and provides breeders with insight into its genetic determinants to enable increased efficiency in breed management.
In conclusion, we have identified markers within KIT, which are likely in linkage disequilibrium with the mutation/s causative of the BEW trait in alpacas. Our study provides a genetic testing tool to enable breeders to select either for or against the haplotypes associated with the BEW phenotype. We will continue the search for the specific mutation/s responsible for the BEW phenotype using next-generation sequencing technologies.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
University of Melbourne Faculty of Science Research Development Grant; Intramural Research Program of the National Cancer Institute, National Institutes of Health. F.J. is the recipient of an Australian Postgraduate Award.
Supplementary Material
Acknowledgments
Sincere and special thanks go to the alpaca breeders who donated samples and veterinarians for donating time to collect samples. Special thanks to Daphne Gregory of Chiverton Alpacas for providing photographs of gray alpacas. Thanks to the Australian Alpaca Association and Alpaca Genomics Australia Pty. Ltd. for their ongoing support for this research and dedication to the genetic improvement of the Australian alpaca stock. Thanks to Professor Michael Goddard for helpful advice on project design. Thanks also to James Kavourakis for assistance with SAS software.
References
- Barsh GS.. 1996. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 12: 299–305 [DOI] [PubMed] [Google Scholar]
- Benson G.. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Bailey E.. 2005. Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mamm Genome. 16: 893–902 [DOI] [PubMed] [Google Scholar]
- Bustamante AV, Maté ML, Zambelli A, Vidal-Rioja L. 2003. Isolation and characterization of 10 polymorphic dinucleotide microsatellite markers for llama and guanaco. Mol Ecol Notes. 3: 68–69 [Google Scholar]
- Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A.. 1988. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 335: 88–89 [DOI] [PubMed] [Google Scholar]
- Cho I-C, Zhong T, Seo B-Y, Jung E-J, Yoo C-K, Kim J-H, Lee J-B, Lim H-T, Kim B-W, Lee J-H, et al. 2011. Whole-genome association study for the roan coat color in an intercrossed pig population between Landrace and Korean native pig. Genes Genom. 33: 17–23 [Google Scholar]
- Cooper MP, Fretwell N, Bailey SJ, Lyons LA.. 2006. White spotting in the domestic cat (Felis catus) maps near KIT on feline chromosome B1. Anim Genet. 37: 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cransberg R, Munyard KA.. 2011. Polymorphisms detected in the TYROSINASE and MATP (SLC45A2) genes did not explain coat colour dilution in a sample of Alpaca (Vicugna pacos). Small Ruminant Res. 95: 92–96 [Google Scholar]
- DeStefano AL, Cupples LA, Arnos KS, Asher JH, Jr, Baldwin CT, Blanton S, Carey ML, da Silva EO, Friedman TB, Greenberg J, et al. 1998. Correlation between Waardenburg syndrome phenotype and genotype in a population of individuals with identified PAX3 mutations. Hum Genet. 102: 499–506 [DOI] [PubMed] [Google Scholar]
- Evdotchenko D.. 2003. New polymorphic microsatellite loci for different camel species. Mol Ecol Notes. 3: 431 [Google Scholar]
- Falush D, Stephens M, Pritchard JK.. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley NL, Bottomley S, Munyard KA.. 2011. Three novel mutations in ASIP associated with black fibre in alpacas (Vicugna pacos). J Agri Sci. 149: 529–538 [Google Scholar]
- Feeley NL, Munyard KA.. 2009. Characterisation of the melanocortin-1 receptor gene in alpaca and identification of possible markers associated with phenotypic variations in colour. Anim Prod Sci. 49: 675–681 [Google Scholar]
- Fleischman RA, Saltman DL, Stastny V, Zneimer S.. 1991. Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc Natl Acad Sci USA. 88: 10885–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L, Tazzoli M, Russo V, Beever J.. 2010. Genetic heterogeneity at the bovine KIT gene in cattle breeds carrying different putative alleles at the spotting locus. Anim Genet. 41: 295–303 [DOI] [PubMed] [Google Scholar]
