Abstract
Organisms rapidly adapt to severe environmental stress by inducing the expression of a wide array of heat shock proteins as part of a larger cellular response program. We have used a genomics approach to identify novel heat shock-induced genes in Saccharomyces cerevisiae. The uncharacterized open reading frame (ORF) YLR251W was found to be required for both metabolism and tolerance of ethanol during heat shock. YLR251W has significant homology to the mammalian peroxisomal membrane protein Mpv17, and Mpv17−/− mice exhibit age-onset glomerulosclerosis, deafness, hypertension, and, ultimately, death by renal failure. Expression of Mpv17 in ylr251wΔ cells complements the 37°C ethanol growth defect, suggesting that these proteins are functional orthologs. We have therefore renamed ORF YLR251W as SYM1 (for “stress-inducible yeast Mpv17”). In contrast to the peroxisomal localization of Mpv17, we find that Sym1 is an integral membrane protein of the inner mitochondrial membrane. In addition, transcriptional profiling of sym1Δ cells uncovered changes in gene expression, including dysregulation of a number of ethanol-repressed genes, exclusively at 37°C relative to wild-type results. Together, these data suggest an important metabolic role for Sym1 in mitochondrial function during heat shock. Furthermore, this study establishes Sym1 as a potential model for understanding the role of Mpv17 in kidney disease and cardiovascular biology.
Exposure of cells to heat shock temperatures results in protein misfolding, growth arrest, and metabolic dysfunction. To counteract these effects, cells respond to temperature changes by activating a highly conserved program of gene expression termed the heat shock response. The products of this response are critical both for protecting the cell from the immediate damage imposed by the stress as well as adjusting structural and metabolic components such that growth can proceed at the elevated temperature (reviewed in references 31 and 52). In concert with other environmental conditions known to threaten cellular viability, such as changes in osmolarity or exposure to DNA-damaging compounds, heat shock induces the transcriptional activation of a subset of genes necessary for survival during exposure to a diverse range of stress conditions, collectively termed the environmental stress response (ESR) (17). The ESR is governed largely by the partially redundant general stress responsive transcription factors Msn2 and Msn4 (17). A second pathway regulated by the heat shock transcription factor responds primarily to the stress of elevated temperature but can also be activated by other protein misfolding agents (52-54). Transcriptional profiling via DNA microarray technology has been critical to determining the relative contributions of the Msn2/4 and heat shock transcription factor pathways as well as providing information pertinent to understanding the total complement of heat shock gene expression (7, 17). Together, these two gene expression pathways account for the majority of heat shock-induced transcription.
The set of stress-inducible genes includes the classic heat shock genes, such as those encoding the Hsp70 chaperones, with established roles in protein refolding (10). The trehalose biosynthetic genes, another well-characterized example, have a metabolic role that correlates with increased resistance to heat shock (14). In addition to serving as a storage carbohydrate during exponential growth, trehalose has been shown to bind and protect partially folded proteins during heat shock (49). The existence of heat shock-inducible isoforms of genes encoding both chaperones (for example, the Hsp90 genes HSP82/HSC82) and metabolic enzymes normally present during non-heat-shock conditions (the glycogen synthase genes GSY2/GSY1) illustrates the cellular requirement for proteins with altered function or stability during conditions of elevated temperature (5, 17). A significant number of genes displaying heat shock induction from published DNA microarray analyses are uncharacterized open reading frames (ORFs) with no known biological role, suggesting that much of the ESR remains unexplored (17). Higher eukaryotes respond to the stress of pathophysiological conditions through activation of pathways sharing remarkable similarity to those activated by heat shock in yeast (31). The high level of functional conservation that exists between components of the lower eukaryotic heat shock response and their homologs in mammalian systems is therefore of considerable interest. Functional analysis of novel stress-induced genes in yeast identified by transcriptional profiling is likely to help elucidate the function of the large number of uncharacterized human stress-induced and disease-related genes.
The Mpv17 protein was first identified as a transgene insertion resulting in severe kidney disease in mice (58). The phenotypes associated with the Mpv17−/− mouse include age-onset glomerulosclerosis, deafness, and hypertension, ultimately leading to complete renal failure (58, 61). Mpv17 shares considerable homology to the poorly understood peroxisomal membrane protein Pmp22 and has likewise been localized to the peroxisomal membrane of mammalian cells (28, 55, 61). Although the molecular function of Mpv17 and other members of the Pmp22 family remains unclear, increased generation of reactive oxygen species (ROS) and lipid peroxidation products in Mpv17−/− mouse embryonic fibroblast cells appears to be a source of tissue damage associated with the glomerulosclerosis phenotype (39, 57, 61). Interestingly, deafness is frequently associated with other human mitochondrial and peroxisomal disorders, and the Mpv17−/− model has proven useful for studying the molecular mechanisms connecting these pathologies (30, 33).
We describe in this report the characterization of the ORF YLR251W, a previously unknown heat shock gene in Saccharomyces cerevisiae. We demonstrate that YLR251W is the yeast functional ortholog of Mpv17 and have renamed the ORF SYM1 (for “stress-inducible yeast MPV17 homolog”). Sym1 may therefore serve as a fungal model for understanding the molecular mechanisms underlying the renal pathologies of Mpv17−/− animals.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study were acquired from the BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 lys2Δ0/LYS2) homozygous diploid knockout collection and resulted from ORF deletions generated by a kanMX4 cassette (Research Genetics/Invitrogen, Grand Island, N.Y.) (18) except for the pex3Δ and pex19 strains, which were obtained from the MATa haploid knockout collection. Haploid gene knockouts of both mating types were obtained by sporulation of the parental knockout strains. Cultures were grown at either 30 or 37°C (heat shock temperature) in rich YP (0.1% yeast extract, 0.2% Bacto Peptone) or synthetic complete (SC) (Qbiogene, Carlsbad, Calif.) medium with carbon sources appropriate to the given experiment (2% glucose, 3% glycerol, 2% ethanol, 2% acetate, 0.1% acetaldehyde). Liquid growth curves of 37°C ethanol-grown cells were obtained by growing cells to logarithmic (log) phase at 30°C prior to dilution and shift to 37°C. Growth phenotype experiments on solid medium were performed as serial dilutions from overnight cultures. Unless otherwise indicated, log-phase cultures were used for all other experiments.
Plasmids.
To construct p416GPDSym1-GFP and p426GPDSym1-HA3, the SYM1 coding region and approximately 650 bp upstream of the ORF were amplified via PCR and cloned into the yeast-Escherichia coli shuttle vectors pRS416GPD (CEN; low copy number) and pRS426GPD (2μm; high copy number) (34). A NotI site was generated internally to the stop codon such that NotI-flanked fragments encoding either green fluorescent protein (GFP) or a triple hemagglutinin (HA) tag could be cloned as carboxyl-terminal fusions (27). To construct p426GPDMpv17, Mpv17 was PCR amplified from an λAct human cDNA library (kindly provided by S. Elledge, Harvard University) with primers complementary to the 5′ and 3′ coding regions of the gene and subsequently cloned into the SmaI site of pRS426GPD (13). The peroxisomal matrix-targeted cyan fluorescent protein (CFP) control (p413GPDCFP-skl) used for fluorescent microscopy in this study was constructed by PCR integration of sequence encoding the amino acid residues serine, lysine, and leucine at the C terminus of CFP and subsequent cloning into pRS413GPD (43). Sequences for all primers are available upon request.
Northern and microarray analysis.
RNA for Northern blot analysis was isolated from a 150-ml YPD culture of strain BY4741 at 0, 15, 30, and 60 min during heat shock with a hot phenol procedure (25). The blots were hybridized with random-prime-labeled DNA fragments generated by PCR corresponding to the entire ORF of PGK1, STI1, and SYM1 per the instructions of the manufacturer (Invitrogen, Carlsbad, Calif.) and subsequently subjected to phosphorimaging analysis (Molecular Dynamics/Amersham, Piscataway, N.J.) for quantitation. RNA for DNA microarray analysis was isolated from YPE cultures of wild-type and sym1Δ cells grown at 30 and 37°C. All RNA-labeling and DNA array manipulations were performed by the University of Texas Southwestern Medical Center at Dallas-Microarray Core Facility (http://microarray.swmed.edu) with a custom-synthesized glass slide microarray consisting of 6,219 yeast genes amplified using a Research Genetics oligonucleotide pair set. Background subtraction and normalization were performed as described previously (15). Only those genes upregulated twofold or more in two duplicate experiments are reported in Table 1. The values shown are the normalized severalfold induction in sym1Δ cells versus wild-type cells for each independent experiment.
