Abstract
Phenotypic switching from the white to the opaque phase is a necessary step for mating in the pathogenic fungus Candida albicans. Suppressing switching during vascular dissemination of the organism may be advantageous, because opaque cells are more susceptible to host defenses. A repressor of white-opaque switching, HBR1 (hemoglobin response gene 1), was identified based on its specific induction following growth in the presence of exogenous hemoglobin. Deletion of a single HBR1 allele allowed opaque phase switching and mating competence, accompanied by a lack of detectable MTL α1 and α2 gene expression and enhanced MTLa1 gene expression. Conversely, overexpression of Hbr1p or exposure to hemoglobin increased MTLα gene expression. The a1/α2 repressed target gene CAG1 was derepressed in the same mutant in a hemoglobin-sensitive manner. Regulation of CAG1 by hemoglobin required an intact MTLa1 gene. Several additional Mtlp targets were perturbed in HBR1 mutants in a manner consistent with commitment to an a mating phenotype, including YEL007w, MFα, HST6, and RAM2. Therefore, Hbr1 is part of a host factor-regulated signaling pathway that controls white-opaque switching and mating in the absence of allelic deletion at the MTL locus.
Candida albicans is both a commensal in the human gastrointestinal tract and a virulent pathogen that colonizes specific host organs and causes disseminated vascular infections (39). Adaptation to each host compartment may require recognition of spatial and temporal cues that induce reversible or heritable changes in phenotype. The latter are accomplished in C. albicans, despite its lacking a meiotic cycle, by nonmeiotic mating between diploid cells (12, 21, 35).
The C. albicans mating-type-like locus (MTL) has a genomic structure that is similar but not identical to that of the Saccharomyces cerevisiae MAT locus (20, 61). Both encode the transcriptional regulators a1, α1, and α2, but control of their mating regulatory circuits differs significantly. Mating in C. albicans is carried out between diploid mating partners, while in S. cerevisiae a and a cells are the products of meiosis. Functional a and α cells in C. albicans have been generated only through directed deletion of MTL genes or loss of an entire chromosome containing an MTL locus (21, 35). Thus, genomic rearrangements at the MTL locus are proposed to be the primary mechanism for generating mating-competent cells. This was supported by the deletion of MTL alleles in some isolates from mammals and clinical specimens. Second, C. albicans possesses a fourth gene in its mating locus, a2 (61). This gene product, as well as the α1 gene product, acts as a positive regulator of some genes required for their respective mating cell-type specificity. Third, a unique morphological change is required for C. albicans mating. Cells must convert from the typical yeast form to an elongated, opaque cell for high-efficiency mating. Opaque cells mate with a 106-fold-higher frequency than white cells (36).
White-opaque switching is one of several known processes that permit reversible changes in cellular morphology without detectable genomic rearrangements (51, 52, 56). In the white-opaque phase transition, cells switch between oval budding cells with smooth cell walls to opaque colonies of elongated cells with surface protrusions known as pimples (3). Opaque cells can be easily distinguished as red colonies on modified Lee's agar containing phloxine B (3). The opaque phenotype in the prototypical switching strain WO-1 (52) results from allelic loss of MTLa1 (31). Consistent with the model of switching regulated through the a1/α2 dimer (22, 36, 55), disruption of either the MTLa1 or the MTLα2 allele results in high-frequency white-opaque phase switching (ca. 10−3 per generation).
White-opaque switching alters phenotypic characteristics both in vivo and in vitro. Opaque cells have altered antigenic and adherence properties (2, 24), are more sensitive than white cells to destruction by neutrophils and oxidants (27), and differentially regulate metabolic genes (29). Switching occurs at some sites of C. albicans infection (28, 53), indicating that it is a normal component of the fungal life cycle within the host. Considering the vulnerability of opaque cells to host defenses, however, a mechanism to selectively suppress switching may offer a survival advantage when C. albicans enters the bloodstream.
The homeodomains of the a1 and α2 proteins are conserved in C. albicans and function similarly to the MAT a1 and α2 proteins in determining cell fate (20). In diploid S. cerevisiae cells, the a1/α2 dimer represses haploid-specific genes (hsg) (17). The α2 gene product acts as a corepressor in the regulation of a genes. The C. albicans a1/α2 dimer represses a subset of those genes repressed by MAT a1/α2 in S. cerevisiae (61). Intriguingly, CAG1, the C. albicans ortholog of the hsg SCG1, which encodes the α-subunit of the mating-specific G-protein complex, can functionally replace its yeast counterpart. The cell type-specific regulation of CAG1 in S. cerevisiae is also consistent with its repression by the Mat a1/α2 dimer in diploid cells (48). Indeed, a transcriptional reporter using the predicted hsg operator sequences from the CAG1 promoter demonstrated that C. albicans has an a1/α2 transcriptional repressor activity that requires the MTLa1 gene (20).
We have now identified a suppressor of white-opaque switching that was isolated based on its specific induction following exposure of cells to hemoglobin. Hemoglobin is a host factor that regulates expression of cell surface receptors for fibronectin, laminin, and fibrinogen (64, 65) through a low-affinity, multivalent hemoglobin receptor (41). Hemoglobin induces increased adhesion to endothelial cells (64), and responsiveness to hemoglobin is conserved in other pathogenic species of the Candida genus (45). Therefore, hemoglobin may be an important environmental signal for pathogenesis of C. albicans in a mammalian host.
To define molecular mechanisms for these phenotypic alterations, we identified genes that are transcriptionally regulated in response to hemoglobin (40, 42). We show here that modulating the expression of one of these genes, HBR1, by disruption of a single allele leads to high-frequency white-opaque switching and mating in a wild-type a/α MTL background. We further show that Hbr1p suppresses phenotypic switching through stimulation of MTLα gene expression and regulation of known Mtl target genes, including CAG1.
MATERIALS AND METHODS
Cell culture conditions.
C. albicans strains were routinely cultured in yeast nitrogen base (YNB) with ammonium sulfate, 2% glucose, low methionine, and appropriate supplements (50) with shaking at 250 rpm at 30°C. Bovine methemoglobin was added to cell cultures at 0.5 mg/ml and was prepared as previously described (41). Modified Lee's medium (8) containing 5 μg of phloxine B (Sigma, St. Louis, Mo.)/ml was used for identification of white and opaque switching cells. Switching frequencies were determined using phloxine B plates as described elsewhere (52).
General nucleic acid manipulations.
Total yeast RNA was prepared using the hot acid phenol method (26). The 5′ end of the HBR1 mRNA was mapped using primer PN10 and primer extension using 50 μg of total RNA from 44807 cells grown with 1 mg of hemoglobin/ml for 4 h (6). Genomic DNA was isolated as described previously (18). Genomic PCR analysis for the presence of MTL genes (47) and λ imm434 (35) used the primers as referenced. All cloned PCR products were sequenced to ensure no errors had occurred during amplification (6). Yeast transformations were carried out by the lithium acetate technique modified for C. albicans (62). The HBR1 DNA sequence from C. albicans strains CAI-4 and B311 was determined by standard methods (6). The C. albicans C9 genomic library (15) was obtained from the NIH AIDS Research and Reference Reagent Program.
Plasmid construction.
A full-length HBR1 gene fragment was synthesized by PCR amplification of the SY1 genomic clone (42) containing CaHMX1 and HBR1, using primers P13 and PN28 and cloned into pCR-Blunt II TOPO (Invitrogen). The cloned fragment was then excised by BamH1-PstI digestion and inserted via identical sites into plasmid pCaDis to form pDisCat4 or into pCaExp to form pExpCat4-9. Plasmids pCaDis and pCaExp have been described previously (11). A transcriptional fusion of the HBR1 promoter and the Renilla luciferase gene was created using Pfx polymerase (Invitrogen) amplification of a 502-bp HBR1 promoter fragment, using primers P24 and P27 with the SY1 genomic clone as a template. This fragment was digested with SalI and cloned into SmaI-SalI-digested pUC19 to create pPT1-UC. The promoter region product was inserted as a PstI-KpnI fragment into PstI-KpnI-cleaved pCRW3 (58) to create pPT502.
Strain construction and gene disruption methods.
