Abstract
The yeast Candida albicans is an opportunistic pathogen that threatens patients with compromised immune systems. Immune cell defenses against C. albicans are complex but typically involve the production of reactive oxygen species and nitrogen radicals such as nitric oxide (NO) that damage the yeast or inhibit its growth. Whether Candida defends itself against NO and the molecules responsible for this defense have yet to be determined. The defense against NO in various bacteria and the yeast Saccharomyces cerevisiae involves an NO-scavenging flavohemoglobin. The C. albicans genome contains three genes encoding flavohemoglobin-related proteins, CaYHB1, CaYHB4, and CaYHB5. To assess their roles in NO metabolism, we constructed strains lacking each of these genes and demonstrated that just one, CaYHB1, is responsible for NO consumption and detoxification. In C. albicans, NO metabolic activity and CaYHB1 mRNA levels are rapidly induced by NO and NO-generating agents. Loss of CaYHB1 increases the sensitivity of C. albicans to NO-mediated growth inhibition. In mice, infections with Candida strains lacking CaYHB1 still resulted in lethality, but virulence was decreased compared to that in wild-type strains. Thus, C. albicans possesses a rapid, specific, and highly inducible NO defense mechanism involving one of three putative flavohemoglobin genes.
The dimorphic fungus Candida albicans causes infections in immunocompromised hosts and is particularly problematic for AIDS and cancer patients. In healthy individuals, phagocytic immune cells such as macrophages (17), monocytes (37, 45), and neutrophils (45) defend against Candida infections by producing several growth inhibitors and cytotoxic compounds, including microbicidal enzymes (41) and reactive oxygen and nitrogen species (50). One potentially powerful weapon against C. albicans is nitric oxide (NO). Macrophages produce high concentrations of this free radical via the action of an inducible NO synthase (36), inhibition of which strongly decreases the candidacidal activity of macrophages (4, 15, 43). Despite the increasing understanding of host immune defenses mounted against this opportunistic pathogen, the means by which C. albicans resists NO or other microbicidal agents is not well understood.
Microbes protect themselves against NO toxicity by using enzymes that convert NO to less toxic molecules. Flavohemoglobin, an NO dioxygenase (NOD) that converts NO to nitrate (26, 29, 55), is found in bacteria and yeasts (7, 58). This enzyme is encoded by a single gene in several different organisms: for example, by hmp in Escherichia coli (49) and by ScYHB1 in Saccharomyces cerevisiae (57). Flavohemoglobin is necessary for virulence of a plant pathogen, the bacterium Erwinia chrysanthemi (18). hmp-negative bacteria are more easily inhibited by NO-releasing compounds and NO (9, 21, 26, 38), and expression of hmp is strongly induced by NO (8, 42). Hmp induction by NO is mediated by a derepression mechanism in which NO inactivates a metal-binding transcription factor, Fnr (10, 42) or Fur (8, 11). In the yeast S. cerevisiae, deletion of ScYHB1 abolishes NO-consuming activity and leads to increased growth inhibition upon exposure to an NO-generating compound. In contrast to bacteria, ScYHB1 expression is not induced by NO (35). The fungal pathogen Cryptococcus neoformans has a single flavohemoglobin gene, FHB1; deletion of this gene blocks NO consumption and attenuates virulence (12).
We searched the recently completed DNA sequence of the C. albicans genome and identified three structural homologues of ScYHB1, which we call CaYHB1, CaYHB4, and CaYHB5. Each gene was individually deleted and tested for an NO defense function. Here we report that one of these genes, CaYHB1, plays a central role in protection of Candida against NO.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae wild-type strain BY4742, along with its isogenic yhb1Δ:KanMX derivative, strain 15887, were obtained from Research Genetics/Invitrogen (Table 1). C. albicans strain RM1000 (his1Δ/his1Δ ura3Δ/ura3Δ) (Table 1) was provided by Jesus Pla (Universidad Complutense de Madrid, Madrid, Spain). Strains were routinely grown in YEPD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose), with 100-μg/ml uridine added for the growth of Ura− Candida strains.
TABLE 1.
Yeast strains used in this study
| Strain type | Genotype | Sourceb |
|---|---|---|
| S. cerevisiae | ||
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | Res. Genetics |
| 15887 | MATα his3Δ leu2Δ lys2Δ ura3Δ yhb1Δ:KanMX | Res. Genetics |
| C. albicans | ||
| CAF2a | ura3Δ::imm434/URA3 | Fonzi and Irwin (19) |
| RM1000 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/ his1Δ::hisG | Alonso-Monge et al. (1) |
| yhb1Δ/YHB1c | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG yhb1Δ::dpl200/YHB1+ | This study |
| yhb1Δ/YHB1c ura3Δ/URA3c | ura3Δ::imm434/URA3+his1Δ::hisG/his1Δ::hisG yhb1Δ::dp1200/YHB1+ | This study |
| yhb1Δ/yhb1Δ His+ Ura+ | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG yhb1Δ::HIS1/yhb1Δ::URA3 | This study |
| yhb1Δ/yhb1Δ | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG yhb1Δ::HIS1/yhb1Δ::dp1200 | This study |
| yhb1Δ/yhb1Δ ura3Δ/URA3c | ura3Δ::imm434/URA3+his1Δ::hisG/his1Δ::hisG yhb1Δ::HIS1/yhb1Δ::dp1200 | This study |
| yhb4Δ/yhb4Δ | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG yhb4Δ::HIS1/yhb4Δ::dp1200 | This study |
| yhb5Δ/yhb5Δ | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG yhb5Δ::HIS1/yhb5Δ::dp1200 | This study |
CAF2 is from reference (19) and is the progenitor of RM1000 (1). The superscript c denotes a heterozygote at that locus in which one wild-type copy of the relevant gene was inserted by transformation back into its native chromosomal location in a homozygous deletion strain. The wild-type gene copy used for transformation was obtained by PCR from genomic DNA of SC5314, the CAF2 progenitor (19), and included DNA flanking the original deletion on each side. Correct insertions into genomic DNA of the transformants were confirmed by PCR analysis.
Res. Genetics, Research Genetics/Invitrogen.
Construction of Candida deletion mutants.