- Gauly M, Vaughan J, Hogreve SK, Erhardt G.. 2005. Brainstem auditory-evoked potential assessment of auditory function and congenital deafness in llamas (Lama glama) and alpacas (L. pacos). J Vet Intern Med. 19: 756–760 [DOI] [PubMed] [Google Scholar]
- Geissler EN, McFarland EC, Russell ES.. 1981. Analysis of pleiotropism at the dominant white-spotting (W) locus of the house mouse: a description of ten new W alleles. Genetics. 97: 337–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler EN, Ryan MA, Housman DE.. 1988. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 55: 185–192 [DOI] [PubMed] [Google Scholar]
- Gruneberg H.. 1936. Grey-lethal, a new mutation in the house mouse. J Hered. 27: 105 [Google Scholar]
- Guridi M, Soret B, Alfonso L, Arana A.. 2011. Single nucleotide polymorphisms in the Melanocortin 1 Receptor gene are linked with lightness of fibre colour in Peruvian Alpaca (Vicugna pacos). Anim Genet. 42: 679–682 [DOI] [PubMed] [Google Scholar]
- Haase B, Brooks SA, Schlumbaum A, Azor PJ, Bailey E, Alaeddine F, Mevissen M, Burger D, Poncet PA, Rieder S, et al. 2007. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 3: e195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintz HF, Van Vleck LD.. 1979. Lethal dominant roan in horses. J Hered. 70: 145–146 [Google Scholar]
- Hofstra RM, Osinga J, Tan-Sindhunata G, Wu Y, Kamsteeg EJ, Stulp RP, van Ravenswaaij-Arts C, Majoor-Krakauer D, Angrist M, Chakravarti A, et al. 1996. A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome). Nat Genet. 12: 445–447 [DOI] [PubMed] [Google Scholar]
- Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A, Yanagisawa M.. 1994. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 79: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Jackson IJ.. 1994. Molecular and developmental genetics of mouse coat color. Annu Rev Genet. 28: 189–217 [DOI] [PubMed] [Google Scholar]
- Johansson Moller M, Chaudhary R, Hellmén E, Höyheim B, Chowdhary B, Andersson L.. 1996. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm Genome. 7: 822–830 [DOI] [PubMed] [Google Scholar]
- Lang KD, Wang Y, Plante Y.. 1996. Fifteen polymorphic dinucleotide microsatellites in llamas and alpacas. Anim Genet. 27: 293 [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN.. 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 33: 177–182 [DOI] [PubMed] [Google Scholar]
- Mariasegaram M, Pullenayegum S, Jahabar Ali M, Shah RS, Penedo MC, Wernery U, Sasse J.. 2002. Isolation and characterization of eight microsatellite markers in Camelus dromedarius and cross-species amplification in C. bactrianus and Lama pacos. Anim Genet. 33: 385–387 [DOI] [PubMed] [Google Scholar]
- Marklund S, Kijas J, Rodriguez-Martinez H, Rönnstrand L, Funa K, Moller M, Lange D, Edfors-Lilja I, Andersson L.. 1998. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 8: 826–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund S, Moller M, Sandberg K, Andersson L.. 1999. Close association between sequence polymorphism in the KIT gene and the roan coat color in horses. Mamm Genome. 10: 283–288 [DOI] [PubMed] [Google Scholar]
- Mau C, Poncet PA, Bucher B, Stranzinger G, Rieder S.. 2004. Genetic mapping of dominant white (W), a homozygous lethal condition in the horse (Equus caballus). J Anim Breed Genet. 121: 374–383 [Google Scholar]
- Mcgregor BA.. 2006. Production, attributes and relative value of alpaca fleeces in southern Australia and implications for industry development. Small Ruminant Res. 61: 93–111 [Google Scholar]
- Merriwether DA, Merriwether AM.. 2003. Alpaca color genetics. Alpacas Magazine. Summer, 2–6
- Munyard KA.. 2011. Inheritance of white colour in alpacas. [cited 2012 Sept 1]. Available from: http://www.alpaca.asn.au/pub/about/info/readings/docs/inheritance_white_colour.pdf
- Newton JM, Wilkie AL, He L, Jordan SA, Metallinos DL, Holmes NG, Jackson IJ, Barsh GS.. 2000. Melanocortin 1 receptor variation in the domestic dog. Mamm Genome. 11: 24–30 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Kasugai T, Ohno K, Morimoto M, Yamazaki M, Dohmae K, Nishimune Y, Kondo K, Kitamura Y.. 1991. Anemia and mast cell depletion in mutant rats that are homozygous at “white spotting (Ws)” locus. Blood. 78: 1936–1941 [PubMed] [Google Scholar]