Strains used in this study are listed in Table 1. HBR1 and MTL gene disruptions were accomplished using the plasmid templates pRS-ARG4 ΔSpe1, pGEM-HIS1, and pGEM-URA3 (63) and the following primer sets: for HBR1, P20 and P21, PN198 and PN199; for MTLa1, PN108 and PN109; for MTLα1, PN110 and PN111 (Table 2). An ARG4 cassette was used to disrupt the first allele of HBR1 in strain BWP17, using primers P20 and P21. A total of 30 Arg+ isolates were identified, and insertion into the HBR1 gene was confirmed in four of these. Strain CAMP 63 was constructed by integration of plasmid pExpCAT4-9 digested at the unique NcoI site into the RP10 gene in strain CAMP R8 and identified by Arg+ Ura+ selection (11). The correct integration site was determined by PCR using primers PN51 and PN202 to detect RP10 integration and primers PN51 and PN52 to detect the presence of the HBR1-MET3 promoter fusion (data not shown). The second HBR1 allele in strain CAMP 63 was targeted by a HIS1 mutation cassette using primers PN198 and PN199, creating strain CAMP 61, or primers P20 and P21, creating CAMP 62. Disruption of the second HBR1 allele was verified by PCR using primers P24 and PN203. URA3 disruption cassettes were used to generate MTLa1 and MTLα1 deletions and were confirmed by Southern analysis using gene-specific DNA probes (data not shown) and by reverse transcription-PCR (RT-PCR) analysis using MTLa1- and MTLα1-specific primers (see above; also see Fig. 4B, lane 1, below). Strain CAMP 35 was constructed using HindIII digestion of pPT502 to direct integration to an ade2 allele of strain Red 3/6 (58).
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype or description | Reference |
|---|---|---|
| CAF2-1 | URA3/ura3::λ imm434 | 14 |
| CAI-4 | ura3::λ imm434/ura3::λ imm434 | 14 |
| CAI-8 | As for CAI-4, except ade2::hisGlade2::hisG | 14 |
| BWP17 | As for CAI-4, except his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 63 |
| CHY477 | ura3/ura3 mtla1::hisG/MTLα1α2 ade2::hisG3URA3hisG/ade2::hisG | 36 |
| MM278 | ura3/ura3 MTLa1/mtlα1::hisG mtlα2::hisG ade2::hisGURA3 hisG/ade2::hisG | 36 |
| CAMP R8 | As for BWP17, except HBR1/hbr1::ARG4 | This study |
| CAMP 43 | As for CAMP-R8, except RP10/rp10::pCaExp-URA3 his1::hisG/his1::pGEM-HIS1 | This study |
| CAMP 45 | As for CAMP-R8, except mtlα1::URA3 his1::hisG/his1::pGEM-HIS1 | This study |
| CAMP 47 | As for CAI-4, except mtlα1::URA3 | This study |
| CAMP 49 | As for CAMP R8, except mtla1::URA3 | This study |
| CAMP 48 | As for CAI-4, except mtla1::URA3 | This study |
| CAMP 51 | As for CAI-8, except mtla1::URA3 | This study |
| CAMP 61 | hbr1::ARG4/hbr1::HIS1 RP10/rp10::pExpCAT4-9-URA3 | This study |
| CAMP 62 | hbr1::ARG4/hbr1::HIS1 RP10/rp10::pExpCAT4-9-URA3 | This study |
| CAMP 63 | As for CAMP-R8, except RP10/rp10::pCaExp-URA3 his1::hisG/his1::pGEM-HIS1 | This study |
| Red 3/6 | As for WO1, except ade2/ade2 | 58 |
TABLE 2.
Primers synthesized for this study
| Primer | Sequence (5′ to 3′) | Use or reference |
|---|---|---|
| PN157 | ATGGCCGTTCTTAGTTGGTGGAGT | Ribosomal RNA |
| PN158 | GTAGTAGCGACGGGCGGTGTG | Pair with PN157 |
| P24 | CTTGCAACGTATCTCTTGGC | HBR1 promoter cloning |
| P27 | GGTTGTCGACGTGATGAGATGTGC | Pair with P24 |
| P26 | AACATTTGTGGTGAACAAGGATG | Actin ORF |
| PN91 | TGATGGTGTTACTCACGTTGTTCC | Pair with P26 |
| PN90 | TGGCTAACTTCAATGTATCTGTTC | Intron, pair with PN91 |
| PN36 | GAGTTGCGTTCAGACACAGC | HBR1 ORF internal |
| PN37 | AAACTTTCGGGTACTTGGACATA | Pair with PN36 |
| PN7 | GAACGATTGTGTGGTCCAG | Phosphoglycerate kinase |
| PN8 | GCAGATTTGACAGCAGCTACC | Pair with PN7 |
| PN10 | GTTCTGTTTGTTGGGAATTCAGC | HBR1 5′ transcript |
| P13 | CGCGCGGATCCATGACAACCATGTCAAGAA | HBR1 gene (5′) |
| PN28 | GGTACCTGCAGATATCTATTGTGCAATATCTTC | HBR1 gene (3′) |
| PN114 | TATATGGGGTAAAGATGACGAT | CAG1 ORF |
| PN115 | AGATAGCCAAAACAAATAAAACAG | Pair with PN114 |
| P20 | TCATTCCCTGAGTTTAGTTTCTCAACTCAATCAAACTCTTGGTAAAGAGACGGTTTTCCCAGT CACGACGTT | HBR1 disruption |
| P21 | AGAGCTTATTCTATCAACGTTTTCATCCATCTCTTCAGCTGTGTCTGAACGCTGTGGAATTGTG AGCGGATA | Pair with P20 |
| PN108 | GTATGTCACCGTGTTTAGCTAATATGATCTTGAATAAAAGAAAACGAATAGTTTTCCCAGTCA CGACGTT | MTLa1 disruption |
| PN109 | GGCTAGGTTGAATTTGAACTTGATTTTGTTTCGTTTGGGTTCCTTCTGTGGAATTGTGAGCGG ATA | Pair with PN108 |
| PN110 | CGAGTACATTCTGGTCGCGATGCTCCAAGAAGAGACACAAGAGAAGTTCAAAAGTTTTCCCA GTCACGACGTT | MTLα1 disruption |
| PN111 | CCAGTCCACAAAATTCAATTTTGCATCAGGAAGAAGTAAATAATTATTGTGTGGAATTGTGAG CGGATA | Pair with PN111 |
| PN198 | CGAAGACAAATTGCTAGACTCGTTAGAGCCTGATTTGGAAAAGGGGGAGTTTTCCCAGTCAC GACGTT | HBR1 disruption |
| PN199 | CAGCTGTGTCTGAACGCAACTCAATTACTATGTCAGGAATATAGCTGTCCTGTGGAATTTGTA GCGGATA | Pair with PN 198 |
| ACT1 | AAGAATTGATTTGGCTGGTAGAGA, TGGCAGAAGATTGAGAAGAAGTTT | qPCR primer pair |
| MTLα1 | AATTAGCGGGATGTTTGGACTCA, CTATCTGGGGCGTTGTATTATCA | qPCR primer pair |
| MTLα2 | ATTATGTTGCAGCAGGATTCA, GATACGGATGGTTCTTGTGTTT | qPCR primer pair |
| CDC36 | GAGCGTCCAGTATAAATCCACCAC, TCAAGACGGGCTCCACATTACTAT | qPCR primer pair |
| HST6 | AAGCTACCGGATGGCGATTAC, AAAACACCGGACTTGATACACCTT | qPCR primer pair |
| CTG1 | AAAAGGGAAGAATTAAGACTACTGG, ATTCTATTTACCCGTTCATCTTCA | qPCR primer pair |
| HBR1 | TGAAATAGCAAAGGAAAGAGACTG, AATATCACAACAATGCCAATCAAC | qPCR primer pair |
| RAM2 | TTTGGCCACCGATAATAC, TTTTTGCCAATGTCTCCA | qPCR primer pair |
| MFα | AGAATCTGCCGTTGAAGC, AGCATCGGCGTTAGCATC | qPCR primer pair |
| MTLa1 | TAATAAAAGGGGAGGAAATAAA, TTGGGAAGGCTAACACC | qPCR primer pair |
| YEL007w | GAGGCGTGCTTATAGTTTCTGG, ACGCTTCTTTTTCTTCTTCTTGTC | qPCR primer pair |
| STE3 | CGACGGGTATTCCCAAGAG, TGCATAACATCGCCAAACTG | qPCR primer pair |
FIG. 4.