Both copies of the entire open reading frame (ORF) of CaYHB1 were deleted in the diploid Candida strain RM1000 (Table 1), using the URA3-dpl200 cassette from pDDB57 (52) to delete the first copy of the gene and the HIS1 cassette from pGEM-HIS1 to replace the second copy (53). Primers YHB1-1 and YHB1-2 (see Table S1 in the supplemental material) were used to amplify each marker gene cassette by PCR and thereby construct a CaYHB1-specific deletion cassette. YHB1-1 contains a 60-nucleotide (nt) sequence homologous to DNA flanking the 5′ end of the CaYHB1 ORF plus 19 nt complementary to one side of the URA3-dpl200 and HIS1 cassettes. YHB1-2 contains a 61-base sequence homologous to DNA flanking CaYHB1 on the 3′ end of the ORF plus 20 nt complementary to the opposite side of the URA3-dpl200 and HIS1 cassettes. In the first round of transformations, deletion cassettes containing URA3-dpl200 were transformed into RM1000 by a modified lithium acetate method (27). Ura+ transformants were selected on YNB/Ura dropout medium (yeast nitrogen base without amino acids [Difco] plus 2% glucose and supplemented with amino acids, but lacking uridine). Deletion transformations were then repeated in subsequent rounds with HIS1-containing cassettes, followed by selection on YNB/Ura His dropout medium (as described above, but also lacking histidine) until a null mutant was identified. Homozygous deletion of CaYHB1 in a specific His+ Ura+ transformant was confirmed by positive PCRs with primer pairs YHB1-3/URA3-1, YHB1-4/URA3-2, YHB1-3/HIS1-1, and YHB1-4/HIS1-2 and a negative PCR with primer pair YHB1-5/YHB1-6 (data not shown). The resulting cayhb1Δ::HIS1/cayhb1Δ::URA3-dpl200 strain was then selected on 5-fluoroorotic acid-containing medium to select against URA3 (5). PCR on genomic DNA from Ura− strains was used to identify a cayhb1Δ::HIS1/cayhb1Δ::dpl200 strain in which the URA3 gene has been removed by recombination between direct repeats of the flanking dpl-200 DNA on the URA3-dpl200 cassette. Final confirmation of cayhb1Δ/cayhb1Δ was shown by the absence of a detectable CaYHB1 mRNA by Northern blot (data not shown). Similar procedures were used to generate the cayhb4Δ/cayhb4Δ and cayhb5Δ/cayhb5Δ deletion mutants (Table 1), except that CaYHB4 mRNA could not be detected even in RM1000.
Complementation of the CaYHB1 deletion.
Reintroduction of CaYHB1 was accomplished through two methods. In the first approach, a complementation plasmid was constructed by using the autonomously replicating plasmid pABSKII (CaARS2 CaURA3), generously provided by B. B. Magee (Department of Genetics, University of Minnesota, Minneapolis). A 3.2-kb region containing the entire CaYHB1 ORF, plus about 1,500 bp upstream and 470 bp downstream of CaYHB1, was amplified from RM1000 genomic DNA through PCR with primers YHB1-9 and YHB1-10 (see Table S1 in the supplemental material). The resulting product was digested with SacI and SpeI and ligated into pABSKII that had been cut with SacI and SpeI. The resultant plasmid (pABSKII CaYHB1) or empty pABSKII was transformed into the cayhb1Δ::HIS1/cayhb1Δ::dpl200 strain; transformants were selected on YNB/Ura dropout medium. For the second reintroduction approach, a 2-kb CaYHB1-containing fragment (including 468 bp upstream and 411 bp downstream of the gene) was created by PCR from RM1000 genomic DNA with primers YHB1-3 and YHB1-4. This PCR product was transformed into the cayhb1Δ::HIS1/cayhb1Δ::URA3-dpl200 strain, and transformants were selected on 5-fluoroorotic acid-containing medium. A cayhb1Δ::dpl200/CaYHB1 strain was obtained, and reintegration of CaYHB1 was confirmed by positive PCRs with YHB1-3/YHB1-4, YHB1-9/YHB1-10, YHB1-3/YHB1-10, and YHB1-4/YHB1-9 and by negative PCRs with YHB1-3/HIS1-1 and YHB1-4/HIS1-2. All transformations were achieved through the lithium acetate method described above.
Cell NO consumption and extract NOD activity measurements.
Fresh cultures were initiated from overnight cultures in YEPD medium plus uridine at A600 = 0.1, with S. cerevisiae grown at 30°C and C. albicans grown at 37°C. Cells were grown in air to an A600 of 0.2 and were then transferred to an atmosphere containing 960 ppm of NO in 21% O2 balanced with N2 or were maintained under air. Maximal gas equilibrium was achieved by shaking sealed 50-ml Erlenmeyer flasks containing a <1:5 ratio of culture volume to flask volume at 275 rpm. Gases were mixed and delivered to the culture flasks from three-way proportioners (Cole-Parmer Instrument Co.) at a rate of 30 ml per min, with gas mixtures passed through a trap containing sodium hydroxide pellets to remove NO·2 and higher oxides of nitrogen formed prior to entering the sealed flasks (24). Cells were exposed for 15, 30, and 60 min for mRNA isolation or for 60 min for NO consumption and NOD activity measurements.
For measurements of NO metabolism by whole cells, culture aliquots were centrifuged, supernatants were aspirated, and cell pellets were washed twice. NO consumption measurements were made with a 2-mm ISO-NOP NO electrode (World Precision Instruments, Sarasota, Fla.) in a 2-ml reaction mixture containing minimal salts glucose medium, cycloheximide, and 2 μM NO in a 37°C, water-thermostatted, glass-stoppered, and magnetically stirred glass reaction vessel (23, 26). For measurements of NOD activity, cell extracts were prepared by harvesting cells by centrifugation, lysing them by addition of Y-PER-S reagent (Pierce), sonicating them, and clarifying them by centrifugation. Clarified extracts were transferred to new tubes and stored at −80°C until assayed. Extract NOD activity measurements were made with the NO electrode in a 2-ml reaction mixture containing 100 mM potassium phosphate (pH 7.0), 0.3 mM EDTA, 100 μM NADH, 0.5-mg/ml Cu,ZnSOD, 1 μM flavin adenine dinucleotide (FAD), and 2 μM NO (22, 23, 25, 26). To avoid interference produced by nonenzymatic decomposition of NO by extracts, rates were determined following a second addition of 2 μM NO. Rates of NO metabolism were determined at 1 μM NO and were corrected for background rates of NO decomposition under otherwise identical conditions. The protein concentration was determined from the A280 of cell extracts by applying a value of 1 mg per ml of protein for an absorbance of 1.
