Abstract
The Western and Eastern species of gorillas (Gorilla gorilla and Gorilla beringei) began diverging in the mid-Pleistocene, but in a complex pattern with ongoing gene flow following their initial split. We sequenced the complete mitochondrial genomes of 1 Eastern and 1 Western gorilla to provide the most accurate date for their mitochondrial divergence, and to analyze patterns of nucleotide substitutions. The most recent common ancestor of these genomes existed about 1.9 million years ago, slightly more recent than that of chimpanzee and bonobo. We in turn use this date as a calibration to reanalyze sequences from the Eastern lowland and mountain gorilla subspecies to estimate their mitochondrial divergence at approximately 380000 years ago. These dates help frame a hypothesis whereby populations became isolated nearly 2 million years ago with restricted maternal gene flow, followed by ongoing male migration until the recent past. This process of divergence with prolonged hybridization occurred against the backdrop of the African Pleistocene, characterized by intense fluctuations in temperature and aridity, while at the same time experiencing tectonic uplifting and consequent shifts in the drainage of major river systems. Interestingly, this same pattern of introgression following divergence and discrepancies between mitochondrial and nuclear loci is seen in fossil hominins from Eurasia, suggesting that such processes may be common in hominids and that living gorillas may provide a useful model for understanding isolation and migration in our extinct relatives.
Key words: hominid, hominoid, mitochondrial genome, molecular clock, primate, speciation
The 3 living genera of African apes (Gorilla, Pan, and Homo) diverged in the late Miocene, beginning approximately 8 or 9 million years ago (Begun 2010; Brunet 2010; Harrison 2010; Wilkinson et al. 2011). The lineage leading to humans produced a radiation of many bipedal species—the exact number of which is an open question—but eventually resulting in only 1 living subspecies of 1 species, Homo sapiens sapiens (Wood and Lonergan 2008). In contrast, our closest living relatives exist as several species and subspecies. Two species of Pan, chimpanzees (Pan troglodytes) and bonobos (Pan paniscus), have been recognized essentially since the initial description of the latter (Coolidge 1933). The divergence between chimpanzees and bonobos, as estimated from complete mitochondrial genome sequences, began about 2 million years ago (Mya), with the radiation of the 4 chimpanzee subspecies beginning about 1 Mya (Stone et al. 2010; Bjork et al. 2011). For many years, gorillas were considered a single species with 3 traditionally recognized subspecies (Groves 1967, 2003). However, beginning in the 1990s they have increasingly been referred to as 2 species: 1) the Western gorilla (Gorilla gorilla) comprising 2 subspecies: the Western lowland gorilla (G. g. gorilla) and the Cross River gorilla (G. g. diehli), and 2) the Eastern gorilla (Gorilla beringei) comprising 2 subspecies: the mountain gorilla (G. b. beringei) and the Eastern lowland gorilla (G. b. graueri) (Groves 2001).
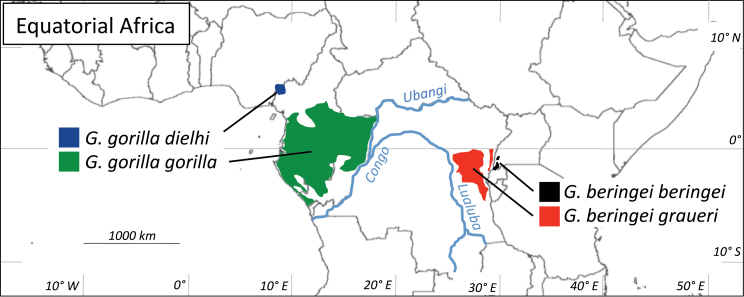
The Eastern and Western gorilla species are separated by approximately 1000 km (Figure 1). Barriers to gene flow between these species include the Congo and Ubangi Rivers and the more open woodland and savanna north of these large rivers. Chimpanzees, however, are not similarly divided, living sympatrically with both gorilla species and in the intervening habitat north of the Congo River system. Discussions of the elevation of the Eastern gorilla to a separate species (G. beringei), rather than a subspecies of G. gorilla, began with mtDNA data indicating that the divergence between these gorilla populations may be as old, or older, than that between the 2 universally recognized species of Pan (Ruvolo et al. 1994; Morell 1994; Garner and Ryder 1996; Groves 1996, 2001, 2003). The estimated dates of gorilla species divergence ranged from 1.3 to 2.7 Mya; with much of the variability among mitochondrial loci attributed to the very short sequences used or varying dating methods (Ruvolo et al. 1994; Garner and Ryder 1996; Jensen-Seaman 2000; Jensen-Seaman et al. 2001; Thalmann et al. 2005). Subsequently, nucDNA sequence data revealed that the mtDNA divergence substantially predated the cessation of gene flow between the Eastern and Western gorillas, potentially with male-mediated gene flow continuing until as recently as 77 kya years ago (Jensen-Seaman et al. 2001, 2003; Kaessmann et al. 2001; Thalmann et al. 2007; Scally et al. 2012). Despite these new data, most authors continue to recognize 2 living species of Gorilla, as we do herein.
Figure 1.
Extant gorilla populations in equatorial Africa and the major rivers that may have restricted gene flow in the past. Although distributions are shown as continuous, gorillas exist primarily in small isolated pockets of remaining forest in West and East Africa. Distributions reproduced from Mitchell and Gonder (2013).
The Eastern lowland and mountain gorilla subspecies of the Eastern Democratic Republic of the Congo and the Albertine Rift can be found living as close as 100 km apart near Lake Edward. They are reciprocally monophyletic at mitochondrial loci, estimated by mtDNA data to have diverged approximately 400 kya with no strong evidence for recent maternal gene flow between these subspecies (Ruvolo et al. 1994; Jensen-Seaman and Kidd 2001; but see Ackermann and Bishop 2010). In West Africa, the Cross River populations are also classified as distinct from other Western gorillas, based on autosomal loci and anatomical data (Sarmiento and Oates 2000; Groves 2001; Thalmann et al. 2011), even though their mitochondrial haplotypes are not monophyletic with respect to other Western gorilla populations (Anthony, Johnson-Bawe, et al. 2007). In general, Western gorilla mitochondrial control region haplotypes fall into 2 major clades (C, D) that can be further subdivided into geographically defined subclades (Clifford et al. 2004). Geographic patterns of genetic diversity suggest a role for Pleistocene forest refugia in driving haplotype structure within Western gorillas and also suggest a role of rivers as significant barriers to gene flow. In particular, the mid-Pleistocene expansion of the Sahara and savanna at the expense of rain forest along with the coeval formation of the Congo River system may have sealed off female-mediated gene flow between what are now G. gorilla and G. beringei (Jensen-Seaman 2000; Thalmann et al. 2007). Subsequent restriction of gorillas to forest refugia likely induced changes in population size and migration in both Eastern and Western gorillas (Jensen-Seaman and Kidd 2001; Clifford et al. 2004; Anthony, Johnson-Bawe, et al. 2007).
