Abstract
Heterochrony, or a change in developmental timing, is an important mechanism of evolutionary change. Historically the concept of heterochrony has focused alternatively on changes in size and shape or changes in developmental sequence, but most have focused on the pattern of change. Few studies have examined changes in the mechanisms that embryos use to actually measure time during development. Recently, evolutionary studies focused on changes in distinct timekeeping mechanisms have appeared, and this review examines two such case studies: the evolution of increased segment number in snakes and the extreme rostral to caudal gradient of developmental maturation in marsupials. In both examples, heterochronic modifications of the somite clock have been important drivers of evolutionary change.
Keywords: heterochrony, marsupial, evolution, developmental timing, snake, somitogenesis, limb, timekeeping mechanisms
1. Introduction
Development consists of a series of events that take place in a highly regulated spatial and temporal context. In most organisms there is a clear directionality to development as later events are commonly contingent on the proper completion of prior events. In animals at least, with a few exceptions such as regeneration and some processes that occur during metamorphosis there is rarely significant reversibility in developmental processes. In multicellular organisms, development proceeds from large scale patterning of the whole organism to events that are increasingly smaller in scale, and more modular and localized as individual parts differentiate and become more specialized. The field of developmental biology largely consists of the study of the mechanisms by which these intricate processes are controlled in space and time.
The process of timing and the role of changes in timing during development have been a strong focus of studies of comparative development and the interaction of development and evolution. The processes of development construct the organism and biologists have looked for ways that changes in developmental processes produce evolutionary change. Historically one kind of developmental change – the change in the timing of events – has received particular focus. Change in the timing of developmental events generally is termed heterochrony. The term heterochrony was initially coined to designate changes in the relative time of developmental processes between ancestors and descendants but in practice heterochrony is applied in a comparative sense to changes among taxa that are related at some level [1].
1.1 Historical perspective
The concept of heterochrony has evolved considerably over the past century. The term “heterochrony” was coined by Haeckel to denote certain deviations from his now discredited Biogenetic Law, which states that ontogeny recapitulates phylogeny. Haeckel believed that during development, an embryo repeated the, “…most important of the form changes which its ancestors traversed during the long and slow course of their paleontological evolution” [Haeckel quoted by 2]. Heterochrony indicated a shift in which a feature appeared at a different time in an organism’s developmental sequence than which it appeared in the sequence of that organism’s phylogeny [3,4]. Haeckel’s definition stands in contrast to the contemporary definition in that it is a comparison of changing ontogenies across phylogeny rather than a comparison of an ontogenetic sequence in an individual species with its presumed evolutionary pattern.
In an effort to refute Haeckel’s concept of recapitulation and join the fields of developmental biology, evolutionary biology and genetics, de Beer showed how timing changes during development could generate diversity among organisms [5–8]. He contended that heterochrony did not require an association with recapitulation; rather, de Beer used heterochrony to denote differences in the ontogenies of related taxa [9]. It is this comparative definition that is the principal one currently in use [3].
While de Beer’s treatment of heterochrony is regarded as a valuable early effort to join development and evolution, Gould made heterochrony a well-known concept in the field of evolutionary biology [9,10]. His work, and that of his colleagues, defined the scope of heterochrony for many years. Gould’s view of heterochrony, largely adopted by evolutionary biologists at the time, was characterized by a re-association of heterochrony with recapitulatory patterns and a focus on relative rates of growth rather than developmental sequences. Gould shifted the emphasis on heterochrony from the relative timing of developmental events to changes in the relationship between size and shape. In the 1980’s and early 1990’s, a surge of heterochrony studies focused nearly exclusively on size and shape changes such that the concept of heterochrony came to be practically synonymous with allometry [3].
In the late 1990’s the application of Gould’s definition of heterochrony was questioned. In certain instances “evolution by heterochrony” was invoked to explain a change in the relative proportion of any structure and often studies were so non-specific as to lack explanatory power. Attention was primarily on size and shape, and size was frequently used as a surrogate for time. In certain circumstances size can be a suitable proxy for age, but in other cases this exchange is inappropriate as rate of development, size and shape can evolve separate from one another [11–18]. Finally, the attention on size and shape restricted studies to global, organismal-level events and later processes in development [19,20]. Numerous variations among closely related species do result from growth heterochrony, but these methods cannot be used to examine a number of the most significant events in development: patterns of gene expression, cell and tissue specification and differentiation, induction and signaling cascades, and the emergence of segmental or regional identity, for example. Heterochronies in events such as these are absolutely critical in producing evolutionary change [3,19,21–25].
1.2 Heterochrony today
The study of heterochrony has been revitalized in the last two decades by a shift in focus from relative growth to relative timing of developmental events, and also an increasing focus on events at molecular and genetic levels. These studies focus on specific elements and increasingly on the underlying developmental mechanisms responsible for evolutionary change. New analytical tools now offer methods to analyze multiple events in many taxa, as well as to test hypotheses in a phylogenetic context. Thus the combination of modern developmental biological approaches using molecular and genetic data, with the renewed approach to heterochrony has brought new explanatory power to classic problems in evolutionary biology. Instead of emphasizing size and shape changes, modern heterochrony studies examine the basis for variation in an array of mechanisms and types of phenotypic modifications. Processes including shifts in critical periods, inductive events, and relative timing of gene expression have been studied; phenomena include patterning mechanisms, the timing of formation of organs and structures, alterations in life history phases, and morphological changes [3].
Most studies of heterochrony do not examine changes in timing mechanisms in the explicit sense–that is, the mechanisms that embryos use to actually measure time. This is partly due to the nature of development: many events in development are simply contingent on the completion of prior events. Within an embryo scheduling is often based on a sequence of events as opposed to clock time. The occurrence of many events depends on induction, cell and tissue interactions, and connections within signaling cascades. It is more appropriate to describe these types of control processes as scheduling rather than timing mechanisms. In addition, there appears to be no one mechanism organisms use for time assessment during development [26–41]. Diverse organisms at different stages of their life history use an array of mechanisms to track developmental time, complicating any evolutionary comparison of changes in timing mechanisms across taxa. However, our understanding of developmental timing mechanisms has increased dramatically in recent years, and heterochrony studies addressing the modification of timing mechanisms are beginning to emerge.