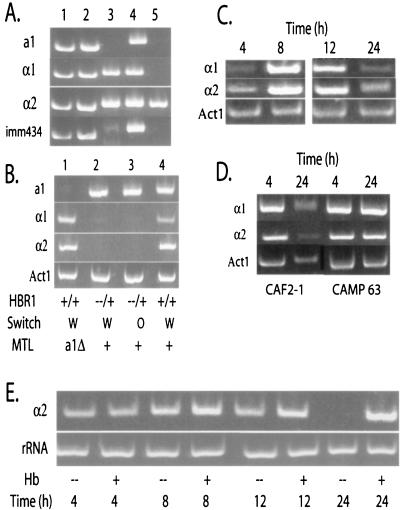
Hbr1p is a positive regulator of MTLα expression. (A) HBR1 heterozygosity does not lead to MTL gene deletions. Results of PCR analysis of genomic DNA using primers specific for MTL genes and for the λimm434 region used in construction of parental strain CAI-4 (14) are shown. Lanes 1 and 2, CAMP43, white and opaque, respectively; lane 3, Red 3/6, white; lane 4, CAI-4, white; lane 5, clinical isolate 156, white. (B) MTL α gene expression is not detectable in the HBR1 heterozygote, as shown by an RT-PCR analysis of RNA isolated from 4-h exponential-phase cells cultured in YNB-glucose medium at 30°C. Gene-specific primers listed on the left were used in PCRs of 26 cycles (MTL genes) or 20 cycles (ACT1). Lane 1, strain CAMP 48; lanes 2 and 3, strain CAMP 43; lane 4, strain CAI-4. (C) MTLα genes are induced only during exponential growth. Results of the RT-PCR analysis of RNA isolated from CAF2-1 cells at the indicated times after transfer of stationary-phase cells to YNB-glucose medium are shown. (D) HBR1 overexpression sustains MTLα expression into early stationary phase. Results of RT-PCR analysis of RNA harvested at 4 and 24 h after transfer of stationary-phase cells to low-methionine medium are shown. CAMP 63 contains an HBR1 copy under the control of the MET3 promoter. The MET3 promoter maintained HBR1 RNA at higher levels in the CAMP 63 strain than in strains containing only the native HBR1 promoter (see Fig. 6B). (E) Hemoglobin can sustain MTLα gene expression into stationary phase. Strain CAF2-1 cells were cultivated in the presence or absence of 0.5 mg of hemoglobin/ml, and RNA was harvested at the indicated times. Primer sets for RT-PCR analysis are listed in the figure. MTLα, 26 cycles; rRNA, 15 cycles.
Essential gene analysis.
To test for the essentiality of HBR1, two isolates each of strains CAMP 63, CAMP 61, and CAMP 62 were grown for 3 days in nonselective medium (YPD with uridine) at 30°C. A total of 109 cells from each strain were plated on 5-fluoroorotic acid agar and incubated at 30°C for 3 days. The number of CAMP 63 URA− colonies per number of cells plated was taken as the frequency of excision of the pExpCat4-9 plasmid occurring spontaneously during unselected growth. The absence of Ura− colonies of strain CAMP 61 and CAMP 62 would indicate that the Arg-His mutations at the HBR1 loci interfere with the essential function of this gene. The presence of colonies would indicate that the gene is not essential.
Quantitative mating analysis.
Opaque cells of strains CAMP R8 (Ade+ Ura−) and CAMP 51 (Ade− Ura+) were identified and isolated from phloxine B plates as described above. Mating between these two strains was carried out using cells grown to an optical density at 600 nm of 1.5 in YPAD medium at 25°C in a 4-h mating procedure as described previously (36). Mating frequency was determined in four independent experiments as follows: (number of Ura+ Ade+ conjugates/(number of Ura+ Ade+ conjugates + total limiting parent).
RT-PCR analysis.
RT-PCR utilized 5 μg of total RNA, and first-strand cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen, Carlsbad, Calif.). Quantification of cDNA by PCR used the following primer sets: P26 and PN91 for ACT1 (both contained within the distal exon); PN36 and PN37 for HBR1; and PN114 and PN115 for CAG1 (Table 2). MTLa1, MTLα1, and MTLα2 (47) and phase-specific WH11, SAP1, and OP4 (36) primers have been described elsewhere. To detect DNA contamination, a PCR primer within the ACT1 intron sequence (P26) and the other within the distal exon (PN90) were used. Cycling parameters were as follows: initial denaturation, 94°C for 2 min; 15 to 26 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min. RT-PCR products were visualized by electrophoresis using 2% agarose ethidium bromide gels (E-Gel; Invitrogen) and digital photography (Kodak, Rochester, N.Y.).
Quantitative real-time RT-PCR (qPCR).
Steady-state mRNA was quantified using real-time SYBR green fluorescence detection using a DNA Engine Opticon I continuous fluorescence detection system (MJ Research, Waltham, Mass.) and the Dynamo SYBR PCR kit (Finnzymes Oy). PCR primers were designed to amplify products of 100 to 180 bp using Primer Select software (DNASTAR, Madison, Wis.) with PCR settings of 95°C for 5 min, 40 cycles of 94°C for 15 s, 51 to 58°C for 15 s, and 72°C for 20 s. Melting curves were generated for each sample at the end of each run to serve as a validation of the purity of the amplified product. Each assay was performed in 96-well plates in duplicate using identical cDNA stocks for each primer set. Typically, for 5 μg of total mRNA reverse transcribed, the total volume of the reaction mixture was taken up to 100 μl with H2O, and 4.5 μl of cDNA was used per well. Each cDNA preparation was normalized using the CDC36 gene as an internal control, and each plate included this control to ensure plate-to-plate uniformity. Each RNA sample was derived from a cell culture grown for 4 h at 30°C and was prepared in an identical manner each time. Batch-to-batch variation of CDC36 was tested for the three strains YJB6284, CAMP 43, and BWP17 grown under these standard conditions and was less than 5%. Ct values ± standard deviations (SD) for a test analysis were as follows (n = 16): 20.83 ± 0.27, 21.00 ± 0.30, and 20.42 ± 0.21. Primer sequences are listed in Table 2.
Renilla luciferase assays.
C. albicans cells were grown at 30°C in minimal YNB medium with ammonium sulfate and 2% glucose. Typically, 5 × 107 cells were harvested, and cell extracts were obtained by glass bead lysis (58). Luminescence was determined using a commercial substrate (Promega, Madison, Wis.), and luciferase activity is reported as light units per 5 × 104 cells.
Scanning electron microscopy.
CAMP 43 opaque cells grown in modified Lee's medium overnight at 25°C were fixed in 4% formaldehyde-2% glutaraldehyde in phosphate-buffered saline at 4°C. Samples on carbon-coated coverslips were fixed with 1% OsO4, dehydrated, and treated with tetramethylsilane (13). After coating with gold-palladium, the cells were observed with a Hitachi S-570 scanning electron microscope operated at 10 kV.
Protein sequence analysis.
Proteins related to the human ortholog of Hbr1 (adrenal gland protein AD-004) were identified by BLINK analysis (http://www.ncbi.nlm.nih.gov) with a cutoff of 100. Sequences for the 19 unique gene products obtained were aligned using CLUSTAL W (60). The aligned sequences were analyzed using the PHYLIP phylogeny programs (J. Felsenstein, PHYLIP Phylogeny Inference package, version 3.5c.). The distance matrix data were analyzed using Fitch-Margoliash and least-squares methods (FITCH program). Sequence data for C. albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida.
Nucleotide sequence accession number.
The nucleotide sequence for HBR1 was deposited in GenBank under accession number AF466197 (release date 1 April 2002).
RESULTS
Identification of HBR1 and its regulation by hemoglobin.
We used random arbitrarily primed PCR to identify C. albicans genes that are transcriptionally altered in cells cultured with hemoglobin (42). One EST obtained from this analysis hybridized to a genomic DNA fragment containing two genes that were induced under these conditions (42) (Fig. 1A). One gene encodes a heme oxygenase (CaHMX1) that catalyzes conversion of heme to α-biliverdin (40). Immediately adjacent to CaHMX1, we identified a second open reading frame that was designated HBR1 (Hb-regulated gene 1) (Fig. 1A).
FIG. 1.