TABLE 1.
Genes upregulated in sym1Δ versus wild-type cells on ethanol at 37°Ca
| Gene | Function | Fold induction | Category |
|---|---|---|---|
| PDC5 | Pyruvate decarboxylase | 14.5, 4.0 | Metabolism |
| PYC2 | Pyruvate carboxylase | 5.1, 2.1 | Metabolism |
| PGI1 | Glucose-6-phosphate isomerase | 2.6, 2.0 | Metabolism |
| SDH2 | Succinate dehydrogenase | 2.6, 2.9 | Metabolism |
| NTH2 | Alpha, alpha-trehalase | 2.4, 2.7 | Metabolism |
| BAP2 | Amino acid transporter | 2.6, 2.1 | Metabolism |
| ACH1 | Acetyl-CoA hydrolase | 2.6, 2.1 | Metabolism |
| SSC1 | Hsp70-mitochondria | 5.5, 2.9 | Heat shock |
| SSA1b | Hsp70-cytosolic | 2.3, 2.3 | Heat shock |
| FKS1 | 1,3-Betaglucan synthase | 5.3, 2.0 | Cell wall |
| PIR1 | Cell wall structural protein | 4.8, 2.5 | Cell wall |
| PMA2 | Plasma membrane ATPase | 4.5, 2.6 | Other |
| YCL042Wc | Uncharacterized ORF | 3.4, 2.1 | Other |
| YAL053W | Uncharacterized ORF | 2.1, 2.1 | Other |
Wild-type and sym1Δ cells were grown overnight at 30°C in glucose medium, shifted to 30°C and grown to mid-logarithmic phase in ethanol as sole carbon source. Cultures were then split and either continued growth at 30°C or shifted to 37°C until growth arrest. RNA was isolated and transcriptional profiles generated comparing the two strains at each temperature. The induction ratios in sym1Δ versus wild-type cells for genes upregulated twofold or more in both replicates are listed.
SSA1 was also independently found as the overlapping predicted ORF YAL004W.
Hypothetical ORF partially overlapping the glucokinase gene GLK1.
Fluorescence microscopy.
Wild-type, sym1Δ, pex3Δ, or pex19Δ cells expressing either Sym1-GFP or CFP-skl were grown overnight (to enhance mitochondrion and peroxisome development) at 30°C in dextrose-containing medium. Mitochondria were specifically detected by staining with MitoTracker Red (Molecular Probes, Eugene, Oreg.) as indicated by the manufacturer. Fluorescence and differential interference contrast images of the stained cells were obtained on an Olympus BX60 microscope using ×100 magnification and captured using a Photometrics charge-coupled device camera.
Subcellular fractionation and membrane association.
Spheroplasting procedures as described elsewhere were used to obtain soluble and membrane pellet fractions from sym1Δ cells expressing Sym1-HA (20). The spheroplasting buffer used for this procedure contained 50 mM Tris (pH 7.5), 3 mM 2-mercaptoethanol, 1.2 M sorbitol, and freshly added protease inhibitors from a cocktail stock to give the following final concentrations: aprotinin, 2 μg/ml; pepstatin A, 2 μg/ml; leupeptin, 1 μg/ml; phenylmethylsulfonyl fluoride (PMSF), 2 mM; and chymostatin, 2 μg/ml. Spheroplasts were lysed with a Dounce homogenizer in buffer containing 5 mM morpholineethanesulfonic acid-KOH (pH 6), 0.6 M sorbitol, 0.5 mM EDTA, and protease inhibitors. Unbroken cells and cellular debris were removed by low-speed centrifugation (800 × g) at 4°C. The low-speed supernatant (total cell extract) was subjected to centrifugation at 16,100 × g to obtain the soluble and membrane pellet fractions. The fractions were analyzed using standard Western immunoblotting procedures after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with antibodies recognizing known soluble (α-PGK1; Molecular Probes) and membrane (α-AAC/porin; this polyclonal antibody recognizes both the general yeast mitochondrial ATP/ADP carrier and outer membrane porin) (gift of C. Koehler, University of California at Los Angeles) proteins and the HA epitope (12CA5; Roche Diagnostics, Indianapolis, Ind.).
For membrane association experiments, crude cell extract isolated by glass bead lysis in TEGN buffer (20 mM Tris-HCl [pH 7.9], 0.5 mM EDTA, 10% glycerol, 50 mM NaCl) was centrifuged at 16,100 × g at 4°C to obtain a membrane pellet. The membrane pellet was resuspended in TEGN and treated with 0.1 M NaCO3, 1% Triton X-100, or TEGN buffer for 30 min on ice. The samples were subsequently centrifuged at 16,100 × g at 4°C to obtain soluble and membrane fractions and analyzed as described above.
Protease protection assay.
A modified version of the protease protection assay of Yaffe (59) was utilized to determine the submitochondrial localization of Sym1. For preparation of the mitochondria, the sym1Δ strain harboring p426GPDSYM1-HA was grown overnight in semisynthetic lactate medium (0.67% yeast-nitrogen base, 1× amino acids, 0.05% yeast extract, 2% lactic acid) to induce mitochondrial proliferation. The culture was diluted and grown in 1 liter of semisynthetic lactate medium at 30°C with vigorous aeration to a final optical density (600 nm) of approximately 1.0. Cells were harvested by centrifugation, resuspended in buffer A (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4), and converted to spheroplasts. Spheroplasts were washed once in buffer B (0.6 M sorbitol, 20 mM potassium morpholineethanesulfonic acid, pH 6.0) and resuspended in 10 ml of buffer B with 1× protease inhibitors. Lysis was achieved through 20 strokes in a chilled Dounce homogenizer, after which unlysed cells were collected by centrifugation at 1,500 × g. The cell pellet was subject to Dounce homogenization again, and after centrifugation the two low-speed supernatants were combined. Intact mitochondria were harvested by centrifugation at 12,000 × g, washed, and resuspended in cold buffer C (0.6 M sorbitol, 20 mM potassium HEPES, pH 7.4) to a final concentration of 25 mg/ml of mitochondrial protein as determined by a Lowry protein assay. Mitochondria were frozen in small aliquots and stored at −80°C.
For the protease protection assay, a sample of frozen mitochondria was diluted to give six equal aliquots of 100 μl. Three were further diluted in 1 ml of buffer C containing either 1 mM PMSF, 100 μg of proteinase K (Roche Diagnostics)/ml, or 100 μg of proteinase K-1% Triton X-100/ml (mitochondria). The other three were diluted in 1 ml of mitoplasting buffer (20 mM potassium HEPES [pH 7.4], 1 mg of bovine serum albumin/ml) containing 1 mM PMSF, 100 μg of proteinase K/ml, or 100 μg of proteinase K-1% Triton X-100/ml (mitoplasts). All six tubes were incubated for 30 min on ice followed by precipitation with 10% trichloroacetic acid. Protein pellets were washed twice with cold acetone to remove acid, dried, and resuspended in SDS-PAGE sample buffer. After SDS-PAGE and transfer to nitrocellulose membrane, proteins were detected using antibodies 12CA5, anti-AAC/porin, and anti-Fis1 (gift of J. Nunnari, University of California at Davis).
Ethanol and heat shock induction analysis.
Heat shock induction was performed at 37°C for 15 min with glucose-grown cultures. Cultures used for ethanol induction experiments were first grown to mid-log phase in glucose prior to growth for 9 h in ethanol. Crude cell extract was obtained from standard glass bead lysis procedures and analyzed as described above. Quantitation of band intensity was performed on scans from film exposures of the enhanced chemiluminescence reaction with Image J (National Institutes of Health).
RESULTS
Identification of a novel gene required for growth on ethanol-containing medium at 37°C.
