Abstract
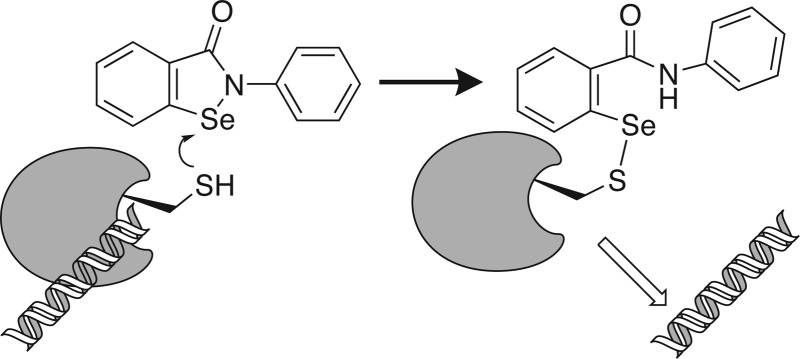
The hepatitis C virus (HCV) nonstructural protein 3 (NS3) is both a protease, which cleaves viral and host proteins, and a helicase that separates nucleic acid strands, using ATP hydrolysis to fuel the reaction. Many antiviral drugs, and compounds in clinical trials, target the NS3 protease, but few helicase inhibitors that function as antivirals have been reported. This study focuses on the analysis of the mechanism by which ebselen (2-phenyl-1,2-benzisoselenazol-3-one), a compound previously shown to be a HCV antiviral agent, inhibits the NS3 helicase. Ebselen inhibited the abilities of NS3 to unwind nucleic acids, to bind nucleic acids, and to hydrolyze ATP, and about 1 μM ebselen was sufficient to inhibit each of these activities by 50%. However, ebselen had no effect on the activity of the NS3 protease, even at 100 times higher ebselen concentrations. At concentrations below 10 μM, the ability of ebselen to inhibit HCV helicase was reversible, but prolonged incubation of HCV helicase with higher ebselen concentrations led to irreversible inhibition and the formation of covalent adducts between ebselen and all 14 cysteines present in HCV helicase. Ebselen analogues with sulfur replacing the selenium were just as potent HCV helicase inhibitors as ebselen, but the length of the linker between the phenyl and benzisoselenazol rings was critical. Modifications of the phenyl ring also affected compound potency over 30-fold, and ebselen was a far more potent helicase inhibitor than other, structurally unrelated, thiol-modifying agents. Ebselen analogues were also more effective antiviral agents, and they were less toxic to hepatocytes than ebselen. Although the above structure–activity relationship studies suggest that ebselen targets a specific site on NS3, we were unable to confirm binding to either the NS3 ATP binding site or nucleic acid binding cleft by examining the effects of ebselen on NS3 proteins lacking key cysteines.
The hepatitis C virus (HCV) is a positive sense RNA virus that causes chronic liver disease in roughly 2% of the world’s population. HCV causes profound morbidity and mortality and is a leading cause of fibrosis, cirrhosis, hepatocellular carcinoma, and liver failure. The HCV RNA genome encodes a single open reading frame that is translated from an internal ribosome entry site (IRES). Host and viral proteases cleave the resulting proteins into structural (core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins. After HCV was first isolated in 1988, numerous academic and industrial laboratories intensely studied each of the HCV proteins as possible drug targets.1 These efforts led to the design of many direct acting antivirals, most of which target the NS3 protease, the NS5B polymerase, or the NS5A RNA binding protein. Three of these NS3 protease inhibitors and one NS5B polymerase inhibitor have been approved to treat HCV. Few inhibitors that act as antivirals have been identified for the other HCV encoded enzymes, namely, the NS2 protease and the NS3 helicase, which is the subject of this study.2,3
The NS3 proteins encoded by HCV and related viruses are the only known proteins that contain both protease and helicase active sites. The NS3 protease function resides in the N-terminal domains, which fold into a cashew-shaped structure, with a serine protease active site in a shallow cleft. The NS3 protease cleaves the NS3–NS4A, NS4A–NS4B, NS4B–NS5A, NS5A–NS5B junctions and some cellular proteins, like the mitochondrial antiviral signaling protein (MAVS)4 and the Toll-like receptor 3 adaptor protein TRIF.5 The NS3 protease is active only when it binds the NS4A protein. The NS3 helicase activity, which unwinds duplex RNA and DNA and RNA/DNA hybrids in a reaction fueled by ATP hydrolysis, resides in the C-terminal domains of NS3. The two N-terminal helicase domains resemble the RecA-like motor domains seen in all other helicases and related nucleic acid translocating motor proteins. The third helicase domain is composed mainly of alpha helices, and it does not resemble domains seen in other related superfamily 2 helicases. ATP binds between the two motor domains,6 and one strand of nucleic acid binds in the cleft that separates the motor domains from the C-terminal helicase domain.7
The NS3 helicase is a remarkably difficult protein to inhibit with small molecules. Most high-throughput screens designed to identify inhibitors of NS3 helicase-catalyzed DNA strand separation identify few inhibitors, and most inhibitors identified in vitro are either toxic or do not act as antivirals in cells. We therefore reasoned that screening collections of compounds that are already known to inhibit HCV replication in cells using an assay designed to detect helicase inhibitors might more easily identify antivirals that target HCV helicase. The assay we chose was a recently reported nucleic acid binding assay that uses fluorescence polarization to find compounds that displace single-stranded DNA (ssDNA) from recombinant truncated NS3 lacking the first 163 amino acids, which encode the protease (called here NS3h).8
We decided to screen the NIH clinical collection because it was recently screened for compounds that inhibit HCV replication in human hepatocytes, and about 17% of the compounds in the collection showed some antiviral activity.9 Gastaminza et al. used the infectious HCV genotype 2a HCV isolate (called JFH1)10 to infect cells in the presence of various compounds in the NIH collection, and they measured the amount of the HCV E2 protein present in each assay using a colorimetric assay. After comparing the amount of E2 present with the amount of cells remaining after compound exposure, as determined by staining with crystal violet, they found 76 nontoxic antivirals that inhibited HCV replication more than 50% out of 446 tested.9
As described below, our screen of the NIH clinical collection identified only three helicase inhibitors, and all but one functioned nonspecifically, meaning that they also prevented an unrelated nucleic acid binding protein from binding the same fluorescent oligonucleotide. The one specific helicase inhibitor in the collection was a selenium-containing compound called ebselen (PubChem CID 3194), which is known to form covalent adducts with cysteines in other protein targets.11,12 In the present study, we also characterize the mechanism of action whereby ebselen inhibits the HCV NS3 helicase. We show that ebselen, and similar compounds where the selenium is replaced by sulfur, can modify all of the cysteines in NS3h. However, its ability to displace NS3h from nucleic acids does not appear to be related to modifications of cysteines in the nucleic acid binding cleft, as one might suspect.
Results
Ebselen Prevents NS3h from Binding Nucleic Acids
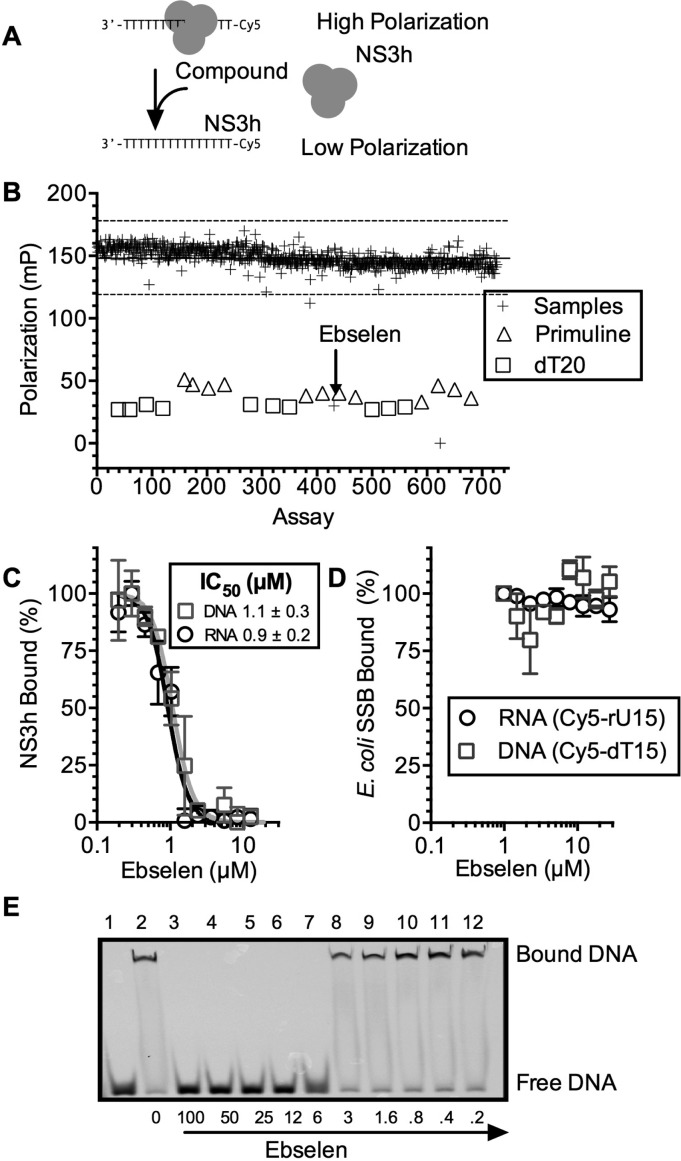
The NIH clinical collection sets 1 (446 compounds) and 2 (281 compounds) contain a diverse variety of compounds that have been used in human clinical trials. Each NIH clinical collection compound was included in a FP-based binding assay to assess its ability to displace HCV NS3h from the oligonucleotide Cy5-dT15 (Figure 1A).8 In the assay, each compound was added to a final concentration of 100 μM to a solution containing 15 nM NS3h and 5 nM Cy5-dT15. The resulting fluorescence polarization was normalized to that observed in negative control assays with DMSO only or positive controls containing 100 μM primuline, which was previously shown to prevent NS3h from binding DNA or RNA.13 In additional positive control assays, an unlabeled oligonucleotide (dT20) was added to a final concentration of 100 nM (Figure 1B).
Figure 1.
