Abstract
There are many similarities between the interactions of environmental protozoa with pathogenic bacterial species and those observed in mammalian macrophages. Since single-celled protozoa predate mammalian hosts, it is likely that interactions in environmental biofilms have selected for many of the bacterial virulence mechanisms responsible for human disease. In order to better understand bacterial-phagocyte interactions, we developed a selection for Acanthamoeba castellanii variants that are more resistant to killing by bacterial pathogens. We identified four amoebal clones that display decreased phagocytosis of bacteria but no difference in uptake of latex beads compared to wild-type amoebae. These amoebal variants display differences in cellular morphology, partial resistance to killing by bacteria, more bactericidal activity, and higher frequencies of lysosome fusion with the bacterial vacuole. Three proteins are present at lower levels in these variants than in wild-type amoebae, and matrix-assisted laser desorption ionization-time of flight mass spectrometry allowed identification of two of them as actin and hsp90. We found that specific inhibitors of hsp90 produce a similar phenotypic effect in macrophages. These data suggest that hsp90 plays a role in phagocytic and, possibly, bactericidal pathways that affect interactions of phagocytic cells with bacteria.
Uptake and degradation of bacterial particles are key functions of phagocytes in the mammalian immune system. Phagocytic environmental protozoa within aquatic biofilms carry out analogous activities in complex ecosystems that contain numerous prokaryotic organisms. Recent data suggest that environmental protozoa, particularly amoebae such as Acanthamoeba castellanii, have played an important role in the evolution of bacterial pathogens of mammals and may represent natural hosts for virulent bacteria (4, 9). A number of different pathogenic species, including Mycobacterium spp. (12), Vibrio cholerae (69), Burkholderia cepacia, Chlamydia pneumoniae, Edwardsiella tarda, Legionella pneumophila (32, 59), Cryptococcus neoformans (66), Listeria monocytogenes, and Francisella tularensis (1), have the ability to survive and replicate in environmental protozoa. Environmental amoebae, including A. castellanii, can undergo a respiratory burst (19), produce oxygen radicals (20), express cell surface receptors (2), and have phagocytic mechanisms (3, 8, 39) that are similar to those of human macrophages.
There are several important steps during the interactions of phagocytic cells with bacterial pathogens, including adherence, phagocytosis, signal transduction, trafficking, and intracellular replication. It is likely that a large number of host cell components are involved in these processes, yet few have been described in great detail. Protozoal models for bacterial-host cell interactions, such as the amoeba A. castellanii, should be very useful for identification of the host cell components involved. As opposed to that of mammalian hosts, the unicellular nature of A. castellanii makes it possible to isolate clones for genetic analyses. Thus, the goal of the present study was to develop methods that would allow identification and characterization of host cell components involved in bacterial-host cell interactions in A. castellanii.
We developed a selection that would allow identification of amoebal variants having defects in pathways necessary for optimal host cell infection by using L. pneumophila, an intracellular pathogen that normally kills amoebae efficiently (11, 52). A total of four A. castellanii clones were isolated that display defects in their interactions with both mycobacteria and legionellae. These variants show decreased uptake of bacteria and enhanced bactericidal activity. Proteomic analyses suggest that decreased levels of hsp90 are responsible for the phenotype of the amoebal variants. In addition, pharmacological studies confirmed that inhibition of hsp90 in murine macrophages produces a similar phagocytic phenotype. Overall, these studies suggest, for the first time, that hsp90 is involved in phagocytosis of or bactericidal activity against bacteria in host cells.
MATERIALS AND METHODS
Cells and culture conditions.
A. castellanii (ATCC 30234) amoebae, originally cultured from a single amoeba (47), were grown to 90% confluency at 23°C in the dark in 75-cm2 tissue culture flasks (Falcon) in PYG broth (12). The amoebae were harvested before use by rapping the flask sharply to bring them into suspension, and the number of viable cells was determined as described previously (12). The MH-S (ATCC CRL-2019) murine alveolar macrophage cell line was maintained at 37°C and 5% CO2 in RPMI 1640 medium with 2 mM l-glutamine (Gibco, Bethesda, Md.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 10 mM HEPES, 1 mM sodium pyruvate, 4.5 g of glucose per liter, 1.5 g of bicarbonate per liter, and 0.05 mM 2-mercaptoethanol. The murine macrophage cell line J774A.1 (ATCC TIB67) was maintained at 37°C and 5% CO2 in high-glucose Dulbecco's modified Eagle medium (Mediatech CELLGRO) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine.
Bacterial strains and growth conditions.
The L. pneumophila strain used for these studies, AA100, is a streptomycin-resistant variant (42) of L. pneumophila serogroup 1 (23). AA100 was grown on BCYE agar (22) for 3 days at 37°C in 5% CO2 as described previously (10). Cultures of M. marinum strain M, a clinical isolate obtained from the skin of a patient (55), and M. smegmatis strain mc2155 (63) were grown in Middlebrook 7H9 broth (Difco) supplemented with 0.5% glycerol, 10% albumin-dextrose complex, and 0.25% Tween 80 at 33 and 37°C for 7 and 3 days, respectively.
Isolation of A. castellanii variants.
A. castellanii was infected with L. pneumophila in a 75-cm2 tissue culture flask by a standard invasion assay protocol as described previously (11). Each flask, containing a monolayer of approximately 107 amoebae, was infected with 109 L. pneumophila amoebae for 30 min at 37°C in high-salt (HS) buffer (43). The monolayer was then washed once with HS buffer, 5 ml of HS buffer containing 100 μg of gentamicin per ml was added, and the mixture was incubated 2 h at 37°C, washed once with HS buffer, and incubated for 4 days at 37°C in 25 ml of HS buffer. The flasks were then washed four times with HS buffer and incubated at 24°C in the dark in 25 ml of PYG broth plus 200 μg of streptomycin per ml, 50 μg of kanamycin per ml, 2 μg of polymyxin B per ml, 100 μg of ampicillin per ml, 25 μg of tetracycline per ml, and 25 μg of chloramphenicol per ml for ∼20 to 30 days. During this period, the monolayers were washed with HS buffer and fresh PYG broth plus antibiotic was added every 5 to 7 days. After this incubation, clones were isolated by limiting dilution (31). The first four rounds of selection were carried out with a single flask, whereas subsequent rounds of selection were done by splitting the output from round 4 into a total of 10 different flasks that were enriched for L. pneumophila-resistant variants independently. A total of 12 successive rounds of selection (4 as a single pool plus 8 independent rounds) were carried out in this manner, and clones were isolated from each round by limiting dilution and characterized for resistance to L. pneumophila infection. Each of the individual clones further characterized in this study was cloned by limiting dilution from a different flask after the final round of selection. The stability of each clone was tested by continuous passage in PYG broth for 6 months, followed by reevaluation of the frequencies of Legionella phagocytosis.
Phagocytosis assays.
Phagocytosis assays were carried out in 24-well tissue culture dishes (Falcon) (10, 14). A. castellanii was seeded into the dishes at 1.5 × 105/well, allowed to adhere overnight at 23°C, washed once with HS buffer, and incubated in 1 ml of HS buffer for 1 h at 37°C, and 3 × 106 bacteria were added. After coincubation with the bacteria for 30 min, the amoebae were washed twice with HS buffer and incubated in HS buffer plus 100 μg of gentamicin per ml (legionellae) or 200 μg of amikacin per ml (mycobacteria) for 2 h at 37°C. The amoeba were then lysed by incubation for 10 min in 1 ml of sterile water, followed by vigorous pipetting. Dilutions were then plated on the appropriate medium to determine CFU counts.
We also used fluorescein isothiocyanate-conjugated latex beads (Molecular Probes, Eugene, Oreg.) and bacteria labeled with Oregon Green 418 (Molecular Probes) as described previously (13) to evaluate the phagocytic phenotype of the amoebal variants and macrophages treated with cellular inhibitors. In these assays, the cells were seeded in eight-well Permanox chamber slides (Nunc) at a density of 104/well and latex beads (1 μm; Molecular Probes) or fluorescent bacteria were added at a ratio of 20 per cell. After 30 min of coincubation, unbound beads or bacteria were removed by extensive washing with HS buffer. The cells were fixed in 4% paraformaldehyde in PBS buffer and mounted in 50% glycerol. The slides were examined under a Nikon TE300 fluorescence microscope with a fluorescein isothiocyanate filter. The percentage of cells that contained latex beads or bacteria and the number of beads or bacteria per cell were then determined by visual counts of 150 cells in random fields for each sample.