1.3 The somite clock
One such timing mechanism that has recently been the focus of heterochrony studies is the somite clock. Somites are transient structures in vertebrate embryos that are the first morphological sign of segmentation; they ultimately give rise to skeletal muscle, cartilage, tendons, endothelial cells, and dermis. Somites “bud off” from the anterior presomitic mesoderm, forming in rostral to caudal sequence. The most commonly referenced model for the mechanism of somitogenesis is that of the “Clock and Wavefront” [27,28,42,43]. The model posits that each cell in the presomitic mesoderm has its own internal clock, which oscillates between permissive and non-permissive states for formation of a segment boundary; cells are coupled so that oscillations within the presomitic mesoderm are synchronized. A wavefront travels rostro-caudal through the presomitic mesoderm, and after it has passed, cells are competent to form a segment boundary. In this fashion, a segment boundary is formed when the clock is in the permissive state and wherever the wavefront happens to be at that time. A large number of studies have detailed the specific molecular components of the Clock and Wavefront; these include members of the Notch, FGF, and Wnt signaling pathways [26–28,44]. While the identity of the clock pacesetter remains unknown, the readout of the clock can be seen in the periodic expression of members of the Notch, Wnt and Fgf signaling pathways in the presomitic mesoderm.
It is important to note that while an array of experimental evidence supports the Clock and Wavefront model, alternative models exist for which there is also experimental evidence, and not all models of the segmentation process rely on a clock mechanism [45,46]. For the purposes of this review, the discussion is framed in terms of the Clock and Wavefront model.
2. Case studies
2.1 Segmentation in snakes
One example of a heterochronic mechanism of evolutionary change comes from the study of segment number in vertebrates, and in particular, work by Gomez and colleagues [47] on somitogenesis in snakes. Among vertebrates the number of vertebrae is constant within a species, but varies widely from species to species, ranging from as few as six in some frogs to several hundred in certain fish and snakes. After investigating a number of possible mechanisms, the authors found that in the evolution of the snake body plan, heterochrony in somitogenesis rate is predominantly responsible for the impressive increase in segment number [47].
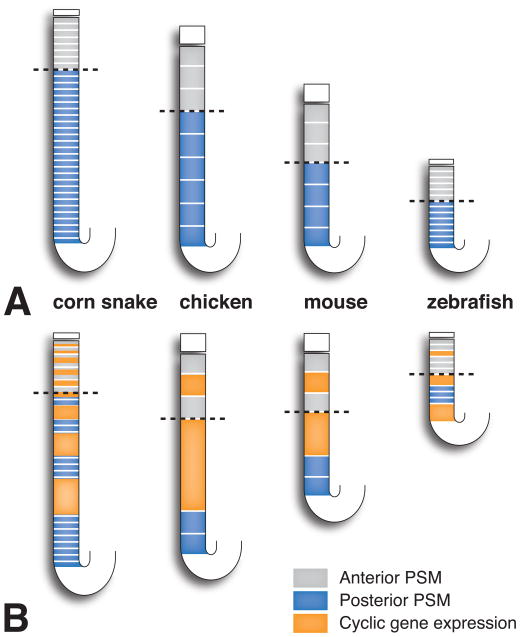
Segment number, and hence vertebral number, depends primarily on two factors–elongation of the body axis and segment size. A greater number of segments can be formed in a longer axis, and more smaller-sized segments can be formed in an axis of the same length [48]. According to the Clock and Wavefront model, segment size is determined by the speed of wavefront regression and the rate of the segmentation clock. Smaller sized somites are formed when either the wavefront regresses more slowly or when the segmentation clock ticks more quickly. In cases in which the segmentation clocks ticks more quickly, the wavefront covers a smaller area of the presomitic mesoderm between the delineation of each somite boundary, leading to more smaller-sized somites (Figure 1).
Figure 1.
Somitogenesis compared between different vertebrate models. In the corn snake, more stripes of cyclic gene expression are seen simultaneously in the presomitic mesoderm and many more smaller-sized somites are formed because the rate of the somite clock is accelerated. Dotted line indicates the position of the determination front. Anterior is to the top, posterior to the bottom. PSM, presomitic mesoderm. Redrawn from [48].
One possibility to explain the dramatic increase in somite number in snakes and other animals with elongated body plans is that axis growth and extension continue for a longer period of time. Modeling of axis growth has shown that the number of cell generations in the presomitic mesoderm needed to produce the snake embryonic axis is approximately 21, compared to 16 and 13 generations in the mouse and chicken axes, respectively [47]. As the number of somites in snakes can be five times or more that of chicken or mouse, number of cell generations of axis growth only partially explains the increase in segment number in snakes.
Another possibility to account for the increase in segment number in snakes is that there are many more smaller-sized somites. Measurements of somite size in chicken, mouse, and snake indicated that snake somites are at a minimum three times smaller than mouse and chicken somites [47]. As discussed above, smaller somites can result from the slowing of determination front regression or acceleration of the somite clock. Gomez and colleagues compared determination front regression in snake and other amniote embryos, but did not find a considerable difference. The authors also compared the time required to form a somite, which is equal to the clock period, among various species. In the corn snake one somite pair forms every 100 min, which is comparable to the 90 min required in chicken. However, the overall developmental rates of snake and chicken are different. Using the time between the formation of conserved developmental landmarks, the study authors determined that the overall developmental rate is three times slower in snake versus chicken, and therefore, the cell cycle is three times slower in the tail bud. The whiptail lizard is much more similar in overall developmental time to the snake but has a 4 hour clock pace, much slower than that of the snake. These results imply that the rate of somitogenesis is accelerated in snake relative to axis growth.
Gomez and colleagues additionally observed within the presomitic mesoderm multiple stripes of dynamic lunatic fringe (LFNG) expression [47]. LNFG is one of the cyclic genes, whose expression is driven by the somite clock. Vertebrates examined previously exhibit only one to three stripes of cyclic gene expression. Mathematical modeling was used to show that the increase in stripe number is explained by the increased clock rate relative to axis growth in snakes compared to other amniotes. Accelerated clock rate causes more traveling waves so that an increased number of stripes are seen concurrently in the presomitic mesoderm. Given this evidence, a relatively accelerated clock rate in the snake was determined to lead to the greatly increased number of smaller-sized segments.