Ebselen specifically inhibits the ability of HCV helicase to bind nucleic acids. (A) FP-based assay to monitor HCV NS3h (or Escherichia coli SSB) binding to a Cy5-labeled oligonucleotide. (B) Ability of compounds in the NIH clinical collection to inhibit NS3h–DNA binding. All compounds were tested at 100 μM. Positive control assays contained primuline (triangles) or dT20 (squares). The solid line represents the mean of all assays, and the dotted lines three standard deviations around the mean. (C) Cy5-dT15–NS3h (squares) or Cy5-rU15–NS3h (circles) complexes were titrated with ebselen (n = 3, normalized mean ± SD). (D) Cy5-dT15–SSB (squares) or Cy5-rU15–SSB (circles) complexes were titrated with ebselen. (E) Electrophoretic mobility shift assay (EMSA). Samples containing the Cy5-dT15 (20 nM), NS3h (200 nM), and various concentrations of ebselen were examined on a 15% native polyacrylamide gel (lanes 3–12). Lane 1, no NS3h; lane 2, no ebselen.
In the FP-based binding assays, only three compounds in the collection inhibited the ability of the protein to bind DNA by more than 3 times the standard deviation observed with all samples (Figure 1B and Table 1). The Z′ factor14 for this screen was 0.91. Only two of these compounds inhibited more than 50%: the DNA topoisomerase II inhibitor, mitoxantrone (CID 5458171), and the hydrogen peroxide scavenger, ebselen (CID 3194). Both inhibited the ability of NS3h to bind either DNA (Cy5-dT15) or RNA (Cy5-rU15) in a concentration-dependent manner (Table 1 and Figure 1C), but only ebselen had no effect on SSB binding to DNA or RNA (Figure 1D). About 1 μM ebselen was needed to reduce the polarization of an NS3h-Cy5-dT15 or NS3h–Cy5-rU15 complex by 50% (IC50) (Figure 1C). The interaction of ebselen with NS3h-bound Cy5-dT20 was also examined using nondenaturing gel electrophoresis to confirm that ebselen displaced NS3h from DNA in an orthogonal assay (Figure 1E).
Table 1. Compounds in the NIH Clinical Collections That Prevented Either HCV NS3h or E. coli SSB from Binding an Oligonucleotide.
| compd | PubChem CID | NS3h (% I)a | SSB (% I)b | HCV (% I)c |
|---|---|---|---|---|
| Mitoxantrone | 5458171 | 125 | 121 | n.d.d |
| Ebselen | 3194 | 99 | 3 | 83 |
| HMS2052E19 | 23581806 | 28 | 95 | <0 |
| Seapuron | 6410757 | 22 | 56 | <0 |
| Spectrum 001824 | 6398970 | 11 | 96 | 5.8 |
| Doxorubicin Hydrochloride | 443939 | 8 | 90 | 100 |
| L 694247 | 23581822 | 3 | 37 | <0 |
| SR 57227A | 131746 | –1 | 133 | 12 |
| Rolitetracycline | 6420073 | –4 | 45 | <0 |
| 5-Nonyloxy-tryptamine | 23581825 | –8 | 67 | 100 |
Percent inhibition of NS3h binding to Cy5-dT15 in the presence of 100 μM of each compound.
Percent inhibition of SSB binding to Cy5-dT15 in the presence of 100 μM of each compound.
Percent inhibition of HCV replication in cells in the presence of 20 μM of each compound. Data are from Gastaminza et al.9
Not determined.
To test if compounds might inhibit NS3h nonspecifically, the same FP-based assay was repeated substituting NS3h with the Escherichia coli single-stranded DNA binding protein (SSB). Nine compounds in the NIH clinical collections inhibited the ability of SSB to bind a Cy5-labeled oligonucleotide (Z′ factor = 0.85, Table 1), including two of the compounds that inhibited NS3h binding. However, ebselen did not inhibit the ability of E. coli SSB to bind Cy5-dT15, consistent with the hypothesis that the interactions seen in the NS3h binding assays were not due to a nonspecific interaction of ebselen with Cy5-dT15 or Cy5-rU15 (Figure 1D). In contrast, mitoxantrone inhibited E. coli SSB–Cy5-dT15 to a similar extent as NS3h, and marginal NS3h inhibitors like HMS2052E19 (PubChem CID 23581806) and seapuron (PubChem CID 6410757) inhibited SSB binding 3–4 times more than NS3h binding. Other compounds in the collection only inhibited SSB binding to DNA (Table 1).
The Ability of Ebselen To Disrupt NS3h–Nucleic Acid Interactions Depends on Reaction Conditions
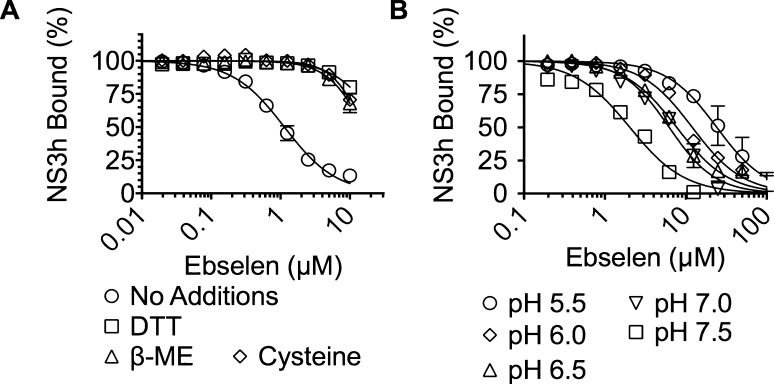
Ebselen is a highly reactive compound known to form selenium–sulfur bonds with reduced thiols in proteins.15 There are 14 cysteines in the NS3h protein used in the above assays and none in E. coli SSB, which could explain the observed specificity. If ebselen functions by modifying cysteines in NS3h, then adding free cysteine or other compounds with reduced thiols should abrogate the ability of ebselen to inhibit HCV helicase. To test this hypothesis, we repeated titrations with ebselen in the presence of 50 μM dithiothreitol (DTT), 50 μM β-mercaptoethanol (β-ME), and 50 μM cysteine. The inhibitory effect of ebselen was abolished in each case, and less than 50% inhibition was observed in the presence of any thiol compound even at the highest concentration of ebselen tested (Figure 2A).
Figure 2.
Effect of thiol compounds and pH on ebselen activity. (A) The Cy5-rU15–NS3h complex was titrated with ebselen in the presence of NS3h alone (black circles), 50 μM DTT (squares), 50 μM β-mercaptoethanol (triangles), or 50 μM cysteine (diamonds). (B) The Cy5-rU15–NS3h complex was titrated with ebselen at pH 5.5 (circles), 6.0 (diamonds), 6.5 (triangles), 7.0 (inverted triangles), and 7.5 (squares). Normalized values are fit to a concentration response equation (n = 3, mean ± SD).
We also examined the effect of pH on the ability of ebselen to inhibit HCV helicase because a cysteine must lose its proton in order for it to act as a nucleophile to react with ebselen. Ebselen was a more potent inhibitor of NS3h–nucleic acid binding at higher pH than at lower pH (Figure 2B), as would be expected if ebselen inhibits NS3h by reacting with cysteines.
Ebselen Reversibly Inhibits NS3h, but Prolonged Incubation of NS3h with Ebselen Leads to Irreversible Inhibition
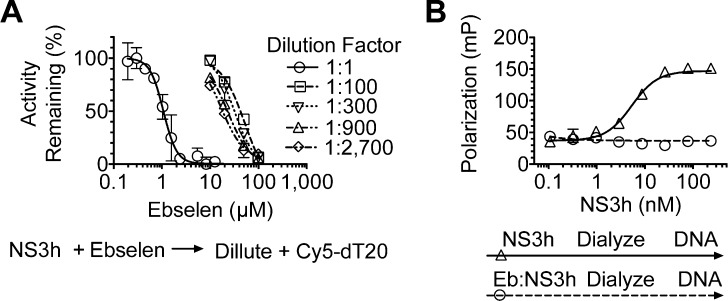
If ebselen functions by covalently modifying cysteine residues, then it could function as an irreversible inhibitor. To test this idea, we incubated 10 μM NS3h with various concentrations of ebselen for 1 h and then diluted the protein 100–100 000-fold while adding it to Cy5-dT15 to determine if the protein retained the ability to bind DNA with a similar affinity as the protein when it was treated with DMSO only. Interestingly, when NS3h was treated with 10 μM ebselen, a concentration capable of displacing all NS3h from DNA or RNA (Figure, 3A), the protein retained an ability to bind DNA after dilution of ebselen to <0.1 μM, suggesting that the reaction was reversible. However, treatments with higher concentrations of ebselen led to progressively less active enzyme upon dilution (Figure 3A).
Figure 3.
Reversibility of ebselen inhibition of a HCV helicase–DNA interaction. (A) NS3h (10 μM) was incubated with the indicated concentrations of ebselen and diluted such that it could be added to Cy5-dT15, FP was measured, and activity remaining was calculated by comparing ebselen containing assays to controls not treated with ebselen. (B) Fluorescence polarization of Cy5-dT15 (5 nM) at different concentrations of NS3h, which was dialyzed following incubation with 100 μM ebselen (circles) or 0.5% v/v DMSO (triangles). Points are the averages of values obtained from two independent assays, and the error bars are the standard deviations.
Even though the highest concentration of ebselen tested (100 μM) was diluted to below its IC50 value seen in concentration–response assays (Figure 1C), it is possible that low amounts of ebselen remaining after dilution might prevent binding. We therefore also removed ebselen after incubation with NS3h by extensively dialyzing the sample. Dialysis, however, did not restore the ability of NS3h to bind DNA (Figure 3B) after the protein was incubated with 100 μM ebselen, suggesting that an irreversible reaction occurred between NS3h and ebselen. The dialyzed NS3h that had been treated with 100 μM ebselen also lost the ability to separate DNA duplexes and hydrolyze ATP (data not shown).