Until recently, the only mitochondrial sequence data available from G. beringei were short sequences from portions of the COII, NADH5, and the first hypervariable region of the D-loop (Ruvolo et al. 1994; Garner and Ryder 1996; Saltonstall et al. 1998; Jensen-Seaman et al. 2003; Thalmann et al. 2005). Short sequences provide unreliable estimates of divergence, especially between closely related taxa. Previous attempts to date the mtDNA split between the extant gorilla species have also been confounded by the presence of nuclear integrations of mitochondrial DNA, or numts (Garner and Ryder 1996; Jensen-Seaman et al. 2004; Anthony, Clifford, et al. 2007). As such, it has been suggested that gorillas are particularly susceptible to inadvertent numt amplification, with 1 solution being to utilize only long-range amplification of large DNA fragments (Thalmann et al. 2004). Recently, long sequence contigs comprising most of the mitochondrial genome were generated with whole-genome next-generation shotgun sequencing of 28 Western gorillas and 2 Eastern gorillas (Prado-Martinez et al. 2013). These sequences do not include the ~1-kb D-loop, and no empirical methods to avoid numts. In order to provide a reliable estimate of the mtDNA divergence between Eastern and Western gorillas, we sequenced the complete mitochondrial genome of 1 Eastern lowland gorilla and compared it to available complete mtDNA sequences of Western lowland gorillas and other hominoids. Furthermore, to help understand the genetic diversity and history of Western gorillas, we sequenced the complete mitochondrial genome of 1 representative of the D3 haplotype clade to complement those available from the C haplogroup.
Materials and Methods
DNA Sequencing
Genomic DNA from 1 wild-born male Eastern lowland gorilla (“M’kubwa”; G. b. graueri) was obtained from the Coriell Cell Repositories, Hamden, NJ (ID PR00206). According to the International Gorilla studbook, this individual was captured in the Eastern Democratic Republic of the Congo in 1953, and died in the Houston Zoo in 2004 (Studbook ID 9907, Niekisch 2011). Genomic DNA from 1 male Western lowland gorilla (“Chipua”; G. gorilla gorilla) was extracted from a lymphoblast cell line, originally obtained from the Coriell Cell Repositories (Coriell ID PR00622 Studbook ID 1419, Niekisch 2011). Using 1ng of these genomic DNAs as templates, the mitochondrial genome was PCR-amplified in 3 overlapping fragments of 7.1, 7.5, and 5.5kb with primer pairs mt10261_F/mt726_R, mt551_F/mt7969_R, and mt7022/mt12400_R, respectively (see Supplementary Table 1 online for primer sequences). PCR volumes were 50 μl and included 0.4mM each dNTP, 0.8 μM each primer, 1.25U LA-Taq polymerase (Takara Bio/Clontech, Mountain View, CA), in 1X manufacturer’s buffer. Following a 2-min denaturation at 94 °C, reactions ran for 35 cycles of 94 °C for 15 s, 60 °C for 15 s, and 72 °C for 10min, with a final extension of 10min at 72 °C. Amplicons were purified with Wizard SV columns (Promega, Madison, WI) and sequenced with multiple primers in both directions (Supplementary Table 1 online) with BigDye v3.1 sequencing kit using an ABI3100 automated sequencer (Applied Biosystems/Life Technologies, Grand Island, NY). The SeqMan program of the LaserGene package (DNA-Star, Madison, WI) was used to edit sequences, create contigs, and produce consensus sequences.
Data Archiving
In accordance with data archiving guidelines (Baker 2013), the assembled and annotated genome sequences have been deposited in GenBank under accession numbers KF914213 and KF914214.
Primate mtDNA Sequence Alignments
The 2 novel gorilla mitochondrial genome sequences obtained in the present study were aligned with whole genome sequences from 2 additional published Western lowland gorilla (G. g. gorilla, GenBank accession numbers NC_001645 and NC_011120), 4 chimpanzees (Pan troglodytes troglodytes, HM068587; P. t. schweinfurthii, HM068591; P. t. verus, HM068593; P. t. ellioti, HM068585), 2 bonobos (Pan paniscus, GU189674 and GU189674), 1 human (Homo sapiens, J01415.2), Neanderthal (H. neanderthalensis, FM865411), 2 orangutans (Pongo abelii, NC_002083; and P. pygmaeus, NC_001646), 1 gibbon (Hylobates lar, NC_002082), and 1 macaque (Macaca mulatta, NC_005943) using the ClustalW online server (Larkin et al. 2007). From the alignment 2 data sets were created. One data set contained the concatenated 12 protein-coding genes located on the heavy strand of the mitochondrial genome (10887 nucleotides), following Raaum et al. (2005), as they are likely to be evolving in a similar fashion. The other data set (15599 nucleotides) contained the entire mitochondrial genomes except the D-loop, which is largely unalignable across ape genera. We also compared our sequences with the nearly complete gorilla mitochondrial genome sequences described by Prado-Martinez et al. (2013).
There is little mitochondrial sequence data available from mountain gorillas, so for dating of the mitochondrial divergence between the Eastern lowland and mountain gorilla subspecies, we used 1 published NADH5 sequence (G. b. beringei, AF240447; Jensen-Seaman et al. 2001), and 12 previously published sequences from the first hypervariable region (HV1) of the D-loop, including those from 2 mountain gorillas (AY530103, AF089820), 2 Eastern lowland gorillas (AF050738, AF187549), 7 Western lowland gorillas (AY530138, AY530141, AY530128, AY530132, AY530118, AY530119, AY530120), and 1 Cross River gorilla (AY530112) (Jensen-Seaman and Kidd 2001; Clifford et al. 2004).
