Abstract
Despite growing interest on the potential use of de-cellularized whole lungs as 3-dimensional scaffolds for ex vivo lung tissue generation, optimal processing including sterilization and storage conditions, are not well defined. Further, it is unclear whether lungs need to be obtained immediately or may be usable even if harvested several days post-mortem, a situation mimicking potential procurement of human lungs from autopsy. We therefore assessed effects of delayed necropsy, prolonged storage (3 and 6 months), and of two commonly utilized sterilization approaches: irradiation or final rinse with peracetic acid, on architecture and extracellular matrix (ECM) protein characteristics of de-cellularized mouse lungs. These different approaches resulted in significant differences in both histologic appearance and in retention of ECM and intracellular proteins as assessed by immunohistochemistry and mass spectrometry. Despite these differences, binding and proliferation of bone marrow-derived mesenchymal stromal cells (MSCs) over a one month period following intratracheal inoculation was similar between experimental conditions. In contrast, significant differences occurred with C10 mouse lung epithelial cells between the different conditions. Therefore, delayed necropsy, duration of scaffold storage, sterilization approach, and cell type used for re-cellularization may significantly impact the usefulness of this biological scaffold-based model of ex-vivo lung tissue regeneration.
Introduction
Increasing interest in the use of de-cellularized complex whole organ scaffolds for ex vivo tissue engineering has provided both opportunity and also unique challenges. Among the unresolved issues which require clarification include defining optimal, organ specific approaches for de-cellularization and for sterilization and storage of de-cellularized organs prior to re-cellularization [1–4]. With respect to trachea and lung, a number of recent publications have comparatively assessed different de-cellularization protocols. Notably, the resulting architecture and extracellular matrix (ECM) protein composition of either trachea or lungs may differ substantially between the different regimens utilized [5–7]. Whether this will subsequently affect re-cellularization of scaffolds and therefore the generation of functional tissue suitable for transplantation, remains unresolved [4,5]. Methods of optimal sterilization and storage have been suggested for trachea [7,8] but not yet clearly delineated for de-cellularized lungs. One further consideration is that of post-mortem time prior to lung harvest and de-cellularization, a practical issue for procurement of human lungs. A number of hours or even days may pass prior to post-mortem tissue harvest. It is unknown at present whether this will affect the suitability of the donor lung for de-cellularization and subsequent re-cellularization.
To address these questions, we assessed architecture and ECM protein content and distribution in mouse lungs obtained following a prolonged postmortem period prior to harvest compared to freshly procured lungs. We also assessed lungs obtained immediately after euthanasia and then subsequently stored after de-cellularization for prolonged periods (3 and 6 months). We further evaluated effects of sterilization using either irradiation or a final rinse with peracetic acid, a commonly used protocol in storage of other biologic scaffolds [9–13]. We then assessed growth of two different cell types, murine bone marrow-derived mesenchymal stromal cells (MSCs) and C10 mouse lung type 2 alveolar epithelial cells, following intratracheal inoculation into the different de-cellularized lungs.
Materials and Methods
Mice
Adult male C57BL/6J mice (8–24 wks, Jackson Laboratories) were maintained at UVM in accordance with institutional and American Association for Accreditation of Laboratory Animal Care standards and review. There was a total of 34 mice used for this experiment.