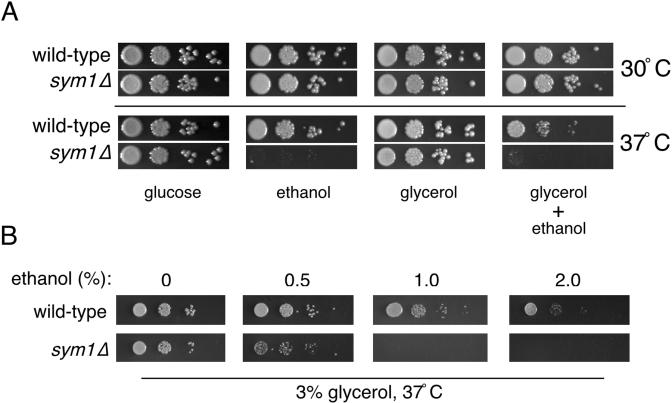
Published DNA microarray data were used to identify approximately 120 previously uncharacterized ORFs displaying a typical heat shock induction profile at 39°C (7, 17). Phenotypic analysis designed to address potential stress-specific growth requirements for this subset of novel ORFs was performed in parallel using homozygous diploid strains obtained from the yeast knockout collection. Growth at both standard (30°C) and heat shock (37°C) temperatures was tested under a wide array of growth conditions, including nonoptimal carbon sources (ethanol-glycerol), oxidative stress (2 mM hydrogen peroxide), osmotic stress (1 M sorbitol), salt stress (1 M NaCl), etc. (40). One ORF, YLR251W (SYM1), was found to be required for growth on the nonfermentable carbon source ethanol specifically at 37°C and was the focus of our present study. As shown in Fig. 1A, wild-type and sym1Δ cells grow equally well on glucose at 30 or 37°C but the sym1Δ strain was unable to grow on ethanol-containing medium at 37°C. Interestingly, no growth inhibition on ethanol was observed for sym1Δ cells at 30°C. We speculated that the observed growth phenotype might reflect a general respiratory defect that would be expected to hinder growth on other nonfermentable carbon sources. However, growth at 37°C on medium containing glycerol as the sole nonfermentable carbon source was unaffected, thus indicating that SYM1 is not required for general respiratory growth. If the inability of sym1Δ cells to grow on ethanol at 37°C is due to a specific defect in ethanol metabolism, this phenotype should be remediated by the addition of glycerol as an alternate carbon source. Interestingly, sym1Δ cells were also unable to grow on medium containing both carbon sources. Growth on medium containing both glucose and ethanol was not inhibited (A. Trott and K. A. Morano, unpublished data), suggesting that the apparent ethanol toxicity was a result of metabolism during respiratory growth.
FIG. 1.
sym1Δ mutant cells are hypersensitive to ethanol and heat shock. (A) Wild-type and sym1Δ strains were plated on SC medium containing glucose (2%), ethanol (2%), glycerol (3%), or glycerol plus ethanol and incubated at 30 or 37°C. Plates with nonfermentable carbon sources were incubated for several days longer than glucose plates due to slower growth rates. (B) Wild-type and sym1Δ strains were plated on SC medium containing glycerol (3%) plus the indicated concentrations of ethanol and grown at 30 and 37°C for the same length of time. Because no growth effects were observed at 30°C, only the 37°C series is shown.
We noted that growth of wild-type cells on glycerol-ethanol medium at 37°C was slower than on either nonfermentable carbon source alone, suggesting that simultaneous metabolism of both compounds may be deleterious during thermal stress (Fig. 1A). To investigate this further, wild-type and sym1Δ cells were plated on medium containing 3% glycerol and increasing concentrations of ethanol ranging from 0 to 2% (Fig. 1B). No growth defects were observed for either strain at 30°C (Trott and Morano, unpublished). However, wild-type cells exhibited progressively poorer growth with increasing ethanol concentrations at 37°C. Importantly, sym1Δ cells displayed hypersensitivity to ethanol at 37°C relative to wild-type results and failed to grow at concentrations above 0.5%. Therefore, metabolism of ethanol at 37°C causes growth impairment in S. cerevisiae and deletion of SYM1 greatly exacerbates this phenotype. It should also be noted that the growth defects are observed at concentrations of ethanol far below those known to cause proteotoxicity, heat shock gene induction, or cell death (6 to 8%) (1, 2, 9).
SYM1 expression is increased during heat shock and growth on ethanol.
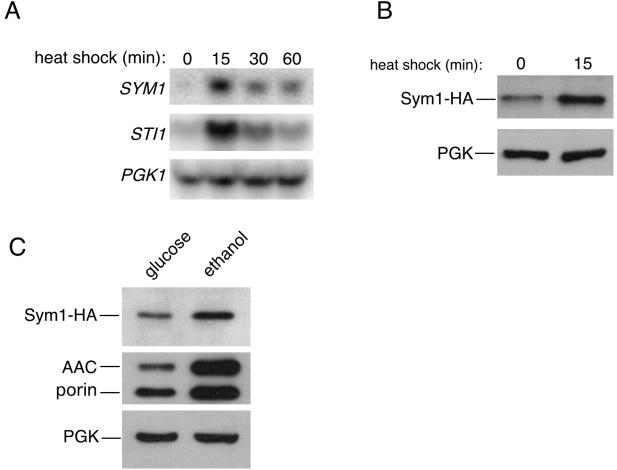
To verify the heat shock induction of SYM1 reported for published microarrays, Northern blot analysis of SYM1 transcript levels was performed over a 1-h heat shock period at 37°C. As shown in Fig. 2A, the SYM1 gene is heat shock induced with an expression profile similar to that of the previously characterized heat shock gene, STI1 (35). Transcription of both genes was strongly induced at the 15-min time point and then fell to levels below the maximum but higher than prestress conditions. The induction ratio of SYM1 was determined by PhosphorImager analysis to be approximately sevenfold. PGK1 levels were determined as a load control for this experiment.
FIG. 2.
SYM1 is stress inducible. (A) A wild-type culture in mid-logarithmic-growth phase was subjected to a 1-h heat shock at 37°C, aliquots were removed at the indicated time points, and total RNA was prepared for Northern analysis as described in Materials and Methods. Radiolabeled probes were used to detect mRNA levels of the indicated genes. STI1 is a well-established heat shock gene, and PGK1 is used as a loading control. (B) Sym1 protein levels after a brief heat shock were examined in the sym1Δ mutant strain transformed with a plasmid expressing SYM1 tagged with a triple-HA epitope at the carboxyl terminus under the control of its own promoter. Equal amounts of mid-logarithmic-phase cells were harvested at 30°C or after 15 min of heat shock at 37°C, protein extracts were prepared, and the levels of Sym1-HA and the loading control PGK were analyzed by Western blotting. (C) Protein extracts were prepared from the same strain used as described for panel B from cells grown in glucose medium or after a 9-h shift to ethanol-containing medium and were analyzed by Western blotting. The polyclonal antibody used to detect the mitochondrial ATP/ADP carrier protein (AAC) also recognizes the mitochondrial outer membrane protein porin.
We next determined whether Sym1 protein levels likewise responded to heat shock. A carboxyl-terminal HA epitope-tagged allele of SYM1 was constructed under the transcriptional control of its own promoter. Expression of Sym1-HA fully complemented the 37°C ethanol phenotype of sym1Δ cells (Trott and Morano, unpublished), verifying that the tag did not interfere with Sym1 function. As anticipated, glucose-grown sym1Δ cells expressing Sym1-HA exhibited an increase in Sym1 levels after 15 min of heat shock at 37°C (Fig. 2B). On the basis of the observed sym1Δ phenotype, we asked whether Sym1-HA protein levels would also increase during growth on ethanol. Glucose-grown sym1Δ cells expressing Sym1-HA were resuspended in ethanol-containing medium for 9 h at 30°C. Western blot analysis of Sym1-HA protein levels in cells grown in both carbon sources indicated that Sym1 exhibited increased expression during respiratory growth on ethanol (Fig. 2C). The induction of Sym1 as determined by scanning densitometry was approximately threefold, in consistency with the observed fivefold increase for AAC and a twofold increase for porin, two mitochondrial proteins whose expression increases during respiration (17). The induction or stabilization of Sym1 in ethanol-containing medium at 30°C suggests a metabolic need for Sym1 during non-heat-shock conditions. Moreover, SYM1 expression and therefore protein availability appears to be coordinated with the conditional requirement for the protein for growth on ethanol at 37°C.
sym1Δ cells undergo reversible growth arrest at 37°C and exhibit an intermediate sensitivity to acetaldehyde.