Ebselen Analogues Lacking Selenium Retain the Ability To Inhibit HCV Helicase and the HCV Subgenomic Replicon
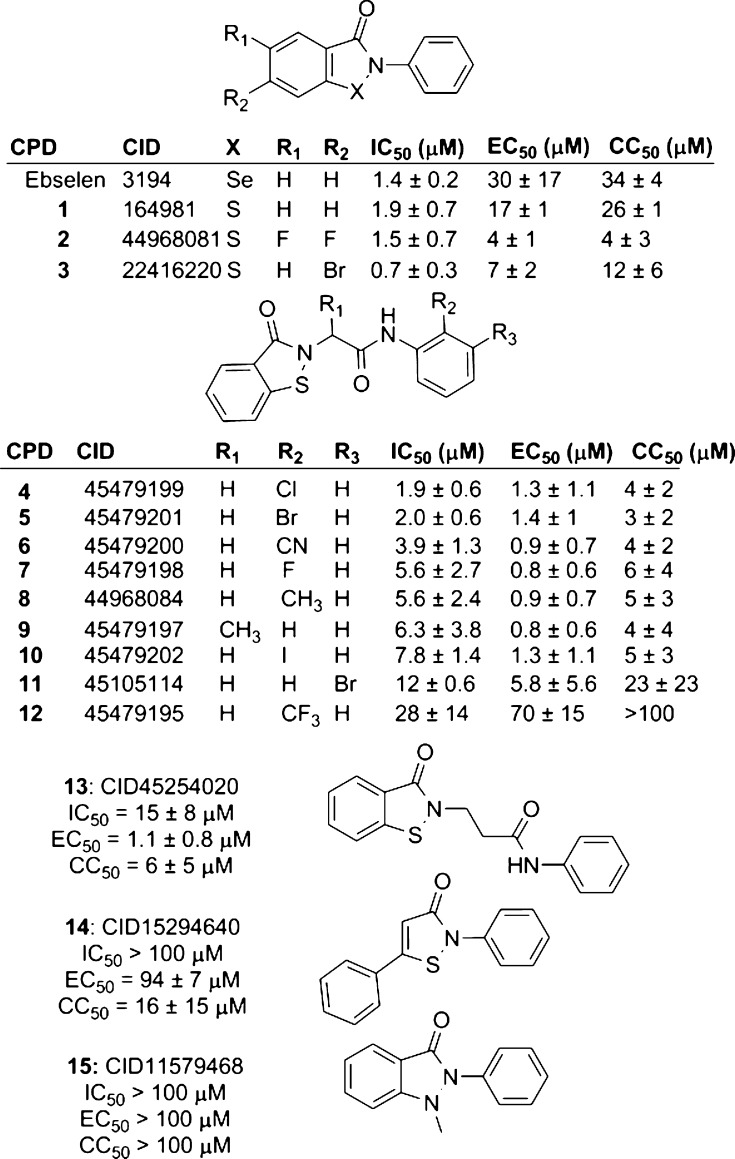
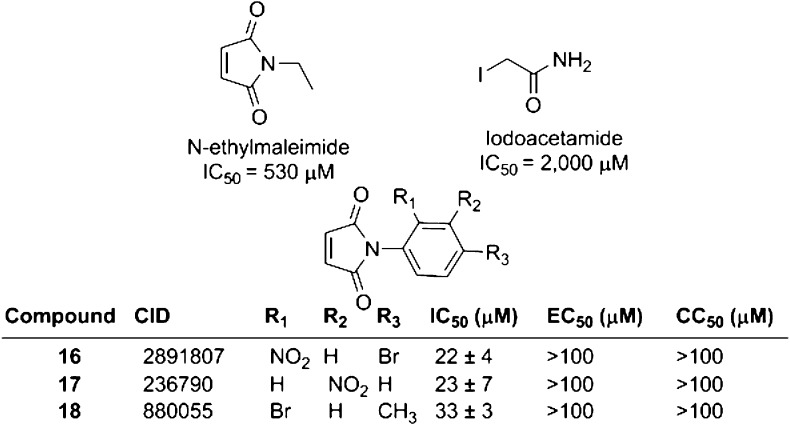
We next set out to determine if ebselen (2-phenyl-1,2-benzisoselenazol-3(2H)-one) makes specific contacts with NS3h by examining the ability of related compounds to inhibit the capacity of HCV helicase to bind DNA. First, we tested the importance of the selenium by replacing it with sulfur (compound 1, aka ebsulfur),16 and we found that the benzisothiazolone derivative was as potent as ebselen. Adding halogens to the benzisothiazolone ring system (compounds 2 and 3) did not affect activity, but other substitutions for the selenium (compound 15) and the disruption of the benzisothiazolone ring system (compound 14) were not tolerated, and both changes resulted in complete loss of activity (Figure 4).
Figure 4.
Structures of ebselen analogues and their ability to inhibit HCV helicase and HCV RNA replication. IC50 values are the concentrations of each compound needed to displace 50% of NS3h from DNA in a FP-based DNA binding assay. EC50 values are the concentrations of each compound needed to decrease Rluc activity (which reflects cellular HCV RNA content) of a subgenomic HCV replicon in Huh7.5 cells by 50%. CC50 values are the concentrations of each compound needed to reduce Huh7.5 cell viability by 50%. Three independent titrations were performed of each of the three assays. Shown are the averages, with uncertainties representing standard deviations.
To probe the role of the 2-phenyl moiety, we first asked whether the length of the linker would affect the activity of ebselen analogues. Peptide linkers (compounds 4–12) were tolerated, but a longer linker (compound 13) led to a 10-fold decrease in the ability to inhibit HCV helicase. Substitutions on the phenyl ring led to a 20-fold effect on potency, with the 2-chlorophenyl derivative (compound 4) being most active and the 2-trifluoromethyl derivative (compound 12) being least active (Figure 4).
Ebselen and its analogues were also tested for their ability to inhibit HCV RNA replication in hepatocytes using a previously described genotype 1b subgenomic replicon system in which cellular Renilla luciferase levels reflect HCV RNA content in cells.13,17 The ability of each compound to decrease the viability of the HCV replicon cell line was also measured (Figure 5). Of note is the observation that many of the new compounds appeared to have a better antiviral potential than that of ebselen. The ebselen concentration needed to reduce cell viability by 50% (CC50) was only slightly higher than the ebselen concentration needed to reduce replicon content by 50% (EC50). Ebsulfur behaved like ebselen, but the halogenated ebsulfur derivatives (2 and 3) were almost 10 times more potent. Both 2 and 3 were still relatively toxic, and they also had a therapeutic index (i.e., CC50/EC50) near 1. In contrast, the peptide derivatives showed a similarly improved antiviral activity, but they were relatively less toxic. Compound 7 had the highest therapeutic index, which was about 6 times higher than that seen with ebselen. The antiviral potential of the peptide derivatives also mimic their ability to inhibit NS3h binding to DNA, with compounds 12, 14, and 15 being the least active in both the in vitro and cell-based assays.
Figure 5.
Ability of other sulfhydryl-modifying agents to inhibit NS3h–DNA binding and HCV replication. IC50, EC50, and CC50 values are as defined in Figure 4. Three independent titrations were performed of each of the three assays. N-ethmaleimide and iodoacetamide were not tested in cell-based assays.
Ebselen Is a More Potent NS3h Inhibitor than Other Thiol-Modifying Compounds
If ebselen inhibits NS3h merely by modifying critical cysteine residues, then similar reagents should also inhibit NS3h similarly. To test that hypothesis, we examined the sensitivity of NS3h to iodoacetamide and N-ethylmaleimide, two common reagents used to covalently modify reduced cysteines. Neither was a potent inhibitor of the ability of NS3h to bind DNA (Figure 5).
We also searched the results of other high-throughput screens that our lab had previously conducted to find compounds that contained a Michael acceptor similar to that present in N-ethylmaleimide, which could react with cysteines in NS3h. The three most potent compounds were found in a screen of a ChemBridge compound library that we previously deposited in PubChem BioAssay (assay identification no. AID 687043). However, these compounds (16–18) were all between 20 and 30 times less potent than ebselen or its most potent analogues, and they had no effect on cellular HCV replicon content or cell viability even at 100 μM, which was the highest concentration tested (Figure 5).
Ebselen Binds NS3h To Covalently Modify All 14 Cysteine Residues
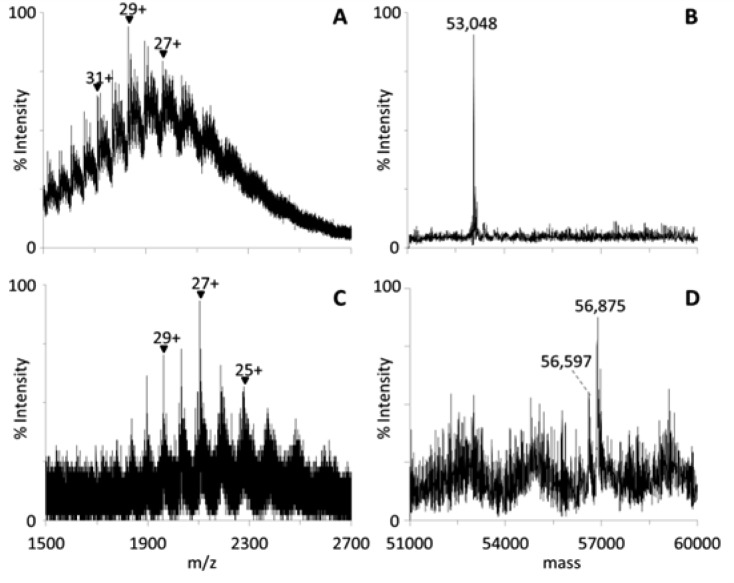
To better understand the nature of possible adducts formed between NS3h and ebselen, we examined NS3h treated with either DMSO or ebselen dissolved in DMSO using electrospray ionization (ESI) mass spectrometry. First, we compared the ESI mass spectra of untreated NS3h with those of DMSO-treated NS3h. The ESI mass spectrum of untreated NS3h showed the same multiply charged protein ions as those in the mass spectrum of DMSO-incubated NS3h (Figure 6A). Deconvolutions of these mass spectra revealed the intact mass of the untreated NS3h protein to be 53 050 Da, and the intact mass of the DMSO-incubated NS3h protein was determined to be 53 048 Da (Figure 6B), showing that DMSO exposure did not significantly alter NS3h.
Figure 6.
Analysis of NS3h–ebselen complexes using mass spectrometry. (A, C) Electrospray ionization mass spectra and (B, D) deconvolutions: (A, B) DMSO-treated NS3h; (C, D) ebselen–NS3h complexes. The peak at 56 875 Da corresponds to the covalent binding of ebselen to all 14 cysteines in NS3h.
In contrast, ESI-MS spectrum of ebselen-treated NS3h (Figure 6C) revealed a complex where the majority of the protein had a mass of 56 875 Da, which is 3827 Da greater than that of untreated NS3h (Figure 6D). Since the average molecular mass of ebselen is ∼274, such a complex has 14 ebselens bound to NS3h, which is the same as the number of cysteines present in NS3h. Other peaks in the deconvoluted spectrum of ebselen-treated NS3h might represent NS3h with fewer covalent modifications. For example, the peak at ∼56 597 Da could correspond to the covalent binding of ebselen to 13 cysteines in NS3h (Figure 6D).