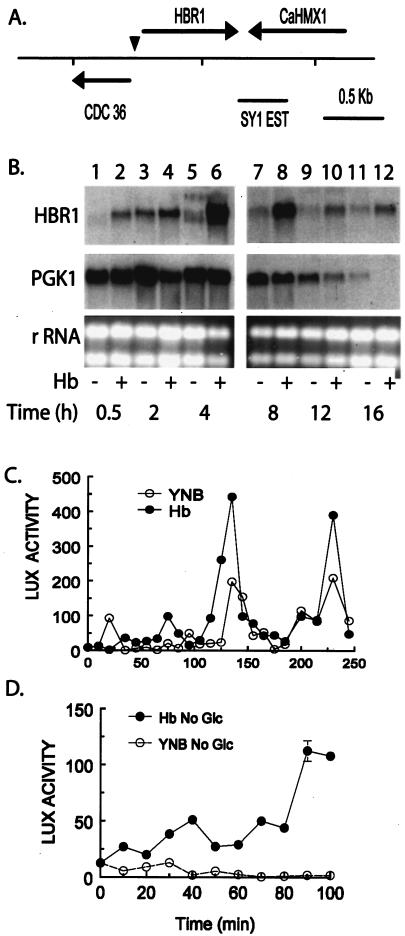
Hemoglobin and growth signals control HBR1 expression. (A) HBR1-CDC36 region in C. albicans. The SY1 EST used to identify the HBR1 genomic clone overlaps the 3′ end of CaHMX1, a predicted heme oxygenase (5, 49). The arrowhead indicates the mapped transcription start. This orientation and positioning of CDC36 with HBR1 is conserved with the ortholog FAP7 in S. cerevisiae. (B) Hemoglobin increases HBR1 basal expression. C. albicans 44807 cells were grown with and without 500 μg of hemoglobin/ml at 30°C, and RNA was isolated for Northern analysis. The same blot was sequentially hybridized with DNA probes of HBR1 and PGK1. rRNA was used as a loading control (ethidium bromide-stained gel). RNA isolated from strain CAF2-1 gave similar results (data not shown). (C) Hemoglobin and glucose stimulate periodic HBR1 transcriptional activity. CAMP 35 cells were transferred to YNB medium containing 2% glucose with (•) or without (○) 500 μg of hemoglobin/ml and sampled at the indicated times for luciferase activity. This assay was repeated four times with similar results each time. (D) Hemoglobin and glucose signaling to HBR1 are separable. CAMP 35 cells were transferred to YNB medium lacking glucose with (•) or without (○) hemoglobin as indicated in panel C. Luciferase activity (LUX) is reported as light units per 5 × 104 cells.
Regulation of HBR1 expression was characterized by Northern analysis of RNA extracted from C. albicans cells grown with and without hemoglobin to different stages of exponential and early stationary phase (Fig. 1B). Expression of HBR1 was minimal in stationary-phase cells. Within 0.5 h after transferring cells from stationary phase into fresh medium, hemoglobin induced HBR1 expression over basal levels, and this was further increased about threefold by 4 h. In separate experiments, steady-state HBR1 mRNA levels determined by qPCR at 4 h following hemoglobin addition were increased twofold relative to cells at the same growth phase without hemoglobin, consistent with the Northern analysis (Table 3).
TABLE 3.
Quantitative RT-PCR analysis of gene expression
| Gene |
Ct (± SD)a for strain
|
|
|---|---|---|
| CAF2-1 | CAMP 43 | |
| ACT1 | 12.9 ± 0.2 | 12.3 ± 0.33 |
| HBR1 | 25.6 ± 0.16 | 26.4 ± 0.44 |
| HBR1/Hbb | 24.1 ± 0.7 | 25.2 ± 0.09 |
| CDC36 (n = 16) | 20.8 ± 0.27 | 20.4 ± 0.21 |
| MTLa1 | 29.5 ± 0.1 | 27.5 ± 0.5 |
| MTLα1 | 26.2 ± 0.16 | >40 |
| MTLα2 | 23.1 ± 0.24 | >40 |
n = 4 except where indicated.
Bovine methemoglobin, 0.5 mg/ml (see Materials and Methods).
Expression levels of both basal and hemoglobin-induced HBR1 mRNA were maximal during the early stages of exponential growth and declined at later times (Fig. 1B). PGK1 mRNA levels were uniform during the period of maximal HBR1 mRNA accumulation and declined to detection limits by 12 h (Fig. 1B) as the cell doubling time increased from 1.4 to 1.8 h (data not shown). Both changes indicated the transition to diauxic growth (1). HBR1 basal mRNA levels also declined during this transition period, but HBR1 steady-state mRNA was maintained by hemoglobin at detectable levels until 16 h (Fig. 1B). Thus, HBR1 expression depends upon cell proliferation, and hemoglobin causes a two- to threefold increase in accumulation of steady-state mRNA levels that persists after basal levels have declined.
The major transcriptional start site of HBR1 was mapped to a C residue at position −149 (data not shown). We analyzed HBR1 transcription by fusing upstream regions with a Renilla luciferase (Rlux) reporter and integrating a single copy in C. albicans cells (58). An Rlux reporter construct containing 502 bp upstream from the predicted HBR1 AUG start codon (strain CAMP 35) showed optimal responses to both proliferation and hemoglobin (data not shown). Following pregrowth for 48 h in glucose-containing medium, transfer of cells into new medium containing glucose but lacking hemoglobin resulted in a burst of HBR1-Rlux reporter activity within 20 min (Fig. 1C). Reporter activity rapidly returned to basal levels but was reproducibly followed by activity peaks at 130 and 230 min as the cells were allowed to grow (Fig. 1C). Transfer to medium lacking glucose did not result in activity above baseline, indicating that glucose is necessary for increased basal transcription (Fig. 1D). Hemoglobin addition increased the magnitude of these peaks 1.5- to 2-fold in the presence of glucose (Fig. 1C), and hemoglobin increased transcription in the presence or absence of glucose (Fig. 1D). These data indicate that HBR1 transcription increases during cell proliferation and that signaling from exogenous hemoglobin (40) also regulates HBR1 transcription.
The HBR1 DNA sequence derived from our genomic clone exactly matched the Stanford genome database sequence (http://www-sequence.stanford.edu/group/candida). The predicted open reading frame consists of 747 bases, encoding a hypothetical protein of 248 amino acids. The protein sequence contains a predicted nucleotide-binding fold (P-loop) at the amino terminus and a consensus SUMO site, but no other functional motifs were identified (Fig. 2). Hbr1p has 68% amino acid identity with S. cerevisiae Fap7p and 62% DNA sequence identity. Although many genes are conserved between these two species, the relative positioning of genes on chromosomes is typically not conserved (25). However, both HBR1 and FAP7 share 5′ untranslated regions with the adjacent gene CDC36 and are divergently transcribed from opposite strands (Fig. 1A). FAP7 is an essential gene in S. cerevisiae (23).
FIG. 2.
Conservation of the predicted amino acid sequences of HBR1 and FAP7. The alignment of predicted protein sequences of Hbr1p and Fap7p is shown. A consensus sequence was generated using DIALIGN 2.2 (37; http://bibiserv.techfak.uni-bielefeld.de/dialign/) with the following eukaryotic orthologs of Hbr1p: Schizosaccharomyces (CAB52884), Candida (AF466197), Saccharomyces (CAA98740), Caenorhabditis (Q09527), Drosophila (AAF58491), Oryctolagus (AAF09498), Homo sapiens (Q9Y3D8), Mus (BAB29612), Anopheles (EAA06472), and Arabidopsis (BAB10972). All uppercase residues were aligned in the analysis. Consensus residues that were highly conserved are indicated below the aligned yeast sequences. Residues 65 to 68 are the SUMO consensus (43).
Hbr1p orthologs were identified in most archaeal and eukaryotic genomes but not in any eubacteria. Evolutionary distances between these apparent Hbr1p orthologs suggested a common ancestry for this protein between eukaryotes and archaea (30). A conserved consensus sequence extending from Leu(118) through Ala(148) was identified in all orthologs (Fig. 2), although no function has been defined for any of these hypothetical proteins apart from Fap7p (23, 43). Hbr1p contains a 56-amino-acid extension containing 54% acidic residues at its C terminus (Fig. 2A). The latter feature is lacking in most Hbr1p orthologs except for a short region in S. cerevisiae Fap7p (Fig. 2). The extension is not a sequencing artifact, because identical sequences were found in C. albicans strains CAI-4 and B311 (data not shown).
Disruption of a single HBR1 allele enables white-opaque switching.