2.2 Heterochrony in segmentation and limb development in marsupials
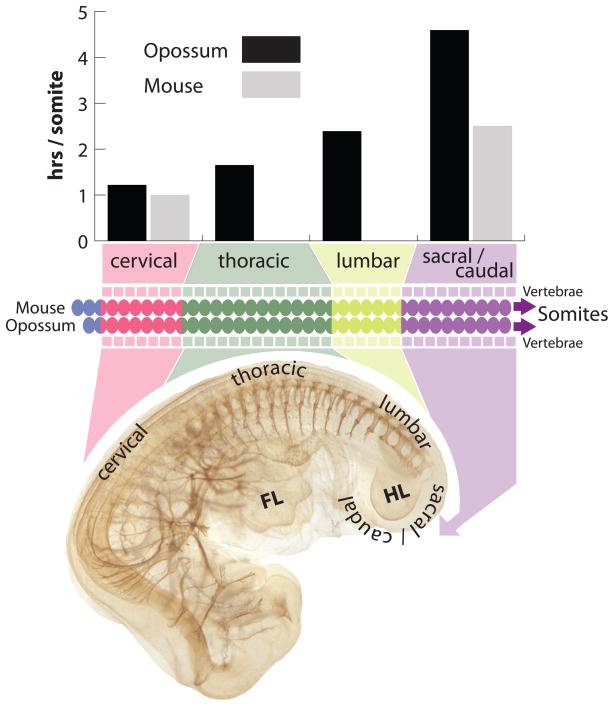
Marsupial and placental mammals are distinguished by reproductive mode where marsupials, relative to eutherian mammals, are born at a highly embryonic state after a short gestation. The altricial newborn must possess sufficient adaptations for independent survival at a stage that is equivalent to a 10–12 week human or 10 day mouse [49]. This embryonic neonate crawls unaided to the teat and completes most development while attached to the teat, nursing. The morphological configuration of the neonate is highly distinctive with an extreme rostral to caudal gradient in development (Figure 2). The forelimbs are well differentiated and relatively large and are used to crawl to the teat directly after birth (Figure 4G). In contrast the hindlimbs are much smaller, comparatively under-developed, and not yet functional. The steep anterior-posterior developmental gradient can also be seen in the degree of chondrification of the axial skeleton (Figure 2). A comparison of cleared skeletal preparations of model marsupial and eutherian mammals shows that vertebrae at cervical and upper thoracic levels are well chondrified at birth in an opossum, while in posterior regions structures are relatively undifferentiated. In contrast, a mouse at a similar stage of development displays much more uniform chondrification along the anterior-posterior axis.
Figure 2.
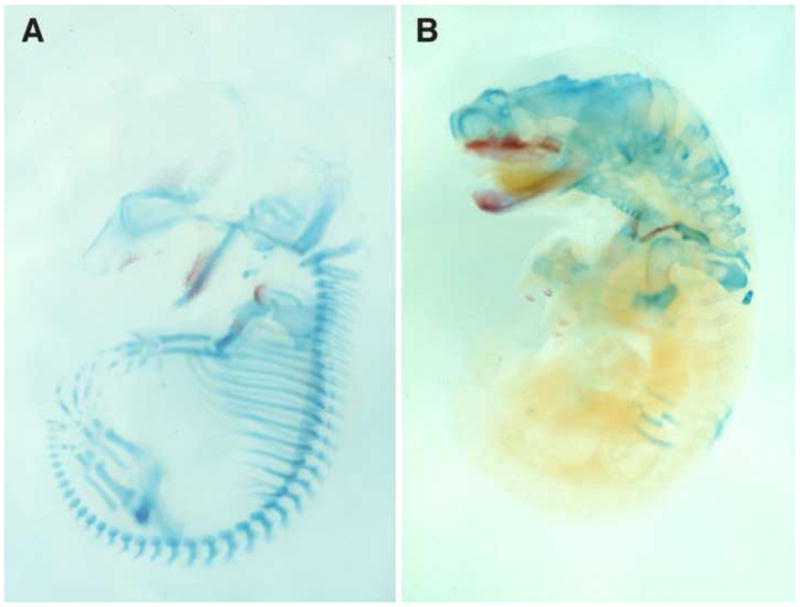
Alcian blue and alizarin red skeletal preparations of an E14 mouse (A) and a two day old opossum (B). Bone is red and cartilage is blue. Modified from [51].
Figure 4.
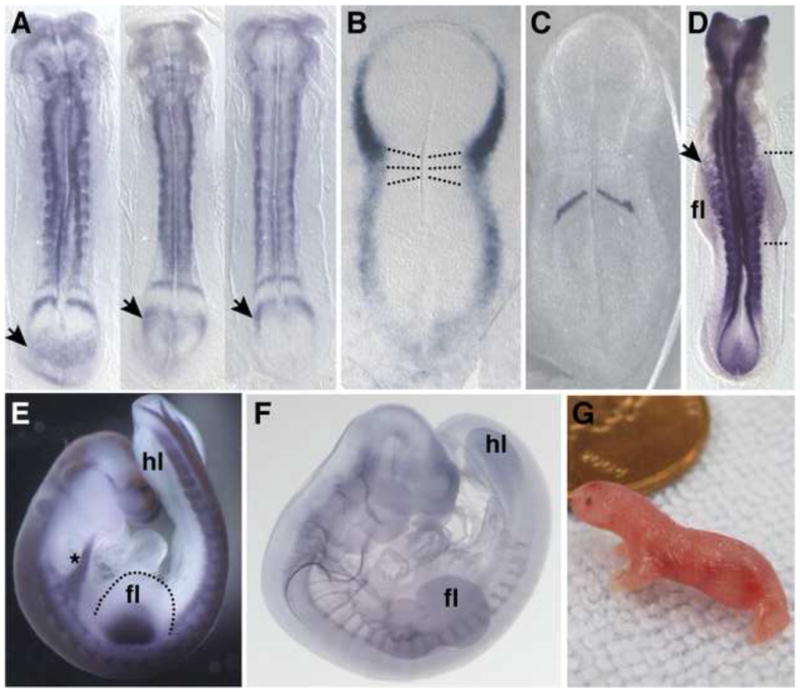
A, stage 24 somite-matched opossum embryos hybridized with in situ probe to Lnfg. Three different phases of cyclic gene expression can be seen in the presomitic mesoderm. B, stage 22 opossum embryo hybridized with Tbx5 probe and shown with first two somites highlighted by dotted lines. At this stage the first somites form and the forelimb field is specified. C, stage 23 opossum embryo hybridized with probe to Mesp2, a gene that defines the future segmental boundary. D, stage 26 opossum embryo hybridized with Pax3 probe. Pax3 positive myocytes can be seen leaving the somites and migrating into the forelimb. Note that more anterior myocytes have migrated further from the somites than more posterior myocytes. Anterior and posterior limits of the forelimb bud are indicated by the dashed lines. E, stage 28 opossum embryo hybridized with Pax3 probe. Asterisk indicates a stream of myocytes en route to the future tongue. Note the difference in the size of the fore- and hindlimb buds. F, opossum embryo stained with antibody to neurofilament. Note the gradient in spinal nerve maturation and outgrowth from rostral to caudal. G, newborn opossum crawling. Anterior is to the top. fl, forelimb; hl, hindlimb.