Northern blot analysis.
Total RNA was isolated from pelleted log-phase culture cells by bead beating with glass beads in the presence of phenol and RNA lysis buffer (0.3 M NaCl, 1 mM EDTA, 10 mM Tris [pH 7.5], 0.2% sodium dodecyl sulfate). Standard electrophoretic techniques, denaturing formaldehyde gels, and blotting procedures were employed (44). DNA probe templates were prepared by PCR from RM1000 genomic DNA by using gene-specific primers (see Table S1 in the supplemental material). These were then radioactively labeled with [α-32P]dATP (ICN Radiochemicals), using the DECAprime II kit (Ambion).
lacZ reporter gene.
Part of CaYHB1 containing the 1.2-kb ORF plus either 998 bp (presumptive full promoter) or 279 bp (minimal promoter) of DNA upstream of the start codon was amplified by PCR from genomic DNA and inserted upstream of Streptococcus thermophilus lacZ in the pAU95 plasmid (48), replacing the 0.5-kb-long HWP1 promoter DNA between KpnI and PstI. The resulting CaYHB1-lacZ plasmids were then cut at a unique PshAI site in the HWP1 3′ untranslated region downstream of lacZ, and the linearized DNAs were each transformed into the Ura− strain RM1000. For each reporter gene plasmid, β-galactosidase assays were then performed on cell extracts (2) of a Ura+-transformed strain that by PCR analysis had correctly integrated the plasmid downstream of the HWP1 locus.
Growth inhibition assays.
Overnight cultures in YEPD medium plus uridine were diluted to A600 = 0.025, grown for 4 h at 30°C, and then inoculated with either a small volume of freshly made solution containing an NO-generating compound or of control solution. When nitrite was added to log-phase Candida cultures in YEPD medium, the initial pH was between 5.5 and 5.8. After 4 h at 30°C, A600 values were recorded and growth during the 4-h period was determined by subtracting the A600 values at t = 0 from final A600 values.
Virulence assays.
The C. albicans strains used were all Ura+ His+, with the URA3 gene present at its normal chromosomal location to avoid any effects on virulence due to misexpression of this gene (3, 6, 46). All animal experiments were performed in accordance with institutional regulations in an Association for Assessment and Accreditation of Laboratory Animal Care-certified facility under the National Institutes of Health guidelines for the care and use of laboratory animals. Female BALB/c mice, 6 to 7 weeks old, obtained from the National Cancer Institute, were housed in groups in bioclean hoods and provided with water and food ad libitum. Eight mice were used for each experimental group. Mice were allowed a 1-week acclimatization period before experiments were started. Cells from the different Candida strains were grown overnight at 37°C in YEPD medium, harvested by centrifugation, washed twice, resuspended in pyrogen-free saline, and counted with a hemocytometer. Each mouse received an intravenous challenge of 0.2 ml containing 5 × 105 cells of the corresponding strain. Confirmation of the number and viability of cells present in the infecting inocula was performed by plate count. Mice were monitored for survival daily after infection, and the days of death were recorded. Survival data and differences between groups were analyzed by using the Kaplan-Meier and log rank tests. P < 0.05 was considered statistically significant.
RESULTS
Flavohemoglobin orthologues in C. albicans.
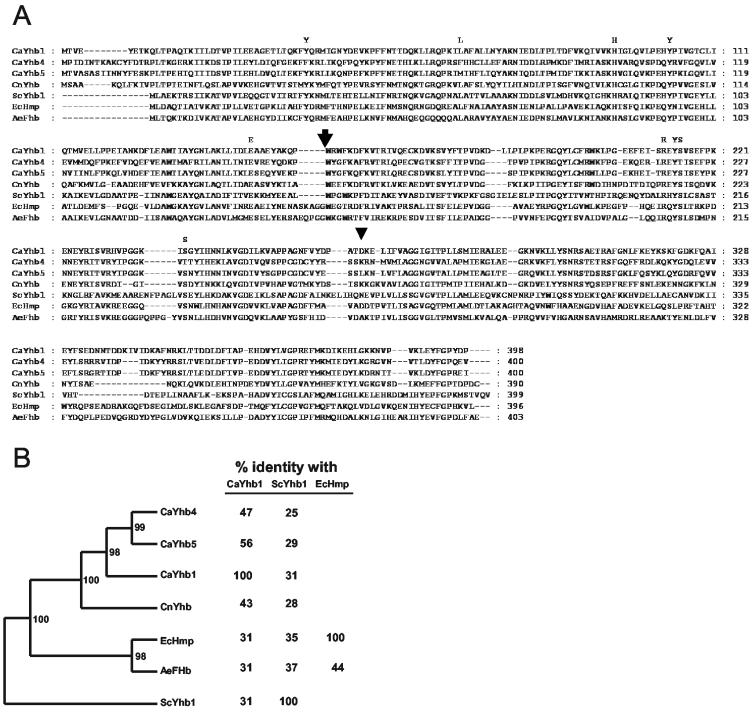
The BLAST program (National Center for Biotechnology Information) was used to search the completed DNA sequence of the C. albicans genome (version 6; http://www-sequence.stanford.edu/group/candida) for sequences related to the Saccharomyces flavohemoglobin gene ScYHB1. This search yielded three structural homologues, two of which are named YHB1 and YHB5 based on contig annotation (YHB1, contig6-2060, nt 7817 to 9013; and YHB5, contig6-2400, nt 9264 to 10467). We refer to them in this work as CaYHB1 and CaYHB5. The third unannotated putative ORF is more closely related to CaYHB5 than CaYHB1 and therefore is referred to as CaYHB4 (contig6-2518, nt 174143 to 175346). The percentages of sequence identity between the predicted Candida flavohemoglobins and the S. cerevisiae flavohemoglobin are as follows: CaYhb1, 31%; CaYhb4, 25%; and CaYhb5, 29% (Fig. 1). The X-ray structures of two bacterial flavohemoglobins show three domains: a globin domain, an FAD-binding domain, and an NAD(P)-binding domain (16, 31). The homology between the three Candida CaYhb proteins and the E. coli flavohemoglobin extends over the whole protein length: i.e., all three domains, rather than being clustered in just one domain.
FIG. 1.