Assays for amoebal growth and killing by L. pneumophila.
The ability of the A. castellanii variants to be killed by L. pneumophila was evaluated by measuring the viability of the amoebae after infection with the bacteria over time. The amoebae were infected with L. pneumophila in the same manner as described for phagocytosis assays. After gentamicin treatment, the amoebae were washed with HS buffer and incubated in HS buffer for 12 h prior to the initial determination of amoebal viability. Viability of amoebae was determined as described previously (12) at various time points. Basically, the amoebae were brought into suspension by pipetting, aliquots were stained with eosin Y (Sigma), and viable amoebae were counted with a hemacytometer (Hausser Scientific). Amoebal growth rates were determined in a similar manner but with incubation in standard PYG growth medium for 10 days and no bacterial infection.
Bactericidal activity of amoebal variants.
Intracellular survival assays were carried out as described previously (12). L. pneumophila was added to a monolayer of 1.5 × 105 amoebae in 24-well tissue culture dishes at a multiplicity of infection (MOI) of 20 and incubated at 37°C for 30 min; the cells were washed three times with HS buffer and lysed at various time points with water. Survival is expressed as the percentage of CFU present at each time point compared to the number present at time zero (30 min), i.e., % survival = (CFU at Tx/CFU at T0) × 100.
Amoebal morphology and lysosomal fusion frequencies.
Light microscopy was carried out with live amoebae seeded at 37°C into a temperature-controlled closed perfusion chamber (Brook Industries) on a Nikon TE300 inverted microscope with differential-interference-contrast (DIC) optics.
Lysosomes were stained, and the frequency of fusion with the bacterial phagosome was determined by transmission electron microscopy essentially as described previously (12). After the amoebae were prelabeled with thorium dioxide (Polysciences), they were infected with L. pneumophila for 30 min. These samples and parallel unlabeled, uninfected amoeba samples for morphology studies were then fixed and prepared for electron microscopy as described previously (6, 12, 49).
Proteomic analysis.
Amoebae were grown to log phase, and 2 × 107 cells were collected by centrifugation at 100 × g for 10 min. The pellets were washed twice with cold phosphate-buffered saline (PBS) and resuspended in 500 μl of lysis buffer (10 mM HEPES, 200 mM NaCl, 2 mM CaCl2, 2.5 mM MgCl2, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail). The suspension was incubated on ice for 30 min and sonicated for 10 s. The lysate was centrifuged at 5,000 × g at 4°C for 20 min to remove debris, and the supernatant was taken as the total protein sample. The concentration of protein was determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). Six hundred micrograms of protein from both wild-type and variant amoebae was then subjected to two-dimensional gel electrophoresis. The first isoelectric focusing (IEF) dimension was performed with immobilized pH gradient gel strips (17 cm, pH 3 to 10, nonlinear; Bio-Rad) and a Bio-Rad IEF cell. The strips were rehydrated with the protein samples in rehydration buffer {7 M urea, 2 M thiourea, 100 mM dithiothreitol (DTT), 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.001% bromophenol blue, 0.2% Bio-Lytes} at 50 V for 12 h in the IEF cell and focused at 10,000 V for a total of 80,000 V · h. Before being loaded on the second dimension, focused immobilized pH gradient strips were equilibrated in buffer I, which contained 6 M urea, 2% sodium dodecyl sulfate, 0.375 M Tris-HCl (pH 8.8), 20% glycerol, and 130 mM DTT, for 15 min and then in equilibration buffer II, in which DTT was replaced with 135 mM iodoacetamide, for another 15 min. The second-dimension electrophoresis was performed with a Bio-Rad Protein II xi apparatus on 10% acrylamide gels. After electrophoresis, the proteins were stained with Coomassie G-250 (Sigma, St. Louis, Mo.). Gel images were captured with a ProImage Analyzer camera and analyzed with Image Analyzer HD software (Genomic Solutions). To identify differences, all spots were compared to at least five reference spots that were consistently similar and well separated in all gels and quadruplicate pairs of independently prepared samples were used for each variant and wild-type amoebae.
MALDI-TOF mass spectrometry.
For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, protein spots of interest were excised from the gel, washed, and analyzed as described previously (28). In brief, gel pieces were washed in 25 mM ammonium bicarbonate (NH4HCO3), pH 8.0, for 30 min, in 50% acetonitrile-25 mM NH4HCO3 for another 30 min, and finally with ultrapure water before complete dehydration in a vacuum centrifuge. The gel pieces were rehydrated with a minimum amount of sequence grade modified porcine trypsin solution containing 0.25 to 0.5 μg of protease, depending on the amount of protein. Digestion was performed at 37°C for 3 to 5 h. Mass spectra of the tryptic digests were acquired on a MALDI-TOF mass spectrometer operating in the reflector mode. A 0.5-ml volume of each digest solution (in 25 mM NH4CO3-10% acetonitrile) was deposited directly onto the sample probe on a dry thin layer of matrix made of α-cyano-4-hydroxy-trans-cinnamic acid mixed with nitrocellulose (4:3 [vol/vol] mixture of a saturated solution of α-cyano-4-hydroxy-trans-cinnamic acid in acetone and a solution consisting of 5 mg of nitrocellulose dissolved in 1 ml of isopropanol-acetone [1:1, vol/vol]). Deposits were washed with 5 ml of 0.1% trifluoroacetic acid before analysis. The peptide mass fingerprint obtained from each protein digested was analyzed with Mascot software (Matrix Science, London, United Kingdom) for protein identification.
Immunoprecipitation and Western analysis.
The same number of cells from wild-type amoeba and the variants were collected and washed once with cold PBS, and cell lysates were prepared as described above. The cell lysates were immunoprecipitated with 10 μg of monoclonal antibody against hsp90 (StressGen Biotechnologies, Victoria, Canada) or α-tubulin (Sigma) while shaking at 4°C overnight. The immunocomplex was then incubated with protein G-Sepharose (Amersham) for 2 h, washed with lysis buffer four times, and fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were then transferred to a nitrocellulose membrane (Poll) with a Bio-Rad transfer apparatus in accordance with the manufacturer's protocols. After incubation with 5% nonfat milk in TTBS (0.05% Tween 20, 20 mM Tris-HCl, pH 7.4) for 60 min, the membrane was washed once with TTBS and incubated with mouse monoclonal antibodies (1:1,000) directed against hsp90 or α-tubulin at room temperature for 1 h. Membranes were washed three times for 10 min and incubated with a 1:3,000 dilution of horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Sigma) for 1 h and detected with the ECL system (Amersham Pharmacia, Piscataway, N.J.). Densitometric analysis was carried out with ImageQuant software (Molecular Dynamics).
Pharmacological inhibitors of hsp90.
The hsp90 inhibitors used in this study, geldanamycin (GA), radicicol (RD), herbimycin A (HB), and novobiocin (NB), were purchased from Sigma. After pretreatment of MH-S murine alveolar macrophages with each of the inhibitors at various concentrations, phagocytosis assays were performed in the same manner as described for amoebae, except that the cells were maintained in RPMI medium and washed with PBS. Intracellular survival assays were carried out with MH-S cells that had been either pretreated with the inhibitors and removed before addition of the bacteria or left in the medium throughout the entire experiment (24 h). For these assays, the cells were infected with bacteria in the same manner as for assays of survival in amoebae. The viability of the cells after treatment with these inhibitors and their solvents was determined with eosin Y and counting in a hemacytometer as described previously (12).
Quantitation of mitochondria in macrophages.
The effects of hsp90 inhibitors on the number of mitochondria in MH-S murine alveolar macrophages was determined with a fluorescence-activated cell sorter. MH-S cells were treated with each inhibitor for 48 h, the cells were incubated with MitoTracker (Molecular Probes) at 20 mM for 30 min, excess dye was washed away, and at least 10,000 cells were analyzed for each sample on a Becton Dickinson FACScan flow cytometer.
Statistical analyses.
Data presented are the means and standard deviations of triplicate samples from a representative experiment. All experiments were repeated at least three times, unless otherwise noted. Significance was determined by analysis of variance. P values of <0.05 were considered significant.