Using the opossum, Monodelphis domestica, as a model marsupial, we have investigated the developmental origins of the steep anterior-posterior gradient, focusing on the forelimb/hindlimb disparity and timing of segmentation [50–52]. Many of the structures discussed above are derived from the somites. Somites give rise to much of the axial skeleton, all postcranial skeletal muscles, including limb muscles, as well as the tongue muscle precursors which migrate from the anterior-most somites. While certain elements of somitogenesis appear to be conserved–gene expression of Clock and Wavefront components and rate of somite maturation along the anterior-posterior axis are consistent with expression reported for other amniotes–two changes in the relative timing of somitogenesis in M. domestica contribute to the characteristic configuration of the marsupial neonate [51]. There exists a four-fold change in the rate of somitogenesis along the anterior-posterior axis (Figure 3). Somitogenesis slows from approximately one hour per somite pair at cervical levels, to at least four hours per somite pair at caudal levels, one of the slowest somitogenesis rates so far reported for any vertebrate. The rate of somitogenesis slows to a greater extent in marsupials than has been previously observed in other amniotes with similar overall rates of development, contributing to the steep anterior-posterior gradient. Additionally, somitogenesis initiates early relative to other events in development, when the embryo is little more than a flat featureless plate, providing anterior somitic derivatives at an earlier stage of development (Figure 4B).
Figure 3.
Somitogenesis rate in opossum decreases 4-fold from rostral to caudal, slowing to greater extent than the caudal rate for mouse. Opossum embryo is stained with antibody to neurofilament to highlight spinal nerves. FL indicates forelimb; HL indicates hindlimb. Mouse data from [97].
Slowing of somitogenesis in marsupials likely involves regulation of the length of the presomitic mesoderm. Under the Clock and Wavefront model, as the somite clock slows, movement of the determination front must also slow in order to maintain consistent somite size. (If the clock were to slow with no compensatory change in determination front progression, somite size would increase, as the wavefront would cover more of the presomitic mesoderm between the delineation of consecutive segment boundaries.) Progression of the determination front depends in part on the rate of addition of new cells to the end of the embryonic axis [53]. As the growth of the axis slows and the length of the presomitic mesoderm decreases, movement of the determination front also slows. Given how dramatically the somite clock slows in marsupials without a corresponding increase in caudal somite size, we predict that in the context of the Clock and Wavefront model, presomitic mesoderm length and rate of axis extension may decrease precociously or to a greater extent than in other amniotes.
We have also investigated the discrepancy at birth in fore- and hindlimb maturation [50]. It was previously known that the forelimb is accelerated from first appearance, while the hindlimb is delayed from the bud stage onward [54–56]. We have shown that during embryonic development the marsupial forelimb is accelerated through a combination of early field specification, early initiation of outgrowth and patterning, and greater tissue allocation [50]. Concomitant with forelimb field expansion and early specification, the number of somites contributing limb muscle myocytes appears to be greater than in other vertebrates, and the migration of myocytes into the forelimb bud occurs earlier. The expression of the Hox genes, known to be involved in limb positioning, has also shifted along the AP axis in correlation with increased limb field size. The acceleration of forelimb development in M. domestica appears to be produced by multiple changes in early limb development. However, because the limb development program is a well integrated network, a complete cascade of events most likely arises from two simple shifts in development: first, an early initiation of the forelimb field, or heterochrony, and second, a shift in the area of the early limb field, or heterotopy. Once the forelimb field is specified early, as evidenced by early initiation of Tbx5 expression (Figure 4B), downstream genes, Fgf10 and then Fgf8, are also expressed early, as Fgf10 is a direct target of Tbx5 [57] and Fgf8 a target of Fgf10 [58]. Limb lateral plate mesoderm induces myocyte migration from the somites to the limb area [59,60]. Therefore, shifts in the timing and extent of these latter events are likely to be a consequence of the early and expanded forelimb field. One heterochronic and one heterotopic change in early limb development may lead to an entire series of shifts in gene expression and cellular behavior which contribute to the precocial forelimb of the marsupial neonate.
The heterochronies of the paraxial mesoderm and of the limbs are likely linked in marsupials. Early initiation of somitogenesis is most likely critical for timely development of the forelimb musculature and somite-derived supporting structures. This theory is supported by a number of observations. Firstly, limb muscle develops from myocytes, which migrate from limb-level somites. In M. domestica forelimb specification, outgrowth, and patterning all occur early relative to other developmental events [50]. Myocyte migration is induced by FGF signaling from the apical ectodermal ridge (AER) of the limb bud [60]. Relative to other events in the M. domestica embryo, the appearance of Fgf8 in the AER is early and myocytes migrate early [50]. Secondly, in M. domestica the anterior-posterior developmental gradient can be observed even within the population of migrating forelimb myocytes. Myocytes originating from more anterior somites begin migration from the somites earlier than myocytes from more posterior somites (Figure 4D). In mouse there is no discernible gradient in timing of anterior to posterior myocyte migration at forelimb level [61]. The presence of this gradient in M. domestica suggests that myocytes migrate as soon as they are mature enough to do so; there exists a constraint on the timing of myocyte maturation and migration. Thirdly, Weisbecker et al. have demonstrated early ossification of anterior axial skeletal elements, including cervical vertebrae, scapula, and ribs–all somite derived structures and points of attachment for limb muscles [62,63]. For these reasons, early initiation of somitogenesis anteriorly likely allows for timely development of the forelimb musculature and somite-derived supporting structures.
Contrary to most expectations, the hindlimb field is also specified and patterned early in marsupials despite the fact that the hindlimb is small and not functional at birth. It is possible that a common developmental mechanism in anterior-posterior patterning couples specification of the limb fields as evidenced by expression of the limb specific T-box genes: once Tbx5 is induced early in the forelimb field, early expression of Tbx4 in the hindlimb field follows. Additionally, downstream genes, Shh and Fgf10, are expressed early in the hindlimb in M. domestica, as would be expected given the model of a tightly integrated developmental module. However, unlike the forelimb, there is no expansion in the relative size of the field. Moreover, when Tbx4 is first expressed, little mesoderm has accumulated at the posterior end of the body. Tbx4 is expressed as cells destined to become the hindlimb mesoderm leave the primitive streak. Outgrowth of the hindlimb cannot be observed until later, after further addition of the caudal somitic and lateral plate mesoderm [50].
These results thus suggest that the disparity in the size and stage of development of the opossum forelimb and hindlimb at birth are not due to changes in the timing of hindlimb specification, but of later events in hindlimb development. It appears that the hindlimb is first specified when there is little cellular material available to build the limb bud and produce subsequent outgrowth. The fact that hindlimb Fgf8 expression is delayed in the opossum (not early relative to mouse) suggests a pause in development after limb field specification. Other studies have also hypothesized a period of developmental “dormancy” at much later stages [55,62]. Differential expression of Igf1 and other growth-related genes in developing opossum limbs suggests that divergent growth rate additionally contributes to the forelimb/hindlimb disparity at birth [64].