Cys Residues in the NS3h Nucleic Acid Binding Cleft Are Not Specifically Targeted by Ebselen
Although all Cys residues in NS3h seem to be covalently modified upon prolonged incubation with saturating concentrations of ebselen, all of these cysteines might not contribute to the mechanism of inhibition. There are four cysteines that line the known NS3h nucleic acid binding cleft, but there are also three near the ATP binding site, which is known to modulate nucleic acid binding.18 To help understand which of these cysteines might be critical to modulate DNA binding (Figure 7A), we examined the effect of ebselen on various NS3 activities and used site-directed mutagenesis to examine proteins lacking certain cysteines.
Figure 7.
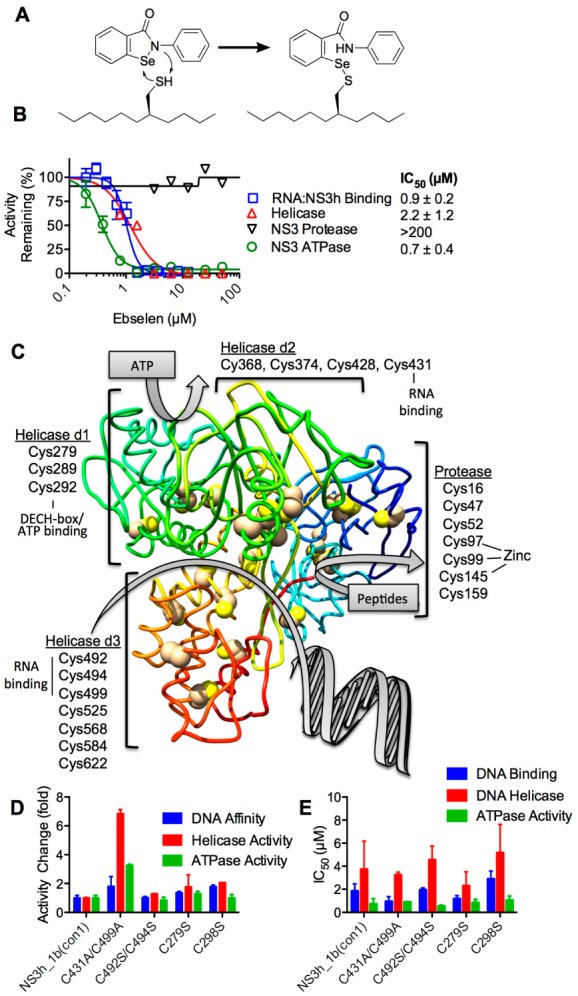
Effects of ebselen on various activities of the NS3 protease/helicase. (A) Schematic for the putative reaction of ebselen with critical cysteine(s) in NS3. (B) Ebselen mainly affects NS3 helicase, not protease, activity. Concentration response of ebselen on RNA binding (squares) as well as ATPase (circles), protease (inverted triangles), and helicase (triangles) activities, reported as normalized activity remaining (n = 2, mean ± SD). (C) Location of cysteines in the HCV NS3 protein. Cys in the helicase domains (d1, d2, and d3) and the protease domain mapped on PDB file 3KQL.6 The NS3 chain is colored as a gradient from the N-terminus (blue) to the C-terminus (red). (D) Relative DNA binding affinity (blue), helicase activity (red), and ATPase activity (green) of NS3h_2a(JFH1) (i.e., C431A, C499A) and site-directed mutants C492S/C494S, C279S, and C289S normalized to that of wild-type NS3h_1b(con1). (E) Ability of ebselen to inhibit the various activities of the different NS3h proteins. IC50 values were calculated from 12-point, 2-fold dilutions starting at 100 μM (n = 3, mean ± SD).
We found that ebselen inhibits NS3-catalyzed DNA unwinding and ATP hydrolysis with a similar potency as it inhibits the NS3-nucleic acid interaction (Figure 7A). However, even at the highest concentrations tested, ebselen had no effect on the ability of full-length NS3 to cleave peptide substrates (Figure 7B). The later result showed that ebselen likely does not completely denature NS3, and the result was somewhat surprising given the fact that there are seven cysteines in the protease domain, including three that coordinate a structural zinc ion (Figure 7C).
To examine the possible roles of various cysteines in the NS3 helicase domains, we focused mainly on studying residues near the known nucleic acid and ATP-binding clefts. The protein that we used in all of the above studies was derived from the con1 strain of HCV genotype 1b.19 Therefore, we first examined how natural genetic variation in HCV might affect the ability of ebselen to inhibit HCV helicase. Only two of these four cysteines are conserved in all HCV genotypes (for an alignment, see Figure 8 of Lam et al.20). In HCV genotype 2a, both Cys431 and Cys499 are alanines. The HCV genotype 2a NS3h (i.e., C431A/C499A) unwound DNA about 6.5 times more rapidly than did the genotype 1b protein, and it bound DNA about twice as tightly (Figure 7D). However, ebselen inhibited all three activities (DNA binding, ATP hydrolysis, and DNA unwinding) of both the genotype 1b and 2a (C431A/C499A) helicases with similar IC50 values (Figure 7E). These data suggest that neither Cys431 nor Cys499 is needed for ebselen to exert its effects.
The other two cysteines in the DNA binding cleft are conserved in all HCV genotypes, and their roles were examined by altering the NS3h_1b(con1) protein using site-directed mutagenesis. The Cys492/Cys494 double mutant had activities that were indistinguishable from those of wild type (Figure 7D), and similar amounts of ebselen were needed to inhibit all three activities of the Cys492/Cys494 double mutant as were needed to inhibit the wild type. Again, these data provide no evidence that cysteine residues in the NS3 DNA binding cleft are targeted by ebselen.
The other cysteine residues that we examined using site-directed mutagenesis were in the known ATP binding cleft (Figure 7C). Two of these residues were altered to serine, but again no differences were observed when either C279S or C289S were compared to the wild-type protein, and neither protein was more or less sensitive to ebselen inhibition in either DNA binding, ATP hydrolysis, or DNA unwinding assays (Figure 7D,E). We also attempted to examine Cys in the conserved helicase signature DECH motif, but a stable protein was not successfully expressed in E. coli. Prior studies with another NS3h construct showed that such a C292S abolishes all NS3h activities,21 suggesting that the sensitivity of such a protein to ebselen would be difficult to study even if it could be expressed and purified in the genetic background studied here.
Discusssion
This study highlights why viral helicases, like the one encoded by HCV, can be extraordinarily difficult drug targets while at the same time it elucidates the mechanism of action of a known HCV antiviral. The results should be helpful to guide the further development of ebselen or related compounds targeting thiols as enzyme inhibitors, molecular probes, or therapeutic agents.
The screens performed here show that antivirals targeting HCV helicase are rare compared to those that attack other antiviral targets. Only one of the 76 compounds in the NIH clinical collection that are known to inhibit HCV replication by more than 50% (at 20 μM) also inhibited HCV helicase.9 Nevertheless, using our approach, we were able to identify one compound that functions both as an antiviral and as a helicase inhibitor. Cell-based screens have since been performed on larger libraries, and they have led to the discovery of drugs such as the NS5A inhibitor daclatasvir (BMS-790052).22 Screening hits from those collections for helicase inhibitors would be the next logical step in this project.
Ebselen was previously identified as an HCV antiviral in another cell-based screen in which Sigma’s library of pharmacologically active compounds (LOPAC) was analyzed for compounds capable of protecting a reporter cells from HCV pathogenesis.23 However, it should be noted that the data here correlate only with an antiviral effect and they do not prove that ebselen’s antiviral activity is solely due to ebselen’s effects on the HCV helicase. We have only ruled out the possibility that ebselen inhibits the NS3 protease function. It might also inhibit the NS5B polymerase, other HCV enzymes, or host factors needed to support HCV replication.
Ebselen clearly forms covalent adducts with cysteines in NS3h, as has been seen previously with other proteins such as the Mycobacterium tuberculosis antigen 85,11Plasmodium falciparum hexokinase,24Trypanosoma brucei hexokinase,25 and diguanylate cyclases.12 Targeting cysteines in NS3h with antiviral agents has been explored before. Kandil et al. reported a rationally designed helicase inhibitor that was intended to react with Cys431 in the RNA-binding cleft.26 The compound was a potent helicase inhibitor (IC50 = 0.26 μM) and showed some antiviral activity in HCV replicon cells,26 but it does not resemble ebselen and instead incorporates a reactive pyrrole.
We had hoped, therefore, to use ebselen as a chemical probe to identify which of the 14 cysteines in NS3h are critical for helicase action. Unfortunately, none of the obvious candidate cysteines seemed to be essential for ebselen to exert its action (Figure 7). We suspect that Cys292 in the DECH-box motif might be needed, but since the protein did not tolerate the site-directed mutation we intended to construct, we were not able to test that hypothesis. Cys292 has been shown by others to be critical for ATP hydrolysis and DNA unwinding.21,27 Coupled with the fact that ATP binding near Cys292 leads to RNA release,6 it is possible that a reaction of ebselen near Cys292 would exert the same effect to displace nucleic acid from the enzyme. Another possibility is that ebselen oxidizes one or more cysteines without covalent modification. Joice et al. recently showed that ebselen inhibits T. brucei hexokinase by oxidizing two cysteines, one of which was revealed to be a key catalytic residue.25
We suspect that inhibition seen with low doses of ebselen does not result from either irreversible adduct formation or oxidation because inhibition with low concentrations of ebselen is reversible (Figure 3). If ebselen functions by covalently modifying cysteine residues, then it could function either as an irreversible or reversible inhibitor. Serafimova et al.28 recently showed that some compounds covalently modify cysteines in a reversible manner, and it is possible that ebselen and its analogues form reversible adducts with NS3h. If this is the case, it might be possible to design chemically tuned electrophiles using this scaffold that are specific for HCV helicase.28
In addition to these new insights into how ebselen interacts with NS3h on a molecular level, the comparisons we report here between ebselen and its structural (Figure 4) and functional (Figure 5) analogues should help to guide the further chemical development of this scaffold, although care would need to be taken in such studies because the chemical reaction of this scaffold could lead to compounds that interfere with assays for undesired reasons.29 The structure–activity relationships identified here reveal also that ebselen is far more potent than other thiol-modifying reagents and that the selenium is not needed for activity. We have also learned that decorating either the phenyl or benzisothiazolone rings systems can alter activity by one or more orders of magnitude. Some of these analogues are also dramatically better antiviral compounds in cells.