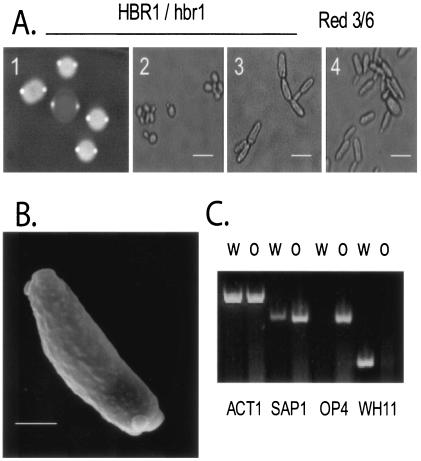
The first allele of HBR1 was mutated using an ARG4 disruption cassette in strain BWP17 and was made prototrophic by the insertion of HIS1 and URA3 genes (see Materials and Methods). The prototrophic HBR1/hbr1 strain CAMP 43, when plated on defined glucose-containing medium, generated colonies containing mixed populations of white and elongated cells resembling opaque cells (3) (data not shown). Growth on phloxine B agar plates to detect switching revealed characteristic red and white colonies that contained opaque and white cells, respectively (Fig. 3A). As shown previously for the prototypical switching strain WO-1 (3), analysis of CAMP 43 opaque cells by scanning electron microscopy revealed the characteristic pimples of opaque cells (Fig. 3B). Thus, the HBR1 heterozygote undergoes phenotypic switching that appears to yield an opaque cell.
FIG. 3.
HBR1 deletion leads to high-frequency white-opaque phase switching. (A) Prototrophic HBR1+/− strain CAMP 43 growing on phloxine B agar plates (frame 1), showing an opaque colony (center) surrounded by white colonies, from microscopic examination of CAMP 43 (frame 2, opaque; frame 3, white), and Red 3/6 (frame 4, opaque) cells. Bar, 10 μm. (B) Scanning electron micrograph of a CAMP 43 opaque cell, illustrating the surface protrusions or pimples characteristic of opaque cells. Bar, 3 μm. (C) Regulation of phase-specific genes in the HBR1 heterozygote was determined by RT-PCR analysis of RNA isolated from 4-h exponential-phase cultures of strain CAMP 43 grown at 30°C in YNB-glucose. W, white; O, opaque; ACT1, actin. Probes for phase-specific transcription were as follows: white, WH11 (59); opaque, OP4 and SAP1 (38).
Because CDC36 and HBR1 are divergently transcribed from a short common promoter region (Fig. 1A), we considered the possibility that the observed switching phenotype of the HBR1 heterozygote could result from interruption of either HBR1 or CDC36 gene function. However, qPCR analysis indicated that CDC36 mRNA levels were typically approximately 30-fold greater than HBR1 levels in wild-type cells, and the HBR1 mutation in CAMP 43 cells did not significantly alter the level of CDC36 mRNA (Table 3). This result was highly reproducible in several HBR1 mutants, indicating that disruption of an HBR1 allele does not interfere with transcription of the adjacent CDC36 gene.
The frequency of the white-opaque transition in CAMP 43 cells was similar to that reported for the prototypical switching strain WO-1 (56), to those determined for an mtla1 mutant in the CAI-4 background (CAMP 48), and independently by us for the WO-1 derivative, Red 3/6 (Table 4). Consistent with a previous report (36), disruption of MTLα1 did not increase switching (Table 4, CAMP 47), but allelic deletion of HBR1 in this background resulted in approximately 10-fold-higher switching than in a wild-type MTL background (Table 4, CAMP 43 and 45). Reintroduction of HBR1 controlled by the MET3 promoter under inducing conditions (CAMP 63) restored the white-opaque transition frequency to that of the parental CAF2-1 strain (Table 4). These data indicated that Hbr1p is a haplo-insufficient repressor of white-opaque phase switching.
TABLE 4.
White-opaque transition frequencies of C. albicans strains
| Strain | Genotype
|
Frequency | |||
|---|---|---|---|---|---|
| HBR1 | MTLα1 | MTLα2 | MTLa1 | ||
| CAMP 43 | −/+ | + | + | + | 3 × 10−3 |
| Red 3/6 | +/+ | + | + | − | 5 × 10−3 |
| CAMP 63 | −/+/+a | + | + | + | <10−5 |
| CAF2-1 | +/+ | + | + | + | <10−5 |
| CAMP 45 | −/+ | − | + | + | 5 × 10−2 |
| CAMP 47 | +/+ | − | + | + | <10−5 |
| CAMP 48 | +/+ | + | + | − | 1.2 × 10−3 |
pDisCat4 integrated as a single copy generating an HBR1 duplication.
HBR1 is an essential gene in C. albicans.
The haplo-insufficient phenotype of the single allelic knockout of HBR1 and the complementation results presented here indicated that HBR1 gene dosage is crucial to proper regulation of the switching phenotype. Our inability to generate a knockout of the second HBR1 allele by standard means (data not shown) prompted us to generate conditionally lethal HBR1 double mutants. These strains (CAMP 61 and CAMP 62) contained deletions of both HBR1 alleles with Hbr1p supplied in trans from a plasmid construct integrated into the neutral RP10 locus (11). They differed only in the primers used to construct the HIS1 mutagenic cassette (see Materials and Methods). The integrated plasmid pExpCAT4-9 in these strains contains HBR1 linked to a URA3 marker (11). Therefore, selection for URA− cells on agar containing 5-fluoroorotic acid (46) should result in the absence of any colonies if HBR1 were necessary for cell survival. A total of 109 cells from two isolates of each strain were tested and compared for the rate of marker excision from strain CAMP 63 lacking the second His insertion (Table 1). CAMP 63 cells produced Ura− colonies at a frequency 2 × 10−7, while the CAMP 61 and CAMP 62 strains produced no colonies. Therefore, the HBR1 double deletion rendered the cells nonviable, indicating that HBR1 is essential for cell survival.
Phase-specific genes confirm white-opaque switching.
Changes in cellular morphology due to the white-opaque transition are accompanied by altered transcription of phase-specific genes, including white phase expression of WH11 and opaque phase expression of OP4 and SAP1 (reviewed in reference 54). RNA was isolated from cultures comprising >95% opaque or white cells, respectively, and analyzed for phase-specific gene expression using RT-PCR. Expression of these three genes in CAMP 43 cells followed the pattern expected for the respective cell types (Fig. 3C). Therefore, the molecular phenotype of the HBR1 mutant is consistent with white and opaque phenotypes.
Hbr1p-regulated switching occurs in the absence of MTL deletions.
White-opaque switching in C. albicans is negatively regulated via the MTL locus (36; reviewed in references 22 and 55). Therefore, switching in the HBR1 heterozygous strain could arise either from rearrangements of this locus or from effects of Hbr1p on MTL gene expression. Genomic PCR analysis demonstrated that both white and opaque cells of strain CAMP 43 retained the full complement of MTL genes, whereas Red 3/6 was deleted for MTLa1 (Fig. 4A, lanes 1 to 3), as previously reported (31). A clinical strain lacking both MTLa1 and MTLα1 alleles was used as an additional control (Fig. 4A, lane 5). The presence of the genomic marker λ imm434 (14) verified that strain CAMP 43 (Fig. 4A, lanes 1 and 2) was a derivative of the parental CAI-4 strain (Fig. 4A, lane 4). Thus, the HBR1 heterozygote functions as a white-opaque switching strain in the presence of all three MTL genes.
Hbr1p regulates MTLα gene expression.
The second possibility to explain switching in the HBR1 heterozygote is that Hbr1p regulates MTL gene expression. We addressed this issue by measuring steady-state levels of MTL RNA by RT-PCR 4 h after transfer of stationary-phase cells to new medium. All three MTL alleles were expressed in the parental strain CAI-4 (Fig. 4B, lane 4), and disruption of the MTLa1 gene did not produce a noticeable effect on MTLα gene expression (Fig. 4B, lane 1). However, MTLα expression could not be detected in either white or opaque Hbr1+/− cells, and MTLa1 appeared to be expressed at a slightly elevated level (Fig. 4B, lanes 2 and 3). Measurement of mRNA levels from CAMP 43 and wild-type cells using qPCR indicated that MTLα gene mRNA expression in CAMP 43 cells was at least 16,000-fold less than in wild-type cells (Table 3). Therefore, Hbr1p is a positive regulator of both MTLα genes, and mutation of one HBR1 allele prevents their expression.