Is there a developmental mechanism that unifies the deceleration of somitogenesis and hindlimb outgrowth? There exists a delay caudally in the production of somites, derived from presomitic mesoderm, as well as a delay in hindlimb outgrowth, derived from lateral plate mesoderm. As the somites and limb buds have different mesodermal origins, the delay in their development is likely linked through a delay in generation of mesoderm from the primitive streak, and later the tailbud. As discussed above, a slowing of somitogenesis strongly suggests that the production of presomitic mesoderm from the end of the axis should slow as well. Slowing of the generation of the entire posterior axis in marsupials may link the delay in maturation of posterior structures of varied mesodermal origin, and remains to be investigated further.
The newborn marsupial does not only exhibit a steep anterior-posterior gradient in the postcranium, but also shows an intricate pattern of heterochrony within the cranial region [1,22,65–71]. In the head elements of the feeding apparatus are accelerated, while the central nervous system is generally delayed in development. Given the demands for elaborated anterior structures and a functional feeding apparatus during the short period from primitive streak to birth (approximately 4.5 days in M. domestica) it has been proposed that an energy trade-off exists to emphasize structures required at birth and de-emphasize non-necessary structures, such as the hindlimbs and pelvis [62,72,73]. Somitogenesis slows posteriorly in opossum relative to mouse [51]. With somitogenesis occurring more slowly, the maturation of posterior somites is delayed, possibly freeing resources for other structures in the embryo. Newborns of different marsupial species with differing lengths of gestation tend to have anterior-posterior developmental gradients that are more or less steep [49,74], supporting the energy trade-off hypothesis. For example, Dasyurus viverrinus (eastern quoll) is ultra-altricial at birth, undergoing organogenesis in just 2.5 days and weighing only 10–15 mg [75]. D. viverrinus and other dasyurid neonates exhibit the steepest anterior-posterior gradients among marsupial newborns, most obvious in the huge difference in the relative degree of development of the fore- and hindlimb [76]. In dasyurids the oral apparatus is also extraordinarily large relative to the brain or sense organs. At the other extreme are the macropodid marsupials (kangaroos and wallabies). Macropus eugenii (Tammar wallaby) goes from primitive streak to birth in approximately 10 days, neonates weigh roughly 370 mg, and the forelimb to hindlimb difference is much less pronounced. The exact constraints that bring about this difference in overall timing among various marsupial species are unknown. It is hypothesized that limited resources, possibly in the form of cellular material and/or metabolic energy, are available to marsupial embryos during gestation, provided by the mother in the oocyte and later through the placenta. During the relatively short time for gestation, metabolic energy expenditure must allow for the development of a functional forelimb at birth. This includes significant soft tissue development–muscle, nerves, and blood vessels–essential for the climb to the teat, and may take place at the metabolic expense of the hindlimb [62]. The shorter the period of gestation in a given marsupial species, the less time these embryos have to build structures necessary for survival at birth, and the less time they have to absorb nutrients through the placenta. Therefore the necessary structures must be accelerated to a greater extent with presumably fewer resources, resulting in the range of developmental gradients observed.
3. Developmental timing mechanisms and the nature of developmental time
In many instances timing appears to simply arise from other properties of developmental processes and regulation (growth, induction, or differentiation), while in other cases timing is explicitly controlled by a molecular mechanism [38]. Thus, a change in the timing of specific events does not necessarily signify a change in a definitive timing mechanism. Heterochrony may result from changes in any number of developmental mechanisms that are not explicitly involved in the regulation of developmental timing. For example, modifications of a regulatory pathway governing differentiation or induction of one structure may delay or accelerate the formation of a dependent tissue or cell type.
Timing mechanisms separate from other fate regulation have been identified using experimental methods in model systems. A number of cell types are able to monitor developmental time or schedule events in embryos. As described above, the somite clock in vertebrate embryos directs the timing of formation of somite boundaries from cells within the presomitic mesoderm [27]. In the central nervous system, timers control specific differentiation programs in retinal neuron precursors, oligodendrocyte precursors, and Drosophila neuroblasts [29–32]. In the zebrafish neural rod, cells contain an intrinsic timer that directs the schedule of epithelial polarization during neural tube formation [33]. Developmental timers also direct global, whole embryo transitions in development, such as the Xenopus midblastula transition, the maternal to zygotic transition in Drosophila and zebrafish, and apoptosis at the onset of gastrulation in Xenopus [34–36,77]. The heterochronic gene network in the worm, Caenorhabditis elegans, specifies temporal cell fate selection using the microRNAs lin-4 and let-7 [37,41,78]. The microRNA let-7 is conserved in sequence and expression across wide evolutionary distance, and the developmental timing role of microRNAs in general appears to be conserved as well [39,41,79–85].
Recent evidence suggests that the molecular oscillator of the segmentation clock may function to provide temporal information in other embryological tissues. Cyclic gene expression associated with the clock commences in the primitive streak as gastrulation is initiated, but expression is not restricted to presomitic mesoderm precursors. Precursors of many different mesodermal tissues experience waves of cyclic expression before they migrate to their individual positions in the embryo [86]. It is unknown whether early exposure to this molecular oscillator plays a role in the timing of differentiation or regionalization of other primitive streak derived tissues [87]. Additionally, Hairy2, a hairy/E(spl) family member which encodes a component of the somitogenesis clock, oscillates in cells at the tip of the chick wing bud [88] and may function in proximo-distal patterning of the wing [89]. Finally, a cell culture experiment performed with several mouse cell lines of different tissue types demonstrated that following exposure to serum shock the hes1 mRNA and protein exhibit a 2 hour oscillation period [90]. These findings suggest that the hairy/E(spl) oscillator is not limited to somitogenesis and that a molecular clock may provide timing cues to cells in many different embryonic tissues, although at tissue-specific time periods [87].
Among developmental timing mechanisms, a distinction can be made between true clock-like, “timekeeping” mechanisms and other scheduling or sequencing processes, which control the timing of specific events, but do not necessarily track time, per se. A timekeeping mechanism tracks increments of time, such an in the case of the somite clock, in which the readout of the clock is the oscillating expression of the cyclic genes and the periodic formation of somite boundaries. For other scheduling mechanisms, changes in the timing pathway may result in heterochrony, but the underlying mechanism is fundamentally different in that time itself is not measured. Scheduling or sequencing mechanisms appear to be quite common in development, while true timekeeping mechanisms are relatively rare. Why is this the case? This fact reflects the nature of development and developmental time: for most processes, what matters most to the embryo is that events occur in the correct order relative to one another, not that the events occur at a specific clock time. Sequencing is sufficient to ensure that development proceeds smoothly, and may even be a more robust mechanism in most situations.