C. albicans contains three predicted flavohemoglobins with similarity to flavohemoglobins from other microbes. (A) Alignment of flavohemoglobins from several microbial genes. CaYhb1, -4, and -5 are the three predicted flavohemoglobins from C. albicans. CnYhb is the flavohemoglobin from Candida norvegensis (34), also known as Pichia norvegensis. ScYhb1 is from Saccharomyces cerevisiae, EcHmp is from Escherichia coli, and AeFhb is from Alcaligenes eutrophus. Amino acids important for binding the coenzymes heme and FAD in the X-ray structure of EcHmp (31) and AeFHb (16) are shown above the aligned sequences. Based on the alignment with the E. coli Hmp sequence (31), the globin domain comprises about the first third of the polypeptide chain, the FAD domain starts after the large arrow, and the NAD(P) domain is after the small arrowhead. Alignments were created with ClustalX and displayed with GeneDoc. (B) Phylogenetic tree of a subset of the microbial flavohemoglobins. The tree is based on an alignment (A) of the sum of the amino acids in these flavohemoglobins and is unrooted. Numbers adjoining the branches indicate bootstrap values (percentages from 500 replicates) based on results of the unweighted pair group method with arithmetic mean (40) calculated by using the PHYLIP program suite (J. Felsenstein; http://evolution.genetics.washington.edu/phylip.html). The different flavohemoglobins are shown together with the percent identity between select pairs.
The three flavohemoglobins show sequence differences at key positions thought to be important for NOD activity and function. Catalytic NO dioxygenation is achieved by O2 binding to the ferrous heme and a rapid reaction of NO with the bound O2 to form NO3− (22, 26, 47). To reduce the ferric ion in heme during repetitive cycles of NO dioxygenation, electrons are univalently transferred from FADH2 to heme, which in turn receives electrons from NAD(P)H. Interestingly, the Tyr B10, which is essential for high O2 affinity and efficient NOD function of the E. coli flavohemoglobin (22), is conserved in the distal heme pocket in CaYhb1 and CaYhb5, but not in CaYhb4 (Fig. 1A). CaYhb4 and CaYhb5 also lack the unique Leu E11 residue that contacts ferric iron in E. coli flavohemoglobin structure (31) and is conserved for NOD activity function (16). Amino acids whose side chains contact FAD (Arg212, Tyr214, Ser215, and Ser238) and amino acids within the proximal region of the heme pocket (His93, Tyr103, and Glu146) are conserved in all isoforms.
The phylogenetic comparison in Fig. 1B shows the closer relatedness of each of the Candida flavohemoglobins to each other than to three flavohemoglobins shown to have an NOD function. The analysis also shows that the flavohemoglobin of Candida norvegensis is more closely related to those from C. albicans than to the others (Fig. 1B).
Constitutive and inducible NO metabolism in C. albicans versus S. cerevisiae.
To investigate the role of the three flavohemoglobin genes in the response of C. albicans to NO, we generated mutant strains for each YHB gene in which both gene copies were deleted. These mutants were generated by replacing each gene copy sequentially in the C. albicans strain RM1000 (his1/his1 ura3/ura3) with a selectable marker (HIS1 or URA3) following the methods of Wilson et al. (52, 53) as detailed in Materials and Methods.
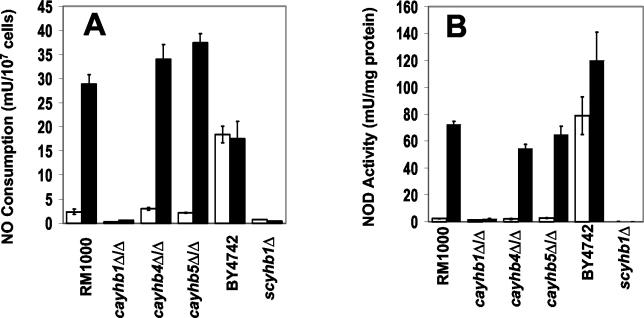
To determine whether the NO consumption rate of C. albicans is induced by exposure to NO, RM1000 cells were treated for 1 h with 960 ppm of gaseous NO (≤2 μM in solution) and compared to untreated cells for NO consumption rate and extract NOD activity. After the 1-h exposure of cells to NO, whole-cell NO consumption and cell extract NOD activities are increased by about 13- and 30-fold, respectively (Fig. 2). The inducible and constitutive NO metabolism by C. albicans is eliminated by the deletion of CaYHB1, but not by deletion of either of the CaYHB4 or CaYHB5 homologues. Thus, the CaYHB1 gene confers the majority of the NO metabolic activity observed under these conditions.
FIG. 2.
Flavohemoglobin gene YHB1 is required for inducible NO metabolism in yeast. (A) NO consumption in yeast exposed either to air only (open bars) or to 960 ppm of gaseous NO in an air mixture (solid bars) for 60 min. (B) NOD activities in naïve and NO-exposed cells. Cells were harvested, extracts were prepared, and NOD activities were measured as described in Materials and Methods. scyhb1Δ indicates strain 15887, and cayhbxΔ/Δ refers to cayhbxΔ::HIS1/cayhbxΔ::dpl200 (Table 1). One milliunit is equivalent to 1 nmol per min. Error bars represent the standard deviation of three independent trials.
Similar to previously reported results (35), intact Saccharomyces cells exhibit NO consumption (Fig. 2A). Furthermore, S. cerevisiae cell extracts contain high levels of NOD activity (Fig. 2B). Consistent with a previous report (35), NO consumption and NOD activity of Saccharomyces are eliminated in a strain containing a deletion of ScYHB1. However, there are clear differences in the NO stress responses of these two yeasts. Basal levels of NO consumption by C. albicans cells are about sevenfold lower than those of S. cerevisiae cells (Fig. 2A). NO exposure induces only a modest 1.5-fold increase in S. cerevisiae cell extract NOD activity and no detectable increase in NO consumption by intact cells (Fig. 2). These small differences between S. cerevisiae extract and cell measurements of NO metabolic activity changes may reflect intracellular substrate or cofactor limits on flavohemoglobin catalysis.
NO induction of YHB mRNA in C. albicans versus S. cerevisiae.