Because Hbr1p appeared to be a limiting factor for MTLα expression in Hbr1+/− cells, we asked whether the same was true during the vegetative growth cycle in an Hbr1+/+ strain. Both MTLα alleles were highly expressed during the exponential growth phase and decreased to basal levels by 24 h (Fig. 4C). This pattern paralleled that of HBR1 expression (Fig. 1B and data not shown). If Hbr1p were limiting for MTLα expression in stationary-phase cells, maintaining high Hbr1p levels into stationary phase should sustain MTLα expression. To test this hypothesis, HBR1 expression was increased directly by expression using a heterologous promoter or indirectly by the addition of hemoglobin to the cultures. As predicted, HBR1 overexpression driven solely by the C. albicans MET3 promoter in strain CAMP 63 sustained MTLα expression at high levels after 24 h, while expression decreased in the parental strain CAF2-1 (Fig. 4D) and was absent in the HBR1 heterozygote (Fig. 4B, lanes 2 and 3 [24-h data not shown]). Addition of hemoglobin to strain CAF2-1, with an intact HBR1 locus, caused only a modest increase in MTLα2 levels at 4 and 12 h (Fig. 4E, lanes 1 to 6). However, hemoglobin sustained MTLα2 expression up to 24 h (Fig. 4E, compare lanes 7 and 8). Therefore, increased MTLα expression in proliferating cells is mediated through Hbr1p, and exposure to hemoglobin sustains MTLα2 expression in early-stationary-phase cells through this pathway.
HBR1 regulates the a1/α2 repressor targets CAG1 and YEL007w.
To confirm that regulation of the MTLα genes by Hbr1 has functional significance, we measured mRNA levels of CAG1, an established direct target of the a1/α2 repressor complex in both C. albicans and S. cerevisiae (called GPA1 in the latter) (20, 48). We used qPCR to measure CAG1 levels and normalized the values to those of an equivalent MTL wild-type strain (CAF-2) grown in YNB with glucose. The CDC36 gene was chosen as an internal standard because its expression levels were comparable to those of the genes under study. In addition, CDC36 expression did not significantly vary under any of the growth conditions or with any of the strains used in the experiments reported here (see Table 3 and Materials and Methods).
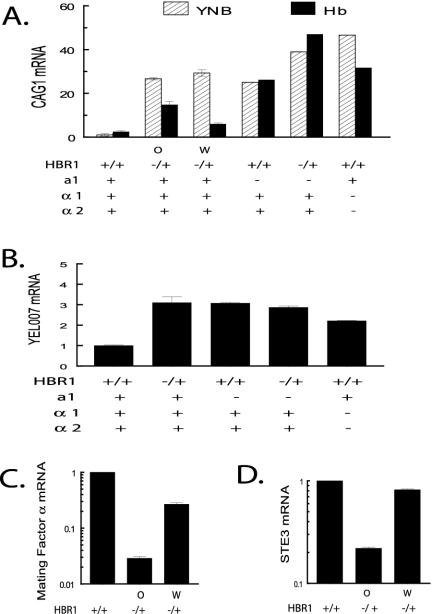
CAMP 43 white and opaque cells displayed 27- to 29-fold derepression of CAG1 when compared with CAF-2 control cells grown under identical conditions (Fig. 5A). If this result were caused by a1/α2 dimer disruption through repression of MTLα genes, then increasing Hbr1p expression induced by hemoglobin should restore MTLα gene expression and repress CAG1. This was confirmed in both white and opaque CAMP 43 cells (Fig. 5A). The difference in the degree of inhibition between white and opaque cells was reproducible, but its basis is unknown. Nevertheless, the reversal of CAG1 derepression was as predicted for increased α2 production as a result of increasing Hbr1 expression. This degree of inhibition is consistent with the twofold increase in HBR1 mRNA levels in CAMP 43 and CAF-2 cells grown with hemoglobin (Table 3).
FIG. 5.
Regulation of a1/α2 repression and α-cell gene targets through HBR1. (A) An a1/α2 target is derepressed in an HBR1 heterozygote. Results of qPCR analysis of RNA isolated from 4-h log-phase cells with and without 0.5 mg of hemoglobin/ml are shown. MTL and HBR1 genotypes as well as white (W) and opaque (O) phenotypes are indicated. CAG1 expression from each strain was corrected for the level of the CDC36 internal standard and then normalized to CAF2-1 levels (arbitrarily set at 1). Strain designations (from left to right): CAF2-1, CAMP 43, CAMP 43, CHY477, CAMP 49, and MM278. This figure represents two separate analyses. Levels are reported as ± the SD. (B). YEL007w expression is derepressed in HBR1 heterozygous cells. qPCR analysis was carried out with the same cDNA preparations as described for panel A. Strain designations are as in panel A, except CAMP 43 opaque cells were not tested. Normalized levels are reported as ± the SD. (C and D) Two α-specific genes are down-regulated in CAMP 43 opaque cells. Mating factor α and STE3 mRNA levels were determined using qPCR with the same cDNA preparations as described for panel A. Levels are reported as ± the SD.
The role of a1/α2 in HBR1-dependent CAG1 regulation was further supported by a comparison of CAG1 mRNA levels from an MTLa1 deletion strain (CHY477) grown with and without hemoglobin. Without hemoglobin, the 25-fold CAG1 derepression was comparable to that of the CAMP 43 cells (Fig. 5A). Hemoglobin did not restore repression of CAG1 in this strain or in a strain containing both HBR1 and MTLa1 mutations (CAMP 49) (Fig. 5A). These results were expected, since a1 is required for a1/α2 repressor function (36). Interestingly, CAMP 49 displayed CAG1 mRNA levels somewhat higher than in either singly mutated strain (Fig. 5A), suggesting that some derepression of CAG1 in the HBR1 mutant may be a1/α2 independent. Nonetheless, these results confirm that hemoglobin- and Hbr1-dependent repression of CAG1 occurs through a1/α2.
Modulation of a1/α2 repressor function by Hbr1 was also confirmed for a recently described gene of unknown function (YEL007w) that is expressed in a and α white cells but not in a/α cells (61). YEL007w mRNA expression was derepressed in the HBR1 heterozygote (CAMP 43) and in MTLa1 and MTLα1,2 deletion strains, as expected for regulation by a1/α2 (Fig. 5B).
The HBR1 mutation represses α cell and the a-cell molecular traits.
The differential regulation of genes required for mating is a primary function of the MTL. Commitment to a specific mating type involves suppression of α-specific genes in a cells and vice versa (16). CAMP 43 cells lack MTLα gene expression and can undergo switching to the mating-competent opaque cell form. This indicated that HBR1 mutant cells display some a-cell attributes, prompting us to further examine genes known to be differentially expressed between a and α cells.
The recently described α mating pheromone gene (MFα) (9, 32) and the a pheromone receptor (STE3) are expressed in functional α cells (61). We found that both of these genes were repressed in CAMP 43 but more strongly in opaque cells, consistent with a transition to an a-cell phenotype (Fig. 5C and D).
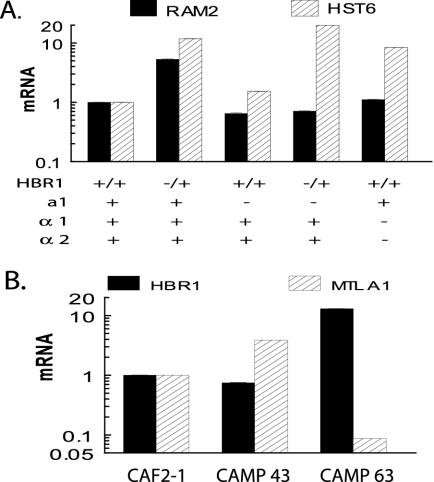
We additionally analyzed the a-cell-specific genes RAM2 and HST6, both of which are required for processing and exporting a-factor. RAM2 encodes the common α subunit of two prenyltransferases (57), and HST6 is the ortholog of the ABC transporter STE6 required for a-factor transport (44) and for a cells to mate (34). RAM2 was induced sixfold in CAMP 43 cells, but not when MTLa1 was deleted either alone or in an HBR1+/− background, nor in an MTLα1,2 double deletion strain (Fig. 6A). These results could be rationalized if Hbr1p were required for a1 to act as a positive regulator of a-specific genes or if an a1 target is necessary for Hbr1p-dependent regulation of RAM2. The former hypothesis is supported by the observed positive regulation of a1 in HBR1+/− cells (Table 3 and Fig. 6B).
FIG. 6.
HBR1 regulates a-specific genes and is a negative regulator of MTL a. (A) Two a-specific genes are regulated through HBR1. qPCR analysis was performed using the same cDNA preparations as in Fig. 5. (B) Hbr1p expression represses MTLa1 expression. Strain CAMP 43 contains only one HBR1 allele, and CAMP 63 contains HBR1 expressed at high levels under the control of the MET3 promoter.
Negative regulation of a1 by HBR1 was confirmed by overexpressing Hbr1p (CAMP 63), which resulted in marked suppression of a1 levels (Fig. 6B). Therefore, Hbr1p may regulate some a-specific genes, such as RAM2, through its effects on a1 expression (Fig. 7).