An alternative or additional explanation for the apparent rarity of developmental clocks is that explicit timekeeping mechanisms may have been difficult to observe in most animals and may actually be more common than previously thought. For example, the heterochronic gene pathway was discovered in C. elegans, in part, because the worm has such well-defined developmental stages, each of which is delineated by a molt. Timing changes between stages were therefore much easier to detect. Developmental stages in vertebrates are not as discrete and each tissue is likely to be regulated independently [38]. In addition, oscillations characteristic of many timers have been difficult to identify. Oscillating gene expression of the somite clock is easier to observe because cells are coupled within the presomitic mesoderm and a large group of cells expresses the cyclic gene concurrently. However in other tissues, oscillators within cells may not be synchronized. Advances in genetics, live cell imaging and biotechnology are likely to improve our ability to identify these single-cell oscillators.
Molecular timers tend to share certain features of their mechanisms. Many timers are oscillators, typically driven by delayed negative feedback loops. The length of the period of oscillations depends on the lengths of delays in the loop. In the case of the segmentation clock, the period of intracellular oscillation is approximately equal to the time required to synthesize and deliver each molecule of mRNA and protein, plus the lifetimes of these molecules. Recent work has demonstrated that the presence of introns within oscillatory genes of the Notch pathway and their associated splicing delays are major determinants of clock period, and thus of somite spacing and number [91,92]. Together, the kinetics of mRNA and protein production and destruction can account for much of the oscillatory period, as mRNA splicing and export are much slower than transcript elongation [93]. Synchronization of oscillations between cells by Delta-Notch coupling also regulates the collective period of the segmentation clock [94]. In C. elegans, rising and falling levels of LIN-42A allow the start and completion, respectively, of larval molts [95]. The period of these molts is undoubtedly influenced by similar delays in transcription, translation, and other molecular processing of gene products. Heterochronic mechanisms of evolutionary change will likely be found through the examination of the critical delays of these oscillatory loops within a comparative context.
Another common mechanism, sometimes referred to as an “hour glass” mechanism, features time measurement using the decay or accumulation of a substance to a threshold level. This type of mechanism has been proposed in the case of the maternal-zygotic transition in Xenopus and zebrafish, in which changing nuclear to cytoplasmic ratio across cell divisions appears to be important in zygotic genome activation [35,96]. In the case of Xenopus laevis, the maternal-zygotic transition requires titration of four DNA replication factors by embryonic nuclei. These proteins are limiting for initiation of replication at increasing nuclear to cytoplasmic ratios. They trigger cell cycle slowing, transcriptional onset, and check-point kinase (Chk1) activation, which becomes transiently activated at the time of the maternal-zygotic transition [96]. Heterochronies in hour glass type mechanisms can be accomplished through shifts in the threshold or the initial levels of critical substances.
We have seen in the past few decades a major expansion in studies of heterochrony in a wide variety of animals and systems. Also the past few decades have witnessed an explosion in our understanding of the underlying mechanisms and also the mechanisms of timing during development. However, thus far there have been few evolutionary studies on heterochrony that integrate these data on timekeeping mechanisms. Most evolutionary studies focus on pattern rather than underlying mechanism. As the specific molecular components of these pathways are identified in model organisms, evodevo studies focusing on the modification of timekeeping mechanisms will undoubtedly become more common.
4. Conclusions
The concept of heterochrony has changed significantly since it was first introduced in the late nineteenth century. Heterochrony studies are now largely focused on changes in developmental sequences and increasingly include events at the molecular level, linking classic questions in evolutionary biology with modern advances in developmental biology. Studies addressing the modification of explicit timing mechanisms are still quite rare. However, our understanding of the basic developmental biology governing timing has grown to the point where a sufficient knowledge base now exists upon which to build a comparative framework and pose evolutionary questions.
Highlights.
Historically heterochrony studies have focused on the pattern of timing changes.
Few studies have examined evolutionary changes in developmental timing mechanisms.
Snakes have more smaller sized somites due to an increase in somitogenesis speed.
The steep AP gradient in marsupial neonates arises from multiple timing changes.
Embryos appear to rely more on “sequence” timing of events rather than “clock” time.
Acknowledgments
The authors wish to thank Olivier Fedrigo for assistance with figures. ALK is supported by NIH NRSA 5F32HD070631-03.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Smith KK. Heterochrony revisited: the evolution of developmental sequences. Biological Journal of the Linnean Society. 2001;73:169–86. [Google Scholar]
- 2.Russell ES. Form and Function. London: John Murry Ltd; 1916. [Google Scholar]
- 3.Smith KK. Time’s arrow: heterochrony and the evolution of development. Int J Dev Biol. 2003;47:613–21. [PubMed] [Google Scholar]
- 4.Richardson MK, Keuck G. Haeckel’s ABC of evolution and development. 2002. [DOI] [PubMed] [Google Scholar]
- 5.De Beer SG. Embryology and evolution. 1930. [Google Scholar]
- 6.De Beer G. Embryos and Ancestors. Oxford: Clarendon Press; 1958. [Google Scholar]
- 7.De Beer G. Embryos and Ancestors. Oxford: Clarendon Press; 1940. [Google Scholar]
- 8.De Beer G. Embryos and ancestors. Oxford: Clarendon Press; 1951. [Google Scholar]
- 9.Gould SJ. Ontogeny and phylogeny. Cambridge, MA: Belknap Press of Harvard University Press; 1977. [Google Scholar]
- 10.Alberch P, Gould SJ, Oster GF, Wake DB. JSTOR: Paleobiology. 1979 Summer;5(3):296–317. [Google Scholar]; Paleobiology. 1979 [Google Scholar]
- 11.Roth VL. How elephants grow: heterochrony and the calibration of developmental stages in some living and fossil species. Journal of Vertebrate Paleontology. 1984;4:126–45. [Google Scholar]
- 12.Emerson SB. JSTOR: The American Naturalist. American Naturalist 1986 Feb;127(2):167–183. [Google Scholar]
- 13.Blackstone NW. Size and time. Systematic Biology. 1987 [Google Scholar]
- 14.Blackstone NW. Allometry and relative growth: pattern and process in evolutionary studies. Systematic Biology. 1987 [Google Scholar]