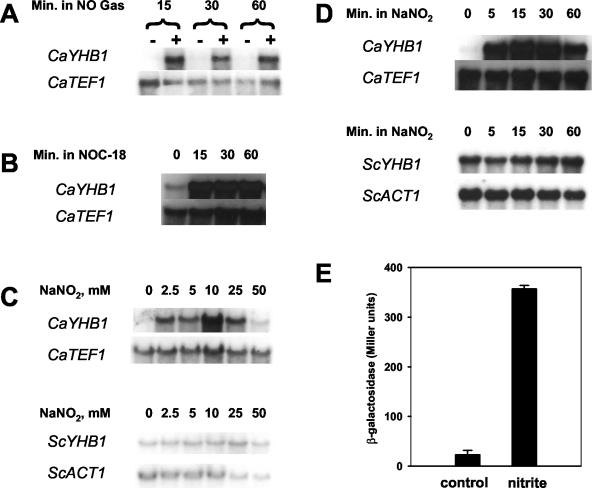
Using a gene-specific probe and RNA from log-phase cultures, Northern blot analysis shows that CaYHB1 mRNA is strongly induced by exposure of Candida to gaseous NO (≤2 μM in solution) (Fig. 3A). The chemical specificity of CaYHB1 induction was probed by addition of hydrogen peroxide or the superoxide-generating drug plumbagin (32), and only gaseous NO exposure strongly induces CaYHB1 expression (data not shown). We also tested the effects of an additional NO source on CaYHB1 expression. A large induction of CaYHB1 mRNA is observed with the NO-releasing chemical NOC-18 (Fig. 3B), but not with its other breakdown product, diethylenetriamine (33) (data not shown). Northern blot analysis shows a detectable mRNA for CaYHB5 but not for CaYHB4 (data not shown). YHB1 mRNA levels were also analyzed in C. albicans or S. cerevisiae cells treated with sodium nitrite, a nitrosating agent and a physiologically relevant NO donor (14). ScYHB1 exhibits little or no change in mRNA levels after exposure of S. cerevisiae to sodium nitrite. In contrast, there is a strong induction of CaYHB1 mRNA after such treatment (Fig. 3C). Induction by nitrite is also observed earlier (5 min) in Candida than in Saccharomyces (15 min) (Fig. 3D). There is no detectable increase in either CaYHB4 or CaYHB5 mRNA after treatment of cells with sodium nitrite. Thus, induction of a single flavohemoglobin gene is observed in both yeasts, although the magnitude and rate of the nitrite-induced changes in YHB1 mRNA levels are much greater in Candida than in Saccharomyces. NO-induced increases in mRNA levels could be mediated by either changes in transcription or mRNA degradation. To test for changes in transcription, a strain was constructed that contains a lacZ reporter gene fused downstream of the CaYHB1 promoter (998 bp) and ORF. In this strain, nitrite induced a large increase in β-galactosidase (Fig. 3E). To test whether the CaYHB1 promoter is required for this induction, a strain was constructed that contains the same reporter gene construct except that the CaYHB1 promoter segment was truncated to just 279 bp upstream of the ORF. In this strain, expression of β-galactosidase in the presence and absence of 10 mM nitrite was 2.3 ± 0.2 and 1.5 ± 0.4 Miller units (mean ± standard deviation), respectively. Together these results show that nitrite induction of flavohemoglobin expression is mediated by an increase in transcription rather than by changes in stability of CaYHB1 mRNA or CaYhb1 protein.
FIG. 3.
CaYHB1 mRNA is induced by NO in C. albicans. (A) CaYHB1 is induced by exposure to 960 ppm of NO gas (≤2 μM in solution). Naïve and NO-exposed cultures were grown as described in Materials and Methods, and cells were harvested at 15, 30, and 60 min, followed by mRNA isolation. (B) Induction of CaYHB1 mRNA by the NO-releasing compound NOC-18. A total of 3 mM NOC-18 was added to exponentially growing yeast (37°C), and cells were collected at 0, 15, 30, and 60 min for RNA extraction. (C) Induction of YHB1 in C. albicans and S. cerevisiae by nitrite. Various concentrations of sodium nitrite (NaNO2) were added to log-phase cultures of the S. cerevisiae strain BY4742 (grown at 30°C) or the C. albicans strain RM1000 (grown at 37°C). After 30 min of treatment, cells were collected and mRNA was extracted. (D) A total of 25 mM NaNO2 was added to exponential cultures of BY4742 (30°C) or RM1000 (37°C), and after 0, 5, 15, 30, and 60 min, cells were harvested for mRNA isolation. ACT1 was probed as a loading control for Saccharomyces RNA; CaTEF1 was the control for Candida samples. (E) Induction of CaYHB1-lacZ by nitrite. A C. albicans strain containing a CaYHB1-lacZ reporter gene plasmid, integrated into a chromosome downstream of the HWP1 gene (see Materials and Methods), was grown in rich YEPD medium with or without 10 mM nitrite at 37°C for 1 h, and β-galactosidase activity in cell extracts was measured as described previously (2).
Protection of C. albicans against NO toxicity.