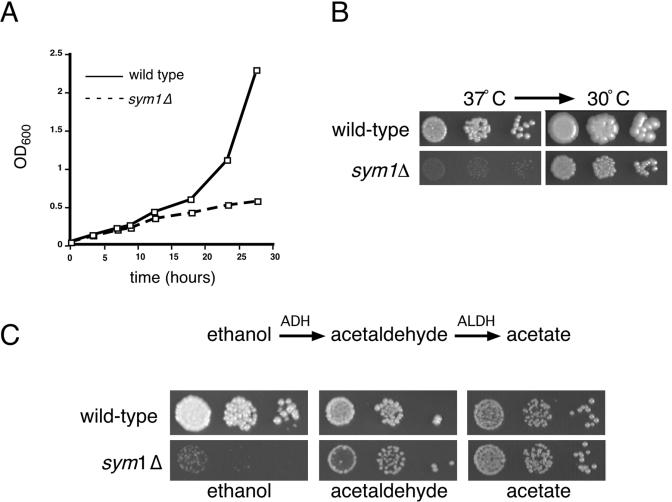
To better understand the nature of the sym1Δ growth phenotype, the growth rates of wild-type and sym1Δ ethanol-grown cells after a shift to 37°C were compared (Fig. 3A). sym1Δ cells are able to replicate for several generations following the 37°C temperature shift. After approximately three generations, sym1Δ cells ceased growth whereas wild-type cells continued to grow exponentially. Microscopic analysis of the growth-arrested sym1Δ culture revealed that the cells arrested asynchronously, with no significant bias toward G1 or G2 phases of the cell cycle. Two scenarios could account for the observed growth defect on ethanol medium; cells may be dying due to ethanol cytotoxicity, or growth may be arrested due to a metabolic imbalance or deficiency. To distinguish between these two possibilities, wild-type and sym1Δ cells were serially diluted onto ethanol plates and incubated at 37°C for 7 days. As shown in Fig. 3B, sym1Δ cells were unable to form robust colonies under these conditions, although microcolonies were observed. The same plate was then shifted to 30°C for several additional days. Both strains resumed growth following shift to 30°C with no detectable loss in CFU. In no case could we detect papillae arising from a background of dead cells in the 37°C microcolonies, illustrating that sym1Δ cells exhibit reversible growth arrest and not cell death at 37°C on ethanol-containing medium.
FIG. 3.
sym1Δ cells show reversible cessation of growth on ethanol during heat shock and differential sensitivity to ethanol catabolic intermediates. (A) Wild-type and sym1Δ cells were grown in YPD medium at 30°C, diluted into YP-2% ethanol medium, and grown to mid-log phase and then rediluted to an optical density at 600 nm (OD600) of 0.1 in YP-2% ethanol, and growth was monitored at 37°C for 28 h. (B) Dilutions of wild-type and sym1Δ cells were spotted onto SC medium containing 2% ethanol and grown for 7 days at 37°C. The same plates were then shifted to 30°C for an additional 3 days. (C) Dilutions of wild-type and sym1Δ cells were spotted onto SC medium containing ethanol (2%), acetaldehyde (0.1%), or acetate (2%) and grown at both 30 and 37°C. Incubation times differed for each carbon source but were identical for both strains. Because no growth defects were observed at 30°C, only the 37°C plates are shown. Note the smaller colony size of the sym1Δ strain and acetaldehyde and the absence of a defect on acetate.
To further address the nature of the sym1Δ sensitivity to ethanol, metabolic intermediates in the ethanol catabolism pathway were tested for their ability to support growth of sym1Δ cells. The utilization of ethanol as a carbon source requires alcohol dehydrogenase (ADH) for the oxidation to acetaldehyde, a potentially toxic intermediate in the pathway that is subsequently oxidized to acetate by acetaldehyde dehydrogenase (ALDH). As shown in Fig. 3C, growth inhibition of sym1Δ cells was observed on 0.1% acetaldehyde relative to the results seen with wild-type cells at 37°C, as evidenced by smaller colony size. However, no reduction in growth rate was seen on acetate-containing medium or on any of the other carbon sources at 30°C. The sensitivity of sym1Δ cells to acetaldehyde is moderate compared to the dramatic phenotype observed for growth on ethanol, suggesting a temperature-specific requirement for SYM1 in the early steps of ethanol metabolism rather than a general defect in metabolism of two-carbon, nonfermentable carbon sources.
SYM1 shares homology and functional similarity to a murine glomerulosclerosis gene.
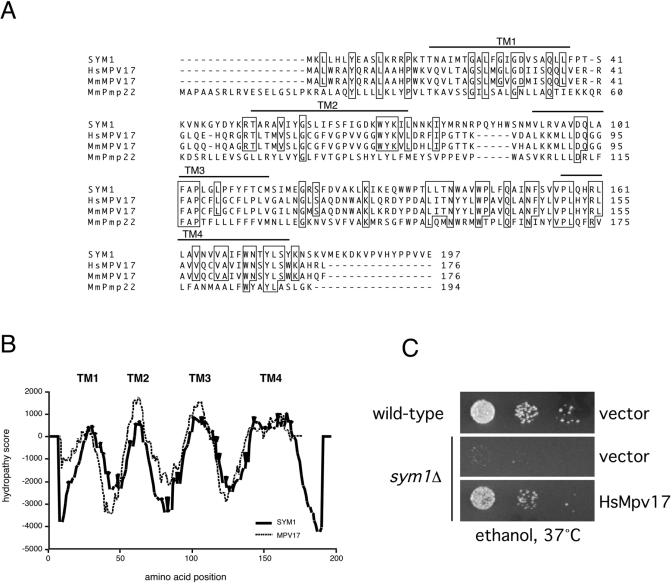
BLAST analysis of the predicted protein sequence of SYM1 identified a family of mammalian peroxisomal membrane proteins with high-level sequence homology (∼48% similarity, ∼32% identity), as shown in Fig. 4A. All are predicted to be small (20- to 22-kDa) integral membrane proteins with four transmembrane domains. SYM1 shares the highest homology with Mpv17, previously identified by insertional mutagenesis as a murine gene whose absence results in progressive, age-onset glomerulosclerosis, hypertension, deafness, and, ultimately, death by renal failure (58). Importantly, the molecular function of Mpv17 is presently unknown. Comparative hydropathy plots of the SYM1 and Mpv17 predicted protein sequences were generated using the program TMPred (23). As shown in Fig. 4B, the hydropathy traces were nearly superimposable, suggesting that in addition to sequence similarity, there is strong conservation of transmembrane domain architecture. To determine whether SYM1 is the yeast functional ortholog of mammalian Mpv17, the Mpv17 coding region was amplified from a human cDNA library by PCR and subsequently cloned and expressed in sym1Δ cells. Expression of human Mpv17 in the sym1Δ strain restored cell growth on ethanol medium at 37°C, thus establishing a functional link between SYM1 and its mammalian homolog (Fig. 4C).
FIG. 4.
SYM1 is the yeast functional ortholog of the kidney disease gene Mpv17. (A) CLUSTAL alignment of the protein coding sequences of SYM1, Homo sapiens Mpv17, Mus musculus Mpv17, and M. musculus Pmp22. Identical residues are boxed. The four predicted transmembrane domains (TM1, TM2, TM3, and TM4) are indicated with bars over the protein sequences. (B) Hydropathy profiles of the SYM1 and Mpv17 protein coding sequences were generated using TMPred. Putative transmembrane domains are indicated. (C) Dilutions of wild-type and sym1Δ cells carrying either pRS426GPD (vector) or pRS426GPDHsMPV17 were spotted onto SC-URA plates containing 2% ethanol as sole carbon source and incubated at 37°C.