- 15.Klingenberg CP, Spence JR. Heterochrony and Allometry: Lessons from the Water Strider Genus Limnoporus. Evolution. 1993;47:1834. doi: 10.1111/j.1558-5646.1993.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 16.Klingenberg CP. Heterochrony and allometry: the analysis of evolutionary change in ontogeny. Biol Rev Camb Philos Soc. 1998;73:79–123. doi: 10.1017/s000632319800512x. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey LR, Sutherland MR. Flawed inference: why size-based tests of heterochronic processes do not work. J Theor Biol. 1995 [Google Scholar]
- 18.Godfrey LR, Sutherland MR. What’s growth got to do with it? Process and product in the evolution of ontogeny. Journal of Human Evolution. 1995 [Google Scholar]
- 19.Raff RA, Wray GA. Heterochrony: Developmental mechanisms and evolutionary results. J Evolution Biol. 1989;2:409–34. [Google Scholar]
- 20.Hall BK. Evolutionary Developmental Biology. 2. Springer; 1999. [Google Scholar]
- 21.Gunter HM, Koppermann C, Meyer A. Revisiting de Beer’s textbook example of heterochrony and jaw elongation in fish: calmodulin expression reflects heterochronic growth, and underlies morphological innovation in the jaws of belonoid fishes. Evodevo. 2014;5:8. doi: 10.1186/2041-9139-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaglia JL, Smith KK. Early differentiation and migration of cranial neural crest in the opossum, Monodelphis domestica. Evol Dev. 2003;5:121–35. doi: 10.1046/j.1525-142x.2003.03019.x. [DOI] [PubMed] [Google Scholar]
- 23.Carleton KL, Spady TC, Streelman JT, Kidd MR, McFarland WN, Loew ER. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biol. 2008;6:22. doi: 10.1186/1741-7007-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–7. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 25.Parsons KJ, Albertson RC. Roles for Bmp4 and CaM1 in shaping the jaw: evo-devo and beyond. Annu Rev Genet. 2009;43:369–88. doi: 10.1146/annurev-genet-102808-114917. [DOI] [PubMed] [Google Scholar]
- 26.Krol AJ, Roellig D, Dequeant M-L, Tassy O, Glynn E, Hattem G, et al. Evolutionary plasticity of segmentation clock networks. Development. 2011;138:2783–92. doi: 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pourquié O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell. 2011;145:650–63. doi: 10.1016/j.cell.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusumi K, May CM, Eckalbar WL. A large-scale view of the evolution of amniote development: insights from somitogenesis in reptiles. Curr Opin Genet Dev. 2013;23:491–7. doi: 10.1016/j.gde.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–18. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 30.Raff M. The mystery of intracellular developmental programmes and timers. Biochem Soc Trans. 2006;34:663–70. doi: 10.1042/BST0340663. [DOI] [PubMed] [Google Scholar]
- 31.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–21. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 32.Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14:823–38. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girdler GC, Araya C, Ren X, Clarke JDW. Developmental time rather than local environment regulates the schedule of epithelial polarization in the zebrafish neural rod. Neural Dev. 2013:8. doi: 10.1186/1749-8104-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc Natl Acad Sci USA. 1996;93:2060–4. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 36.Hensey C, Gautier J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech Dev. 1997;69:183–95. doi: 10.1016/s0925-4773(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 37.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–16. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 38.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–34. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 39.Moss EG. More intriguing roles for mammalian homologs of worm heterochronic genes. Cell Cycle. 2009:8. [Google Scholar]
- 40.Raff M. Intracellular developmental timers. Cold Spring Harbor Symposia on Quantitative Biology. 2007 doi: 10.1101/sqb.2007.72.007. [DOI] [PubMed] [Google Scholar]
- 41.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–7. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–76. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- 43.Aulehla A, Herrmann BG. Segmentation in vertebrates: clock and gradient finally joined. Genes Dev. 2004;18:2060–7. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- 44.Dequeant M-L, Pourquie O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9:370–82. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- 45.Dias AS, De Almeida I, Belmonte JM, Glazier JA, Stern CD. Somites without a clock. Science. 2014;343:791–5. doi: 10.1126/science.1247575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauschke VM, Tsiairis CD, François P, Aulehla A. Scaling of embryonic patterning based on phase-gradient encoding. Nature. 2013;493:101–5. doi: 10.1038/nature11804. [DOI] [PubMed] [Google Scholar]
- 47.Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquié O. Control of segment number in vertebrate embryos. Nature. 2008;454:335–9. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- 48.Gomez C, Pourquié O. Developmental control of segment numbers in vertebrates. J Exp Zool Part B. 2009;312:533–44. doi: 10.1002/jez.b.21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes LR, Hall SL. Structural adaptations of the newborn marsupial. In: Tyndale-Biscoe H, Janssens PA, editors. The Developing Marsupial. New York: Springer-Verlag; 1988. pp. 8–27. [Google Scholar]
- 50.Keyte AL, Smith KK. Developmental origins of precocial forelimbs in marsupial neonates. Development. 2010;137:4283–94. doi: 10.1242/dev.049445. [DOI] [PubMed] [Google Scholar]
- 51.Keyte A, Smith KK. Heterochrony in somitogenesis rate in a model marsupial, Monodelphis domestica. Evol Dev. 2012;14:93–103. doi: 10.1111/j.1525-142X.2011.00524.x. [DOI] [PubMed] [Google Scholar]
- 52.Keyte AL, Smith KK. Opossum (Monodelphis domestica): A Marsupial Development Model. CSH Protoc. 2008;2008 doi: 10.1101/pdb.emo104. pdb.emo104. [DOI] [PubMed] [Google Scholar]
- 53.Dubrulle J, Pourquié O. Coupling segmentation to axis formation. Development. 2004;131:5783–93. doi: 10.1242/dev.01519. [DOI] [PubMed] [Google Scholar]
- 54.McCrady E. The Embryology of the Opossum. Philadelphia: Wistar Institute of Anatomy and Biology; 1938. [Google Scholar]
- 55.Bininda-Emonds ORP, Jeffery JE, Sánchez-Villagra MR, Hanken J, Colbert M, Pieau C, et al. Forelimb-hindlimb developmental timing changes across tetrapod phylogeny. BMC Evol Biol. 2007;7:182. doi: 10.1186/1471-2148-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sears KE. Differences in the timing of prechondrogenic limb development in mammals: the marsupial-placental dichotomy resolved. Evolution. 2009;63:2193–200. doi: 10.1111/j.1558-5646.2009.00690.x. [DOI] [PubMed] [Google Scholar]