RESULTS
Isolation of stable A. castellanii variants.
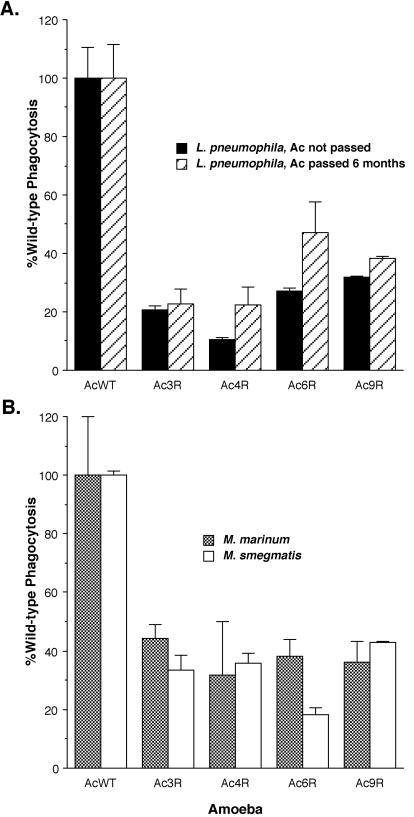
In order to better understand the host side of bacterium-host cell interactions, we took advantage of the single-celled nature of the environmental amoeba A. castellanii. When A. castellanii is infected with L. pneumophila at an MOI of 100 for 4 days, the entire monolayer of >107 amoebae is completely destroyed (11). Considering this information, it might be possible to use the amoebicidal activity of L. pneumophila as a direct selection for amoebae that have acquired changes in host cell components that play a role during interactions with bacteria. To test this hypothesis, we infected amoebae under conditions in which they would be almost entirely killed by L. pneumophila and grew those amoebae that survived this treatment. We carried out multiple infections in this manner for up to 12 rounds of selection. Individual amoebae were cloned from each round of selection by limiting dilution. Clones from the first through the fourth rounds were evaluated by comparing their frequencies of phagocytosis of L. pneumophila to that of wild-type amoebae. No significant differences were observed between the wild type and clones from these early rounds of selection (data not shown). However, four clones from the 12th round of selection, designated Ac3R, Ac4R, Ac6R, and Ac9R, were identified that displayed a decreased level of bacterial phagocytosis (P < 0.025; Fig. 1A). Although these data are for a ratio of bacteria per amoeba (MOI) of 20, we tested multiple MOIs ranging from 20 to 1 and all MOIs displayed the same level of decreased phagocytosis (data not shown). We examined the stability of the variants by continuous passage in the laboratory for more than 6 months, followed by reexamination of the phagocytic phenotype (Fig. 1A). These variants displayed a stable phagocytic phenotype after continuous laboratory passage. In addition, none of the variants grew at a rate different from that of wild-type amoebae (P > 0.1) in standard PYG laboratory medium (data not shown).
FIG. 1.
Phagocytosis of L. pneumophila (A) or M. smegmatis and M. marinum (B) by A. castellanii (Ac) variants calculated as the percentage of that displayed by wild-type amoebae (AcWT) under the same conditions. Phagocytosis assays were carried out by coincubating the bacteria with cells for 30 min and evaluating the number of bacterial CFU that are intracellular at the end of this time period. Amoebal variants (Ac3R, Ac4R, Ac6R, and Ac9R) were characterized for L. pneumophila phagocytosis immediately after cloning by limiting dilution and after passage during continuous culture for 6 months. The data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. The data shown are representative of at least three independent experiments.
Amoebal variants are defective in phagocytosis of mycobacteria.
We examined the specificity of the phagocytic defect with mycobacteria as phagocytic particles. We chose both pathogenic and nonpathogenic mycobacterial species, Mycobacterium marinum and Mycobacterium smegmatis, respectively, for these studies. The amoebal variants were defective for phagocytosis of both M. smegmatis and M. marinum at levels similar to that observed for L. pneumophila (Fig. 1B). These observations suggest that the defect present in these amoebae is relatively broad and likely to affect a number of bacterial species, since mycobacteria (gram positive) and legionellae (gram negative) are very distantly related.
Amoebal variants are morphologically different from the wild type.
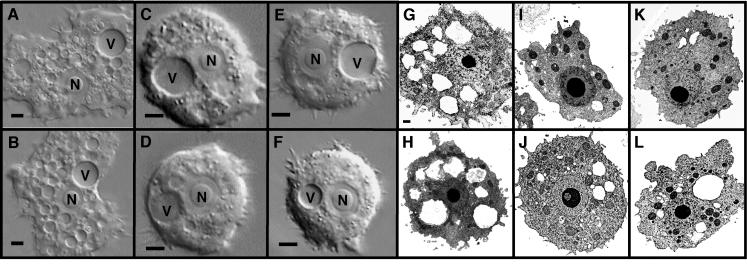
To further characterize these amoebal variants, we examined their morphology by light and electron microscopy. Light microscopy demonstrated that the variants contain fewer intracellular vacuoles and have a more rounded shape than wild-type amoebae (Fig. 2). Electron microscopy confirmed the relative paucity of intracellular vacuoles in the variants and suggested that the number of mitochondria is greater in the variants than in the wild type (Fig. 2). We quantitated the number of vacuoles and mitochondria in both the variants and the wild type. The number of intracellular vacuoles is significantly lower in all variants than in the wild type, and the number of mitochondria is significantly higher in Ac3R and Ac9R (Table 1). The morphological differences observed are stable over 6 months of passage in the laboratory. These observations suggest that the amoebal variants carry differences that affect more than one cellular characteristic and, as a result, might affect all phagocytic events.
FIG. 2.
Morphology of wild-type A. castellanii (A, B, G, H) and amoebal variants Ac3R (C, I), Ac4R (E, K), Ac6R (D, J), and Ac9R (F, L) by DIC microscopy (A to F) and transmission electron microscopy (G to L). The A. castellanii nucleus (N) and contractile vacuole (V) are visible in each DIC panel. The bar in each DIC panel is 10 μm. The bar in panel G is 1 μm and applies to panels G to L. Electron-dense particles in panels G to L are mostly mitochondria with a single nucleus and nucleolus visible. Electron-transparent regions are vacuolar compartments with the contractile vacuole commonly appearing as the largest vacuole.
TABLE 1.
Structural characteristics of the amoebae used in this study
| Amoebae | No. of vacuoles/cella | No. of mitochondria/cell |
|---|---|---|
| Wild type | 7.8 ± 2.5 | 9.2 ± 2.7 |
| Ac3R | 4.5 ± 1.9b | 12 ± 4.3b |
| Ac4R | 4.4 ± 2.2b | 11 ± 3.5 |
| Ac6R | 4.1 ± 2.2b | 11 ± 5.3 |
| Ac9R | 4.5 ± 2.1b | 15 ± 4.4b |
| 6-mo wild type | 6.1 ± 1.5 | 9.3 ± 2.6 |
| 6-mo Ac3R | 4.2 ± 1.2b | 14.4 ± 3.3b |
| 6-mo Ac4R | 4.8 ± 1.2b | 11.4 ± 2.7 |
| 6-mo Ac6R | 5.1 ± 1.5b | 12.0 ± 3.1 |
| 6-mo Ac9R | 4.8 ± 1.4b | 14.8 ± 2.9b |
Number of food vacuoles (excluding contractile vacuole) per amoeba in two counts of 50 cells each from amoebal variants and the wild type immediately after isolation and following passage in the laboratory for 6 months. The results are the means ± standard deviations. The data are representative of at least two independent experiments.
Significantly different (P < 0.05) from the result for wild-type A. castellanii.
Amoebal variants are not defective for phagocytosis of latex beads.
To test whether the phagocytic defect is due to a general cellular defect, rather than specific to bacterial uptake, we examined the ability of these amoebae to take up latex beads. Although all of the amoebal variants displayed reduced uptake of fluorescent bacteria by microscopy, both the number of beads per cell and the percentage of cells that phagocytosed at least one bead were the same for all mutants as for wild-type amoebae (Table 2). Although the mean values for variant Ac9R appear somewhat lower than for the wild type, this difference is not significant (P > 0.2). These observations suggest that the phagocytic phenotype of these variants is at least somewhat specific to their interactions with bacteria, rather than affecting uptake of all phagocytic particles, and is not solely the result of rapid intracellular killing, since enumeration of fluorescent bacterial particles phagocytosed is not dependent on bacterial viability.