Protein-Coding Genes, tRNA, and rRNA Analysis
For analysis of individual genes, we used the mitochondrial genomes from 1 human (J01415.2), 1 chimpanzee (HM068587), 1 bonobo (GU189674), along with our 2 novel gorilla mtDNA genome sequences. The protein coding, tRNA, and rRNA genes, as well as conceptually translated amino acid sequences, were aligned using ClustalW (Larkin et al. 2007). Genetic distances, transition and transversion rates, and rate heterogeneity parameters were estimated using MEGA v5.2.2 (Tamura et al. 2011). Pairwise K a/K s values between chimpanzee and bonobo, and Eastern and Western gorilla were calculated using “seqinr” package (Charif and Lobry 2007) implemented in R v3.0.2. The difference in sequence divergence between chimpanzee–bonobo on one hand, and Western–Eastern gorillas on the other, was evaluated by creating 1000 bootstrap pseudoreplicates of each pairwise sequence alignment with the 12-gene data set using the SEQBOOT and DNADIST programs of the PHYLIP v3.395 package (Felsenstein 1993).
Phylogenetic Analysis
For dating, we used a Bayesian Markov Chain Monte Carlo approach (MCMC) implemented in BEAST v1.7.5 (Drummond and Rambaut 2007). The BEAST input file was generated using BEAUTi v1.7.5 (Drummond and Rambaut 2007). An uncorrelated lognormal relaxed molecular clock was used to allow evolutionary rate to vary from branch to branch. This approach has been previously shown to provide better estimate of time to most recent common ancestor (tMRCA) over the strict molecular clock that does not allow the evolutionary rate to vary among branches (Drummond et al. 2006). The SRD06 model of nucleotide substitution was used to partition the nucleotide data by codon position, so that the third codon position can differ from position 1 and 2 (Shapiro et al. 2006). The rate of evolution was calibrated using lognormally distributed priors as described in Raaum et al. (2005) with lognormal means of zero and lognormal standard deviation of 0.56. As discussed by several authors (Ho 2007; Bjork et al. 2011), lognormally distributed priors perform better than both normally distributed and exponentially distributed priors, when using fossil calibration points for dating the tree as it will sample values from the more distant past more frequently than recent values. We offset these distributions at the internal node of human–chimpanzee split by 5 Myr to ensure that the median values of the distribution approximates the expected 6 Mya split (as per Bjork et al. 2011). The same offset point was used for the NADH5 phylogeny construction.
MCMC simulation ran for 10 million generations for the 12-gene and whole mtDNA genome data sets, discarding 10% of the generations as “burnin.” The maximum clade credibility (MCC) tree was identified and annotated using TreeAnnotator v1.7.5 (Drummond and Rambaut 2007). Nodes with posterior probabilities exceeding 90% (P > 0.9) were used for tree building. The MCC tree generated by TreeAnnotator v1.7.5 was visualized and dated using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree). Tracer v1.5 (Rambaut and Drummond 2009) was used to summarize the posterior estimates of the various parameters sampled by the Markov Chain. tMRCA means, medians, 95% highest posterior density (95% HPD) intervals (all in Myr), and effective sample sizes (ESS) were calculated using Tracer.
For maximum likelihood and maximum parsimony phylogenetic reconstruction, the ClustalW alignments of both data sets were used with 1000 bootstrapped replicates, using MEGA v5.2.2 (Tamura et al. 2011). A model test was first performed using jModelTest v2.1.4 ( Guindon and Gascuel 2003; Darriba et al. 2012) to determine the best fitting nucleotide substitution model for each data set, using Akaike’s information criteria with correction (AICc) and Bayesian information criteria (BIC) values.
Results
DNA Sequencing
Complete mitochondrial genome sequences were obtained for 1 Eastern lowland gorilla (“M’kubwa,” G. b. graueri) and 1 Western lowland gorilla (“Chipua,” G. g. gorilla), and deposited in GenBank (accession numbers KF914213 and KF914214, respectively). The exact number of cytosines within the first hypervariable region (HV1) of the D-loop could not be determined for the Western lowland gorilla, presumably due to polymerase stutter or possible heteroplasmy. The Western lowland gorilla sequence was therefore submitted as …C8TTC10… in this region and annotated as undetermined, following previous publications (Garner and Ryder 1996; Thalmann et al. 2005). Similarly in a poly-C stretch in the second hypervariable region (HV2), the number of cytosines is submitted as 11 in our Western lowland gorilla and 12 in the Eastern gorilla, with some uncertainty. No nucleotide positions appeared to be heteroplasmic, although rigorous testing for this possibility was not done. All sequences appear to derive from authentic mitochondrial DNA, not numts, in that no apparently heterozygous sites were observed, nor were any premature stop codons or indels seen within protein-coding portions. We also note that the PCR amplicons generated to reconstruct the whole mtDNA genome in this study are 2 to 3 times larger than the largest gorilla numts reported in a recent comprehensive search of the gorilla genome for numts (Soto-Calderón 2012), making amplification on numts unlikely. Phylogenetic analysis of the novel HV1 sequences along with all previously published gorilla HV1 sequences confirms that wild-born M’kubwa is an Eastern lowland gorilla belonging to haplogroup B and that the mtDNA of Chipua belongs to the D3 haplogroup (Supplementary Figure 1 online), following the HV1 haplotype nomenclature of Anthony, Johnson-Bawe, et al. (2007). We observed that 1 of the 2 previously published complete genomes from G. gorilla, NC_001645 (Horai et al. 1995) appears to be a composite sequence derived from more than 1 individual, with its D-loop clearly derived from haplogroup D, while the bulk of its genome is much more similar to the other published G. gorilla sequence from haplogroup C (NC_011120; Xu and Arnason 1996). Therefore this sequence, NC_001645, was not used in any subsequent analyses.