Another S. cerevisiae predicted ORF named YOR292C also exhibits substantial homology to the Mpv17/Pmp22 family. A yor292cΔ knockout strain was tested for growth defects on nonfermentable carbon sources at both 30 and 37°C, with no observable defect (Trott and Morano, unpublished). Moreover, a sym1Δ yor292cΔ double-mutant strain was constructed, and cells of this strain displayed no additional ethanol sensitivity relative to sym1Δ cells (Trott and Morano, unpublished). Together, the sequence analysis and functional complementation results strongly support assignment of SYM1 as the functional ortholog of Mpv17 in S. cerevisiae.
Sym1 is an integral membrane protein of the mitochondrial inner membrane.
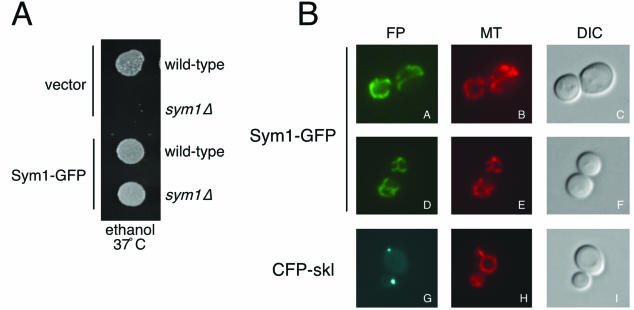
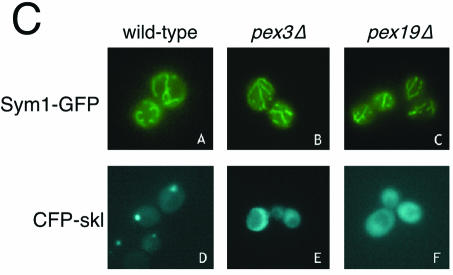
Mpv17 localizes to the peroxisome in murine cells (61). Given the sequence and functional similarity of Sym1 to Mpv17, we predicted that Sym1 would likewise reside in the peroxisome in yeast. To address this question, a carboxyl-terminal fusion of GFP to Sym1 was constructed. Sym1-GFP was found to fully complement the ethanol growth phenotype of sym1Δ cells, indicating that the addition of GFP most likely did not disrupt targeting and function of the Sym1 protein (Fig. 5A). To visualize peroxisomes, a version of CFP was constructed using PCR to add the amino acid sequence ser-lys-leu, a well-established peroxisomal matrix-targeting signal, to the carboxy terminus (CFP-skl) and was expressed in sym1Δ (8, 16, 50). Cells expressing Sym1-GFP or CFP-skl were examined by fluorescence microscopy as shown in Fig. 5B. Sym1-GFP was localized to a reticulated network distinct from the punctate fluorescence observed with CFP-skl. Because the Sym1-GFP pattern resembled that observed with mitochondrial proteins, cells expressing Sym1-GFP were stained with MitoTracker Red, a fluorescent dye that specifically stains charged mitochondria (Molecular Probes) (21). The MitoTracker Red pattern was identical to that observed with Sym1-GFP (compare panel A to panel B and panel D to panel E). To further demonstrate that Sym1 was localized predominantly to a mitochondrial compartment, we expressed both the Sym1-GFP fusion and the CFP-skl peroxisomal marker in pex3Δ and pex19Δ cells. These genes are required for peroxisome biogenesis and have been demonstrated to mislocalize both peroxisomal membrane and matrix proteins to the cytoplasm (22). As shown in Fig. 5C, the localization pattern of Sym1-GFP remained unchanged in pex3Δ and pex19Δ cells relative to wild-type results (panels B and C versus panel A). In contrast, the punctate peroxisomal signal observed in wild-type cells with CFP-skl became diffuse throughout the cytoplasm in the pex mutants (panels E and F). On the basis of these live cell observations made using a fully functional GFP-tagged allele, we therefore conclude that Sym1 is a mitochondrial protein in yeast.
FIG. 5.
Sym1 is localized to the mitochondrial network. (A) Wild-type or sym1Δ cells carrying pRS416GPD (vector) or pRS416GPDSym1-GFP were spotted onto SC-URA plates containing 2% ethanol as sole carbon source and incubated at 37°C. (B) sym1Δ cells carrying either pRS416GPDSym1-GFP or pRS416GPDCFP-skl were grown overnight in selective medium and stained with the mitochondrial dye MitoTracker Red. Cells were photographed under epifluorescence illumination with the appropriate filters to detect the fluorescent protein fusions (FP) and MitoTracker signal (MT) or using differential interference contrast optics (DIC). (C) Wild-type, pex3Δ, and pex19Δ cells transformed with either pRS416GPDSym1-GFP or pRS413GPDCFP-skl were grown overnight in selective medium and photographed as described above.
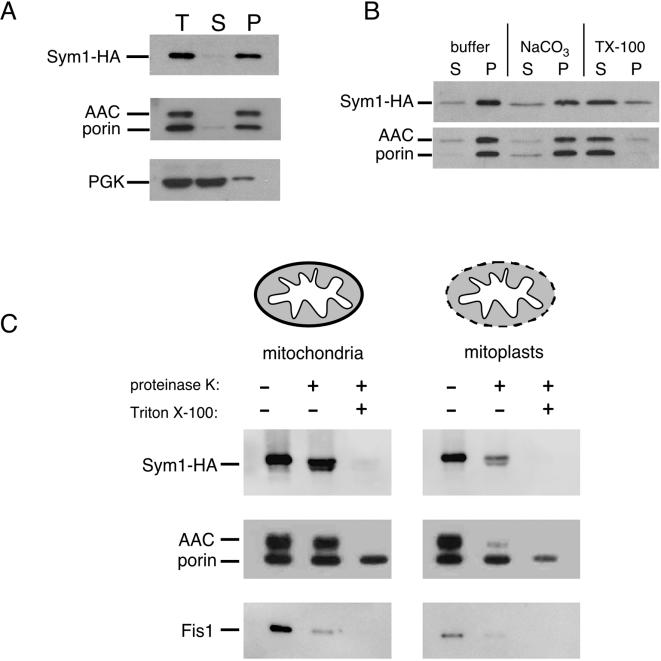
The strong similarity of Sym1 to members of the Pmp22/Mpv17 family of integral membrane proteins and the predicted hydropathy suggested that Sym1 would likewise be an integral membrane protein. To address whether Sym1-HA was associated with the soluble or insoluble (membrane) fraction of a cellular extract, a fractionation experiment using the lysate of spheroplasted cells was performed (Fig. 6A). More than 95% of Sym1-HA cofractionated with the mitochondrial membrane proteins porin and AAC after medium-speed centrifugation at 16,100 × g, while only a small percentage remained in the supernatant with the soluble cytoplasmic protein phosphoglycerate kinase (PGK), verifying that Sym1 behaved as a membrane-associated protein. To verify the predicted integral membrane nature of the protein, medium-speed (16,100 × g) membrane fractions were treated with buffer, 1% Triton X-100, or 0.1 M sodium carbonate and supernatant and pellet fractions were generated by centrifugation. The solubility of Sym1 and known integral membrane proteins porin and AAC was determined by Western blotting (Fig. 6B). As expected for an integral membrane protein and as demonstrated by blotting for AAC-porin, Sym1 was solubilized after treatment with Triton X-100 but remained insoluble with sodium carbonate treatment, which only removes peripheral membrane proteins. These data indicate that Sym1, like other investigated members of the Pmp22/Mpv17 family, is an integral membrane protein.
FIG. 6.
Sym1 is an integral membrane protein of the mitochondrial inner membrane. (A) sym1Δ cells expressing the Sym1-HA fusion were grown in SC-URA, harvested, spheroplasted, and lysed by Dounce homogenization. The sample was split into two equal aliquots, one of which was held as the total cell extract (T); the other was subjected to centrifugation at 16,100 × g, and both supernatant (S) and membrane pellet (P) fractions were isolated. Equivalent amounts of all fractions were subjected to SDS-PAGE and Western blotting to identify the indicated proteins. (B) sym1Δ cells expressing the Sym1-HA fusion were grown in SC-URA, harvested, and lysed using glass beads. A medium-speed membrane pellet was generated by centrifugation at 16,100 × g. The membrane pellet was split into thirds and treated with buffer, 0.1 M NaCO3, or 1% Triton X-100, and the aliquots were again centrifuged at 16,100 × g to isolate membrane pellet (P) and supernatant (S) fractions. SDS-PAGE and Western blotting was carried out as described for panel A. (C) Mitochondria were isolated for the protease protection assay as described in Materials and Methods. Mitochondria were split into equal aliquots and diluted into osmotically supportive buffer (mitochondria) or nonsupportive buffer (mitoplasts), which selectively ruptures the outer membrane. These samples were further diluted into three equivalent aliquots and treated with buffer, proteinase K (100 μg/ml), or proteinase K-1% Triton X-100. Equivalent amounts of all samples were resolved with SDS-PAGE and subjected to Western blotting with antisera as described previously. Fis1 was used as an outer membrane protein control.