TABLE 2.
Phagocytosis of latex beads and bacteria by amoebae
| Amoebae | % Phagocytosisa
|
No. of particles/amoeba
|
||
|---|---|---|---|---|
| Beads | Bacteria | Beads | Bacteria | |
| Wild type | 90 ± 16 | 71 ± 5.3 | 3.4 ± 2.5 | 3.2 ± 0.1 |
| Ac3R | 92 ± 15 | 37 ± 3.1b | 2.9 ± 2.1 | 2.0 ± 0.1b |
| Ac4R | 91 ± 17 | 37 ± 9.4b | 3.4 ± 2.4 | 3.0 ± 0.2 |
| Ac6R | 91 ± 19 | 53 ± 9.8b | 3.4 ± 2.3 | 2.7 ± 0.2b |
| Ac9R | 78 ± 19 | 55 ± 10b | 2.2 ± 1.6 | 2.7 ± 0.2b |
Percentage of amoebae that contained at least one bacterium (L. pneumophila) or bead as determined by fluorescence microscopy. The results are the means ± standard deviations for three counts of 50 amoebae. The data are representative of at least two independent experiments.
Significantly different (P < 0.05) from the result for wild-type A. castellanii.
Amoebal variants are resistant to killing by L. pneumophila.
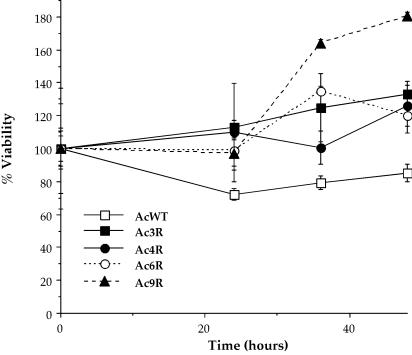
We infected each of the amoebal variants with L. pneumophila for 30 min and compared their subsequent viability over time (Fig. 3). All of the amoebal variants were more resistant to killing by L. pneumophila than were wild-type amoebae (P < 0.01). Thus, these variants display an L. pneumophila-resistant phenotype, possibly resulting from decreased uptake of the bacteria or increased bactericidal activity.
FIG. 3.
Percent viability of the A. castellanii wild type (AcWT) and variant clones Ac3R, Ac4R, Ac6R, and Ac9R after infection with L. pneumophila. For viability studies, the amoebae were first infected with bacteria in the same manner as for phagocytosis assays, followed by removal of extracellular bacteria and determination of amoebal viability at various time points. Time zero is 12 h after addition of bacteria. The viability of each amoeba was arbitrarily set to 100% at the first time point. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. The data shown are representative of at least three independent experiments.
Amoebal variants display enhanced bactericidal activity.
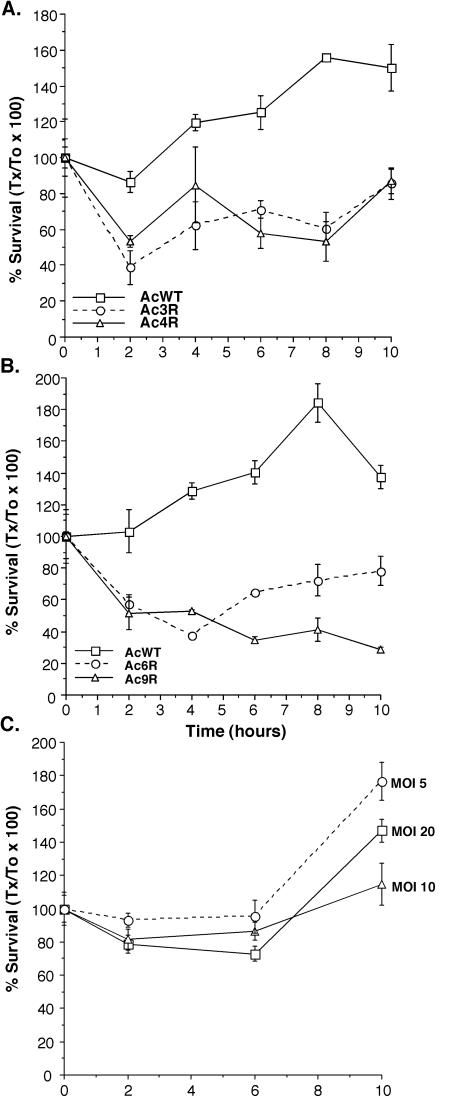
We monitored the intracellular survival of L. pneumophila in wild-type amoebae and the variants over time. All amoebal variants displayed higher (P < 0.01) bactericidal activity than did wild-type amoebae (Fig. 4). Normally, by 10 h after infection at least some bacterial replication is observable in wild-type amoebae, but replication is not observed with each of the variants until later and the bacteria are killed at higher levels early during infection. In order to determine whether the lower levels of phagocytosis leading to fewer intracellular bacteria is the reason for the increased ability of the amoebae to kill them, we tested survival and replication at a lower MOI than 20 bacteria per amoeba, which is the standard condition, in wild-type amoebae (Fig. 4). On the basis of these data, there is no correlation between the numbers of bacteria phagocytosed and their ability to survive and replicate in wild-type amoebae. These observations suggest that there are differences in how the amoebal variants interact with bacteria at early time points during signal transduction, phagocytosis, and/or trafficking.
FIG. 4.
Survival of L. pneumophila in wild-type (AcWT) A. castellanii and amoebal variants Ac3R and Ac4R (A) or Ac6R and Ac9R (B) and in wild-type amoebae when different MOIs were used on the basis of the ratio of the number of bacteria available for phagocytosis per amoeba (C). Amoebae were infected in the same manner as for phagocytosis assays, and the number of intracellular bacterial CFU was determined at various time points (Tx) relative to time zero (T0, 30 min). Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. The data shown are representative of at least three independent experiments.
Amoebal variants circumvent inhibition of lysosomal fusion by L. pneumophila.
Inhibition of lysosomal fusion by L. pneumophila is a key factor in their ability to survive and replicate in macrophages (33) and amoebae (7). Since we observed a decrease in the ability of L. pneumophila to survive in the amoebal variants, differences in trafficking of the bacterial phagosome may be at least partially responsible. We investigated this possibility by transmission electron microscopy with thorium dioxide as a marker for the lysosomal compartment (Fig. 5). The frequency of lysosomal fusion with the L. pneumophila vacuole in the variants was significantly higher (P < 0.02) than for wild-type A. castellanii (Table 3). These data suggest that one of the factors involved in the enhanced bactericidal activity of the amoebal variants is a decreased ability of L. pneumophila to inhibit phagosome-lysosome fusion in these cells.
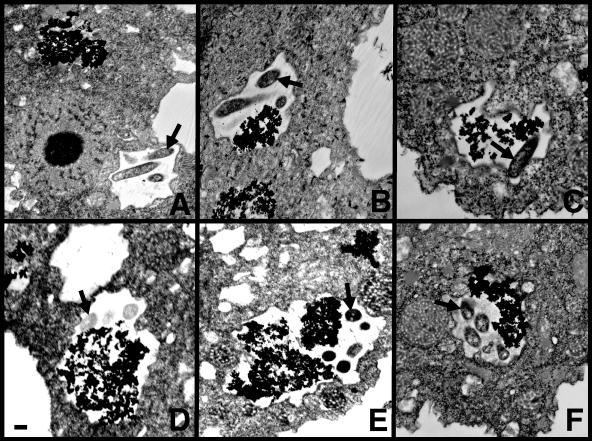
FIG. 5.
Transmission electron micrographs of L. pneumophila vacuoles that have not fused (A) and that have fused (B to F) with lysosomes in thorium dioxide-labeled wild-type A. castellanii (A, D) and clones Ac3R (B), Ac4R (C), Ac6R (E), and Ac9R (F) at 30 min postinfection. Granular, electron-dense material is thorium dioxide that has accumulated in lysosomes. After the amoebae were prelabeled with thorium dioxide and the excess label was removed, the amoebae were infected with bacteria in the same manner as for phagocytosis assays prior to fixation at various time points and preparation for electron microscopy. When thorium dioxide is present in the same intracellular compartment with bacteria, they are considered to have fused with lysosomes. The bar in the bottom left of panel D is 1 μm and applies to all of the panels. Arrows indicate bacteria in each panel.