- Nocka K, Tan JC, Chiu E, Chu TY, Ray P, Traktman P, Besmer P.. 1990. Molecular bases of dominant negative and loss of function mutations at the murine c-kit/white spotting locus: W37, Wv, W41 and W. EMBO J. 9: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul E.. 1999. Theory of colour inheritance in alpacas. Alpacas Australia. 28
- Paul E.. 2006. Colour review Adelaide, Australia: Australian Alpaca Association National Conference; [Google Scholar]
- Peakall R, Smouse PE.. 2006. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 6: 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo MC, Caetano AR, Cordova K.. 1999. Eight microsatellite markers for South American camelids. Anim Genet. 30: 166–167 [DOI] [PubMed] [Google Scholar]
- Penedo MCT.. 1998. Microsatellite markers for South American camelids. Anim Genet. 29:411–412 [PubMed] [Google Scholar]
- Powell AJ, Moss MJ, Tree LT, Roeder BL, Carleton CL, Campbell E, Kooyman DL.. 2008. Characterization of the effect of Melanocortin 1 Receptor, a member of the hair color genetic locus, in alpaca (Lama pacos) fleece color differentiation. Small Ruminant Res. 79: 183–187 [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N.. 2010. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA.. 1999. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 65: 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P.. 2000. Association mapping in structured populations. Am J Hum Genet. 67: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulos WL, Hutt FB.. 1969. Lethal dominant white in horses. J Hered. 60: 59–63 [DOI] [PubMed] [Google Scholar]
- Reinsch N, Thomsen H, Xu N, Brink M, Looft C, Kalm E, Brockmann GA, Grupe S, Kühn C, Schwerin M, et al. 1999. A QTL for the degree of spotting in cattle shows synteny with the KIT locus on chromosome 6. J Hered. 90: 629–634 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ.. 2000. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz SA, Misener S.. (eds.) Bioinformatics methods and protocols: methods in molecular biology NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Russell ES.. 1979. Hereditary anemias of the mouse: a review for geneticists. In: Caspari EW.. (ed.) Advances in Genetics. New York: Academic Press; [PubMed] [Google Scholar]
- Sánchez-Martín M, Rodríguez-García A, Pérez-Losada J, Sagrera A, Read AP, Sánchez-García I.. 2002. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet. 11: 3231–3236 [DOI] [PubMed] [Google Scholar]
- Santschi EM, Purdy AK, Valberg SJ, Vrotsos PD, Kaese H, Mickelson JR.. 1998. Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm Genome. 9: 306–309 [DOI] [PubMed] [Google Scholar]
- Sarno RJ, David VA, Franklin WL, O’Brien SJ, Johnson WE.. 2000. Development of microsatellite markers in the guanaco, Lama guanicoe: utility for South American camelids. Mol Ecol. 9: 1922–1924 [DOI] [PubMed] [Google Scholar]
- Schuelke M.. 2000. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 18: 233–234 [DOI] [PubMed] [Google Scholar]
- Sherwin WB, Jabot F, Rush R, Rossetto M.. 2006. Measurement of biological information with applications from genes to landscapes. Mol Ecol. 15: 2857–2869 [DOI] [PubMed] [Google Scholar]
- Silvers WK.. 1979. The coat colors of mice: a model for gene action and interaction New York: Springer-Verlag; [Google Scholar]
- Southard-Smith EM, Angrist M, Ellison JS, Agarwala R, Baxevanis AD, Chakravarti A, Pavan WJ.. 1999. The Sox10(Dom) mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 9: 215–225 [PubMed] [Google Scholar]
- Tachibana M, Kobayashi Y, Matsushima Y.. 2003. Mouse models for four types of Waardenburg syndrome. Pigment Cell Res. 16: 448–454 [DOI] [PubMed] [Google Scholar]
- Valbonesi A, Apaza N, La Manna V, Gonzales ML, Huanca T, Renieri C.. 2011. Inheritance of white, black and brown coat colours in alpaca (Vicuna pacos). Small Ruminant Research. 99: 16–19 [Google Scholar]
- Yajima I, Sato S, Kimura T, Yasumoto K, Shibahara S, Goding CR, Yamamoto H.. 1999. An L1 element intronic insertion in the black-eyed white (Mitfmi-bw) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum Mol Genet. 8: 1431–1441 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.