Dating Mitochondrial Divergence
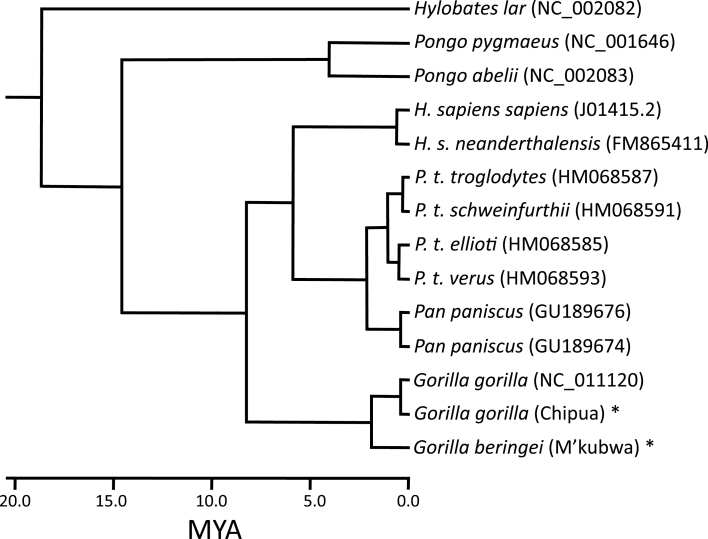
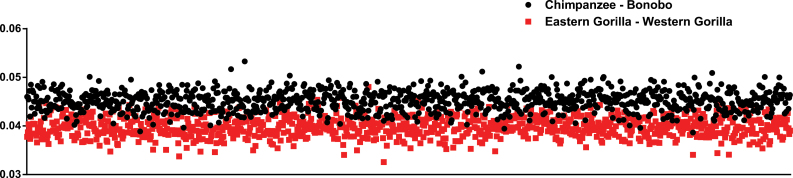
Both the 12-gene data set and the complete mtDNA sequence data set (see Methods) produced identical tree topologies, with the combined Bayesian posterior probability of 1.000 (Figure 2). The maximum parsimony tree and the maximum likelihood tree generated under the GTR+I+G model with either data set has an identical topology to the Bayesian tree, with all nodes supported in 99% or more of bootstrap replicates (not shown). We estimate, using a Bayesian MCMC approach, that the divergence between the mitochondrial genomes of the gorilla species occurred nearly 2 million years ago, slightly more recently than the chimpanzee–bonobo split, calibrated with a human–chimpanzee divergence of 6 Mya (Table 1). In a gene-by-gene comparison using the 13 protein-coding genes, 8 of the Pan species splits are older than the Gorilla species splits (Table 2). Of the 1000 bootstrapped replicate pairwise sequence alignments created for the Pan and for the Gorilla species using the 12-gene data set, 97.1% exhibit a greater divergence between chimp and bonobo than between the Eastern and Western gorillas (Figure 3). The deepest split within Western gorillas is between our novel Chipua sequence and previously published genomes, occurring relatively recently, less than 400000 years ago. Estimated divergence dates from the complete mtDNA genome (excluding the D-loop) are very similar to the 12-gene data set (Table 1).
Figure 2.
Phylogeny of hominoid mitochondria, based on the 12 protein-coding genes of the heavy strand, with dates inferred using a Bayesian MCMC approach with the human–chimpanzee divergence offset by 5 Myr to approximate a 6 Mya date for the human–chimpanzee split. Novel genome sequences are indicated with an asterisk.
Table 1.
Dates with confidence intervals
| 12 Heavy strand genes | Whole mthNA minus D-loop | |||
|---|---|---|---|---|
| Taxon divergence | tMRCAa | 95% HPDb | tMRCAa | 95% HPDb |
| Old World monkey–Hominoid | 32.215 | 25.288–40.411 | 32.535 | 25.339–40.758 |
| Gibbon–Hominid | 19.068 | 15.423–23.432 | 19.280 | 15.314–23.507 |
| Pongo–African Apes | 14.853 | 12.053–18.163 | 14.537 | 11.657–17.763 |
| Pongo pygmaeus–P. abelii | 4.090 | 3.192–5.213 | 3.989 | 3.015–5.089 |
| Gorilla–Homo/Pan | 8.396 | 7.063–10.192 | 8.280 | 6.919–10.003 |
| Homo–Pan | 5.983 | 5.200–7.058 | 5.982 | 5.197–7.082 |
| Pan troglodytes–P. paniscus | 2.163 | 1.742–2.691 | 2.172 | 1.715–2.679 |
| P. t. troglodytes/P. t. schweinfurthii–P. t. verus/P. t. ellioti | 1.054 | 0.824–1.330 | 1.027 | 0.803–1.304 |
| Gorilla gorilla–G. beringei | 1.900 | 1.456–2.397 | 1.895 | 1.438–2.391 |
| Deepest root within Western gorilla | 0.370 | 0.258–0.494 | 0.404 | 0.284–0.531 |
| Human–Neanderthal | 0.591 | 0.428–0.762 | 0.587 | 0.430–0.758 |
aTime to most recent common ancestor, in Myr.
b95% highest posterior density.
Table 2.
Chimpanzee/bonobo versus Eastern/Western Gorilla genetic distance and pairwise K a/K s values
| Locusa | Chimp–Bonobo distanceb | Western–Eastern gorilla distanceb | Chimp–Bonobo K a/K s | Western–Eastern K a/K s |
|---|---|---|---|---|
| ATPase6 | 0.040 | 0.054 | 0.317 | 0.321 |
| ATPase8 | 0.018 | 0.015 | 0.300 | 0.408 |
| COI | 0.031 | 0.021 | 0.078 | 0.075 |
| COII | 0.028 | 0.033 | 0.000 | 0.116 |
| COIII | 0.042 | 0.031 | 0.118 | 0.170 |
| Cytb | 0.050 | 0.050 | 0.206 | 0.060 |
| NADH1 | 0.051 | 0.040 | 0.091 | 0.144 |
| NADH2 | 0.049 | 0.038 | 0.151 | 0.189 |
| NADH3 | 0.027 | 0.036 | 0.143 | 0.313 |
| NADH4 | 0.042 | 0.037 | 0.113 | 0.211 |
| NADH4L | 0.045 | 0.033 | 0.052 | 0.035 |
| NADH5 | 0.062 | 0.058 | 0.253 | 0.173 |
| NADH6 | 0.049 | 0.047 | 0.106 | 0.000 |
| tRNAs | 0.018 | 0.016 | — | — |
| 12S rRNA | 0.014 | 0.008 | — | — |
| 16S rRNA | 0.024 | 0.025 | — | — |
| D-Loop | 0.105 | 0.103 | — | — |
aGenetic distance results were calculated separately for each gene, with the exception of the 22 tRNAs, which were concatenated into 1 sequence for analysis.
bGenetic distance calculated with MEGA v5.2.2.