TABLE 3.
Fusion of L. pneumophila vacuoles with lysosomes
| Amoebae | % Fusiona
|
|
|---|---|---|
| 30 min | 120 min | |
| Wild type | 31 ± 5 | 46 ± 6 |
| Ac3R | 57 ± 2b | 64 ± 4b |
| Ac4R | 63 ± 7b | 68 ± 6b |
| Ac6R | 51 ± 2b | 72 ± 3b |
| Ac9R | 55 ± 1b | 62 ± 5b |
Percentage of bacterial (L. pneumophila) vacuoles containing thorium. The results are the means ± standard deviations for three counts of 50 amoebae in different sections of the same preparation. The data are representative of at least two independent experiments.
Significantly different (P < 0.02) from the result for wild-type A. castellanii.
Amoebal variants have a limited number of protein differences.
Since the amoebal variants differ from the wild type in a number of phenotypic characteristics, we carried out proteomic analysis to identify any molecular differences from wild-type amoebae. Analysis of total proteins from the variants resulted in the identification of three to six proteins that are absent and three to four that are at lower levels in each of the variants compared to wild-type amoebae. Although all of these differences were consistent over four sets of gels run in parallel, three proteins were consistently at lower levels or absent in all four of these variants (Fig. 6). These observations suggest that some or all of these three proteins are important for the phenotypic characteristics of the amoebal variants.
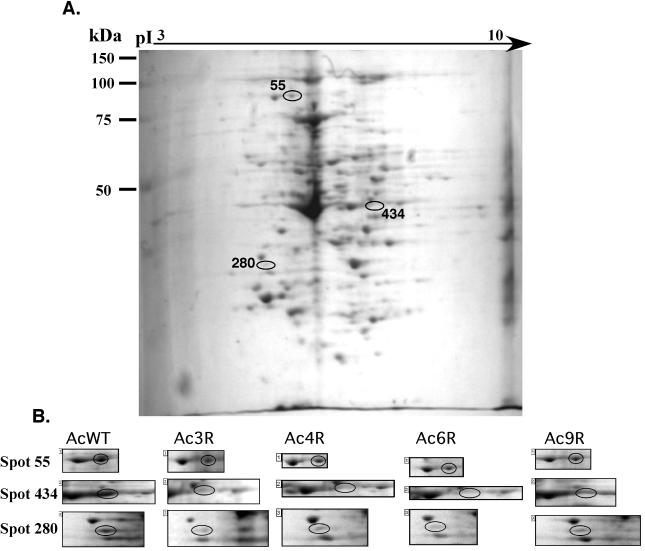
FIG. 6.
Proteomic analysis of differences between wild-type (AcWT) and variant amoebae. Example of a single Coomassie-stained two-dimensional polyacrylamide gel electrophoresis gel of total proteins from amoebal variant Ac3R (A) and comparative analysis (B) of spots 55, 280, and 434, which were found in all variants at lower levels than in the wild type. During this analysis, all spots on each gel were compared to a minimum of five reference spots that were consistently present and clearly separated in all gels. Once differences were found, they were reexamined in a minimum of three independent experiments and quadruplicate pairs of gels to evaluate whether the differences were consistent.
Hsp90 is down-regulated in all of the amoebal variants.
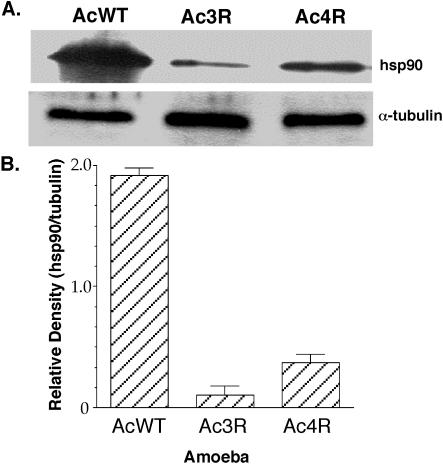
Each of these spots was excised and subjected to MALDI-TOF mass spectrometry in order to determine the identity of the protein that it represents. Two of these spots matched, at very high significance, actin and hsp90, and the third spot displayed some similarity to cyclophilin D but did not reach significance (Table 4). The presence of low levels of hsp90 in the amoebal variants was confirmed by Western analysis of proteins immunoprecipitated with an anti-hsp90 monoclonal antibody (Fig. 7). Identical volumes of the same amoebal lysate were also immunoprecipitated with α-tubulin monoclonal antibody as a control for protein loading differences. Densitometric analysis of the relative levels of hsp90 compared to α-tubulin demonstrated that all amoebal variants have between 5- and 20-fold lower levels of hsp90 than the wild type.
TABLE 4.
MALDI-TOF analysis of differentially expressed proteins
| Spot no. | Gene | Molecular mass (kDa)
|
No. of peptidesa | Scoreb | Sequencec | |
|---|---|---|---|---|---|---|
| Estimated | Actual | |||||
| 55 | gi:1708316 | 85 | 80 | 8 | 242 | ELISNASDALDK |
| hsp90 | HFSVEGQLEFK | |||||
| GIVDSEDLPLNISR | ||||||
| 280 | gi:5739196 | 20 | 19 | 3 | 15 | QGNTPLGR |
| Cyclophilin D | IGCLSMANAGK | |||||
| 434 | gi:71629 | 40 | 42 | 12 | 437 | IIAPPER |
| Actin | AGFAGDDAPR | |||||
| GYSFTTTAER | ||||||
| DSYVGDEAQSKR | ||||||
| IWHHTFYNELR | ||||||
| EEYDESGPSIVHR | ||||||
| SYELPDGQVITIGNER | ||||||
| VAPEEHPVLLTEAPLNPK | ||||||
Number of peptides that matched by Mascot (www.matrixscience.com) on the spectra obtained from each protein spot.
Scores of >23 are considered significant (P < 0.05).
Amino acid sequences of unique peptide matches.
FIG. 7.
Levels of hsp90 or α-tubulin in wild-type (AcWT) and variant (Ac3R and Ac4R) amoebae as determined by Western analysis of identical volumes of the same amoebal lysate immunoprecipitated with anti-hsp90 or α-tubulin monoclonal antibody (A) and densitometric analysis of the levels of hsp90 calculated relative to the α-tubulin present in each amoebal lysate (B). The data shown are representative of at least three independent experiments.
Inhibition of hsp90 in macrophages increases mitochondrial numbers and bacterial phagocytosis and survival.