Lung De-cellularization
Mice were euthanized by lethal intraperitoneal injection of sodium pentobarbital in accordance with accepted AAALAC standards. After opening the chest, the trachea was cannulated with a blunted 18 gauge Luer-lock syringe, the thymus was removed and discarded, and the heart-lung bloc was harvested. The lungs underwent de-cellularization and were subsequently stored for defined periods of time or underwent specific sterilization techniques. The lungs were de-cellularized under sterile conditions according to previously published protocols [5,6,14–16]. Each step listed was accomplished by both rinsing through the trachea and also by perfusing solutions through the right ventricle. Lungs were washed in de-ionized (DI) water containing 5X penicillin/streptomycin (from 100X stock, Cellgro) for one hour at 4°C. 3mL of the DI water solution was injected through the cannulated trachea to rinse the lung, which was allowed to deflate before repeating the tracheal rinse four additional times. A DI water rinse of 15 cc total volume was repeated through the vasculature by injection through the right ventricle. 3cc of 0.1% Triton-X (Sigma) and 5X pen/strep in DI water was then infused through both the trachea and the right ventricle, and the lungs were submerged in Triton-X solution and incubated at 4° C for 24 hours. The following day, the lungs were removed from the Triton solution and rinsed with DI/pen-strep as described above. 3cc of 2% sodium deoxycholate (Sigma) and 1X pen/strep in DI water were then infused through the trachea and right ventricle and the lungs incubated in this solution at 4° C fo r 24 hours. The lungs were then removed from the sodium deoxycholate solution and rinsed with DI water as described above. 3cc of 1M NaCl (USB) and 5X pen/strep in de-ionized water were then infused through the trachea and right ventricle and the lungs incubated in the solution for 1 hour at room temperature (~25° C). The lungs were then removed from the NaCl solution and rinsed with DI water as described above. 3cc of 30ug/mL porcine pancreatic DNase (Sigma), 1.3 mM MgSO4 (Sigma), 2mM CaCl2 (Sigma), 5X Pen/Strepin DI water were then infused through the trachea and right ventricle and the lungs incubated in the solution for 1 hour at room temperature. Finally the lungs were removed from the DNase solution and rinsed with 5x pen/strep (Cellgro) in 1x PBS as described above for the DI solution rinses. Lungs were stored in PBS/pen-strep solution at 4° C until utilized.
To assess effects of prolonged storage of the de-cellularized lungs, lungs were stored in sterile PBS with 5X pen/strep at 4°C for either 3 or 6 months prior to assessment. To assess effects of two different sterilization approaches, one set of de-cellularized whole lungs was rinsed three times, through both the trachea and right ventricle, with 15mL of a 0.1% peracetic acid in 4% ethanol solution and then incubated in this solution for two hours prior to assessment [9,10]. Another set of 6 de-cellularized whole lungs was irradiated for 12 minutes at a constant dose of 5 Gy/minute using a RadSource 2000 Biological Irradiator prior to assessment. To assess effects of delayed lung harvest, 6 mice were euthanized and then kept at 4°C for 72 hours prior to necropsy and removal of the heart-lung bloc for subsequent de-cellularization and re-cellularization.
Lung Histology
De-cellularized lungs were fixed (20 cm H2O) with 4% paraformaldehyde for 20 minutes at room temperature, embedded in paraffin, and 5-μm sections mounted on glass slides. Following deparaffinization, sections were stained with hematoxylin & eosin, Verhoeff’s Van Gieson (EVG), Masson’s Trichrome, or Alcian Blue, and were assessed by standard light microscopy [5,14].
Immunohistochemical (IHC) Staining
Standard deparaffinization was performed with three separate 10 min incubations of xylenes, followed by rehydration in a descending series of ethanols, and finally in water. Antigen retrieval was performed by heating tissue in 1x sodium citrate buffer (Dako, Carpentaria, CA) at 98°C for 20 minutes followed by a brief 20 minutes cool at room temperature. Tissue sections were permeabilized in 0.1% Triton-X solution (Sigma Aldrich, St. Louis, MO) for 15 minutes. Triton-X was removed with two 10 minute washes in 1% BSA solution. Blocking was performed with 10% goat serum for 60 minutes. After blocking, primary antibody was added and tissue sections were incubated overnight at 4°C in a humidified chamber. Tissues were washed three times with 1% BSA solution for 5 minutes each. Secondary antibody was added and incubated for 60 min at room temperature in a dark humidified chamber. Tissues were again washed three times in 1% BSA solution for 5 minutes each in the dark. DAPI nuclear stain was added for 5 minutes at room temperature in the dark followed by 2 washes in 1% BSA solution for 5 minutes each. The sections were finally mounted in Aqua Polymount (Lerner Laboratories, Pittsburg, PA). Primary antibodies used were: Purified Mouse Anti-Fibronectin monoclonal (610077 – 1:100 – BD Transduction Laboratories), Laminin antibody polyclonal (ab11575 – 1:100 – Abcam), Rabbit polyclonal to alpha elastin (ab21607 – 1:100 – Abcam), Smooth muscle myosin heavy chain 2 polyclonal (ab53219 – 1:100 – Abcam), Collagen I polyclonal (ab292 – 1:100 – Abcam), Ki67 Proliferation marker polyclonal (ab16667 - 1:50 - Abcam), Cleaved Caspase-3 polyclonal (Asp175 – 1:100 – Cell Signaling Technology), Mouse clone anti-human Actin polyclonal (1A4 - 1:10,000 - Dako via FAHC). Secondary antibodies used: Alexa Fluor 568 goat anti-rabbit IgG (H+L) (1:500, Invitrogen), Alexa Fluor 568 F(ab′)2 fragment of goat anti-mouse IgG (H+L) (1:500, Invitrogen) [5,14].