Ebselen has been studied for many years as a possible drug to treat cerebral ischemia and stroke. It is effective and well tolerated in animal models.30 In cells, ebselen reduces hydrogen peroxide by mimicking the activity of the selenoenzyme glutathione peroxidase. It is possible that ebselen’s antioxidant activity might abrogate or mask any direct antiviral effect such that the compound is only a modest HCV inhibitor.9,23 Supporting this notion is our observation that far more ebselen needs to be added to cells to inhibit HCV replication than is needed to inhibit HCV helicase in vitro (Figure 4) and the fact that Chockalingam et al. reported a similarly modest effect of ebselen on HCV RNA replication.23
The diverse effects of ebselen in cells likely result from the selenium moiety, and the toxicity of the selenium has also been a concern limiting ebselen drug development. In our HCV systems, the selenium does not appear to be needed either for activity in vitro or in cells (Figure 4), and by eliminating the selenium, we have been able to design compounds that are more potent and selective antivirals (Figure 4). The best of the new compounds synthesized here (compounds 4–7) are similarly potent in vitro, but they are considerably more active and selective in cell-based assays (Figure 4), suggesting that they are more specific antivirals and have fewer off-target effects than ebselen. While the relative toxicity of these new compounds to hepatocytes still limits their utility at this time, we have shown that modifications to the scaffold can improve the efficacy in cells more than 20-fold and the therapeutic index (CC50/EC50) by more than 5-fold.
Whether potent HCV helicase inhibitors will be ever needed as drugs is also a subject that is open for debate. Many other promising HCV antivirals have been discovered, and with the FDA approval of direct-acting antivirals like the protease inhibitors telaprevir,31 boceprevir,32 and simeprevir,33 and the NS5B polymerase inhibitor sofosbuvir,34 nontoxic, interferon-free treatments capable of curing most HCV patients are in sight. So, additional drug candidates might not be needed at this stage in HCV drug development. On the other hand, HCV helicase inhibitors like ebselen and its analogues studied here might be useful probes to study why RNA viruses encode helicases like NS3. It is also not understood why a helicase and a protease are combined in the same polypeptide, and potent specific helicase inhibitors active in cells might help to solve this and other important biological questions.
Methods
Materials
The truncated C-terminally His-tagged NS3 helicase lacking the protease domain (NS3h) was purified as described by Hanson et al.,35 and the full-length single chain NS3-4A protein used for protease assays (Figure 7) was purified as described by Frick et al.36 All experiments, other than those in Figure 7, were performed with NS3h isolated from the con1 strain of HCV genotype 1b, aka NS3h_1b(con1), GenBank AB114136.19
NS3h from HCV genotype 2a(JFH1), which carries the C431A and C499A substitutions (GenBank AJ238799), was used to examine the roles of Cys431 and Cys499 (Figure 7). The C492S/C494S double mutant was prepared from a plasmid expressing wild-type NS3h_1b(con1) using the primers 5′-TCC TCG GTT CTG AGC GAG AGC TAT GAC GCG G-3′ and 5′-CCG CGT CAT AGC TCT CGC TCA GAA CCG AGG A-3′ and the QuikChange II kit (Agilent Technologies). The same kit and plasmid was used to construct genes encoding C279S with primers 5′- GCC GAC GGT GGT AGC TCT GGG GG-3′ and 5′-CCC CCA GAG CTA CCA CCGTCG GC-3′, and C289S using primers 5′-GGG CGC CTA TGA CAT CAT AAT AAGTGA TGA GTG CC-3′ and 5′-GGC ACT CAT CAC TTA TTA TGA TGT CAT AGG CGC CC-3′. All substitutions were verified by sequencing (Genewiz, South Plainfield, NJ) before expression and purification.
DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Helicase substrates were prepared by combining DNA oligonucleotides at a 1:1 molar ratio to a concentration of 50 μM in 10 mM Tris HCl, pH 8.0, heating them to 95 °C for 5 min, and allowing them to cool to RT over 3 h.
The 446 compounds in NIH clinical collection set 1 and the 281 compounds in NIH clinical collection set 2 (http://www.nihclinicalcollection.com/) were obtained from Evotec Inc. (South San Francisco, CA). Ebselen was from Sigma (E-3520), and synthesis of ebsulfur (compound 1) has been described previously.37
Instant JChem was used for structure database management, search, and prediction, Instant JChem 6.0 (2013), ChemAxon (http://www.chemaxon.com).
General Analytical Chemistry
1H and 13C NMR spectra were recorded on a Bruker AM 400 spectrometer (operating at 400 and 101 MHz, respectively) or a Bruker AVIII spectrometer (operating at 500 and 126 MHz, respectively) in CDCl3 or DMSO-d6 with 0.03% TMS as an internal standard. The chemical shifts (δ) reported are given in parts per million (ppm), and the coupling constants (J) are in Hertz (Hz). The spin multiplicities are reported as s = singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublet, ddd = doublet of doublet of doublet, dt = doublet of triplet, td = triplet of doublet, and m = multiplet. The LC–MS analysis was performed on an Agilent 1200 RRL HPLC system with photodiode array UV detection and an Agilent 6224 TOF mass spectrometer. The chromatographic method utilized the following parameters: a Waters Acquity BEH C-18 2.1 × 50 mm, 1.7 μm column; UV detection wavelength = 214 nm; flow rate = 0.4 mL/min; gradient = 5–100% MeCN over 3 min with a hold of 0.8 min at 100% MeCN; the aqueous mobile phase contained 0.15% NH4OH. The mass spectrometer utilized the following parameters: an Agilent multimode source which simultaneously acquires ESI+/APCI+; a reference mass solution consisting of purine and hexakis(1H,1H,3H-tetrafluoropropoxy) phosphazine; and a makeup solvent of 90:10:0.1 MeOH/H2O/HCO2H, which was introduced to the LC flow prior to the source to assist ionization. Melting points were determined on a Stanford Research Systems OptiMelt apparatus. Flash chromatography separations were carried out using a Teledyne Isco CombiFlash Rf 200 purification system with silica gel columns. Mass-directed fractionation separations were carried out using an Agilent 1200 HPLC system with photodiode array UV detection and an Agilent 6120 mass spectrometer. The chromatographic method utilized a Waters XBridge C-18, 19 × 150 mm, 5 μm column; UV detection wavelength = 214 nm; flow rate = 20 mL/min; focused gradient = 5–100% MeCN; the aqueous mobile phase contained 0.15% NH4OH.
Compound Synthesis
2-(Benzylthio)-4-bromobenzoic Acid
Using a vial, 4-bromo-2-fluorobenzoic acid (0.622 g, 2.84 mmol) and Cs2CO3 (1.854 g, 5.69 mmol) were dissolved in NMP (3 mL), and the mixture was briefly heated at 120 °C for 3 min. After cooling to RT, BnSH (0.40 mL, 3.4 mmol) was added to the mixture, and the mixture was reheated at 120 °C for 3 h. The reaction mixture was cooled to RT, diluted with water (25 mL), and quenched with 1 M aq. HCl (20 mL). The product was extracted with EtOAc (25 mL), washed with brine (2 × 10 mL), and dried with MgSO4 to give the title compound (0.857 g, 93%) as a peach-colored solid. It was used in the next step without further purification.
2-(Benzylthio)-4-bromo-N-phenylbenzamide
A catalytic amount of DMF (4 drops) was added to a crude mixture of 2-(benzylthio)-4-bromobenzoic acid (0.82 g, 2.54 mmol) in DCM (10 mL), and the mixture was cooled to 0 °C. Neat oxalyl chloride (0.35 mL, 4.1 mmol) was added to the mixture. After stirring at 0 °C for 20 min, the mixture was stirred at RT for 1.4 h and subsequently added dropwise to a freshly prepared mixture of aniline (255 μL, 2.80 mmol) and DIPEA (0.50 mL, 2.9 mmol) in DCM (4 mL) at 0 °C. After stirring at 0 °C for 2 min, the mixture was stirred at RT for 1 h. The product was concentrated and purified using flash chromatography (0–100% DCM/hexanes) to give the title compound (0.714 g, 71%) as an off-white solid. 1H NMR (400 MHz, CDCl3) δ 8.42 (s, 1H), 7.67 (d, J = 8.3 Hz, 1H), 7.61–7.53 (m, 3H), 7.46 (dd, J = 8.3, 2.0 Hz, 1H), 7.39–7.33 (m, 2H), 7.26–7.22 (m, 3H), 7.20–7.13 (m, 3H), 4.11 (s, 2H).
6-Bromo-2-phenylbenzo[d]isothiazol-3(2H)-one (3: CID22416220)
Sulfuryl chloride (23 μL, 0.28 mmol) was added to a solution of 2-(benzylthio)-4-bromo-N-phenylbenzamide (86 mg, 0.22 mmol) in DCM (3 mL). After stirring at 23 °C for 13 h, the volatiles were removed, and the remaining residue was purified using flash chromatography (0–30% EtOAc/hexanes) followed by mass-directed fractionation to give 3 (12 mg, 18%) as a white solid. mp 202–205 °C. 1H NMR (400 MHz, CDCl3) δ 7.96 (dd, J = 8.4, 0.6 Hz, 1H), 7.77 (dd, J = 1.6, 0.6 Hz, 1H), 7.70–7.66 (m, 2H), 7.57 (dd, J = 8.4, 1.6 Hz, 1H), 7.51–7.45 (m, 2H), 7.37–7.31 (m, 1H). LC–MS: tR = 3.46 min, purity = 100%. HRMS m/z calcd for C13H9BrNOS [M + H]+, 307.9568; found, 307.9545.
5,6-Difluoro-2-phenylbenzo[d]isothiazol-3(2H)-one (2: CID44968081)
Following general procedure A (Scheme 1), a complex mixture of all three of the possible (-F, -F, -SBn) regioisomers of 2-(benzylthio)-4,5-difluoro-N-phenylbenzamide (1.79 g, 5.04 mmol) was used to produce 2 (0.442 g, 33%) as a white solid. mp 185–188 °C. 1H NMR (400 MHz, CDCl3) δ 7.90 (dd, J = 9.1, 7.4 Hz, 1H), 7.69–7.63 (m, 2H), 7.52–7.45 (m, 2H), 7.40 (dd, J = 8.9, 6.3 Hz, 1H), 7.38–7.32 (m, 1H). LC–MS: tR = 3.27 min, purity = 91%. HRMS m/z calcd for C13H8F2NOS [M + H]+, 264.0289; found, 264.0281.
Scheme 1. General Procedure A.
Representative synthesis of 3 (CID22416220).