- 57.Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, et al. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–33. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, et al. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–65. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi K, Ozawa E. Myogenic cell migration from somites is induced by tissue contact with medial region of the presumptive limb mesoderm in chick embryos. Development. 1995;121:661–9. doi: 10.1242/dev.121.3.661. [DOI] [PubMed] [Google Scholar]
- 60.Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, et al. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev Cell. 2003;5:379–90. doi: 10.1016/s1534-5807(03)00263-6. [DOI] [PubMed] [Google Scholar]
- 61.Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–12. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 62.Weisbecker V, Goswami A, Wroe S, Sánchez-Villagra MR. Ossification heterochrony in the therian postcranial skeleton and the marsupial-placental dichotomy. Evolution. 2008;62:2027–41. doi: 10.1111/j.1558-5646.2008.00424.x. [DOI] [PubMed] [Google Scholar]
- 63.Huang R, Christ B, Patel K. Regulation of scapula development. Anat Embryol. 2006;211 (Suppl 1):65–71. doi: 10.1007/s00429-006-0126-9. [DOI] [PubMed] [Google Scholar]
- 64.Sears KE, Patel A, Hubler M, Cao X, Vandeberg JL, Zhong S. Disparate Igf1 expression and growth in the fore- and hind limbs of a marsupial mammal (Monodelphis domestica) J Exp Zool Part B. 2012;318:279–93. doi: 10.1002/jez.b.22444. [DOI] [PubMed] [Google Scholar]
- 65.Clark CT, Smith KK. Cranial osteogenesis in Monodelphis domestica (Didelphidae) and Macropus eugenii (Macropodidae) J Morphol. 1993;215:119–49. doi: 10.1002/jmor.1052150203. [DOI] [PubMed] [Google Scholar]
- 66.Smith KK. Development of craniofacial musculature in Monodelphis domestica (Marsupialia, Didelphidae) J Morphol. 1994;222:149–73. doi: 10.1002/jmor.1052220204. [DOI] [PubMed] [Google Scholar]
- 67.Smith KK. Integration of Craniofacial Structures during Development in Mammals. Am Zool. 1996;36:70–9. [Google Scholar]
- 68.Smith KK. Comparative Patterns of Craniofacial Development in Eutherian and Metatherian Mammals. Evolution. 1997;51:1663–78. doi: 10.1111/j.1558-5646.1997.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 69.Smith KK, Vaglia J. Early development of the cranial neural crest, neural tube and paraxial mesoderm in marsupials. Am Zool. 2001;41:1589–9. [Google Scholar]
- 70.Smith KK. Sequence heterochrony and the evolution of development. J Morphol. 2002;252:82–97. doi: 10.1002/jmor.10014. [DOI] [PubMed] [Google Scholar]
- 71.Nunn CL, Smith KK. Statistical analyses of developmental sequences: the craniofacial region in marsupial and placental mammals. Am Nat. 1998;152:82–101. doi: 10.1086/286151. [DOI] [PubMed] [Google Scholar]
- 72.Müller F. Zum Vergleich der Ontogenesen von Didelphis virginiana und Mesocricetus auratus. Revue Suisse De Zoologie. 1967;74:607–13. [Google Scholar]
- 73.Smith KK. Comparative patterns of craniofacial development in eutherian and metatherian mammals. Evolution. 1997;51:1663–78. doi: 10.1111/j.1558-5646.1997.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 74.Smith KK. Craniofacial development in marsupial mammals: developmental origins of evolutionary change. Dev Dyn. 2006;235:1181–93. doi: 10.1002/dvdy.20676. [DOI] [PubMed] [Google Scholar]
- 75.Tyndale-Biscoe CH, Renfree M. Reproductive Physiology of Marsupials. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- 76*.Hill JP, Hill WC. The growth-stages of the pouch-young of the Native Cat (Dasyurus viverrinus) together with observations on the anatomy of the new-born young. 1955 The Transactions of the Zoological Society of …. [Google Scholar]
- 77.Newport J, Kirschner M. A Major Developmental Transition in Early Xenopus-Embryos.1. Characterization and Timing of Cellular-Changes at the Midblastula. Stage Cell. 1982;30:675–86. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 78.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 79.Pasquinelli AE, McCoy A, Jiménez E, Saló E, Ruvkun G, Martindale MQ, et al. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution? Evol Dev. 2003;5:372–8. doi: 10.1046/j.1525-142x.2003.03044.x. [DOI] [PubMed] [Google Scholar]
- 80.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 81.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 82.Banerjee D, Kwok A, Lin S-Y, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 2005;8:287–95. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 83.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–50. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sokol NS, Xu P, Jan Y-N, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–6. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 86.Jouve C, Iimura T, Pourquié O. Onset of the segmentation clock in the chick embryo: evidence for oscillations in the somite precursors in the primitive streak. Development. 2002;129:1107–17. doi: 10.1242/dev.129.5.1107. [DOI] [PubMed] [Google Scholar]
- 87.Palmeirim I, Rodrigues S, Dale JK, Maroto M. Development on time. Adv Exp Med Biol. 2008;641:62–71. doi: 10.1007/978-0-387-09794-7_5. [DOI] [PubMed] [Google Scholar]
- 88.Pascoal S, Carvalho CR, Rodríguez-León J, Delfini M-C, Duprez D, Thorsteindottir S, et al. A molecular clock operates during chick autopod proximal-distal outgrowth. Journal of Molecular Biology. 2007;368:303–9. doi: 10.1016/j.jmb.2007.01.089. [DOI] [PubMed] [Google Scholar]
- 89.Towers M, Tickle C. Growing models of vertebrate limb development. Development. 2009;136:179–90. doi: 10.1242/dev.024158. [DOI] [PubMed] [Google Scholar]
- 90.Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–3. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 91.Takashima Y, Ohtsuka T, González A, Miyachi H, Kageyama R. Intronic delay is essential for oscillatory expression in the segmentation clock. Proc Natl Acad Sci USA. 2011;108:3300–5. doi: 10.1073/pnas.1014418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanisch A, Holder MV, Choorapoikayil S, Gajewski M, Oezbudak EM, Lewis J. The elongation rate of RNA polymerase II in zebrafish and its significance in the somite segmentation clock. Development. 2013;140:444–53. doi: 10.1242/dev.077230. [DOI] [PubMed] [Google Scholar]
- 93.Hoyle NP, Ish-Horowicz D. Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proc Natl Acad Sci USA. 2013;110:E4316–24. doi: 10.1073/pnas.1308811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herrgen L, Ares S, Morelli LG, Schroeter C, Juelicher F, Oates AC. Intercellular Coupling Regulates the Period of the Segmentation Clock. Curr Biol. 2010;20:1244–53. doi: 10.1016/j.cub.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 95.Monsalve GC, Van Buskirk C, Frand AR. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr Biol. 2011;21:2033–45. doi: 10.1016/j.cub.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 96.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–6. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.TAM P. The Control of Somitogenesis in Mouse Embryos. J Embryol Exp Morphol. 1981;65:103–28. [PubMed] [Google Scholar]