Mass Spectrometry
Samples (approximately 1 cm3 and 130 mg for each sample) obtained from the right lower lobe of the various lungs were processed according to standard protocol [5, 14] and dried separately in a SpeedVac. Each dried sample was then suspended in 40 μL of 100 mM ammonium bicarbonate (NH4HCO3)/50 mM dithiothreitol and placed at 56°C for 1 h. After cooling, 5 μL of 500 mM iodoacetamide in 100 mM NH4HCO3 was added, and the solution was incubated for 30 min at room temperature in the dark. The sample was then dried in a SpeedVac, suspended in 50 μL of trypsin solution (10 ng/mL) in 50mM NH4HCO3, and incubated overnight at 37°C. Five microliters of 10% formic acid was then added to stop the digestion. The sample was then centrifuged at 14,000 g, and 15 μL of supernatant was desalted using a C18 ZipTip (P10; Millipore Corporation) according to the manufacturer’s protocol. The ZipTip eluate was then dried again, and then reconstituted in 20 uL 0.1% formic acid and 2% acetonitrile. Six μL of each digest was loaded directly onto a 100 um × 120 mm fused silica microcapillary column packed with MAGIC C18 (5 μm particle size, 20 nm pore size, Michrom Bioresources) at a flow rate of 500 nL/min, and peptides were separated by a gradient comprising 3 – 60% ACN/0.1% formic acid in 45 min. The peptides were introduced into a linear ion trap (LTQ)-Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanospray ionization source. Mass spectrometry data was acquired in a data-dependent acquisition mode, in which an Orbitrap survey scan from m/z 400–2000 (resolution: 30,000 FWHM at m/z 400) was paralleled by 10 LTQ MS/MS scans of the most abundant ions.
The product ion spectra were searched against the IPI mouse database (v. 3.75) using SEQUEST (Bioworks 3.3.1, Thermo Fisher Scientific). Search parameters allowed mass tolerances of 2.0 Da and 1.4 Da for precursor and fragment ions, respectively, and variable modifications for oxidized methionine (+15.9949 amu) and carboxyamidomethylated cysteine (+57.021464 amu). After applying filters of cross correlation XCorr [1.9, 2.5, 3.8 for peptide charge states of +1, +2, and +3, respectively], delta correlation (ΔCN) (> 0.1) and peptide probability (< 1e-3), protein identifications were ranked by number of peptides identified. Proteins that were identified by two or more peptides in each of the replicates were compiled and the average number of peptides identified from multiple replicates for each protein was presented in Supplementary Table 1.
Cells and Cell Inoculation
Mesenchymal stromal cells (MSCs) derived from bone marrow of adult male C57BL/6 mice were obtained from the NCRR/NIH Center for Preparation and Distribution of Adult Stem Cells at Texas A and M University [17]. Purity was determined by expression of Sca-1, CD106, CD29, absence of CD11b, CD11c, CD34, and CD45 expression, and the ability to differentiate into osteoblasts, chondrocytes and adipocytes in vitro [17]. MSCs were cultured on cell-culture treated plastic at 37°C and 5% CO2 in MSC basal med ium consisting of Iscove’s Modification of Dulbecco’s Medium supplemented with 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Fisher), 10% fetal bovine serum (Atlanta Biologicals) and 10% horse serum (HS, Invitrogen). Cells were used at passage 9 or lower and maintained in culture at confluency no greater than 70%. C10 mouse lung type 2 alveolar epithelial cells were obtained courtesy of Matthew Poynter Ph.D., University of Vermont and cultured under standard conditions [18]. Only the left lobe of the de-cellularized lungs was used for seeding. To ensure this, the right lobes were tied off using sterile suture under sterile conditions, and the right lobes were then removed. To seed the de-cellularized lungs, a solution of 3% low-melting temperature SeaPrep Agarose (Cambrex) in PBS was warmed until the agarose melted. 1×106 MSCs or C10 cells suspended in 1 mL MSC or C10 basal media, respectively, were mixed with 1ml of the low melting agarose and the 2mL cell suspension injected through the cannulated trachea into the remaining left lung. The inoculated lung was then incubated for 30 minutes at 4°C until the agarose h ardened. The lobe was sliced with a sterile razor blade to yield transverse sections of approximately 1mm in thickness. Each slice was placed in a well of a sterile 24 well dish, covered with sterile cell media and placed in a standard tissue culture incubator at 37°C until agarose melted out of the tissue. The lungs were then submerged overnight in basal MSC or C10 media at 37°C and 5% CO2. The next day, medium was changed to fresh basal medium and subsequently medium was changed every other day [5]. Individual slices were harvested at 1, 3, 7, 14, 21, and 28 days post-inoculation, fixed for 10 minutes at room temperature in 4% paraformaldehyde, and mounted 5 μm paraffin sections assessed by H and E staining for presence and distribution of the inoculated cells.