Figure 3.
The mitochondrial genetic distances between Gorilla species compared to that of the Pan species, of 1000 bootstrapped replicates of the 12-gene data set.
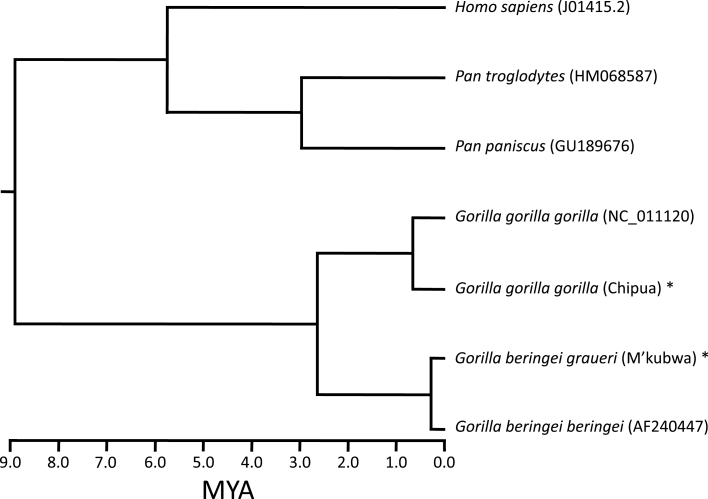
With no source of high-quality genomic DNA from a mountain gorilla, we were not able to amplify long-range PCR products in order to sequence the complete mitochondrial genome of this subspecies. Instead, we used a previously published sequence from the mountain gorilla NADH5 gene (AF240447) along with our novel sequences and a Bayesian MCMC approach to estimate the time of the Eastern lowland (G. b. graueri) and mountain (G. b. beringei) gorilla divergence at 0.378 (0.04–0.864) Mya, slightly more recent than the deepest split within G. gorilla (Figure 4). The dates obtained from HV1 were substantially higher than NADH5; when calibrated with an Eastern–Western gorilla divergence of 2 Mya, the deepest split within Western gorillas is 1.775 Mya (1.176–2.599 Mya; median 1.685 Mya) and the G. graueri versus G. beringei split time is 0.929 Mya (0.764–2.028; median 0.803 Mya). Although the estimates of the Eastern lowland and mountain gorilla mitochondrial divergence time differs depending on loci and calibration node, we note that in all circumstances the deepest split with Western gorillas is deeper than the split between the 2 Eastern gorilla subspecies.
Figure 4.
MCC tree from NADH5 gene sequence, with the human–chimpanzee divergence offset by 5 Myr to approximate a 6 Mya date for this divergence. The tMRCA of the 2 subspecies of G. beringei is estimated to be approximately 380 kya.
Twelve of the 13 protein-coding genes differs by at least 1 predicted amino acid between our Eastern lowland gorilla and our Western lowland gorilla for the gene-by-gene comparisons, although only 10 of these genes contain an apparent fixed difference, where all 3 Western gorilla mitochondrial genomes code for a different amino acid than the single Eastern lowland gorilla genome. With only 1 complete G. beringei sequence it is not possible to determine if such differences are polymorphic within this species. All proteins appear to be evolving under purifying selection in general, in that all gene-wide K a/K s values are less than 1 (Table 2). This is also true for each chimpanzee–bonobo pairwise comparison. Of the 22 tRNA genes, 9 are identical between all G. gorilla and G. graueri, with 21 substitutions found among the remaining 13 tRNA genes.
Integration of Previously Published Mitochondrial Genome Data
There are presently more than 200 gorilla sequences of the first hypervariable region (HV1) of the D-loop submitted to GenBank; mitochondrial haplogroups have traditionally been defined based on HV1 sequence in gorillas (Clifford et al. 2004). Recently, 29 nearly complete gorilla mitochondrial genome sequences have also been described, containing the entire coding sequence of the genome but lacking the ~1-kb D-loop (Supplemental Information online of Prado-Martinez et al. 2013). We therefore took this opportunity to merge these data sets by creating a phylogenetic tree of these 29 genome sequences, along with the 2 novel complete mitochondrial genome sequences of the present paper and the 1 previously reported G. gorilla sequence (NC_011120), excluding the D-loop. Of the 29 gorillas sequenced by Prado-Martinez et al. (2013) we had previously sequenced the HV1 region from 14 of them using long-range PCR to avoid numts (unpublished data), and could infer an additional individual’s haplotype as we had sequenced her wild-born mother. With about half of the 29 gorillas directly haplogrouped we could provisionally, but confidently, assign the others to an existing haplogroup (Supplementary Figure 2 online).
We compared our complete mitochondrial genome sequence of M’kubwa (G. beringei), generated with long-range PCR and dideoxy dye terminator sequencing to the nearly complete sequence generated by Prado-Martinez et al. (2013) from the same individual, but using short-read whole-genome shotgun sequencing of total genomic DNA. The sequences are identical at every nucleotide, excluding the ~1-kb D-loop not present in the latter sequence.
Discussion
The sequence of the Eastern gorilla (G. beringei) completes an important data set, as it is the last living hominid species to have its mitochondrial genome completely sequenced. In addition, the haplogroup D3 Western gorilla mitochondrial genome sequence further extends the genetic diversity sampled in this species by adding the most divergent complete mitochondrial sequence in G. gorilla. We note the importance of providing this sequence in making estimates of the amount of mtDNA diversity in this species, since 1 of the 2 previously described mitochondrial genome sequences is apparently a composite produced from at least 2 individuals, 1 of whom is from haplogroup C and 1 from haplogroup D. The use of large amplicons successfully avoided inadvertent PCR amplification of numts, a common difficulty in the analysis of mitochondri Anthony al DNA in gorillas (Jensen-Seaman et al. 2004; Thalmann et al. 2004; Anthony, Clifford, et al. 2007). With the exception of 2 stretches of cytosines in the control region, all bases were called with complete confidence. We believe that our estimate of the tMRCA of the gorilla species’ mitochondrial genomes using a Bayesian MCMC provides the most accurate estimate to date, as it is the first to be based on complete genome sequence. We do recognize, however, that such estimates are dependent on calibration from an imperfect fossil record and a number of assumptions regarding the nature of the nucleotide substitutions. Only short mitochondrial sequences from a few loci are available from the mountain gorilla, and so at this time the divergence estimates between G. b. beringei and G. b. graueri remain tentative.