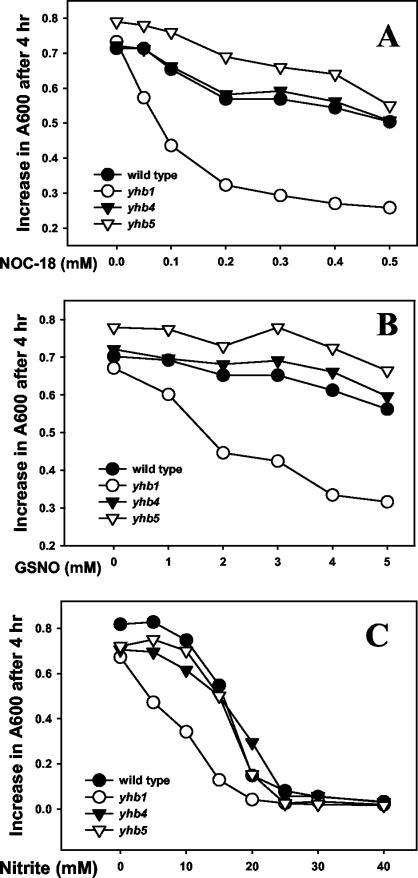
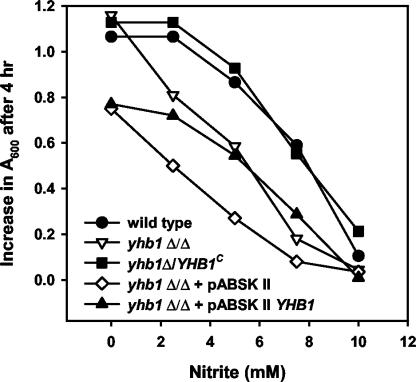
To test whether CaYHB1, CaYHB4, or CaYHB5 protects C. albicans from NO toxicity, we compared the CaYHB deletion strains and the parental strain (RM1000) for sensitivity to NO-releasing compounds and nitrite. Growth of the cayhb1Δ/cayhb1Δ, cayhb4Δ/cayhb4Δ, or cayhb5Δ/cayhb5Δ strain is similar to that of the parent strain RM1000 in rich YEPD medium under aerated conditions. Growth of the CaYHB1 deletion strain is potently inhibited by the NO-generating compounds NOC-18 and S-nitrosoglutathione (Fig. 4A and B). In comparison, neither NO-generating compound significantly inhibits the growth of RM1000 or the CaYHB4 or CaYHB5 deletion strain. The results are similar to those previously reported for wild-type C. albicans strains treated with these agents (51). In contrast, sodium nitrite inhibits the growth of all strains tested, but the cayhb1Δ/cayhb1Δ strain consistently shows greater sensitivity to nitrite (Fig. 4C). Furthermore, nitrite resistance was restored by introducing an intact CaYHB1 gene into the cayhb1Δ/cayhb1Δ strain, either by transformation with a plasmid (pABSKII CaYHB1) expressing CaYHB1 or by insertion of the intact CaYHB1 gene into the original locus (Fig. 5). These findings demonstrate that CaYHB1, but not CaYHB4 or CaYHB5, protects C. albicans against NO toxicity.
FIG. 4.
Deletion of CaYHB1 increases sensitivity to NO-generating compounds and to nitrite. Growth of wild-type (RM1000; solid circles), cayhb1Δ/cayhb1Δ (open circles), cayhb4Δ/cayhb4Δ (solid triangles), or cayhb5Δ/cayhb5Δ (open triangles) yeast cultures was measured following treatment of log-phase cultures with control solvent or NOC-18 (A), S-nitrosoglutathione (GSNO) (B), or sodium nitrite (NaNO2) (C) at the indicated concentrations. After 4 h of treatment, growth was calculated from the change in turbidity at 600 nm (A600t = 4 −A600t = 0). Data are from representative experiments that were repeated at least two times with similar results.
FIG. 5.
Reinsertion of CaYHB1 complements sensitivity of the cayhb1Δ/cayhb1Δ strain to sodium nitrite. Growth of cultures of the wild-type strain (RM1000; solid circles) or mutant strains of the types cayhb1Δ::HIS1/cayhb1Δ::dpl200 (open triangles), cayhb1Δ::dpl200/CaYHB1 (solid squares), cayhb1Δ::HIS1/cayhb1Δ::dpl200 transformed with empty plasmid pABSKII (open diamonds), or cayhb1Δ::HIS1/cayhb1Δ::dpl200 transformed with complementation plasmid pABSKII CaYHB1 (solid triangles) was determined following treatment with the indicated concentrations of sodium nitrite for 4 h. Growth was determined from the change in turbidity at 600 nm. The RM1000, cayhb1Δ/Δ, and cayhb1Δ/+ strains were each grown in YEPD medium supplemented with uridine, while the strains containing plasmid pABSKII or pABSKII CaYHB1 were grown in YEPD medium without uridine to maintain selection for the URA3-containing plasmid. The exact reason for slower growth of the latter cultures is not known, but it may reflect a low activity of the plasmid URA3 gene. Results are representative of two or more experiments.
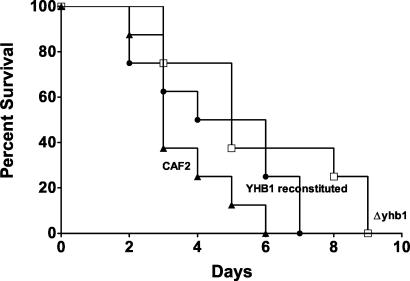
Analysis of virulence of CaYHB1 deletion mutant C. albicans strain, using a murine model.
The effect of CaYHB1 in systemic candidiasis was evaluated by comparing the survival curves of mice given isogenic C. albicans strains, including the homozygous cayhb1/cayhb1, reconstituted CaYHB1/cayhb1, and wild-type (CAF-2) strains in a mouse model of hematogenously disseminated candidiasis. Experiments revealed that, although infections with the cayhb1 mutant still resulted in lethality in this model (all animals died within less than 10 days), its virulence was somewhat attenuated and/or delayed compared to that exhibited by CAF-2 (Fig. 6, median survival times of 5 versus 3 days; P = 0.0294). Reintroduction of one copy of the gene at its original locus partially restored virulence and resulted in survival curves that were intermediate between those generated with the homozygous mutant and those generated with CAF-2 (P = 0.1651 versus CAF-2).
FIG. 6.
C. albicans strains lacking CaYHB1 show attenuated virulence. The His+ Ura+ C. albicans strains used were the CAF2, Δyhb1 (yhb1Δ/yhb1Δ ura3Δ/URA3c), and YHB1 reconstituted (yhb1Δ/YHB1c ura3Δ/URA3c) strains shown. Tail vein injection with Candida was performed on day 0.
DISCUSSION
In C. albicans, removal of NO is dependent upon the presence of a single flavohemoglobin gene, CaYHB1. CaYHB1 is induced by micromolar NO, confers NOD activity, and protects cells against NO toxicity. Our results also indicate that the three Candida flavohemoglobins are not functionally equivalent. Thus, CaYHB5 mRNA is expressed, but in contrast to CaYHB1, is not induced by exposure to NO or NO-generating agents. As for CaYHB4, we were unable to detect its mRNA under either normal or inducing conditions. Furthermore, neither the cayhb4Δ/cayhb4Δ strain nor the cayhb5Δ/cayhb5Δ strain displays NO sensitivity or growth impairment in aerated YEPD medium. Our preliminary data show that Candida strains lacking CaYHB5 show a very small reduction in virulence (data not shown), although the significance of this finding awaits further analysis of the enzymatic function of this flavohemoglobin. These data demonstrate that CaYHB1 is the only flavohemoglobin with a detectable role in NO metabolism and detoxification. Nevertheless, it remains possible that NO metabolic functions of CaYHB4 or CaYHB5 are expressed under specialized growth conditions or during specific stages of differentiation.