Establishing Sym1 as a mitochondrial membrane protein provoked investigation into whether Sym1 was localized to the inner or outer membrane of mitochondria. To determine in which membrane Sym1 resides, a protease protection assay was performed. Preparations of enriched mitochondria with intact or disrupted outer membranes (mitoplasts) were treated with proteinase K in the presence or absence of Triton X-100, as shown in Fig. 6C. Outer mitochondrial membrane proteins are degraded by proteinase K treatment in both intact mitochondria and mitoplasts, as demonstrated by the results seen with the outer membrane protein Fis1, while inner mitochondrial membrane proteins are sensitive to proteinase K only in mitoplast preparations, as demonstrated by results seen with the inner membrane protein, AAC (32, 36). Triton X-100 treatment results in sensitivity of both inner and outer mitochondrial membrane proteins to protease degradation due to disruption of both mitochondrial membranes. Sym1-HA exhibited a significant increase in proteinase K sensitivity treatment in mitoplasts when compared to the results seen with intact mitochondria in a manner similar to the inner membrane localization of AAC. We repeatedly observed a minor, faster-migrating species of Sym1-HA in our proteinase K treatments. Because the HA epitope is located at the carboxyl terminus, a likely explanation for this additional band is cleavage of a hypersensitive amino-terminal region, perhaps due to minute amounts of exogenously added protease accessing Sym1-HA. Another possibility is that portions of Sym1, especially the amino terminus, may extend into or through the outer membrane. However, the bulk of Sym1-HA was protected from protease in intact mitochondria while Fis1 was digested, whereas levels of both proteins and AAC were dramatically reduced in protease-treated mitoplasts. Taken together, these data strongly suggest that unlike members of the peroxisome resident Mpv17/Pmp22 family, the Sym1 protein in yeast localizes to the mitochondrial inner membrane, likely as a polytopic integral membrane protein.
sym1Δ cells exhibit altered transcript levels of several metabolic genes.
The nature of the growth defect of sym1Δ cells on ethanol-containing medium, as well as its localization in the mitochondria, suggests a possible role for Sym1 in respiration-linked metabolic processes at elevated temperature. To explore this hypothesis on a global scale, DNA microarray analysis was performed using RNA extracted from wild-type and sym1Δ cells grown in ethanol at 30 and 37°C. Importantly, there was no significant difference in the transcript profiles of the two strains at 30°C (Trott and Morano, unpublished). Additionally, essentially no genes were downregulated in the sym1Δ strain relative to the wild-type results at 37°C. However, transcriptional profiling revealed moderately increased expression of a number of genes in sym1Δ cells at 37°C relative to wild-type results. The transcript levels of genes displaying a twofold or higher increase in two repeated arrays in comparison to wild-type results were considered to be significant and are listed by functional category in Table 1. The largest class of genes identified in our analysis is metabolic genes, suggesting metabolic deregulation in sym1Δ cells. Interestingly, many of these genes, including PDC5, PGI1, PMA2, FKS1, and PIR1, are not normally responsive to ethanol growth or heat shock (17, 24). In addition, the upregulation of both cytosolic and mitochondrial Hsp70 molecular chaperone genes, both previously known to be involved in mitochondrial protein translocation, suggests that sym1Δ cells may be experiencing some form of organellar stress (11, 12).
DISCUSSION
The combined effects of ethanol and heat on cellular growth have been previously established as detrimental factors leading to a significant decrease in growth rate or viability (46, 47, 54). These effects are likely due to the denaturing properties of both heat and high concentrations of ethanol on the structure and stability of both lipids and proteins, leading to a synergistic effect compared to the results with heat or ethanol alone. To date, analysis of cellular pathways responsible for adaptation and growth under these conditions has focused on the role played by stress-specific response pathways and their induced genes. Our identification of Sym1, a previously unknown mitochondrial heat shock protein required for growth on ethanol at elevated temperatures, establishes a novel role for heat shock proteins in ethanol metabolism at ethanol concentrations well below established toxicity levels.
The ability of sym1Δ cells to grow at 37°C on other nonfermentable carbon sources such as glycerol argues against a general respiratory block imposed by growth at the restrictive temperature. Examples of respiratory defects apparent only during growth at 37°C include deletions of genes encoding components of the electron transport chain (QCR6) and the outer membrane pore protein porin (POR1) (4, 60). The observed ability of sym1Δ mutants to resume growth at 30°C on ethanol-containing medium after extensive exposure to heat suggests that SYM1 is not required for maintenance of mitochondrial DNA integrity during 37°C growth and that extensive mitochondrial damage does not occur. The ethanol growth phenotype of sym1Δ cells is not ameliorated by the addition of glycerol, although growth at 37°C on glycerol alone occurs at wild-type rates. These findings imply that a “dominant” metabolic block ensues in sym1Δ cells during ethanol consumption. This conclusion is further supported by the observation that wild-type cells likewise experience a reduction in growth rate on glycerol in the presence of ethanol and heat (Fig. 1B). Furthermore, sym1Δ cells exhibit a growth rate comparable to the wild-type rate on medium containing both glucose and ethanol. Growth under these conditions is likely to depend primarily on fermentative pathways, with a concomitant reduction in respiratory gene expression and therefore in ethanol metabolism. The suppression of ethanol metabolism by glucose may therefore be sufficient to block growth inhibition by ethanol in wild-type and sym1Δ cells. In support of this hypothesis, we have begun preliminary characterization of high-copy-number suppressors of the sym1Δ 37°C phenotype and have isolated genes that when overexpressed increase protein kinase A (PKA) activity, which is known to repress expression of respiratory genes (41, 51).
Analysis of sym1Δ growth on acetaldehyde and acetate, both intermediate carbon sources in the ethanol metabolic pathway, indicates a temperature-specific defect in the utilization of both ethanol and acetaldehyde, the substrates of the alcohol (ADH) and ALDH families, respectively. The conversion of ethanol to acetyl-coenzyme A occurs predominantly in the cytosol, although both ADH and ALDH isoforms are found in the mitochondria as well (6). Although no examples of altered ADH or ALDH expression leading to a toxicity phenotype similar to that of sym1Δ have been reported, changes in the levels or activity of either enzyme could potentially lead to alterations in levels of the intermediates of these pathways. For example, either increased ADH activity or decreased ALDH activity would lead to an increase in the levels of the toxic intermediate acetaldehyde, which could potentially result in growth inhibition on medium containing either ethanol or acetaldehyde as the carbon source. Determination of cellular or subcellular levels of ethanol and acetaldehyde in wild-type and sym1Δ cells is likely to be hampered by the fact that both compounds are highly diffusible throughout the cell and quickly equilibrate upon cell disruption (19).
The DNA microarray data displaying altered levels of a number of genes, including metabolic enzymes, in sym1Δ cells grown on ethanol at 37°C suggest a metabolic defect. It is not clear whether the basis for the observed gene deregulation is a cause or consequence of the growth phenotype. However, the increased expression of several metabolic genes, such as PDC5, PGI1, and PMA2, normally repressed under respiratory conditions is particularly intriguing. For example, PDC5 encodes pyruvate decarboxylase, which is required for the decarboxylation of pyruvate to ethanol during fermentative growth, and is strongly repressed during respiratory growth (24, 45). Derepression of PDC5 therefore signifies a defect in carbon source sensing or respiratory gene repression in sym1Δ cells at elevated temperature. Alterations in the levels of metabolic enzymes in sym1Δ cells may result in a buildup of toxic intermediates during growth on ethanol at 37°C. Metabolism of glycerol under these conditions could also be negatively affected by such intermediates, providing a potential explanation for the lack of growth of sym1Δ cells on glycerol medium supplemented with ethanol but not on glycerol medium alone. Although a more detailed analysis of the specific genes showing altered expression in sym1Δ cells is required, two conclusions can be drawn from the microarray data. First, sym1Δ cells do not display gross changes in gene expression that might be expected if the cells were experiencing substantial oxidative stress, protein unfolding, or other cellular damage. Second, the observed changes in gene expression only occur at 37°C in ethanol medium, in keeping with the lack of a detectable growth phenotype at 30°C. These data further support the hypothesis that SYM1 is required for a metabolic or growth process that is only markedly impaired when cells are growing using ethanol as a carbon source concomitant with the duress of heat shock.