As noted previously, the divergence of the Eastern and Western gorilla species’ mitochondrial genomes substantially predated the complete cessation of nuclear gene flow. Evidence for this comes from the observation that numts closely related to both Eastern and Western mtDNA haplotypes are found in both the Eastern and Western gorilla species (Jensen-Seaman et al. 2004; Thalmann et al. 2005; Anthony, Clifford, et al. 2007; Soto-Calderón 2012). Also, the genetic distances and divergence dates estimated from autosomal, X chromosomal, and Y chromosomal loci are consistently much less than those from mitochondrial genes (Burrows and Ryder 1997; Jensen-Seaman et al. 2001, 2003; Kaessmann et al. 2001; Altheide 2002; Scally et al. 2012). While the actual estimated divergence dates depend on a variety of factors that may differ among genetic systems including mutation rate, population size, and selection—along with assumptions of models and calibrations—this strong discrepancy between mitochondrial and nuclear loci is seen most clearly when these same loci are compared in other hominid species, such as chimpanzees (Jensen-Seaman et al. 2001; Mailund et al. 2012). One explanation is that long distance male-mediated gene flow persisted much longer than female gene flow as gorilla populations became isolated. This is consistent with the described dispersal patterns of gorillas where females tend to emigrate from their natal group to quickly join a neighboring group, whereas male gorillas may spend years traveling very long distances before establishing or taking over a reproductive group (Harcourt 1978; Douadi et al. 2007; Inoue et al. 2013). There is theoretical support for this sex-biased model (Birky et al. 1989) and empirical support seen in other primate species with strong female philopatry (Evans et al. 2010). However, a recent study failed to detect any significant sex-bias in genetic structure of Western gorillas across large distances (Fünfstück et al. 2014). An alternative explanation is simply that mitochondrial DNA is a single locus, and like any other locus may not be representative of the majority of genetic loci. We also recognize the possibility that selection, in the form of mito-nuclear incompatibility, could restrict mitochondrial introgression in the presence of nuclear gene flow (e.g., Trier et al. 2014).
Our estimated tMRCA of 1.9 Mya is an estimate of the time of cessation of female-mediated gene flow between populations that would ultimately become the Eastern and Western gorilla species. Perhaps the habitat fragmentation of the early to mid-Pleistocene created islands of forest refugia in central Africa between which males would travel, but females would not. The available paths of migration between West Africa and the Eastern populations could have been further reduced by the formation of the Congo/Ubangi River system around this same time, restricting the swath of suitable forest habitat north of these rivers and south of the savanna (Figure 1; Groves 1971; Beadle 1981; Stankiewicz and de Wit 2006; Schultheiβ et al. 2014). It is in this context that we note that the mitochondrial divergence in gorillas began around the same time as that between chimpanzees and bonobos, and much earlier than the deepest split within common chimpanzees (Table 1). It is therefore intriguing to speculate that perhaps the species-level splits in both Gorilla and Pan had a common cause. Although aridity induced habitat fragmentation is commonly invoked to explain Pleistocene allopatric speciation events, equatorial Africa may have been experiencing a wetter, warmer phase around 1.8 Mya (Trauth et al. 2009), suggesting that changes in river drainage may be an equally likely cause, especially considering the effects of major rivers on the patterns of genetic variation in extant Western lowland gorillas (Anthony, Johnson-Bawe, et al. 2007; Fünfstück et al. 2014). What is certain is that the Pleistocene was a time of tremendous climatic fluctuation in Africa, and renewed tectonic activity along the East African Rift, leading to numerous events that may have initiated speciation in gorillas, and perhaps chimpanzees as well.
Historically, following a major revision of taxonomy by Groves, gorillas were considered a single species with 3 subspecies (Groves 1967, 2003). Beginning in the mid-1990s, the consensus of the field began to change with many authors recognizing 2 gorilla species, G. gorilla and G. beringei. This rethinking was explicitly triggered by the observation of what appeared to be a surprisingly ancient split between the Western and Eastern gorilla populations, based solely on mitochondrial DNA sequence (Ruvolo et al. 1994; Morell 1994; Garner and Ryder 1996; Groves 1996, 2001, 2003). Interestingly, the average sequence divergence between Western and Eastern gorillas was reported to be larger than that between the universally recognized chimpanzee and bonobo species at both the COII gene and the HV1 (Ruvolo et al. 1994; Garner and Ryder 1996). Our results show a clear advantage in utilizing the complete mtDNA sequence, which reveal that the chimpanzee–bonobo mitochondrial genomes are actually slightly more divergent overall than the Eastern–Western gorilla genomes (Figure 3), and that the COII gene, by chance, is among the most aberrant loci (Table 2). Although we now recognize that mtDNA does not provide a complete picture of the events surrounding the genetic isolation of G. gorilla and G. beringei, it is unlikely that the consensus opinion will revert to a single species taxonomy, nor do we advocate as such. The picture that is now emerging is that the ancestral gorilla populations began to separate nearly 2 million years ago, based on our dating of the mitochondrial divergence at 1.9 Mya as well as the dating of the average nuclear sequence divergence between Eastern and Western gorilla genomes at 1.75 Mya (Scally et al. 2012), or somewhat more recent (0.9–1.6 Mya; Thalmann et al. 2007). Following this initial split, gene flow continued among these populations until about 100 kya, perhaps predominantly via male migration. The overall degree of anatomical and molecular differentiation between Eastern and Western gorillas is clearly greater than between any chimpanzee subspecies, and on par with other sister species pairs in primates (Groves 2001). The fact that the genetic evidence refutes a simple clean break as predicted by vicariance-driven allopatric speciation does not negate the overall distinctiveness of the 2 currently recognized gorilla species. We note that another recent analysis has placed the initial gorilla divergence to be much more recent (300–500 kya), based solely on nuclear data (Mailund et al. 2012). Our hope is that the 2 new complete mitochondrial genome sequences described herein, along with previously published great ape mtDNA sequences, can be combined with whole nuclear genome data to develop an isolation–migration model that simultaneously considers both genomes as a means to fully understand the speciation dynamics of gorillas.