Interestingly, the results suggest a low predictive value of phylogenetic analyses for an NOD function. CaYhb1 is more closely related to CaYhb4 and CaYhb5 than ScYhb1 or bacterial flavohemoglobins (Fig. 1B). Yet, CaYhb1, but not CaYhb4 or CaYhb5, functions in NO detoxification. The presence of the active site residues Tyr B10 and Leu E11 appears to be a strong indicator of a NOD function. On the other hand, structurally diverse and distantly related hemoglobins and myoglobins also show NOD activity and function but show different active site structures (13, 54), thus cautioning against an exclusion of an NO detoxification function based solely upon structure analysis. Clearly, evidence for significant NO metabolism or induction of activity in response to NO stress is required to establish an NOD function.
Unlike in other organisms, the inducibility of CaYHB1 observed in C. albicans appears to be a specific response to NO stress. The hmp gene in E. coli bacteria is induced not only by NO stress, but also by paraquat, which produces both superoxide and hydrogen peroxide stress (39). Although C. albicans CaYHB1 transcript levels are increased by NO generators, as well as authentic NO, CaYHB1 mRNA levels do not change upon treatment of Candida with sublethal levels of hydrogen peroxide or the superoxide-generating compound plumbagin (data not shown). These differences between bacterial and Candida flavohemoglobin expression suggest unique regulatory mechanisms. Our data show that transcriptional regulation is responsible for at least part of the NO induction of CaYHB1. To our knowledge, this is the first study to show NO-regulated expression of a physiologically relevant gene target in fungi, so there is very little knowledge so far about potential transcription factors that could mediate this response. We are currently analyzing the CaYHB1 promoter to identify the cis-acting DNA sequence; the data obtained so far suggest the involvement of multiple DNA elements.
As a commensal organism and opportunistic pathogen, C. albicans is likely to be exposed to NO in a variety of concentrations and locations. For example, NO is produced by endothelia (20), various epithelia (28, 30, 56), and macrophages (50). In the oral cavity, where Candida is commonly found, NO is also produced by bacteria and by abiotic acidification of nitrite (14). Because of its growth-inhibiting (cytostatic) effects (51), NO released by these sources is likely to limit the domain of Candida. Yet in our experiments on virulence using a murine tail vein injection model, the mutant strain lacking CaYHB1 still shows virulence, although that virulence is less than that of the control wild-type strain or the CaYHB1-complemented mutant. These results are similar to those obtained in the fungal pathogen C. neoformans (12), showing that flavohemoglobin is not absolutely required for virulence when the fungus is directly injected into the bloodstream. However, because it seems likely that NO concentrations will vary widely in different parts of the body, future analysis using other types of virulence-testing models, including mucosal infection routes, will be needed to fully understand the role of CaYHB1 in C. albicans infection in humans.
Supplementary Material
Acknowledgments
We are grateful to Jesus Pla, Aaron Mitchell, and B. B. Magee, who supplied strains and plasmids.
This study was supported by NSF grant MCB 0091236 to M.C.G., NIH grant GM 65090 to P.R.G., NSF IGERT grant DGE 0114264 to W.C., and NIH training grant 5 T32 GM 08362 to B.D.U. J.L.L.-R. is the recipient of a New Investigator Award in Molecular Pathogenic Mycology from the Burroughs Wellcome Fund.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alonso-Monge, R., F. Navarro-Garcia, G. Molero, R. Diez-Orejas, M. Gustin, J. Pla, M. Sanchez, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. Green Publishing Associates and Wiley-Interscience, New York, N.Y.
- 3.Bain, J. M., C. Stubberfield, and N. A. Gow. 2001. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol. Lett. 204:323-328. [DOI] [PubMed] [Google Scholar]
- 4.Blasi, E., L. Pitzurra, M. Puliti, A. R. Chimienti, R. Mazzolla, R. Barluzzi, and F. Bistoni. 1995. Differential susceptibility of yeast and hyphal forms of Candida albicans to macrophage-derived nitrogen-containing compounds. Infect. Immun. 63:1806-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, S., M. H. Nguyen, Z. Zhang, H. Jia, M. Handfield, and C. J. Clancy. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71:6101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1994. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J. Biol. Chem. 269:7349-7354. [PubMed] [Google Scholar]
- 8.Crawford, M. J., and D. E. Goldberg. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273:34028-34032. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, M. J., and D. E. Goldberg. 1998. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543-12547. [DOI] [PubMed] [Google Scholar]
- 10.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Autreaux, B., D. Touati, B. Bersch, J. M. Latour, and I. Michaud-Soret. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. USA 99:16619-16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jesus-Berrios, M., L. Liu, J. C. Nussbaum, G. M. Cox, J. S. Stamler, and J. Heitman. 2003. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13:1963-1968. [DOI] [PubMed] [Google Scholar]
- 13.Dou, Y., D. H. Maillett, R. F. Eich, and J. S. Olson. 2002. Myoglobin as a model system for designing heme protein based blood substitutes. Biophys. Chem. 98:127-148. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, C., H. Dougall, P. Johnston, S. Green, R. Brogan, C. Leifert, L. Smith, M. Golden, and N. Benjamin. 1995. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat. Med. 1:546-551. [DOI] [PubMed] [Google Scholar]
- 15.Elahi, S., G. Pang, R. B. Ashman, and R. Clancy. 2001. Nitric oxide-enhanced resistance to oral candidiasis. Immunology 104:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ermler, U., R. A. Siddiqui, R. Cramm, and B. Friedrich. 1995. Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution. EMBO J. 14:6067-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evron, R. 1980. In vitro phagocytosis of Candida albicans by peritoneal mouse macrophages. Infect. Immun. 28:963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Favey, S., G. Labesse, V. Vouille, and M. Boccara. 1995. Flavohaemoglobin HmpX: a new pathogenicity determinant in Erwinia chrysanthemi strain 3937. Microbiology 141:863-871. [DOI] [PubMed] [Google Scholar]