An intriguing aspect of this investigation is the functional orthology between Sym1 and Mpv17, previously shown to be essential for maintaining normal kidney function in the mouse. We have established conservation of function between Sym1 and Mpv17 through the complementation of the sym1Δ ethanol defect by expression of human Mpv17. Interestingly, the mitochondrial localization of Sym1 in yeast differs strikingly from the reported peroxisomal localization of Mpv17 and suggests a compartmentalization divergence between members of this protein family. Several lines of evidence suggest that Sym1 functions predominantly in the mitochondrial compartment in yeast. First, the fully functional Sym1-GFP fusion protein exhibited the classic reticulated network of mitochondria, demonstrating complete overlap with the mitochondrion-specific dye MitoTracker. Second, this localization pattern was unaffected in pex mutant strains that block peroxisome biogenesis (22). Third, a recently published proteomic analysis of S. cerevisiae mitochondria identified Sym1 as one of 750 proteins localized to this organelle by highly purified mitochondria and tandem mass spectrometry (48). Lastly, a similar proteomic analysis of yeast peroxisomal membranes failed to identify Sym1 (44). While we cannot definitively exclude the possibility that a minor subpopulation of Sym1 resides in the peroxisomal membrane, we think it highly likely that Sym1 carries out its essential stress and respiratory functions in the mitochondrion.
Two scenarios can be envisioned to explain how Mpv17 and Sym1 can be functional orthologs in different organelles. Mpv17, like Sym1, may localize to the yeast mitochondrial inner membrane, suggesting that the relevant pathway in which both proteins operate may reside in the mitochondria in yeast. Alternatively, Mpv17 may retain its peroxisomal targeting determinants in yeast and may be able to carry out its mechanistic role from that membrane. There is precedent for members of the same protein family being differentially localized to distinct organelles in fungi: the Candida boidinii peroxisomal membrane protein PMP47 is highly homologous to the large family of mitochondrial nucleotide carrier proteins (29). Additionally, S. cerevisiae possesses two citrate synthase genes, CIT1 and CIT2, encoding proteins targeted to mitochondria and peroxisomes, respectively (26, 42). We have attempted to determine the subcellular localization of Mpv17 expressed in yeast but have been unable to generate an epitope-tagged Mpv17 allele that retains function as assayed by complementation (Trott and Morano, unpublished). Although little is known about the function of the members of the Pmp22/Mpv17 family, a permeability channel activity was found to be reconstituted in liposomes with peroxisomal membrane fractions from rat liver enriched with a 22-kDa protein (56). Moreover, two reports identified a channel in plant peroxisomes with properties distinct from a classic porin (37, 38). Little mechanistic information exists for the Mpv17 protein except for a putative link with ROS metabolism and upregulation of the matrix metalloproteinase gene MMP-2 (3, 39, 57, 61). We have found no evidence for increased ROS or oxidative damage in sym1Δ cells (Trott and Morano, unpublished). We favor the hypothesis that Sym1, possibly in a multimeric structure, possesses a pore or channel-like activity in the mitochondrial inner membrane required for the transport of small molecules into or out of the mitochondrial matrix under heat shock conditions. This activity, which is required in mammals for peroxisomal metabolic processes, may have diverged evolutionarily from a yeast mitochondrial metabolic process. The absence of the pore during ethanol metabolism at elevated temperatures could result in the inhibition of enzymatic reactions or signaling events required for cell proliferation. The lack of mechanistic information for the Pmp22/Mpv17 protein family, and the demonstrated importance of Mpv17 as one of the few mouse models of renal disease, makes the yeast Sym1 protein an attractive surrogate for structure-function studies.
Acknowledgments
We thank Carla Koehler, Jodi Nunnari, Richard Rachubinski, and Joel Goodman for antibodies and Stephen Elledge for the human cDNA library. We are indebted to Franco Vizeacoumar, Richard Rachubinski, Jesus Eraso, and James McNew for helpful discussions. We also thank William Margolin for use of his fluorescence microscopy facility.
This work was supported by the American Heart Association (Beginning Grant-in-Aid 0160113Y) and grant MBC-103134 from the American Cancer Society. K. Morano is an American Cancer Society Research Scholar.
REFERENCES
- 1.Aguilera, A., and T. Benitez. 1986. Ethanol-sensitive mutants of Saccharomyces cerevisiae. Arch. Microbiol. 143:337-344. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, A., and T. Benitez. 1985. Role of mitochondria in ethanol tolerance of Saccharomyces cerevisiae. Arch. Microbiol. 142:389-392. [DOI] [PubMed] [Google Scholar]
- 3.Binder, C. J., H. Weiher, M. Exner, and D. Kerjaschki. 1999. Glomerular overproduction of oxygen radicals in mpv17 gene-inactivated mice causes podocyte foot process flattening and proteinuria: a model of steroid-resistant nephrosis sensitive to radical scavenger therapy. Am. J. Pathol. 154:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blachly-Dyson, E., S. Peng, M. Colombini, and M. Forte. 1990. Selectivity changes in site-directed mutants of the VDAC ion channel: structural implications. Science 247:1233-1236. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boubekeur, S., N. Camougrand, O. Bunoust, M. Rigoulet, and B. Guerin. 2001. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 268:5057-5065. [DOI] [PubMed] [Google Scholar]
- 7.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves, R. S., P. Herrero, I. Ordiz, M. Angeles del Brio, and F. Moreno. 1997. Isocitrate lyase localisation in Saccharomyces cerevisiae cells. Gene 198:165-169. [DOI] [PubMed] [Google Scholar]
- 9.Costa, V., E. Reis, A. Quintanilha, and P. Moradas-Ferreira. 1993. Acquisition of ethanol tolerance in Saccharomyces cerevisiae: the key role of the mitochondrial superoxide dismutase. Arch. Biochem. Biophys. 300:608-614. [DOI] [PubMed] [Google Scholar]
- 10.Craig, E. A., B. K. Baxter, J. Becker, J. Halladay, and T. Ziegelhoffer. 1994. Cytosolic hsp70s of Saccharomyces cerevisiae: roles in protein synthesis, protein translocation, proteolysis, and regulation, p. 31-52. In R. I. Morimoto, A. Tissieres, and C. Georgopolous (ed.), The biology of heat shock proteins and molecular chaperones, vol. 26. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 11.Craig, E. A., B. D. Gambill, and R. J. Nelson. 1993. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57:402-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig, E. A., J. Kramer, J. Shilling, M. Werner-Washburne, S. Holmes, J. Kosic-Smithers, and C. M. Nicolet. 1989. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol. Cell. Biol. 9:3000-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elledge, S. J., J. T. Mulligan, S. W. Ramer, M. Spottswood, and R. W. Davis. 1991. Lambda yes: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc. Natl. Acad. Sci. USA 88:1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, B., R. S. Haltiwanger, and B. Futcher. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein, C. B., J. A. Waddle, W. Hale IV, V. Dave, J. Thornton, T. L. Macatee, H. R. Garner, and R. A. Butow. 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdmann, R., and S. J. Gould. 2002. Visualization and purification of peroxisomes. Methods Enzymol. 351:365-381. [DOI] [PubMed] [Google Scholar]