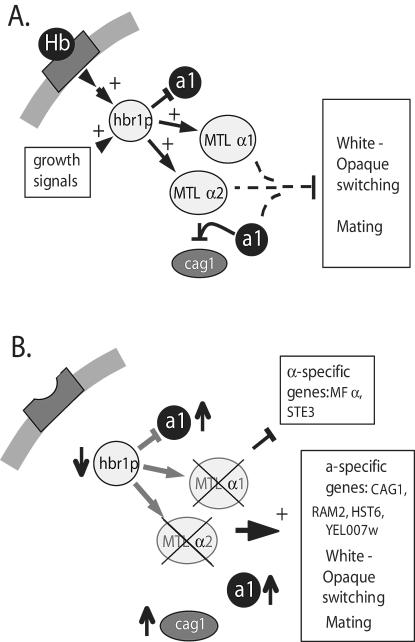
FIG. 7.
Model for the role of HBR1 in MTL gene regulation. (A) Hbr1p expression is stimulated by growth and by signals from the host factor hemoglobin. Normal cellular levels of Hbr1p support MTLα1 and MTLα2 expression and moderately repress MTLα1. Both alleles of HBR1 are necessary to maintain MTLα gene expression and functional levels of the a1/α2 repressor. According to current models (36), this repressor limits expression of CAG1, white-opaque phase switching, and mating. (B) Limiting Hbr1p by removing stimuli controlling its expression or by allelic deletion represses MTLα1 and MTLα2 gene expression. These conditions derepress genes regulated by a1 α2, permit white-opaque phenotypic switching, increase expression of a-cell-specific genes, and decrease α-cell-specific genes. Through this process whereby MTLα1 and MTLα2 expression is down-regulated, the cells acquire a cell characteristics and are capable of mating. Thus, HBR1 regulation of MTL genes is a mechanism that can allow mating without the deletion of MTL genes.
HST6 expression was also substantially elevated in an HBR1+/− strain (Fig. 6A). Like RAM2, it was not induced in the a1 deletion, showing that elevated expression does not result from loss of a1 α2 repression. However, HST6 differed from RAM2 in that deleting one allele of HBR1 in the strain lacking a1 elevated HST6 expression to the same extent as in CAMP 43 (Fig. 6A). Therefore, a1 is not required for this response to Hbr1. Furthermore, deletion of α1 α2 was sufficient to elevate HST6 expression, a response not seen for RAM2 (Fig. 6A). This suggests that HST6 may be regulated by Hbr1 through its effects on α1 or α2. Although the mechanisms for regulation of MTL targets are clearly divergent, these results consistently show that loss of Hbr1 expression results in a gene expression profile typical of an a cell (Fig. 7).
HBR1 heterozygotes function as mating-competent a cells.
Hbr1p regulation of MTLα genes and the targets mentioned above suggested that reducing Hbr1p dosage may permit opaque cells of this strain to mate as a cells. To test this, opaque HBR1 heterozygotes (CAMP R8; Ura− His− Ade+) were crossed with opaque MTLa null cells (CAMP 51; Ade− Ura+) using the quantitative filter mating procedure as described previously (36) (see Table 1 for complete strain descriptions). Based on four independent crosses, Ura+ Ade+ colonies were isolated at frequencies of (3.7 ± 1.0) × 10−3 on selective Arg− Ura− medium. Neither parental strain survived on this selective medium, indicating that spontaneous reversion to prototrophy was undetectable. This demonstrated that the HBR1 heterozygote behaved like an a cell in an MTLa/MTLα HBR1/hbr1 × mtla1/MTLα HBR1/HBR1 strain cross. Therefore, mating is regulated by Hbr1 and can also occur in the absence of chromosomal deletions in the MTL locus.
DISCUSSION
The C. albicans MTL locus (20) is a central regulator of phenotypic phase switching and mating in vitro (35, 36) and in vivo (21). However, mating competency in C. albicans has been previously achieved only through allelic deletion of MTLa1 or MTLα2 (21, 35, 36). Our data demonstrate for the first time that phenotypic phase switching and mating can occur in cells containing an intact MTL locus. We show that the MTLα genes are positively regulated by Hbr1p and that loss of a single HBR1 allele is sufficient to allow white-opaque switching and mating without MTL gene rearrangement. HBR1 in turn is regulated by proliferation signals and exposure of cells to exogenous hemoglobin (Fig. 7). Therefore, both the availability of nutrients and hemoglobin in the local environment may be important regulators of mating for C. albicans in its mammalian host.
The haplo-insufficient phenotype of HBR1 for repressing white-opaque switching suggests that Hbr1p plays a pivotal role in the control of MTLα gene expression. The white-to-opaque transition in HBR1 heterozygotes occurred with a frequency comparable to that of MTLa1 or MTLα2 deletion strains (36, 54) (Table 4). Morphological characteristics and gene expression patterns in the opaque cells were indistinguishable from those generated by MTL deletion. HBR1 mutants formed cells with the oblong shape and dimensions typical of opaque cells (52), possessed the characteristic cell surface pimples (3), and appropriately regulated phase-specific genes (38, 59) (Fig. 3). Reexpression of HBR1 using the MET3 promoter suppressed switching, confirming that the HBR1 deletion caused the increased white-to-opaque switching frequency. The increased switching in HBR1/hbr1 cells results from loss of positive regulation of MTLα gene expression by Hbr1p. Consistent with this model, we demonstrated loss of a1/α2 repressor activity for two known targets, CAG1 and YEL007w (20, 48, 61), in an HBR1+/− strain.
Hbr1p positively regulates MTLα gene expression and is situated upstream of the switch event, but we do not know whether Hbr1p directly or indirectly induces MTLα expression. The two MATα genes in S. cerevisiae are divergently transcribed from a common promoter region and are separated by less than 300 bp (4). The C. albicans MTLα genes are separated by more than 5 kbp (20), suggesting a divergence in regulatory mechanisms.
Hbr1p possesses a predicted P-loop but lacks known DNA-binding motifs (Fig. 2). The Hbr1p ortholog, Fap7p, was identified in a screen for mutants that failed to activate a Gal1-LacZ reporter by a Gal4p-Skn7p hybrid transcription factor in response to oxidative stress (23). This mutant contained a P-loop mutation (G19S), indicating that the P-loop is an essential structural feature for some Fap7p functions. In this mutant, interaction with the transcription factor Skn7p was disrupted, although the cells remained viable. Consistent with this result, Fap7p was found to interact with Skn7p in a two-hybrid assay and was nuclear localized (23). Skn7p has been shown to couple environmental inputs such as oxidative stress to intracellular signaling (reviewed in reference 19). However, H2O2 inhibited transcription from an HBR1-luciferase reporter construct (data not shown), which is inconsistent with an Skn7p interaction under oxidative stress. Nevertheless, this does not preclude interactions of Hbr1p with Skn7p under other metabolic circumstances.
The ability of Hbr1 to regulate mating is also not sufficient to explain why HBR1 and the yeast ortholog FAP7 are essential genes. Other critical functions may depend on these genes. FAP7 was recently identified in a screen for mutants defective in noncoding RNA processing (43), suggesting that it has additional functions in S. cerevisiae.
HBR1 transcription is rapidly induced in response to hemoglobin exposure. The haplo-insufficiency of HBR1 for MTLα expression and sensitivity of CAG1 in the HBR1 heterozygote to hemoglobin addition shows that the MTL signaling pathway is quite sensitive to regulation by this host factor. Hemoglobin also maintains MTLα2 levels into stationary phase in cells with an intact HBR1 locus, demonstrating that this pathway allows hemoglobin to regulate the MTL locus in wild-type C. albicans. Hemoglobin αβ dimers, released via erythrocyte lysis, signal to HBR1 through an unidentified hemoglobin receptor (41). Although the acute phase protein haptoglobin can sequester hemoglobin αβ dimers (7, 10), many pathogenic Candida species exhibit hemolytic activity that could release sufficient hemoglobin to overwhelm this protective protein (33). Thus, hemoglobin exposure may occur when the organism establishes a disseminated infection and is proximal to erythrocytes in a site with low-shear flow. Sensitivity of this pathway to hemoglobin may be modulated by cell proliferation signals that also regulate HBR1 and, thereby, MTLα expression. Fungal cells in the exponential growth phase would express α genes and, therefore, the a1/α2 repressor independently of hemoglobin, but a1/α2 levels would decrease in the postexponential phase. In the latter case, MTLα expression would be more sensitive to hemoglobin stimulation and could restore repressor function to inhibit phase switching in stationary phase.