The data from our study provide the most accurate dates of mitochondrial lineage divergence in gorillas, both within the diverse Western species, as well as between the Western and Eastern species. The study of gorilla mitochondrial DNA variation based on whole genomes has lagged behind that of chimpanzees and bonobos (Stone et al. 2010; Bjork et al. 2011), and we hope that the present paper is a first step toward remedying that paucity of data. Combining the mitochondrial data and recent whole nuclear genome sequences from gorillas with realistic estimates of migration rates and distances of both sexes could be used to develop and test more complex models of ape speciation processes (Ackermann and Bishop 2010; Mailund et al. 2012). Finally, we note that many of the same complexities seen in extant great apes may have also occurred in extinct hominins such as Neanderthals, Denisovans, and the hominins from Sima de los Huesos, where anatomical and molecular data reveal a complex pattern of isolation and migration, potentially the result of hybridization between subspecies or species, sex-biased gene flow, incomplete linage sorting, mitochondrial paraphyly, and geographically structured variation (Krause et al. 2010; Reich et al. 2010; Meyer et al. 2012, 2014; Prüfer et al. 2014). As such, models of great ape divergence could in turn be used to inform scenarios to explain Eurasian hominin demographic evolution whose migratory behavior and demographic structure cannot be directly observed.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Institute for General Medicine at the National Institutes of Health (R15 GM073682-01 to N.M.A.).
Supplementary Material
References
- Ackermann RR, Bishop JM. 2010. Morphological and molecular evidence reveals recent hybridization between gorilla taxa. Evolution. 64:271–290 [DOI] [PubMed] [Google Scholar]
- Altheide TK. 2002. Comparative population genetics of the Hominoidea: an investigation of locus-specific and genome-wide influence [PhD dissertation]. Tucson, AZ: University of Arizona [Google Scholar]
- Anthony NM, Clifford SL, Bawe-Johnson M, Abernethy KA, Bruford MW, Wickings EJ. 2007. Distinguishing gorilla mitochondrial sequences from nuclear integrations and PCR recombinants: guidelines for their diagnosis in complex sequence databases. Mol Phylogenet Evol. 43:553–566 [DOI] [PubMed] [Google Scholar]
- Anthony NM, Johnson-Bawe M, Jeffery K, Clifford SL, Abernethy KA, Tutin CE, Lahm SA, White LJ, Utley JF, Wickings EJ, et al. 2007. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc Natl Acad Sci USA. 104:20432–20436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Beadle LC. 1981. The inland waters of tropical Africa. London: Longman [Google Scholar]
- Begun DR. 2010. Miocene hominids and the origins of the African apes and humans. Ann Rev Anthropol. 39:67–84 [Google Scholar]
- Birky CW, Jr, Fuerst P, Maruyama T. 1989. Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics. 121:613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork A, Liu W, Wertheim JO, Hahn BH, Worobey M. 2011. Evolutionary history of chimpanzees inferred from complete mitochondrial genomes. Mol Biol Evol. 28:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet M. 2010. Two new Mio-Pliocene Chadian hominids enlighten Charles Darwin’s 1871 prediction. Philos Trans R Soc Lond B Biol Sci. 365:3315–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows W, Ryder OA. 1997. Y-chromosome variation in great apes. Nature. 385:125–126 [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. New York: Springer Verlag. p. 207–232 [Google Scholar]
- Clifford SL, Anthony NM, Bawe-Johnson M, Abernethy KA, Tutin CE, White LJ, Bermejo M, Goldsmith ML, McFarland K, Jeffery KJ, et al. 2004. Mitochondrial DNA phylogeography of western lowland gorillas (Gorilla gorilla gorilla). Mol Ecol. 13:1551–1565, 1567 [DOI] [PubMed] [Google Scholar]
- Coolidge HJ, Jr. 1933. Pan paniscus. Pigmy chimpanzee from south of the Congo River. Am J Phys Anthopol. 18:1–59 [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douadi MI, Gatti S, Levrero F, Duhamel G, Bermejo M, Vallet D, Menard N, Petit EJ. 2007. Sex-biased dispersal in western lowland gorillas (Gorilla gorilla gorilla). Mol Ecol. 16:2247–2259 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Pin L, Melnick DJ, Wright SI. 2010. Sex-linked inheritance in macaque monkeys: implications for effective population size and dispersal to Sulawesi. Genetics. 185:923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Available from: http://evolution.genetics.washington.edu/phylip.html
- Fünfstück T, Arandjelovic M, Morgan DB, Sanz C, Breuer T, Stokes EJ, Reed P, Olson SH, Cameron K, Ondzie A, et al. 2014. The genetic population structure of wild western lowland gorillas (Gorilla gorilla gorilla) living in continuous rain forest. Am J Primatol. Advance Access published April 3, 2014, 10.1002/ajp.22274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner KJ, Ryder OA. 1996. Mitochondrial DNA diversity in gorillas. Mol Phylogenet Evol. 6:39–48 [DOI] [PubMed] [Google Scholar]
- Groves CP. 1967. Ecology and taxonomy of the gorilla. Nature. 213:890–893 [DOI] [PubMed] [Google Scholar]
- Groves CP. 1971. Distribution and place of origin of the gorilla. Man. 6:44–51 [Google Scholar]
- Groves CP. 1996. Do we need to update the taxonomy of gorillas? Gorilla J. June:3–4 [Google Scholar]
- Groves CP. 2001. Primate taxonomy. Washington and London: Smithsonian Institution Press [Google Scholar]
- Groves CP. 2003. A history of gorilla taxonomy. In: Taylor AB, Goldsmith ML, editors. Gorilla biology: a multidisciplinary perspective. Cambridge: Cambridge University Press. p. 15–34 [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- Harcourt AH. 1978. Strategies of emigration and transfer by primates, with particular reference to gorillas. Z Tierpsychol. 48:401–420 [DOI] [PubMed] [Google Scholar]
- Harrison T. 2010. Anthropology. Apes among the tangled branches of human origins. Science. 327:532–534 [DOI] [PubMed] [Google Scholar]
- Ho SYW. 2007. Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol. 38:409–414 [Google Scholar]
- Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. 1995. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci USA. 92:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue E, Akomo-Okoue EF, Ando C, Iwata Y, Judai M, Fujita S, Hongo S, Nze-Nkogue C, Inoue-Murayama M, Yamagiwa J. 2013. Male genetic structure and paternity in western lowland gorillas (Gorilla gorilla gorilla). Am J Phys Anthropol. 151:583–588 [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI. 2000. Evolutionary genetics of gorillas [PhD dissertation]. New Haven (CT): Yale University [Google Scholar]
- Jensen-Seaman MI, Kidd KK. 2001. Mitochondrial DNA variation and biogeography of eastern gorillas. Mol Ecol. 10:2241–2247 [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Deinard AS, Kidd KK. 2001. Modern African ape populations as genetic and demographic models of the last common ancestor of humans, chimpanzees, and gorillas. J Hered. 92:475–480 [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Deinard AS, Kidd KK. 2003. Mitochondrial and nuclear DNA estimates of divergence between western and eastern gorillas. In: Taylor ML, Goldsmith AB, editors. Gorilla biology: a multidisciplinary perspective. Cambridge: Cambridge University Press; p. 247–268 [Google Scholar]
- Jensen-Seaman MI, Sarmiento EE, Deinard AS, Kidd KK. 2004. Nuclear integrations of mitochondrial DNA in gorillas. Am J Primatol. 63:139–147 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S. 2001. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet. 27:155–156 [DOI] [PubMed] [Google Scholar]
- Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Pääbo S. 2010. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464:894–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- Mailund T, Halager AE, Westergaard M, Dutheil JY, Munch K, Andersen LN, Lunter G, Prüfer K, Scally A, Hobolth A, et al. 2012. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet. 8:e1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Fu Q, Aximu-Petri A, Glocke I, Nickel B, Arsuaga JL, Martínez I, Gracia A, de Castro JM, Carbonell E, et al. 2014. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 505:403–406 [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science. 338:222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MW, Gonder MK. 2013. Primate speciation: a case study of African apes. Nat Edu Know. 4:1 [Google Scholar]
- Morell V. 1994. Will primate genetics split one gorilla into two? Science. 265:1661. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature. 499:471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. 2005. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J Hum Evol. 48:237–257 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. 2009. Tracer v1.5. Available from: http://beast.bio.ed.ac.uk/Tracer
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo M, Pan D, Zehr S, Goldberg T, Disotell TR, von Dornum M. 1994. Gene trees and hominoid phylogeny. Proc Natl Acad Sci USA. 91:8900–8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K, Amato G, Powell J. 1998. Mitochondrial DNA variability in Grauer’s gorillas of Kahuzi-Biega National Park. J Hered. 89(2):129–135 [DOI] [PubMed] [Google Scholar]
- Sarmiento EE, Oates JF. 2000. The Cross River gorillas: a distinct subspecies, Gorilla gorilla diehli Matschie 1904. Am Mus Novitates. 3304:1–55 [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature. 483:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß R, Van Bocxlaer B, Riedel F, von Rintelen T, Albrecht C. 2014. Disjunct distributions of freshwater snails testify to a central role of the Congo system in shaping biogeographical patterns in Africa. BMC Evol Biol. 14:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 23:7–9 [DOI] [PubMed] [Google Scholar]
- Soto-Calderón ID. 2012. Evolution of nuclear integrations of the mitochondrial genome in Great Apes and their potential as molecular markers [PhD dissertation]. [New Orleans (LA)]: University of New Orleans [Google Scholar]
- Stankiewicz J, de Wit MJ. 2006. A proposed drainage model for Central Africa—did the Congo flow east? J Afr Earth Sci. 44:75–84 [Google Scholar]
- Stone AC, Battistuzzi FU, Kubatko LS, Perry GH, Jr, Trudeau E, Lin H, Kumar S. 2010. More reliable estimates of divergence times in Pan using complete mtDNA sequences and accounting for population structure. Philos Trans R Soc Lond B Biol Sci. 365:3277–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann O, Hebler J, Poinar HN, Pääbo S, Vigilant L. 2004. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol Ecol. 13:321–335 [DOI] [PubMed] [Google Scholar]
- Thalmann O, Fischer A, Lankester F, Pääbo S, Vigilant L. 2007. The complex evolutionary history of gorillas: insights from genomic data. Mol Biol Evol. 24:146–158 [DOI] [PubMed] [Google Scholar]
- Thalmann O, Serre D, Hofreiter M, Lukas D, Eriksson J, Vigilant L. 2005. Nuclear insertions help and hinder inference of the evolutionary history of gorilla mtDNA. Mol Ecol. 14:179–188 [DOI] [PubMed] [Google Scholar]
- Thalmann O, Wegmann D, Spitzner M, Arandjelovic M, Guschanski K, Leuenberger C, Bergl RA, Vigilant L. 2011. Historical sampling reveals dramatic demographic changes in western gorilla populations. BMC Evol Biol. 11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauth MH, Larrasoaña JC, Mudelsee M. 2009. Trends, rhythms and events in Plio-Pleistocene African climate. Quat Sci Rev. 28:399–411 [Google Scholar]
- Trier CN, Hermansen JS, Sætre GP, Bailey RI. 2014. Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid Italian sparrow and its parent species. PLoS Genet. 10:e1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RD, Steiper ME, Soligo C, Martin RD, Yang Z, Tavaré S. 2011. Dating primate divergences through an integrated analysis of palaeontological and molecular data. Syst Biol. 60:16–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood B, Lonergan N. 2008. The hominin fossil record: taxa, grades and clades. J Anat. 212:354–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Arnason U. 1996. A complete sequence of the mitochondrial genome of the western lowland gorilla. Mol Biol Evol. 13:691–698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.