- 17.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 19.Guijarro, J. M., and R. Lagunas. 1984. Saccharomyces cerevisiae does not accumulate ethanol against a concentration gradient. J. Bacteriol. 160:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding, T. M., K. A. Morano, S. V. Scott, and D. J. Klionsky. 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmerhorst, E. J., P. Breeuwer, W. van't Hof, E. Walgreen-Weterings, L. C. Oomen, E. C. Veerman, A. V. Amerongen, and T. Abee. 1999. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274:7286-7291. [DOI] [PubMed] [Google Scholar]
- 22.Hettema, E. H., W. Girzalsky, M. van Den Berg, R. Erdmann, and B. Distel. 2000. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann, K., and W. Stoffel. 1993. TMbase-a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 24.Hohmann, S. 1993. Characterisation of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 241:657-666. [DOI] [PubMed] [Google Scholar]
- 25.Koch, K. A., and D. J. Thiele. 1996. Autoactivation by a Candida glabrata copper metalloregulatory transcription factor requires critical minor groove interactions. Mol. Cell. Biol. 16:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewin, A. S., V. Hines, and G. M. Small. 1990. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol. Cell. Biol. 10:1399-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, X. D., P. C. Liu, N. Santoro, and D. J. Thiele. 1997. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 16:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luers, G. H., D. M. Otte, S. Subramani, and T. Franz. 2001. Genomic organization, chromosomal localization and tissue specific expression of the murine pxmp2 gene encoding the 22 kDa peroxisomal membrane protein (Pmp22). Gene 272:45-50. [DOI] [PubMed] [Google Scholar]
- 29.McCammon, M. T., C. A. Dowds, K. Orth, C. R. Moomaw, C. A. Slaughter, and J. M. Goodman. 1990. Sorting of peroxisomal membrane protein Pmp47 from Candida boidinii into peroxisomal membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265:20098-20105. [PubMed] [Google Scholar]
- 30.Meyer zum Gottesberge, A. M., A. Reuter, and H. Weiher. 1996. Inner ear defect similar to Alport's syndrome in the glomerulosclerosis mouse model Mpv17. Eur. Arch. Oto-Rhino-Laryngol. 253:470-474. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto, R. I. 1993. Cells in stress: transcriptional activation of heat shock genes. Science 259:1409-1410. [DOI] [PubMed] [Google Scholar]
- 32.Mozdy, A. D., J. M. McCaffery, and J. M. Shaw. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller, M., J. W. Smolders, A. M. Meyer zum Gottesberge, A. Reuter, R. M. Zwacka, H. Weiher, and R. Klinke. 1997. Loss of auditory function in transgenic mpv17-deficient mice. Hear. Res. 114:259-263. [DOI] [PubMed] [Google Scholar]
- 34.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 35.Nicolet, C. M., and E. A. Craig. 1989. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9:3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfanner, N., and W. Neupert. 1987. Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem. 262:7528-7536. [PubMed] [Google Scholar]
- 37.Reumann, S., E. Maier, R. Benz, and H. W. Heldt. 1995. The membrane of leaf peroxisomes contains a porin-like channel. J. Biol. Chem. 270:17559-17565. [DOI] [PubMed] [Google Scholar]
- 38.Reumann, S., E. Maier, H. W. Heldt, and R. Benz. 1998. Permeability properties of the porin of spinach leaf peroxisomes. Eur. J. Biochem. 251:359-366. [DOI] [PubMed] [Google Scholar]
- 39.Reuter, A., A. Nestl, R. M. Zwacka, J. Tuckermann, R. Waldherr, E. M. Wagner, M. Hoyhtya, A. M. Meyer zum Gottesberge, P. Angel, and H. Weiher. 1998. Expression of the recessive glomerulosclerosis gene MPV17 regulates MMP-2 expression in fibroblasts, the kidney, and the inner ear of mice. Mol. Biol. Cell 9:1675-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieger, K. J., A. Kaniak, J. Y. Coppee, G. Aljinovic, A. Baudin-Baillieu, G. Orlowska, R. Gromadka, O. Groudinsky, J. P. Di Rago, and P. P. Slonimski. 1997. Large-scale phenotypic analysis—the pilot project on yeast chromosome III. Yeast 13:1547-1562. [DOI] [PubMed] [Google Scholar]
- 41.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenkrantz, M., T. Alam, K.-S. Kim, B. J. Clark, P. A. Srere, and L. P. Guarente. 1986. Mitochondrial and nonmitochondrial citrate synthases in Saccharomyces cerevisiae are encoded by distinct homologous genes. Mol. Cell. Biol. 6:4509-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai, Y., A. Koller, L. K. Rangell, G. A. Keller, and S. Subramani. 1998. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer, H., K. Nau, A. Sickmann, R. Erdmann, and H. E. Meyer. 2001. Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22:2955-2968. [DOI] [PubMed] [Google Scholar]
- 45.Seeboth, P. G., K. Bohnsack, and C. P. Hollenberg. 1990. Pdc10 mutants of Saccharomyces cerevisiae give evidence for an additional structural PDC gene: cloning of PDC5, a gene homologous to PDC1. J. Bacteriol. 172:678-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, F. 1959. The effects of elevated temperatures on yeast: II. Induction of respiratory-deficient mutants. J. Cell. Comp. Physiol. 54:37-52. [DOI] [PubMed] [Google Scholar]
- 47.Sherman, F. 1956. The heat inactivation and production of cytochrome deficiency in yeast. Exp. Cell Res. 11:659-670. [DOI] [PubMed] [Google Scholar]
- 48.Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schonfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100:13207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer, M. A., and S. Lindquist. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1:639-648. [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thevelein, J. M., L. Cauwenberg, S. Colombo, J. H. De Winde, M. Donation, F. Dumortier, L. Kraakman, K. Lemaire, P. Ma, D. Nauwelaers, F. Rolland, A. Teunissen, P. Van Dijck, M. Versele, S. Wera, and J. Winderickx. 2000. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26:819-825. [DOI] [PubMed] [Google Scholar]
- 52.Trott, A., and K. A. Morano. 2003. The yeast response to heat shock, p. 71-119. In S. Hohmann and P. W. H. Mager (ed.), Yeast stress responses, vol. 1. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 53.Trotter, E. W., L. Berenfeld, S. A. Krause, G. A. Petsko, and J. V. Gray. 2001. Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trotter, E. W., C. M. Kao, L. Berenfeld, D. Botstein, G. A. Petsko, and J. V. Gray. 2002. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 277:44817-44825. [DOI] [PubMed] [Google Scholar]
- 55.Tugal, H. B., M. Pool, and A. Baker. 1999. Arabidopsis 22-kilodalton peroxisomal membrane protein. Nucleotide sequence analysis and biochemical characterization. Plant Physiol. 120:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Veldhoven, P. P., W. W. Just, and G. P. Mannaerts. 1987. Permeability of the peroxisomal membrane to cofactors of beta-oxidation. Evidence for the presence of a pore-forming protein. J. Biol. Chem. 262:4310-4318. [PubMed] [Google Scholar]
- 57.Wagner, G., K. Stettmaier, W. Bors, H. Sies, E. M. Wagner, A. Reuter, and H. Weiher. 2001. Enhanced gamma-glutamyl transpeptidase expression and superoxide production in MPV17−/− glomerulosclerosis mice. Biol. Chem. 382:1019-1025. [DOI] [PubMed] [Google Scholar]
- 58.Weiher, H., T. Noda, D. A. Gray, A. H. Sharpe, and R. Jaenisch. 1990. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell 62:425-434. [DOI] [PubMed] [Google Scholar]
- 59.Yaffe, M. 1991. Analysis of mitochondrial function and assembly. Methods Enzymol. 194:627-643. [DOI] [PubMed] [Google Scholar]
- 60.Yang, M., and B. L. Trumpower. 1994. Deletion of QCR6, the gene encoding subunit six of the mitochondrial cytochrome bc1 complex, blocks maturation of cytochrome c1, and causes temperature-sensitive petite growth in Saccharomyces cerevisiae. J. Biol. Chem. 269:1270-1275. [PubMed] [Google Scholar]
- 61.Zwacka, R. M., A. Reuter, E. Pfaff, J. Moll, K. Gorgas, M. Karasawa, and H. Weiher. 1994. The glomerulosclerosis gene MPV17 encodes a peroxisomal protein producing reactive oxygen species. EMBO J. 13:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]