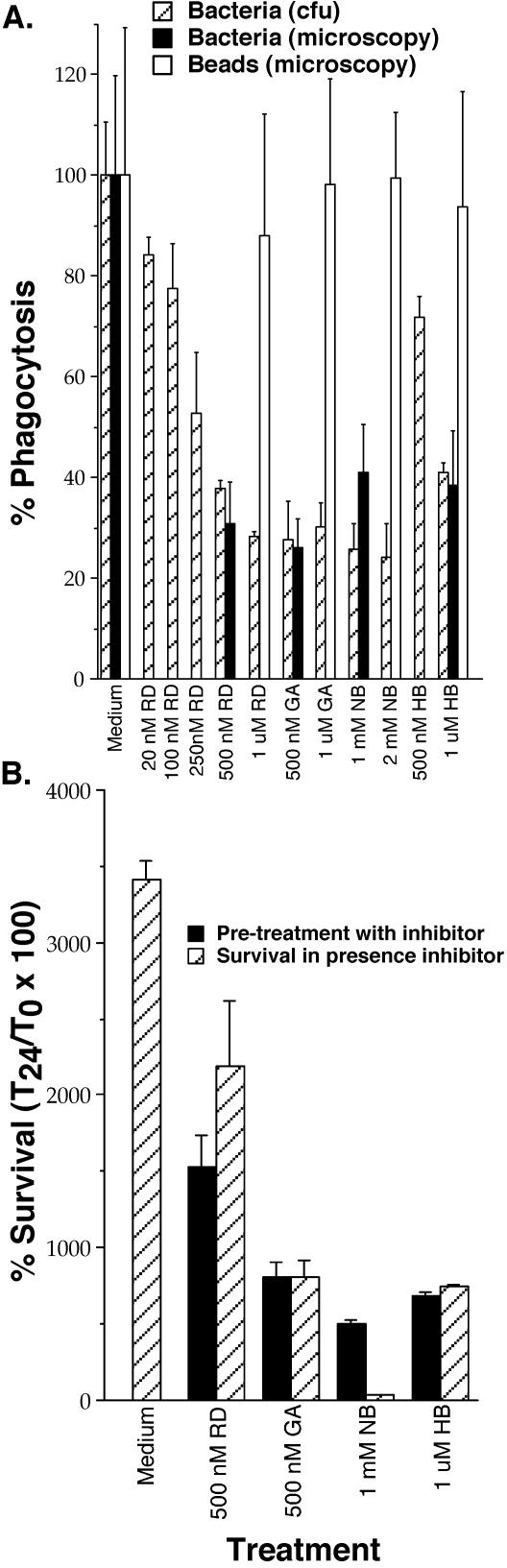
Since hsp90 is thought to interact with both actin (36, 48) and cyclophilins (68, 72), as well as a number of other cellular proteins (53), it is possible that this protein is a key factor in the phenotypic characteristics of the variants. A number of specific inhibitors exist for hsp90 in mammalian cells, allowing us to test whether this protein plays a role in phagocytosis of bacteria. However, since none of these inhibitors produced the expected effects in amoebae (data not shown), we conducted inhibitor studies with mammalian cells. These inhibitors produced similar results with both J774A.1 (data not shown) and MH-S murine macrophage cell lines. We found that all inhibitors of hsp90 significantly increased fluorescent staining with the mitochondrion-specific dye MitoTracker (Table 5). In addition, these inhibitors decreased phagocytosis of L. pneumophila in a dose-dependent manner whether viable intracellular bacteria or microscopy was used to determine the number of particles phagocytosed, yet the inhibitors did not reduce the phagocytosis of latex beads (Fig. 8A). Furthermore, intracellular survival and replication of L. pneumophila were reduced in macrophages pretreated with hsp90 inhibitors prior to infection (Fig. 8B). To ensure that the inhibitors did not directly inhibit the growth of the bacteria, we examined survival and replication of L. pneumophila when the inhibitors were present during bacterial interactions with macrophages. Most of the inhibitors, with the exception of NB, produced similarly reduced levels of L. pneumophila survival and replication either when they were used to pretreat the cells or when they were present throughout the experiment (Fig. 8B). When NB was present throughout the experiment, no bacterial replication was observed, suggesting that this inhibitor affects bacterial replication and pretreatment of the macrophages may have some degree of inhibitory effect on the bacteria themselves. None of these inhibitors or their solvents, at the concentrations and time points used, affected macrophage or bacterial viability (data not shown). These observations support a role for hsp90 in cellular pathways that are important for uptake and/or killing of bacteria in phagocytic cells.
TABLE 5.
Mitochondria in macrophages treated with hsp90 inhibitors
| Treatment | Mitochondrial fluorescencea |
|---|---|
| Medium | 67.7 ± 0.7 |
| 500 nM RD | 80.4 ± 1.6b |
| 500 nM GA | 101.2 ± 3.8b |
| 1 mM NB | 83.0 ± 2.5b |
| 1 μM HB | 114.1 ± 0.0b |
Mean fluorescence ± standard deviation of 10,000 cells as measured by flow cytometry after staining of hsp90 inhibitor-treated MH-S murine alveolar macrophages with the mitochondrion-specific fluorescent marker MitoTracker. The data are representative of at least two independent experiments. The inhibitors used were RD, GA, NB and HB.
Significantly different (P < 0.01) from the result for medium.
FIG. 8.
Phagocytosis of L. pneumophila (Bacteria) or 1-μm latex beads (Beads) by MH-S murine alveolar macrophages (A) and survival and replication of L. pneumophila over 24 h within these same cells (B). Assays were carried out in the presence and absence (Medium) of various concentrations of specific hsp90 inhibitors. Phagocytosis assays for bacteria [Bacteria (cfu)] were carried out in the same manner as described for amoebae and by evaluating the number of bacterial CFU that were intracellular at the end of this time period. Phagocytosis of bacteria [Bacteria (microscopy)] and beads [Beads (microscopy)] was quantitated by fluorescence microscopy to determine the percentage of cells that contained at least one particle for three counts of 50 amoebae. Survival and replication assays were carried out by first pretreating the cells with the inhibitor and either removing it by washing prior to infection (Pretreatment with inhibitor) or pretreatment followed by infection and maintenance of the presence of the inhibitor throughout the remainder of the experiment (Survival in presence inhibitor). Survival and replication were evaluated at 24 h postinfection (T24) relative to time zero (T0, 30 min). The inhibitors used were RD, GA, NB, and HB. Phagocytosis was calculated as the percentage of that displayed in the absence of any inhibitor. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. The data shown are representative of at least three independent experiments.
DISCUSSION
Our selection strategy has allowed the isolation of four A. castellanii variants that display lower levels of bacterial phagocytosis than wild-type amoebae. Potential mechanisms that might lead to this phenotype include loss or alteration of a receptor, modification of signaling pathways, cytoskeletal changes affecting phagocytosis, and increased bactericidal activity. Each of these clones displays differences in multiple stages during its interactions with bacteria. However, through the use of two-dimensional polyacrylamide gel electrophoresis we were able to evaluate differences in the protein profiles of these clones and identify those that differ from wild-type amoebae. On the basis of our data, hsp90 appears to be important for the observed phenotypic traits of these variants. hsp90 is very conserved across species and is an essential stress protein in eukaryotic cells (38, 74). Since hsp90 has truly pleiotropic activities (53), it is possible that lower levels of this single protein are responsible for all of the differences between these variants and wild-type amoebae.
Although these are the first A. castellanii variants identified that affect interactions with L. pneumophila, another model system for L. pneumophila host-pathogen interactions has been used for analysis of host factors involved, i.e., Dictyostelium discoideum (30, 64). Interestingly, most of the D. discoideum mutants examined that were considered good candidates for playing a role in host-pathogen interactions increased rather than decreased the susceptibility of D. discoideum. Only a GB mutant, which is defective for all types of trimeric G-protein signaling, displayed a slightly reduced ability to support growth of L. pneumophila (64). Thus, the current strategy of enriching for bacterium-resistant variants has allowed the identification of a second host factor involved in host cell-L. pneumophila interactions without the cumbersome task of screening a large number of mutants with changes in candidate genes of interest. Possibly, the enrichment method developed in the present study could be applied to D. discoideum or pools of D. discoideum mutants. Certainly, this procedure could be improved by conducting mutagenesis of the amoebae, to increase the frequency at which variants would be found, and enriching for variants from a number of independent pools, to prevent the isolation of siblings, which was most likely responsible for our observation that all variants displayed decreased levels of hsp90. One limitation of this approach with D. discoideum is that L. pneumophila does not infect and kill D. discoideum (30, 64) as efficiently as it does A. castellanii. Furthermore, although D. discoideum has many similarities to other environmental protozoa, there is no evidence that it can serve as a natural host for L. pneumophila, possibly because D. discoideum prefers soil habitats (5) and L. pneumophila prefers aquatic environments (24, 25). Thus, the fact that A. castellanii is a natural host for L. pneumophila may prove to be a valuable asset for further analysis of host-pathogen interactions.
The A. castellanii variants we have isolated display decreased vacuolation, and two of them have an increased number of mitochondria. Previous observations of macrophages suggest that an increased number of mitochondria correlate with activation of macrophages (15, 16) and greater resistance to infection (35). In macrophages, resistance to infection may result from an enhanced ability to carry out oxidative phosphorylation during a respiratory burst (50). A respiratory burst results in secretion of extracellular ATP (61, 70), which is known to increase the production of reactive oxygen and nitrogen species (44, 61, 70). Interestingly, a role for hsp90 in control of superoxide production in host cells has recently been suggested (65). Further characterization of these variants may provide insight into the role of hsp90 in bactericidal mechanisms, similar to those found in mammalian macrophages.