Statistical Analyses
Heat maps for the natural log of unique peptide hits for each positively identified protein in the mass spectrometric analyses of lungs de-cellularized under each experimental condition were generated using the ‘pheatmap’ package for ‘R’ statistical software version 2.15.1. Two group comparisons were done using the raw peptide counts (ie. non log transformed) using the non-parametric exact permutation test with p<0.05 considered statistically significant [19]. This non-parametric equivalent of the t-test was used due to the non-normality of the data, small sample sizes per group, and to directly compare biologically relevant conditions. As a measure of agreement/concordance between lung lobe sample replicates, non-parametric Spearman correlations were also done with concordance considered significant at p<0.05 [19]. The exact permutation tests and correlations were done using SAS statistical software, version 9.2.
Results
Comparison of de-cellularized mouse lungs: architecture and ECM composition
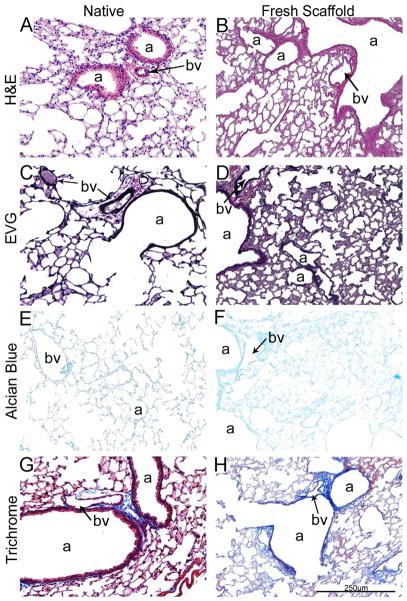
Histologic evaluation with H&E, Verhoeff’s Van Gieson (EVG), and Masson’s trichrome stains demonstrates, as we and others have previously shown [5,6,14–16,20–24], that freshly de-cellularized lungs maintain the architecture of the extracellular matrix compared to native lung (Figure 1A). Glycosaminoglycans (GAGs) were less evident by Alcian Blue staining in freshly de-cellularized lungs, likely representing, in large part, loss of cell-associated GAGs during the de-cellularization process as previously described by us and others (Figure 1A, Panels E and F) [5,14,16].
Figure 1.
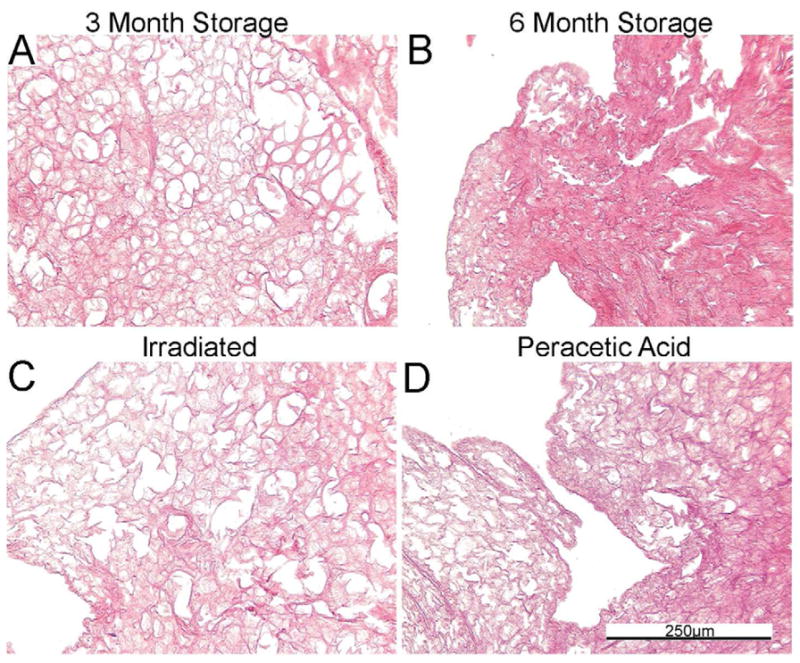
Figure 1A: De-cellularization of freshly obtained mouse lungs maintains the native architecture.