N-(2-Chlorophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (4: CID45479199)
A stock solution of sodium benzoisothiazololate was freshly prepared containing benzoisothiazol-3(2H)-one (0.25 M) and NaOH (0.25 M) in (8:1) EtOH/H2O. Another solution of 2-bromo-N-(2-chlorophenyl)acetamide (249 mg, 1.00 mmol) in EtOH (1 mL) was prepared, and to this solution was added the 0.25 M stock solution of sodium benzoisothiazololate (3 mL, 0.75 mmol). The reaction mixture was stirred at RT for 29 h. The mixture was concentrated, redissolved in DCM (8 mL), and washed with saturated aq. NaHCO3 (2 × 3 mL). The product was purified using flash chromatography (0–10% i-PrOH/DCM) to give 4 (166 mg, 69%) as an off-white solid. mp 200–202 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.92 (s, 1H), 7.99 (dt, J = 8.2, 0.9 Hz, 1H), 7.91 (dt, J = 7.9, 1.0 Hz, 1H), 7.75 (dd, J = 8.1, 1.6 Hz, 1H), 7.70 (ddd, J = 8.3, 7.1, 1.3 Hz, 1H), 7.51 (dd, J = 8.0, 1.5 Hz, 1H), 7.45 (ddd, J = 8.0, 7.2, 1.0 Hz, 1H), 7.33 (td, J = 7.8, 1.5 Hz, 1H), 7.20 (td, J = 7.7, 1.6 Hz, 1H), 4.80 (s, 2H). 13C NMR (126 MHz, DMSO) δ 166.1, 165.0, 141.5, 134.4, 132.0, 129.6, 127.5, 126.5, 126.2, 125.8, 125.7, 125.4, 123.4, 121.8, 45.9. LC–MS: tR = 2.94 min, purity = 97%. HRMS m/z calcd for C15H12ClN2O2S [M + H]+, 319.0303; found, 319.0309.
N-(2-Bromophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (5: CID45479201)
Following general procedure B (Scheme 2), 0.25 M sodium benzoisothiazololate (3 mL, 0.75 mmol) and 2-bromo-N-(2-bromophenyl)acetamide (352 mg, 1.20 mmol) were used to produce 5 (69 mg, 25%) as an off-white solid. mp 202–204 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.85 (s, 1H), 7.99 (dt, J = 8.2, 0.8 Hz, 1H), 7.90 (ddd, J = 7.8, 1.3, 0.7 Hz, 1H), 7.74–7.61 (m, 3H), 7.46 (ddd, J = 8.0, 7.2, 1.0 Hz, 1H), 7.38 (td, J = 7.9, 1.5 Hz, 1H), 7.16 (td, J = 7.9, 1.7 Hz, 1H), 4.76 (s, 2H). LC–MS: tR = 2.96 min, purity = 100%. HRMS m/z calcd for C15H12BrN2O2S [M + H]+, 364.9782; found, 364.9772.
Scheme 2. General Procedure B.
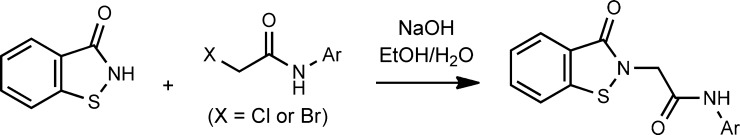
Representative synthesis of 4 (CID45479199).
N-(2-Cyanophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (6: CID45479200)
Following general procedure B, 0.25 M sodium benzoisothiazololate (3 mL, 0.75 mmol) and 2-bromo-N-(2-cyanophenyl)acetamide (287 mg, 1.20 mmol) were used to produce 6 (46 mg, 20%) as an off-white solid. mp 223–225 °C. 1H NMR (400 MHz, DMSO-d6) δ 10.55 (s, 1H), 8.00 (dt, J = 8.2, 0.9 Hz, 1H), 7.91 (ddd, J = 7.9, 1.3, 0.8 Hz, 1H), 7.84 (ddd, J = 7.8, 1.6, 0.6 Hz, 1H), 7.74–7.68 (m, 2H), 7.65 (ddd, J = 8.3, 1.4, 0.5 Hz, 1H), 7.46 (ddd, J = 8.0, 7.2, 1.0 Hz, 1H), 7.37 (ddd, J = 7.8, 7.2, 1.4 Hz, 1H), 4.78 (s, 2H). LC–MS: tR = 2.67 min, purity = 96%. HRMS m/z calcd for C16H12N3O2S [M + H]+, 310.0645; found, 310.0648.
N-(2-Fluorophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (7: CID45479198)
Following general procedure B (Scheme 2), 0.25 M sodium benzoisothiazololate (3 mL, 0.75 mmol) and 2-bromo-N-(2-fluorophenyl)acetamide (232 mg, 1.00 mmol) were used to produce 7 (170 mg, 75%) as an off-white solid. mp 207–209 °C. 1H NMR (400 MHz, CDCl3) δ 8.63 (s, 1H), 8.24 (t, J = 7.9 Hz, 1H), 8.11 (ddd, J = 7.9, 1.3, 0.7 Hz, 1H), 7.68 (ddd, J = 8.3, 7.1, 1.3 Hz, 1H), 7.60 (dt, J = 8.2, 0.9 Hz, 1H), 7.46 (ddd, J = 8.0, 7.1, 1.0 Hz, 1H), 7.14–7.02 (m, 3H), 4.67 (s, 2H). LC–MS: tR = 2.83 min, purity = 99%. HRMS m/z calcd for C15H12FN2O2S [M + H]+, 303.0598; found, 303.0599.
2-(3-Oxobenzo[d]isothiazol-2(3H)-yl)-N-(o-tolyl)acetamide (8: CID44968084)
Following general procedure B, benzoisothiazol-3(2H)-one (66 mg, 0.44 mmol) and 2-bromo-N-(o-tolyl)acetamide (100 mg, 0.44 mmol) were used to produce 8 (7 mg, 5%) as a white solid. mp 221–223 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.66 (s, 1H), 7.99 (dt, J = 8.2, 0.9 Hz, 1H), 7.90 (dt, J = 7.9, 1.0 Hz, 1H), 7.70 (ddd, J = 8.3, 7.2, 1.3 Hz, 1H), 7.45 (ddd, J = 7.9, 7.2, 1.0 Hz, 1H), 7.40 (dd, J = 7.9, 1.4 Hz, 1H), 7.24–7.07 (m, 3H), 4.72 (s, 2H), 2.23 (s, 3H). LC–MS: tR = 2.83 min, purity = 100%. HRMS m/z calcd for C16H15N2O2S [M + H]+, 299.0849; found, 299.0843.
2-(3-Oxobenzo[d]isothiazol-2(3H)-yl)-N-phenylpropanamide (9: CID45479197)
Following general procedure A, 2-(benzylthio)-N-(1-oxo-1-(phenylamino)propan-2-yl)benzamide (94 mg, 0.24 mmol) was used to produce 9 (67 mg, 93%) as a white solid. mp 209–211 °C. 1H NMR (400 MHz, CDCl3) δ 8.95 (s, 1H), 8.05 (ddd, J = 7.9, 1.2, 0.7 Hz, 1H), 7.64 (ddd, J = 8.2, 7.0, 1.2 Hz, 1H), 7.60–7.53 (m, 3H), 7.42 (ddd, J = 8.0, 7.0, 1.2 Hz, 1H), 7.32–7.26 (m, 2H), 7.11–7.05 (m, 1H), 5.51 (q, J = 7.0 Hz, 1H), 1.74 (d, J = 7.0 Hz, 3H). LC–MS: tR = 3.02 min, purity = 100%. HRMS m/z calcd for C16H15N2O2S [M + H]+, 299.0849; found, 299.0843.
N-(2-Iodophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (10: CID45479202)
Following general procedure B (Scheme 2), 0.25 M sodium benzoisothiazololate (3 mL, 0.75 mmol) and 2-bromo-N-(2-iodophenyl)acetamide (408 mg, 1.20 mmol) were used to produce 10 (82 mg, 27%) as a tan solid. mp 218–220 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.80 (s, 1H), 7.99 (dt, J = 8.2, 0.9 Hz, 1H), 7.93–7.87 (m, 2H), 7.71 (ddd, J = 8.3, 7.1, 1.3 Hz, 1H), 7.49–7.43 (m, 2H), 7.40 (td, J = 7.7, 1.5 Hz, 1H), 7.00 (td, J = 7.8, 1.7 Hz, 1H), 4.73 (s, 2H). LC–MS: tR = 2.98 min, purity = 97%. HRMS m/z calcd for C15H12IN2O2S [M + H]+, 410.9659; found, 410.9656.
N-(3-Bromophenyl)-2-(3-oxobenzo[d]isothiazol-2(3H)-yl)acetamide (11: CID45105114)
Following general procedure B, benzoisothiazol-3(2H)-one (42 mg, 0.28 mmol) and 2-bromo-N-(3-bromophenyl)acetamide (100 mg, 0.34 mmol) were used to produce 11 (16 mg, 16%) as a white solid. 1H NMR (400 MHz, DMSO-d6) δ 10.51 (s, 1H), 8.00 (dt, J = 8.2, 0.9 Hz, 1H), 7.93 (t, J = 1.9 Hz, 1H), 7.89 (ddd, J = 7.9, 1.2, 0.7 Hz, 1H), 7.72 (ddd, J = 8.3, 7.1, 1.3 Hz, 1H), 7.51–7.43 (m, 2H), 7.33–7.25 (m, 2H), 4.69 (s, 2H). LC–MS: tR = 3.09 min, purity = 100%. HRMS m/z calcd for C15H12BrN2O2S [M + H]+, 364.9782; found, 364.9767.
2-(3-Oxobenzo[d]isothiazol-2(3H)-yl)-N-(2-(trifluoromethyl)phenyl)acetamide (12: CID45479195)
Following general procedure B (except that the reaction was heated at 40 °C for 43 h and the product was purified by mass-directed fractionation), benzoisothiazol-3(2H)-one (75 mg, 0.50 mmol) and 2-chloro-N-(2-(trifluoromethyl)phenyl)acetamide (141 mg, 0.59 mmol) were used to produce 12 (5 mg, 3%) as a clear, colorless oil. 1H NMR (400 MHz, CDCl3) δ 8.82 (s, 1H), 8.46 (d, J = 8.3 Hz, 1H), 8.00 (dt, J = 8.1, 1.0 Hz, 1H), 7.83 (dt, J = 8.2, 0.9 Hz, 1H), 7.66–7.56 (m, 3H), 7.49 (ddd, J = 8.0, 7.0, 1.0 Hz, 1H), 7.29–7.23 (m, 1H), 5.23 (s, 2H). LC–MS: tR = 3.51 min, purity = 100%. HRMS m/z calcd for C16H12F3N2O2S [M + H]+, 353.0566; found, 353.0559.