Signaling events that affect intracellular trafficking and bactericidal activity may be triggered by bacteria binding to different host cell receptors. hsp90 is known to interact with a number of different cell surface receptors, including scavenger receptors (45) and steroid receptors (21). It has also been suggested that hsp90 itself can serve as a receptor for either lipopolysaccharide (71) or specific bacterial proteins (34). In addition, hsp90 plays an important role in a number of receptor-mediated signal transduction pathways (53, 57). This concept fits well with the observation that phagocytosis of latex beads is triggered by mechanical contact rather than a receptor-mediated pathway (73). Effects on receptors or signal transduction may be responsible for the bactericidal phenotype, but it is possible that trafficking of the bacterial vacuole is affected directly considering that hsp90 can play an important role in receptor trafficking (18, 27). Further analysis of these variants should allow us to differentiate between these possibilities.
Although all of the variants display lower levels of hsp90, they show slightly different phenotypic characteristics and have lower levels of other proteins as well. There are two simple explanations for the differences in phenotype and differences in protein profile. First, the levels of hsp90 vary from two- to fivefold less in the amoebal variants. Since hsp90 can affect the turnover of proteins that it interacts with (17, 26, 46, 60), it is conceivable that the levels of other hsp90-associated proteins would be affected differently depending on the levels of hsp90 present. This possibility is supported by the fact that both actin (36, 48) and cyclophilins (68, 72) have been previously shown to interact with hsp90. Possibly, hsp90 is involved in modification of actin, since only actin migrating at a specific pI is at lower levels in the variants than in the wild type and at least three forms of actin are normally present in eukaryotic cells (56, 67). Second, since multiple rounds of selection were carried out, it is possible that secondary mutations were acquired that are involved in some of the more subtle differences between the variants. Characterization of the other protein differences and either reconstruction of the variants through molecular means or complementation analyses should help to differentiate between these possibilities.
Although known inhibitors of hsp90 in mammalian cells had no effect in amoebae, it is likely that this result is due to differences between the amoebal and mammalian proteins, transport of the inhibitors, or other aspects of amoebal physiology. GA and the closely related compound HB are actinomycete fermentation products of the benzoquinone ansamycin class (62). Ansamycin antibiotics such as GA and HB bind at the ATP-binding site in the N-terminal domain of hsp90 and disrupt the ATPase activity of hsp90, which is essential for its chaperone activity in vivo (29, 51, 54, 58). Although RD has no significant structural similarity to GA or HB, it also binds to the N-terminal nucleotide-binding domain of hsp90 and inhibits the inherent ATPase activity of hsp90 (58). However, RD shows a greater affinity for the nucleotide binding site than does GA. Unlike GA, RD causes no significant conformational changes (58). The binding site for NB lies within the C-terminal region of hsp90 (40, 41). The second C-terminal ATP-binding site is responsible for autophosphorylation of hsp90 (37). GA does not interfere with autophosphorylation of hsp90 and hence does not bind to the C-terminal ATP-binding site (37). Since all of these inhibitors differ with respect to the mechanism of hsp90 inhibition and specific chemical structure, it is unlikely that nonspecific effects are responsible for the observed phenotypic changes in macrophages. It is compelling that hsp90 inhibitors produce a phagocytic defect in macrophages similar to that observed in amoebae. Furthermore, hsp90 inhibitors also reduce bacterial survival and replication in macrophages, as well as increase the numbers of mitochondria. On the basis of these data, it is unlikely that the other protein differences observed play as important a role in the phenotypic characteristics of the amoebal variants as hsp90.
Overall, these studies demonstrate, for the first time, a role for hsp90 in the interactions of phagocytic cells with bacteria. This protein may influence the mechanisms of phagocytosis, bactericidal activity, or a combination of these cellular processes. Since there are direct links between the mechanism of phagocytosis and subsequent intracellular events, it may be difficult to differentiate between these possibilities. However, careful examination of phagocytic and signal transduction pathways in the variants and the wild type should provide insight into the role of hsp90. Thus, the observation that hsp90 plays a role in early bacterium-phagocyte interactions is likely to assist in elucidation of the cellular pathways involved.
Acknowledgments
We thank Gerald McLaughlin, Gautam Sarath, and Philip Stahl for useful discussions and Nancy Caceras for technical assistance.
This work was supported by grants AI40165 and AI47866 from the National Institutes of Health.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, P. G., and Dawidowicz. 1990. Phagocytosis in Acanthamoeba. I. A mannose receptor is responsible for the binding and phagocytosis of yeast. J. Cell. Physiol. 145:508-513. [DOI] [PubMed] [Google Scholar]
- 3.Allen, P. G., and E. A. Dawidowicz. 1990. Phagocytosis in Acanthamoeba. II. Soluble and insoluble mannose-rich ligands stimulate phosphoinositide metabolism. J. Cell. Physiol. 145:514-521. [DOI] [PubMed] [Google Scholar]
- 4.Barker, J., and M. R. W. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 5.Bonner, J. T. 1944. A descriptive study of the development of the slime mold Dictyostelium discoideum. Am. J. Bot. 31:175-182. [Google Scholar]
- 6.Bowers, B., and E. D. Korn. 1968. The fine structure of Acanthamoeba castellanii. I. The trophozoite. J. Cell Biol. 39:95-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, R. C., H. Bass, and J. P. Coombs. 1975. Carbohydrate binding proteins involved in phagocytosis by Acanthamoeba. Nature 254:434-435. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, J. D. 1999. Exploring a novel perspective on pathogenic relationships. Trends Microbiol. 7:96-98. [DOI] [PubMed] [Google Scholar]
- 10.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, S. L., L. Yan, M. Littman, M. M. Samrakandi, and J. D. Cirillo. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667-1677. [DOI] [PubMed] [Google Scholar]
- 14.Cirillo, S. L. G., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohn, Z. A., and B. Benson. 1964. The differentiation of mononuclear phagocytes: morphology, cytochemistry and biochemistry. J. Exp. Med. 121:153-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohn, Z. A., J. G. Hirsch, and M. E. Fedorko. 1965. The in vitro differentiation of mononuclear phagocytes. IV. The ultrastructure of macrophage differentiation in the peritoneal cavity and in culture. J. Exp. Med. 123:747-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 18.Czar, M. J., R. H. Lyons, M. J. Welsh, J. M. Renoir, and W. B. Pratt. 1995. Evidence that the FK506-binding immunophilin heat shock protein 56 is required for trafficking of the glucocorticoid receptor from the cytoplasm to the nucleus. Mol. Endocrinol. 9:1549-1560. [DOI] [PubMed] [Google Scholar]
- 19.Davies, B., L. S. Chattings, and S. W. Edwards. 1991. Superoxide generation during phagocytosis by Acanthamoeba castellanii: similarities to the respiratory burst of immune phagocytes. J. Gen. Microbiol. 137:705-710. [Google Scholar]
- 20.Davies, B., and S. W. Edwards. 1991. Chemiluminescence and superoxide production in Acanthamoeba castellanii: free radicals generated during oxidative stress. J. Gen. Microbiol. 137:1021-1027. [Google Scholar]
- 21.Denis, M., and J. A. Gustafsson. 1989. The Mr approximately 90,000 heat shock protein: an important modulator of ligand and DNA-binding properties of the glucocorticoid receptor. Cancer Res. 49:2275s-2281s. [PubMed] [Google Scholar]
- 22.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fliermans, C. B., W. B. Cherry, L. H. Orrison, and L. Thacker. 1979. Isolation of Legionella pneumophila from nonepidemic-related aquatic habitats. Appl. Environ. Microbiol. 37:1239-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita, N., S. Sato, A. Ishida, and T. Tsuruo. 2002. Involvement of Hsp90 in signaling and stability of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 277:10346-10353. [DOI] [PubMed] [Google Scholar]