Representative photomicrographs comparing native (control) mouse lung and freshly de-cellularized mouse lungs are depicted. a = airways, bv = blood vessels. N = 6 for each condition. Original magnification 200x.
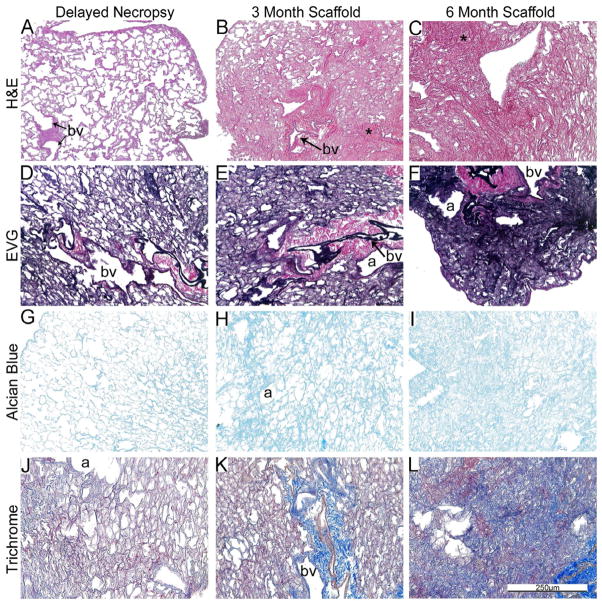
Figure 1B: Delayed necropsy results in preservation of native architecture whereas storage for 3-month and 6-months results in progressive atelectasis and loss of normal architecture. Representative photomicrographs depict the effects of delayed necropsy and of 3 and 6 month storage on freshly de-cellularized mouse lungs. * = atelectatic tissue, a = airways, bv = blood vessels. N = 6 for delayed necropsy & 3-month scaffolds, N = 4 for 6-month scaffolds. Original magnification 200x.
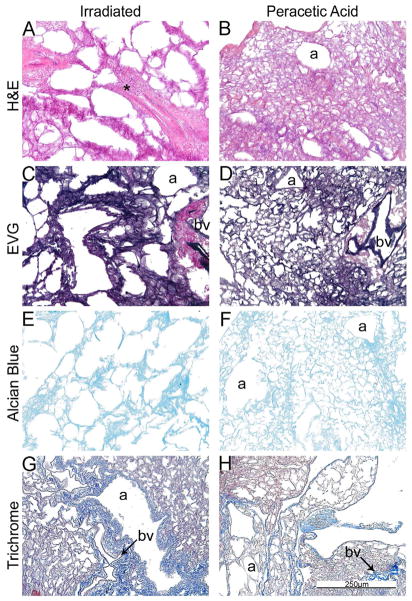
Figure 1C: Peracetic acid treatment preserves while irradiation destroys de-cellularized lung tissue architecture. Representative photomicrographs demonstrate the architecture of irradiated and peracetic acid-treated freshly de-cellularized lungs. * = abnormal architecture due to irradiation treatment, a = airways, bv = blood vessels. N = 6 for each condition. Original magnification 200x.
Figure 1D: Inflation of sterilized or stored de-cellularized lungs restores native architecture. Representative photomicrographs demonstrate that atelectasis of stored lungs can be reversed. a = airways, bv = blood vessels. N = 6 for each condition. Original magnification 200x.
Comparison of the delayed necropsy storage, 3-month, and 6-month storage conditions demonstrated both similarities and differences. Overall, delayed necropsy appeared to minimally affect the histologic appearance and overall presence of collagens (Trichrome), elastin (EVG), and GAGs (Alcian Blue) (Figure 1B, Panels A, D, G, J). Following 3 months of storage, scattered areas of apparent atelectasis (areas marked with an asterisk), were observed particularly in central versus more peripheral regions of the de-cellularized lungs (Figure 1B, Panels B, E, H, K). Following 6-months storage, the de-cellularized lungs were markedly atelectatic, showing very little resemblance to native or freshly de-cellularized lungs (Figure 1B, Panels C, F, I, L).