3-(3-Oxobenzo[d]isothiazol-2(3H)-yl)-N-phenylpropanamide (13: CID45254020)
Following general procedure A (Scheme 1), 2-(benzylthio)-N-(3-oxo-3-(phenylamino)propyl)benzamide (52 mg, 0.13 mmol) was used to produce 13 (32 mg, 82%) as a white solid. mp 193–195 °C. 1H NMR (400 MHz, CDCl3) δ 8.48 (s, 1H), 7.95 (dt, J = 8.0, 1.0 Hz, 1H), 7.62–7.56 (m, 3H), 7.52 (dt, J = 8.2, 0.9 Hz, 1H), 7.35 (ddd, J = 8.0, 7.0, 1.0 Hz, 1H), 7.33–7.27 (m, 2H), 7.13–7.06 (m, 1H), 4.29 (t, J = 6.1 Hz, 2H), 2.89 (t, J = 6.1 Hz, 2H). LC–MS: tR = 2.82 min, purity = 100%. HRMS m/z calcd for C16H15N2O2S [M + H]+, 299.0849; found, 299.0899.
2,5-Diphenylisothiazol-3(2H)-one (14: CID15294640)
Following a previously reported procedure,3814 (12% yield over 4 steps) was isolated as clear, colorless crystals. mp 92–94 °C. 1H NMR (400 MHz, CDCl3) δ 7.67–7.63 (m, 2H), 7.56–7.43 (m, 7H), 7.35–7.29 (m, 1H), 6.57 (s, 1H). 13C NMR (101 MHz, CDCl3) δ 167.8, 156.1, 136.9, 131.3, 130.1, 129.5, 129.5, 127.3, 126.0, 124.6, 110.7. LC–MS: tR = 3.28 min, purity = 100%. HRMS m/z calcd for C15H12NOS [M + H]+, 254.0634; found, 254.0641.
1-Methyl-2-phenyl-1H-indazol-3(2H)-one (15: CID11579468)
Following a previously reported procedure,3915 (15% yield over 2 steps) was isolated as white crystals. mp 87–89 °C. 1H and 13C NMR matched the reported data.39 LC–MS: tR = 2.93 min, purity = 100%. HRMS m/z calcd for C14H13N2O [M + H]+, 225.1022; found, 225.1032.
Fluorescence Polarization-Based Nucleic Acid-Binding Assays
Screening assays were executed as described by Mukherjee et al.8 DMSO or compound dissolved in DMSO was added to each assay by pin transfer such that the final concentration of DMSO was 1% (v/v) in each assay. Polarization was monitored with a TECAN Infinite M1000 PRO multimode microplate reader by exciting at 635 nm (5 nm bandwidth) and measuring total fluorescence intensity, parallel, and perpendicular polarized light at 667 nm (20 nm bandwidth). G-factors were calculated from wells with Cy5-dT15 alone. Inhibition (%) was calculated by normalizing data to values obtained with positive controls (200 nM dT20 or 100 μM primuline) and negative controls (DMSO only). For RNA-binding experiments, Cy5-dT15 was substituted with 5′-Cy5-rUrUrUrUrUrUrUrUrUrUrUrUrUrUrU-3′ (Cy5-rU15).
The relative Kd of each protein for Cy5-dT20 was determined by titrating Cy5-dT20 with each protein and fitting the data to a standard Hill equation, as described recently.40 Relative affinities were normalized (Figure 7B,C) by converting Kd’s to association constants (Ka) and dividing by the average of the Ka values obtained for NS3h_1b(con1).
Electrophoretic Mobility Shift Assay
Binding assays containing 25 mM MOPS, pH 7.5, 1.25 mM MgCl2, 20 nM Cy5-dT15, and 200 nM NS3h_1b were incubated 5 min at RT. Following addition of indicated concentrations of ebselen, the binding reactions were incubated another 5 min at 23 °C. A 15% polyacrylamide Tris Borate EDTA (TBE) gel was prerun at 4 °C for 30 min at 100 V. Ten microliters of each sample was loaded onto the gel. The gel was run 5 min at 200 V to allow samples to enter gel and then 60 min at 100 V at 4 °C. The gel was scanned on a BioRad Molecular Imager FX phosphorimager.
HCV Subgenomic Replicon Assays
Huh-7.5 cells (Dr. Charles Rice, Rockefeller University) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone), 1× nonessential amino acids, and 100 U/mL of each of penicillin and streptomycin (Invitrogen). Huh-7.5 cells containing stable HCV Con1/1b Rluc subgenomic replicons were maintained in the above growth medium supplemented with 0.25 mg mL–1 Geneticin (Invitrogen). Stable HCV Con1/1b subgenomic replicon (HCV Rluc) Huh-7.5 cells were isolated as previously described.13,17 To assess the ability of each compound to inhibit HCV replication, Huh-7.5 cells harboring HCV Rluc subgenomic replicons were seeded at 10 × 103 cells per well in 96-well plates and incubated 4 to 5 h to allow the cells to attach to the plate. Two-fold serial compound dilutions were made in dimethyl sulfoxide (DMSO), diluted into media, such that the DMSO final concentration was 0.5% after adding dilutions to cells. Compounds and cells were incubated at 37 °C in 5% CO2. After 3 days, the medium was removed by aspiration, and the cells were washed with 1× PBS. Cellular Renilla luciferase content was measured using the Renilla luciferase assay kit (Promega). Cells were lysed by adding 60 μL of Promega lysis buffer, and luciferase activity was measured by injecting 50 μL of Promega’s Renilla luciferase substrate into 50 μL of lysate and reading luminescence for 5 s with a FLUOstar Omega microplate reader (BMG Labtech, Germany).
Cell Viability Assays
To assess compound effects on Huh-7.5 cell viability, cells were plated and treated as above, and the effect of compound on cell viability was tested using the CellTiter-Glo luminescent cell viability kit (Promega) following the manufacturer’s instructions. Briefly, at the end of the 3 day incubation, the medium was aspirated and the cells were washed with growth medium; then, an equal volume of growth medium and CellTiter-Glo reagent was added and incubated at RT on an orbital shaker. The plate was allowed to sit at RT for 30 min, and the luciferase activity was measured for 5 s using a FLUOstar Omega microplate reader (BMG Labtech).
Helicase (DNA Unwinding) Assays
Molecular beacon-based helicase assays (MBHAs) were performed as described previously except that DTT was not included in buffers.35,41 Assays contained 25 mM MOPS, pH 6.5, 1.25 mM MgCl2, 5% DMSO, 5 μg/mL BSA, 0.01% (v/v) Tween-20, 5 nM substrate, 12.5 nM NS3h, and 1 mM ATP. The partially duplex DNA substrates used in helicase assays consisted of a 45-mer bottom strand 5′-GCT CCC CGT TCA TCG ATT GGG GAG CTT TTT TTT TTT TTT TTT TTT-3′ and a 25-mer top strand 5′-Cy5-GCT CCC CAA TCG ATG AAC GGG GAG C-IAbRQSp-3. Data were analyzed as described before.35 Specific activities for each protein were determined by performing assays at various protein concentrations and determining the slope of the linear range of a plot of rate vs protein concentration. Relative activity changes (Figure 7D) were determined by dividing by the specific activity of N3h_1b(con1).
ATPase Assays
Assays were assembled in 27 μL in clear 96-well microtiter plates (Corning Inc., catalog no. 9017) and initiated by adding 3 μL of ATP such that the final reactions contained 15 nM NS3h, 1 mM ATP, 25 mM MOPS, pH 6.5, 1.25 mM MgCl2, 5% DMSO, 50 μg/mL BSA, and 0.01% Tween-20 and indicated concentrations of ebselen. After 15 min at 23 °C, reactions were terminated by adding 200 μL of a solution containing 0.034% (w/v) Malachite Green, 1 N HCl, 1% ammonium molybdate, and 0.025% Tween-20, followed within 10 s by addition of 25 μL of 35% (w/v) sodium citrate. After 20 min at 23 °C, an absorbance at 630 nm was read in a Varioskan multimodal reader (ThermoFisher, Inc.). Phosphate released was determined from a standard curve after subtracting A630 values obtained in a reaction lacking NS3h. Specific activities for each protein were determined by performing assays at various protein concentrations and determining the slope of the linear range of a plot of rate vs protein concentration. Relative activity changes (Figure 7D) were determined by dividing by the specific activity of N3h_1b(con1).
Protease Assays
All protease assays were carried out using the 5-carboxyfluorescein-labeled substrate from the AnaSpec Enzolyte 520 protease assay kit (AnaSpec, San Jose, CA). Each assay contained 10 nM scNS3-NS4A, 5% DMSO, and 1× AnaSpec HCV protease assay buffer (no DTT was added). Assays were carried out in a total volume of 20 μL in black 384-well plates with fluorescence at 520 nm measured using a BMG FLUOstar Omega (BMG Labtech) multimode reader. Reactions were performed with 7 concentrations of a 2-fold dilution series of ebselen (in duplicate) starting at 100 μM.
Electrospray Ionization (ESI) Mass Spectrometry (MS)
The protein samples were analyzed using a Waters ESI Q-TOF Micro mass spectrometer. All data were acquired and analyzed using MassLynx software. Three different protein samples were analyzed. The first sample was untreated NS3h in 10 mM ammonium acetate (pH 7.5). It was dialyzed against 10 mM ammonium acetate using centrifugal concentrators with three changes of buffer. Afterward, it was diluted to a concentration of 0.15 mg mL–1 using 50:50 (v/v) acetonitrile/water containing 0.2% formic acid for ESI-MS analysis. The second sample was NS3h protein incubated with 100 μM ebselen in 10 mM ammonium acetate (pH 7.5) containing 1% DMSO for 1 h at RT. It was also dialyzed against 10 mM ammonium acetate with three changes of buffer. Afterward, it was diluted to a concentration of 0.15 mg mL–1 using 50:50 (v/v) acetonitrile/water containing 0.2% formic acid for ESI-MS analysis. The third sample was the NS3h protein in 10 mM ammonium acetate (pH 7.5) incubated with 1% DMSO for 1 h at RT. This sample was also dialyzed against 10 mM ammonium acetate with three changes of buffer. Afterward, it was diluted to a concentration of 0.10 mg mL–1 using 50:50 (v/v) acetonitrile/water containing 0.1% formic acid. All ESI-MS analyses were performed in positive ion mode using a nano-ESI source with a capillary voltage of 3 kV and a flow rate of 0.7 μL/min. All spectra were acquired for 10 min.