- 19.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furchgott, R. F., and P. M. Vanhoutte. 1989. Endothelium-derived relaxing and contracting factors. FASEB J. 3:2007-2018. [PubMed] [Google Scholar]
- 21.Gardner, A. M., and P. R. Gardner. 2002. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J. Biol. Chem. 277:8166-8171. [DOI] [PubMed] [Google Scholar]
- 22.Gardner, A. M., L. A. Martin, P. R. Gardner, Y. Dou, and J. S. Olson. 2000. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J. Biol. Chem. 275:12581-12589. [DOI] [PubMed] [Google Scholar]
- 23.Gardner, P. R., G. Costantino, and A. L. Salzman. 1998. Constitutive and adaptive detoxification of nitric oxide in Escherichia coli. Role of nitric-oxide dioxygenase in the protection of aconitase. J. Biol. Chem. 273:26528-26533. [DOI] [PubMed] [Google Scholar]
- 24.Gardner, P. R., G. Costantino, C. Szabo, and A. L. Salzman. 1997. Nitric oxide sensitivity of the aconitases. J. Biol. Chem. 272:25071-25076. [DOI] [PubMed] [Google Scholar]
- 25.Gardner, P. R., A. M. Gardner, L. A. Martin, Y. Dou, T. Li, J. S. Olson, H. Zhu, and A. F. Riggs. 2000. Nitric-oxide dioxygenase activity and function of flavohemoglobins: sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 275:31581-31587. [DOI] [PubMed] [Google Scholar]
- 26.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:10378-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 28.Gookin, J. L., J. M. Rhoads, and R. A. Argenzio. 2002. Inducible nitric oxide synthase mediates early epithelial repair of porcine ileum. Am. J. Physiol. Gastrointest. Liver Physiol. 283:G157-G168. [DOI] [PubMed] [Google Scholar]
- 29.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:14100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman, R. A., G. Zhang, N. C. Nussler, S. L. Gleixner, H. R. Ford, R. L. Simmons, and S. C. Watkins. 1997. Constitutive expression of inducible nitric oxide synthase in the mouse ileal mucosa. Am. J. Physiol. 272:G383-G392. [DOI] [PubMed] [Google Scholar]
- 31.Ilari, A., A. Bonamore, A. Farina, K. A. Johnson, and A. Boffi. 2002. The X-ray structure of ferric Escherichia coli flavohemoglobin reveals an unexpected geometry of the distal heme pocket. J. Biol. Chem. 277:23725-23732. [DOI] [PubMed] [Google Scholar]
- 32.Jamieson, D. J., D. W. Stephen, and E. C. Terriere. 1996. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 138:83-88. [DOI] [PubMed] [Google Scholar]
- 33.Keefer, L. K., R. W. Nims, K. M. Davies, and D. A. Wink. 1996. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol. 268:281-293. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi, G., T. Nakamura, H. Ohmachi, A. Matsuoka, T. Ochiai, and K. Shikama. 2002. Yeast flavohemoglobin from Candida norvegensis. Its structural, spectral, and stability properties. J. Biol. Chem. 277:42540-42548. [DOI] [PubMed] [Google Scholar]
- 35.Liu, L., M. Zeng, A. Hausladen, J. Heitman, and J. S. Stamler. 2000. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 97:4672-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 37.Marodi, L., H. M. Korchak, and R. B. Johnston, Jr. 1991. Mechanisms of host defense against Candida species. I. Phagocytosis by monocytes and monocyte-derived macrophages. J. Immunol. 146:2783-2789. [PubMed] [Google Scholar]
- 38.Membrillo-Hernandez, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274:748-754. [DOI] [PubMed] [Google Scholar]
- 39.Membrillo-Hernández, J., S. O. Kim, G. M. Cook, and R. K. Poole. 1997. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by σS. J. Bacteriol. 179:3164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michener, C. D., and R. R. Sokal. 1957. A quantitative approach to a problem in classification. Evolution 11:130-162. [Google Scholar]
- 41.Peterson, E. M., and R. A. Calderone. 1978. Inhibition of specific amino acid uptake in Candida albicans by lysosomal extracts from rabbit alveolar macrophages. Infect. Immun. 21:506-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole, R. K., M. F. Anjum, J. Membrillo-Hernandez, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rementeria, A., R. Garcia-Tobalina, and M. J. Sevilla. 1995. Nitric oxide-dependent killing of Candida albicans by murine peritoneal cells during an experimental infection. FEMS Immunol. Med. Microbiol. 11:157-162. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schuit, K. E. 1979. Phagocytosis and intracellular killing of pathogenic yeasts by human monocytes and neutrophils. Infect. Immun. 24:932-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staab, J. F., and P. Sundstrom. 2003. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 11:69-73. [DOI] [PubMed] [Google Scholar]
- 47.Stevanin, T. M., N. Ioannidis, C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J. Biol. Chem. 275:35868-35875. [DOI] [PubMed] [Google Scholar]
- 48.Uhl, M. A., and A. D. Johnson. 2001. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147:1189-1195. [DOI] [PubMed] [Google Scholar]
- 49.Vasudevan, S. G., W. L. Armarego, D. C. Shaw, P. E. Lilley, N. E. Dixon, and R. K. Poole. 1991. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 226:49-58. [DOI] [PubMed] [Google Scholar]
- 50.Vázquez-Torres, A., and E. Balish. 1997. Macrophages in resistance to candidiasis. Microbiol. Mol. Biol. Rev. 61:170-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vazquez-Torres, A., J. Jones-Carson, and E. Balish. 1995. Nitric oxide production does not directly increase macrophage candidacidal activity. Infect. Immun. 63:1142-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wittenberg, J. B., M. Bolognesi, B. A. Wittenberg, and M. Guertin. 2002. Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J. Biol. Chem. 277:871-874. [DOI] [PubMed] [Google Scholar]
- 55.Wu, G., L. M. Wainwright, and R. K. Poole. 2003. Microbial globins. Adv. Microb. Physiol. 47:255-310. [DOI] [PubMed] [Google Scholar]
- 56.Zhan, X., D. Li, and R. A. Johns. 2003. Expression of endothelial nitric oxide synthase in ciliated epithelia of rats. J. Histochem. Cytochem. 51:81-87. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, X. J., D. Raitt, P. V. Burke, A. S. Clewell, K. E. Kwast, and R. O. Poyton. 1996. Function and expression of flavohemoglobin in Saccharomyces cerevisiae. Evidence for a role in the oxidative stress response. J. Biol. Chem. 271:25131-25138. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, H., and A. F. Riggs. 1992. Yeast flavohemoglobin is an ancient protein related to globins and a reductase family. Proc. Natl. Acad. Sci. USA 89:5015-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.