Peracetic acid treated lungs had a similar overall appearance to native or freshly de-cellularized lungs although some central regions appeared slightly more atelectatic (Figure 1C, Panels B, D, F, H). In contrast, irradiated lungs demonstrated a grossly abnormal appearance with a scattered heterogenous pattern of thickened, fused appearing alveolar septa, and large emphysematous-appearing alveolar spaces (marked with an asterisk) (Figure 1C, Panels A, C, E, G).
To assess whether some of the observed changes reflected atelectasis or other reversible changes, de-cellularized lungs treated using each of the different storage and sterilization conditions were re-inflated with low melting point agarose, utilizing the same approach used for intratracheal inoculations previously discussed [5,14,15]. As depicted in Figure 1D, the lung architecture of de-cellularized lungs obtained from the 3 month storage, peracetic acid, and even to some degree the irradiated lungs, following re-inflation better resembled native or freshly de-cellularized lungs. In contrast, no significant improvement was observed in the 6 month storage lungs.
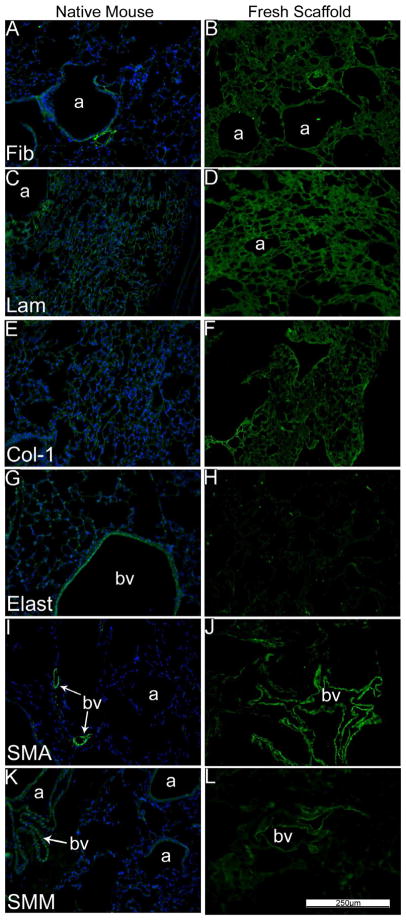
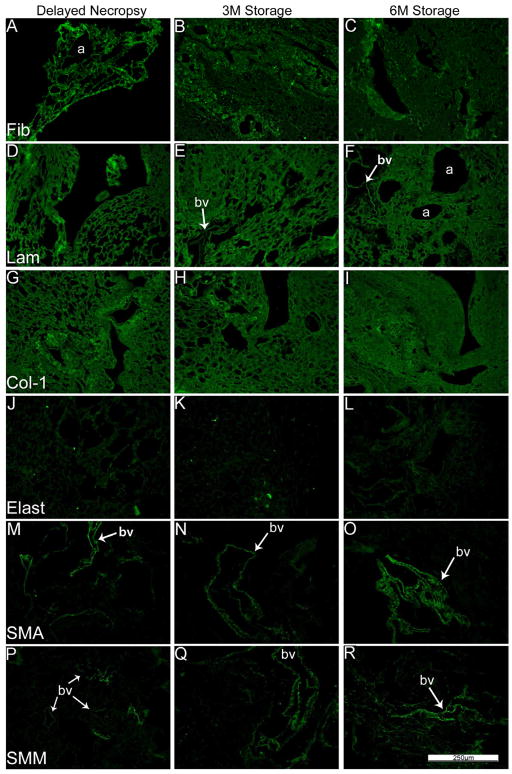
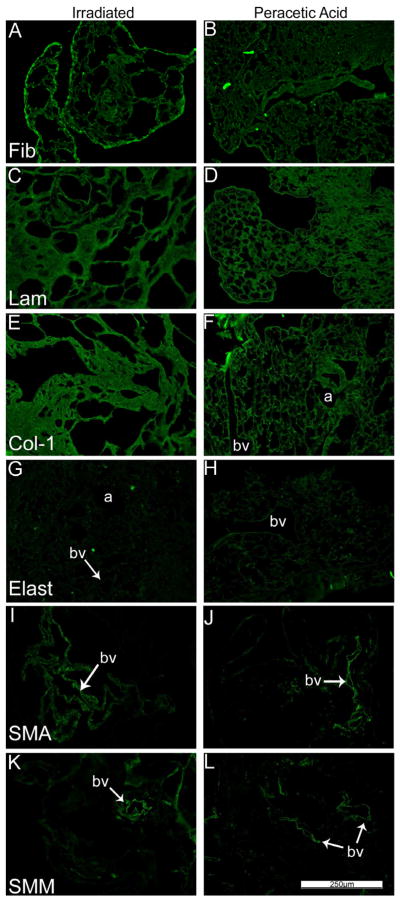
As we and others have previously demonstrated using specific immunohistochemical staining for selected ECM proteins, type 1 collagen and laminin were largely retained in freshly de-cellularized lungs whereas elastin was significantly decreased (Figure 2A, Panels C–H, Control antibody staining is shown in Supplemental Figure 1). Fibronectin was largely retained but became fragmented in appearance [5,14]. Furthermore, some cellular proteins, including smooth muscle actin & smooth muscle myosin were also largely retained in freshly de-cellularized lungs (Figure 2A, Panels A, B, I–L) [5,14]. Neither delayed necropsy, 3 or 6 month storage, nor peracetic acid treatment had any obvious effect on the visual appearance of staining for these proteins although the 6 month storage lungs remained markedly abnormal appearing (Figures 2B, C). In irradiated lungs, staining for laminin and collagen-1 appeared to be more intense, likely due to the agglomeration and thickened tissue. Fibronectin, smooth muscle actin, and smooth muscle myosin appeared to be present in the same patterns and intensities as in native or freshly de-cellularized lungs.
Figure 2.
Figure 2A: Extracellular matrix proteins are largely preserved in freshly de-cellularized lungs.