Acknowledgments
We would like to thank W. Shadrick, D. Vuong, L. Favrot, D. Ronning, P. Porubsky, and K. Noon for valuable technical assistance and helpful discussions during the course of this study. This work was supported, in whole or in part, by National Institutes of Health grants R01 AI088001 (to D.N.F) and U54 HG005031 (to J. Aubé, Molecular Libraries Probe Production Centers Network, University of Kansas Specialized Chemistry Center) and by a Research Growth Initiative Award from the UWM Research Foundation (to D.N.F).
Glossary
Abbreviations
- Cy5
cyanine 5
- DMSO
dimethyl sulfoxide
- FP
fluorescence polarization
- ESI
electrospray ionization
- HCV
hepatitis C virus
- NS3h
nonstructural protein 2 lacking the protease domain
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- De Clercq E. (2007) The design of drugs for HIV and HCV. Nat. Rev. Drug Discovery 6, 1001–1018. [DOI] [PubMed] [Google Scholar]
- Frick D. N. (2007) The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 9, 1–20. [PMC free article] [PubMed] [Google Scholar]
- Belon C. A., and Frick D. N. (2011) Hepatitis C virus NS3 helicase inhibitors, in Hepatitis C: Antiviral Drug Discovery and Development (He Y., and Tan S. L., Eds.( pp 237–256, Caister Academic Press, Norfolk, UK. [Google Scholar]
- Lin R.; Lacoste J.; Nakhaei P.; Sun Q.; Yang L.; Paz S.; Wilkinson P.; Julkunen I.; Vitour D.; Meurs E.; Hiscott J. (2006) Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80, 6072–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Foy E.; Ferreon J. C.; Nakamura M.; Ferreon A. C.; Ikeda M.; Ray S. C.; Gale M. J.; Lemon S. M. (2005) Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U.S.A. 102, 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M.; Rice C. M. (2010) Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 107, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L.; Morgenstern K. A.; Griffith J. P.; Dwyer M. D.; Thomson J. A.; Murcko M. A.; Lin C.; Caron P. R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100. [DOI] [PubMed] [Google Scholar]
- Mukherjee S.; Hanson A. M.; Shadrick W. R.; Ndjomou J.; Sweeney N. L.; Hernandez J. J.; Bartczak D.; Li K.; Frankowski K. J.; Heck J. A.; Arnold L. A.; Schoenen F. J.; Frick D. N. (2012) Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays. Nucleic Acids Res. 40, 8607–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P.; Whitten-Bauer C.; Chisari F. V. (2010) Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. U.S.A. 107, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J.; Gastaminza P.; Cheng G.; Kapadia S.; Kato T.; Burton D. R.; Wieland S. F.; Uprichard S. L.; Wakita T.; Chisari F. V. (2005) Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrot L.; Grzegorzewicz A. E.; Lajiness D. H.; Marvin R. K.; Boucau J.; Isailovic D.; Jackson M.; Ronning D. R. (2013) Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat. Commun. 4, 2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman O. J.; Orr M. W.; Wang Y.; Lee V. T. (2014) High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem. Biol. 9, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndjomou J.; Kolli R.; Mukherjee S.; Shadrick W. R.; Hanson A. M.; Sweeney N. L.; Bartczak D.; Li K.; Frankowski K. J.; Schoenen F. J.; Frick D. N. (2012) Fluorescent primuline derivatives inhibit hepatitis C virus NS3-catalyzed RNA unwinding, peptide hydrolysis and viral replicase formation. Antiviral Res. 96, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. H.; Chung T. D.; Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Schewe T. (1995) Molecular actions of ebselen—an antiinflammatory antioxidant. Gen. Pharmacol. 26, 1153–1169. [DOI] [PubMed] [Google Scholar]
- Lu J.; Vodnala S. K.; Gustavsson A. L.; Gustafsson T. N.; Sjoberg B.; Johansson H. A.; Kumar S.; Tjernberg A.; Engman L.; Rottenberg M. E.; Holmgren A. (2013) Ebsulfur is a benzisothiazolone cytocidal inhibitor targeting the trypanothione reductase of Trypanosoma brucei. J. Biol. Chem. 288, 27456–27468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Frankowski K. J.; Belon C. A.; Neuenswander B.; Ndjomou J.; Hanson A. M.; Shanahan M. A.; Schoenen F. J.; Blagg B. S.; Aube J.; Frick D. N. (2012) Optimization of potent hepatitis C virus NS3 helicase inhibitors isolated from the yellow dyes thioflavine S and primuline. J. Med. Chem. 55, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. K.; Gurjar M. M.; Patel S. S. (2003) ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278, 23311–23316. [DOI] [PubMed] [Google Scholar]
- Belon C. A.; Frick D. N. (2009) Fuel specificity of the hepatitis C virus NS3 helicase. J. Mol. Biol. 388, 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A. M.; Keeney D.; Eckert P. Q.; Frick D. N. (2003) Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 77, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. W.; Kim J.; Gwack Y.; Han J. H.; Choe J. (1997) Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 71, 9400–9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M.; Nettles R. E.; Belema M.; Snyder L. B.; Nguyen V. N.; Fridell R. A.; Serrano-Wu M. H.; Langley D. R.; Sun J. H.; O’Boyle D. R.; Lemm J. A.; Wang C.; Knipe J. O.; Chien C.; Colonno R. J.; Grasela D. M.; Meanwell N. A.; Hamann L. G. (2010) Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam K.; Simeon R. L.; Rice C. M.; Chen Z. (2010) A cell protection screen reveals potent inhibitors of multiple stages of the hepatitis C virus life cycle. Proc. Natl. Acad. Sci. U.S.A. 107, 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. T.; Walker D. M.; Drew M. E.; Mitchell W. G.; Dao K.; Schroeder C. E.; Flaherty D. P.; Weiner W. S.; Golden J. E.; Morris J. C. (2013) Interrogating a hexokinase-selected small-molecule library for inhibitors of Plasmodium falciparum hexokinase. Antimicrob. Agents Chemother. 57, 3731–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joice A. C.; Harris M. T.; Kahney E. W.; Dodson H. C.; Maselli A. G.; Whitehead D. C.; Morris J. C. (2013) Exploring the mode of action of ebselen in Trypanosoma brucei hexokinase inhibition. Int. J. Parasitol.: Drugs Drug Resist. 3, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandil S.; Biondaro S.; Vlachakis D.; Cummins A. C.; Coluccia A.; Berry C.; Leyssen P.; Neyts J.; Brancale A. (2009) Discovery of a novel HCV helicase inhibitor by a de novo drug design approach. Bioorg. Med. Chem. Lett. 19, 2935–2937. [DOI] [PubMed] [Google Scholar]
- Wardell A. D.; Errington W.; Ciaramella G.; Merson J.; McGarvey M. J. (1999) Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J. Gen. Virol. 80, 701–709. [DOI] [PubMed] [Google Scholar]
- Serafimova I. M.; Pufall M. A.; Krishnan S.; Duda K.; Cohen M. S.; Maglathlin R. L.; McFarland J. M.; Miller R. M.; Frodin M.; Taunton J. (2012) Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol. 8, 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B. (2010) Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2, 1529–1546. [DOI] [PubMed] [Google Scholar]
- Parnham M. J.; Sies H. (2013) The early research and development of ebselen. Biochem. Pharmacol. 86, 1248–1253. [DOI] [PubMed] [Google Scholar]
- Zeuzem S.; Andreone P.; Pol S.; Lawitz E.; Diago M.; Roberts S.; Focaccia R.; Younossi Z.; Foster G. R.; Horban A.; Ferenci P.; Nevens F.; Mullhaupt B.; Pockros P.; Terg R.; Shouval D.; van Hoek B.; Weiland O.; Van Heeswijk R.; De Meyer S.; Luo D.; Boogaerts G.; Polo R.; Picchio G.; Beumont M. (2011) Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 364, 2417–2428. [DOI] [PubMed] [Google Scholar]
- Bacon B. R.; Gordon S. C.; Lawitz E.; Marcellin P.; Vierling J. M.; Zeuzem S.; Poordad F.; Goodman Z. D.; Sings H. L.; Boparai N.; Burroughs M.; Brass C. A.; Albrecht J. K.; Esteban R. (2011) Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 364, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist A.; Samuelsson B.; Johansson P. O.; Cummings M. D.; Lenz O.; Raboisson P.; Simmen K.; Vendeville S.; de Kock H.; Nilsson M.; Horvath A.; Kalmeijer R.; de la Rosa G.; Beumont-Mauviel M. (2014) Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J. Med. Chem. 57, 1673–1693. [DOI] [PubMed] [Google Scholar]
- Lawitz E.; Mangia A.; Wyles D.; Rodriguez-Torres M.; Hassanein T.; Gordon S. C.; Schultz M.; Davis M. N.; Kayali Z.; Reddy K. R.; Jacobson I. M.; Kowdley K. V.; Nyberg L.; Subramanian G. M.; Hyland R. H.; Arterburn S.; Jiang D.; McNally J.; Brainard D.; Symonds W. T.; McHutchison J. G.; Sheikh A. M.; Younossi Z.; Gane E. J. (2013) Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 368, 1878–1887. [DOI] [PubMed] [Google Scholar]
- Hanson A. M.; Hernandez J. J.; Shadrick W. R.; Frick D. N. (2012) Identification and analysis of inhibitors targeting the hepatitis C virus NS3 helicase. Methods Enzymol. 511, 463–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D. N.; Ginzburg O.; Lam A. M. (2010) A method to simultaneously monitor hepatitis C virus NS3 helicase and protease activities. Methods Mol. Biol. 587, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engman L.; Hallberg A. (1989) Expedient synthesis of ebselen and related compounds. J. Org. Chem. 54, 2964–2966. [Google Scholar]
- Wright S. W.; Petraitis J. J.; Freimark B.; Giannaras J. V.; Pratta M. A.; Sherk S. R.; Williams J. M.; Magolda R. L.; Arner E. C. (1996) 2,5-Diarylisothiazolone: novel inhibitors of cytokine-induced cartilage destruction. Bioorg. Med. Chem. 4, 851–858. [DOI] [PubMed] [Google Scholar]
- Correa A.; Tellitu I.; Dominguez E.; Sanmartin R. (2006) Novel alternative for the N–N bond formation through a PIFA-mediated oxidative cyclization and its application to the synthesis of indazol-3-ones. J. Org. Chem. 71, 3501–3505. [DOI] [PubMed] [Google Scholar]
- Shadrick W. R.; Mukherjee S.; Hanson A. M.; Sweeney N. L.; Frick D. N. (2013) Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry 52, 6151–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belon C. A.; Frick D. N. (2008) Monitoring helicase activity with molecular beacons. BioTechniques 45, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]