In summary, we show here that one of the genes regulated by hemoglobin suppresses the phenotypic switch required for mating in C. albicans. We previously demonstrated that hemoglobin alters the adhesive phenotype of C. albicans by inducing binding to several host matrix proteins (65), but this is the first direct evidence that hemoglobin regulates expression of genes known to be expressed during infection and mating. Importantly, we have identified HBR1 as an essential gene that is regulated in response to a defined environmental cue in the vascular compartment of a mammalian host. In addition to regulating mating, HBR1 presumably controls other important signaling pathways that may explain its requirement for vegetative growth.
Acknowledgments
We thank Peter E. Sudbery, Judith Berman, Aaron P. Mitchell, Mathew Miller, Alexander Johnson, and William Fonzi for providing reagents; Michael Sein and Mark Chao for technical assistance; and Kunio Nagashima and Michelle Gignac for the scanning electron microscopy analysis.
REFERENCES
- 1.Alloush, H. M., J. L. Lopez-Ribot, B. J. Masten, and W. L. Chaffin. 1997. 3-Phosphoglycerate kinase: a glycolytic enzyme protein present in the cell wall of Candida albicans. Microbiology 143:321-330. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J., R. Mihalik, and D. R. Soll. 1990. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J. Bacteriol. 172:224-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. M., and D. R. Soll. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Astell, C. R., L. Ahlstrom-Jonasson, M. Smith, K. Tatchell, K. A. Nasmyth, and B. D. Hall. 1981. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 27:15-23. [DOI] [PubMed] [Google Scholar]
- 5.Auclair, K., H. W. Huang, P. Moenne-Loccoz, and P. R. Ortiz de Montellano. 2003. Cloning and expression of a heme binding protein from the genome of Saccharomyces cerevisiae. Protein Expr. Purif. 28:340-349. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 7.Baumann, H., and J. Gauldie. 1994. The acute phase response. Immunol. Today 15:74-80. [DOI] [PubMed] [Google Scholar]
- 8.Bedell, G. W., and D. R. Soll. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun. 26:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett, R. J., M. A. Uhl, M. G. Miller, and A. D. Johnson. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 23:8189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman, B. H. 1993. Haptoglobin. In B. H. Bowman (ed.), Hepatic plasma proteins, vol. 149. Academic Press, San Diego, Calif.
- 11.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 12.De Backer, M. D., P. T. Magee, and J. Pla. 2000. Recent developments in molecular genetics of Candida albicans. Annu. Rev. Microbiol. 54:463-498. [DOI] [PubMed] [Google Scholar]
- 13.Dey, S., T. S. Basu Baul, B. Roy, and D. Dey. 1989. A new rapid method of air-drying for scanning electron microscopy using tetramethylsilane. J. Microsc. 156:259-261. [Google Scholar]
- 14.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshorn, A. K., and S. Scherer. 1989. Genetic analysis of prototrophic natural variants of Candida albicans. Genetics 123:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herskowitz, I. 1987. A master regulatory locus that determines cell specialization in yeast. Harvey Lect. 81:67-92. [PubMed] [Google Scholar]
- 17.Herskowitz, I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342:749-757. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 21.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, A. 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1:106-116. [DOI] [PubMed] [Google Scholar]
- 23.Juhnke, H., C. Charizanis, F. Latifi, B. Krems, and K. D. Entian. 2000. The essential protein fap7 is involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 35:936-948. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, M. J., A. L. Rogers, L. R. Hanselmen, D. R. Soll, and R. J. Yancey, Jr. 1988. Variation in adhesion and cell surface hydrophobicity in Candida albicans white and opaque phenotypes. Mycopathologia 102:149-156. [DOI] [PubMed] [Google Scholar]
- 25.Keogh, R. S., C. Seoighe, and K. H. Wolfe. 1998. Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi. Yeast 14:443-457. [DOI] [PubMed] [Google Scholar]
- 26.Köhrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA, p. 398-405. In C. Guthrie and G. R. Fink (ed.), Yeast genetics and molecular biology, vol. 194. Academic Press, San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 27.Kolotila, M. P., and R. D. Diamond. 1990. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect. Immun. 58:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvaal, C., S. A. Lachke, T. Srikantha, K. Daniels, J. McCoy, and D. R. Soll. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan, C.-Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leipe, D. D., E. V. Koonin, and L. Aravind. 2003. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 333:781-815. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, G., L. P. Samaranayake, and J. Y. Yau. 2001. Candida species exhibit differential in vitro hemolytic activities. J. Clin. Microbiol. 39:2971-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magee, B. B., M. Legrand, A. M. Alarco, M. Raymond, and P. T. Magee. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46:1345-1351. [DOI] [PubMed] [Google Scholar]
- 35.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 36.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 37.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 38.Morrow, B., T. Srikantha, J. Anderson, and D. R. Soll. 1993. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect. Immun. 61:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, England.
- 40.Pendrak, M. L., M. P. Chao, S. S. Yan, and D. D. Roberts. 2004. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J. Biol. Chem. 279:3426-3433. [DOI] [PubMed] [Google Scholar]
- 41.Pendrak, M. L., H. C. Krutzsch, and D. D. Roberts. 2000. Structural requirements for hemoglobin to induce fibronectin receptor expression in Candida albicans. Biochemistry 39:16110-16118. [DOI] [PubMed] [Google Scholar]
- 42.Pendrak, M. L., S. S. Yan, and D. D. Roberts. Sensing the host environment: recognition of hemoglobin by the pathogenic yeast Candida albicans. Arch. Biochem. Biophys., in press. [DOI] [PubMed]
- 43.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 44.Raymond, M., D. Dignard, A. M. Alarco, N. Mainville, B. B. Magee, and D. Y. Thomas. 1998. A Ste6p/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in Saccharomyces cerevisiae. Mol. Microbiol. 27:587-598. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues, R. G., S. Yan, T. J. Walsh, and D. D. Roberts. 1998. Hemoglobin differentially induces binding of Candida, Trichosporon, and Saccharomyces species to fibronectin. J. Infect. Dis. 178:497-502. [DOI] [PubMed] [Google Scholar]
- 46.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 47.Rustad, T. R., D. A. Stevens, M. A. Pfaller, and T. C. White. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061-1072. [DOI] [PubMed] [Google Scholar]
- 48.Sadhu, C., D. Hoekstra, M. J. McEachern, S. I. Reed, and J. B. Hicks. 1992. A G-protein alpha subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-alpha 2 repressor. Mol. Cell. Biol. 12:1977-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos, R., N. Buisson, S. Knight, A. Dancis, J.-M. Camadro, and E. Lesuisse. 2003. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology 149:579-588. [DOI] [PubMed] [Google Scholar]
- 50.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 191:3-21. [DOI] [PubMed] [Google Scholar]
- 51.Slutsky, B., J. Buffo, and D. R. Soll. 1985. High-frequency switching of colony morphology in Candida albicans. Science 230:666-669. [DOI] [PubMed] [Google Scholar]
- 52.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soll, D. R. 1992. High-frequency switching in Candida albicans. Clin. Microbiol. Rev. 5:183-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soll, D. R. 2001. Phenotypic switching, p. 123-142. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 55.Soll, D. R., S. R. Lockhart, K. J. Daniels, R. Zhao, and D. Wessels. 2004. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. Bioessays 26:10-20. [DOI] [PubMed] [Google Scholar]
- 56.Soll, D. R., B. Morrow, and T. Srikantha. 1993. High-frequency phenotypic switching in Candida albicans. Trends Genet. 9:61-65. [DOI] [PubMed] [Google Scholar]
- 57.Song, J. L., and T. C. White. 2003. RAM2: an essential gene in the prenylation pathway of Candida albicans. Microbiology 149:249-259. [DOI] [PubMed] [Google Scholar]
- 58.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 62.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42:339-343. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan, S., E. Negre, J. A. Cashel, N. Guo, C. A. Lyman, T. J. Walsh, and D. D. Roberts. 1996. Specific induction of fibronectin binding activity by hemoglobin in Candida albicans grown in defined media. Infect. Immun. 64:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan, S., R. G. Rodrigues, D. Cahn-Hidalgo, T. J. Walsh, and D. D. Roberts. 1998. Hemoglobin induces binding of several extracellular matrix proteins to Candida albicans. Identification of a common receptor for fibronectin, fibrinogen, and laminin. J. Biol. Chem. 273:5638-5644. [DOI] [PubMed] [Google Scholar]