- 27.Galigniana, M. D., C. Radanyi, J. M. Renoir, P. R. Housley, and W. B. Pratt. 2001. Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 276:14884-14889. [DOI] [PubMed] [Google Scholar]
- 28.Garin, J., R. Diez, S. Kieffer, J. F. Dermine, S. Duclos, E. Gagnon, R. Sadoul, C. Rondeau, and M. Desjardins. 2001. The phagosome proteome: insight into phagosome functions. J. Cell Biol. 152:165-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grenert, J. P., W. P. Sullivan, P. Fadden, T. A. Haystead, J. Clark, E. Mimnaugh, H. Krutzsch, H. J. Ochel, T. W. Schulte, E. Sausville, L. M. Neckers, and D. O. Toft. 1997. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J. Biol. Chem. 272:23843-23850. [DOI] [PubMed] [Google Scholar]
- 30.Hagele, S., R. Kohler, H. Merkert, M. Schleicher, J. Hacker, and M. Steinert. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol. 2:165-171. [DOI] [PubMed] [Google Scholar]
- 31.Harlow, E. L. D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Henke, M., and K. M. Seidel. 1986. Association between Legionella pneumophila and amoebae in water. Isr. J. Med. Sci. 22:690-695. [PubMed] [Google Scholar]
- 33.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, S., Y. C. Song, A. Emili, P. M. Sherman, and V. L. Chan. 2003. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90α and triggers signalling pathways leading to the activation of NF-κB and p38 MAP kinase in epithelial cells. Cell Microbiol. 5:165-174. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, G., W. C. Van Voorhis, E. N. Sarno, N. Nogueira, and Z. A. Cohn. 1983. The cutaneous infiltrates of leprosy. A transmission electron microscopy study. J. Exp. Med. 158:1145-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyasu, S., E. Nishida, T. Kadowaki, F. Matsuzaki, K. Iida, F. Harada, M. Kasuga, H. Sakai, and I. Yahara. 1986. Two mammalian heat shock proteins, HSP90 and HSP100, are actin-binding proteins. Proc. Natl. Acad. Sci. USA 83:8054-8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langer, T., H. Schlatter, and H. Fasold. 2002. Evidence that the novobiocin-sensitive ATP-binding site of the heat shock protein 90 (hsp90) is necessary for its autophosphorylation. Cell Biol. Int. 26:653-657. [DOI] [PubMed] [Google Scholar]
- 38.Lindquist, S., and E. A. Craig. 1988. The heat-shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 39.Lock, R., L. Öhman, and C. Dahlgren. 1987. Phagocytic recognition mechanisms in human granulocytes and Acanthamoeba castellanii using type 1 fimbriated Escherichia coli as phagocytic prey. FEMS Microbiol. Lett. 44:135-140. [Google Scholar]
- 40.Marcu, M. G., A. Chadli, I. Bouhouche, M. Catelli, and L. M. Neckers. 2000. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J. Biol. Chem. 275:37181-37186. [DOI] [PubMed] [Google Scholar]
- 41.Marcu, M. G., T. W. Schulte, and L. Neckers. 2000. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J. Natl. Cancer Inst. 92:242-248. [DOI] [PubMed] [Google Scholar]
- 42.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 43.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, J. K., F. R. Livingston, E. Gozal, M. Torres, and H. J. Forman. 1993. Stimulation of the rat alveolar macrophage respiratory burst by extracellular adenine nucleotides. Am. J. Respir. Cell Mol. Biol. 9:505-510. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura, T., J. Hinagata, T. Tanaka, T. Imanishi, Y. Wada, T. Kodama, and T. Doi. 2002. HSP90, HSP70, and GAPDH directly interact with the cytoplasmic domain of macrophage scavenger receptors. Biochem. Biophys. Res. Commun. 290:858-864. [DOI] [PubMed] [Google Scholar]
- 46.Neckers, L., T. W. Schulte, and E. Mimnaugh. 1999. Geldanamycin as a potential anti-cancer agent: its molecular target and biochemical activity. Investig. New Drugs 17:361-373. [DOI] [PubMed] [Google Scholar]
- 47.Neff, R. J. 1957. Purification, axenic cultivation, and description of a soil amoeba, Acanthamoeba sp. J. Protozool. 4:176-182. [Google Scholar]
- 48.Nishida, E., S. Koyasu, H. Sakai, and I. Yahara. 1986. Calmodulin-regulated binding of the 90-kDa heat shock protein to actin filaments. J. Biol. Chem. 261:16033-16036. [PubMed] [Google Scholar]
- 49.Niszl, I. A., and M. B. Markus. 1989. Processing of free-living amoebae for transmission electron microscopy. Stain Tech. 64:259-260. [DOI] [PubMed] [Google Scholar]
- 50.Oren, R., A. E. Farnham, K. Saito, E. Milofsky, and M. L. Karnovsky. 1963. Metabolic patterns in three types of phagocytizing cells. J. Cell Biol. 17:487-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panaretou, B., C. Prodromou, S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polesky, A. H., J. T. D. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111-133. [DOI] [PubMed] [Google Scholar]
- 54.Prodromou, C., S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65-75. [DOI] [PubMed] [Google Scholar]
- 55.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsby, M. L., G. S. Makowski, and E. A. Khairallah. 1994. Differential detergent fractionation of isolated hepatocytes: biochemical, immunochemical and two-dimensional gel electrophoresis characterization of cytoskeletal and noncytoskeletal compartments. Electrophoresis 15:265-277. [DOI] [PubMed] [Google Scholar]
- 57.Richter, K., and J. Buchner. 2001. Hsp90: chaperoning signal transduction. J. Cell. Physiol. 188:281-290. [DOI] [PubMed] [Google Scholar]
- 58.Roe, S. M., C. Prodromou, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 42:260-266. [DOI] [PubMed] [Google Scholar]
- 59.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoeba. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider, C., L. Sepp-Lorenzino, E. Nimmesgern, O. Ouerfelli, S. Danishefsky, N. Rosen, and F. U. Hartl. 1996. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. USA 93:14536-14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikora, A., J. Liu, C. Brosnan, G. Buell, I. Chessel, and B. R. Bloom. 1999. Cutting edge: purinergic signaling regulates radical-mediated bacterial killing mechanisms in macrophages through a P2X7-independent mechanism. J. Immunol. 163:558-561. [PubMed] [Google Scholar]
- 62.Smith, D. F., L. Whitesell, S. C. Nair, S. Chen, V. Prapapanich, and R. A. Rimerman. 1995. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol. Cell. Biol. 15:6804-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 64.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sreedhar, A. S., K. Mihaly, B. Pato, T. Schnaider, A. Stetak, K. Kis-Petik, J. Fidy, T. Simonics, A. Maraz, and P. Csermely. 2003. Hsp90 inhibition accelerates cell lysis: anti-Hsp90 ribozyme reveals a complex mechanism of Hsp90 inhibitors involving both superoxide- and Hsp90-dependent events. J. Biol. Chem. 278:35231-35240. [DOI] [PubMed]
- 66.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sussman, D. J., E. Y. Lai, and C. Fulton. 1984. Rapid disappearance of translatable actin mRNA during cell differentiation in Naegleria. J. Biol. Chem. 259:7355-7360. [PubMed] [Google Scholar]
- 68.Tesic, M., J. A. Marsh, S. B. Cullinan, and R. F. Gaber. 2003. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 278:32692-32701. [DOI] [PubMed]
- 69.Thom, S., D. Warhurst, and B. S. Drasar. 1992. Association of Vibrio cholerae with fresh water amoebae. J. Med. Microbiol. 36:303-306. [DOI] [PubMed] [Google Scholar]
- 70.Tonetti, M., L. Sturla, T. Bistolfi, U. Benatti, and A. De Flora. 1994. Extracellular ATP potentiates nitric oxide synthase expression induced by lipopolysaccharide in RAW 264.7 murine macrophages. Biochem. Biophys. Res. Commun. 203:430-435. [DOI] [PubMed] [Google Scholar]
- 71.Triantafilou, K., M. Triantafilou, S. Ladha, A. Mackie, R. L. Dedrick, N. Fernandez, and R. Cherry. 2001. Fluorescence recovery after photobleaching reveals that LPS rapidly transfers from CD14 to hsp70 and hsp90 on the cell membrane. J. Cell Sci. 114:2535-2545. [DOI] [PubMed] [Google Scholar]
- 72.Ward, B. K., R. K. Allan, D. Mok, S. E. Temple, P. Taylor, J. Dornan, P. J. Mark, D. J. Shaw, P. Kumar, M. D. Walkinshaw, and T. Ratajczak. 2002. A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90. J. Biol. Chem. 277:40799-40809. [DOI] [PubMed] [Google Scholar]
- 73.Weisman, R. A., and E. D. Korn. 1967. Phagocytosis of latex beads by Acanthamoeba. I. Biochemical properties. Biochemistry 6:485-497. [DOI] [PubMed] [Google Scholar]
- 74.Young, J. C., I. Moarefi, and F. U. Hartl. 2001. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]