Representative photomicrographs comparing native (control)mouse lung and freshly de-cellularized mouse lungs are depicted. Nuclear DAPI staining is depicted in blue, and the stain(s) of interest are depicted in green in each respective panel. LAM = laminin, COL-1 = type I collagen, ELAST = elastin, FIB = fibronectin, SMA = smooth muscle actin, SMM = smooth muscle myosin. a = airways, bv = blood vessels. N = 6 for each condition. Original magnification 200x.
Figure 2B: Delayed necropsy, 3-month, and 6-month de-cell storage preserve most ECM proteins in the mouse lung. ECM proteins Fibronectin, laminin, collagen-1, elastin, and cellular smooth muscle actin, and smooth muscle myosin proteins were compared between the three permutations. Nuclear DAPI staining is depicted in blue, and the stain of interest is depicted in green in each respective panel. LAM = laminin, COL-1 = type I collagen, ELAST = elastin, FIB = fibronectin, SMA = smooth muscle actin, SMM = smooth muscle myosin. a = airways, bv = blood vessels. N = 6 for delayed necropsy & 3-month scaffolds, N = 4 for 6-month scaffolds. Original magnification 200x.
Figure 2C: Irradiation and peracetic acid treatments preserve most ECM proteins in de-cellularized mouse lung. Photomicrographs show the presence of fibronectin, laminin, collagen-1, elastin, smooth muscle actin, and smooth muscle myosin in the irradiated and peracetic acid-rinsed de-cellularized mouse lungs. Nuclear DAPI staining is depicted in blue, and the stain(s) of interest are depicted in green in each respective panel. Lam = laminin, Col-1 = type I collagen, Elast = elastin, Fib = fibronectin, SMA = smooth muscle actin, SMM = smooth muscle myosin. a = airways, bv = blood vessels. N = 6 for each condition. Original magnification 200x.
Assessment of residual protein content and composition in de-cellularized mouse lungs
Mass spectrometry-based approaches were utilized to detect differences in residual protein content under the different storage and sterilization procedures. Freshly de-cellularized right lower lobes were used as controls. All IPI accession numbers were manually searched in the UniProtKB/Swiss Prot database (http://www.uniprot.org/help/about). If a protein was matched to more than one category, its predominant subcellular location was used for functional grouping. Protein identification was assigned based on two or more unique peptide hits within the same sample, not across samples. The full range of data presented in Supplemental Table 1 is an average of the peptide hits for each experimental grouping. A value of one occurs when a sample had two or more peptide hits in a sample with 1 or less in the other samples. One such example is utrophin. In two young lung samples, two unique peptide hits for utrophin were detected in a single lung sample but 1 or less was detected in four of the samples. Thus the average (2+2+0+0+0+0) is 0.67 or 1.
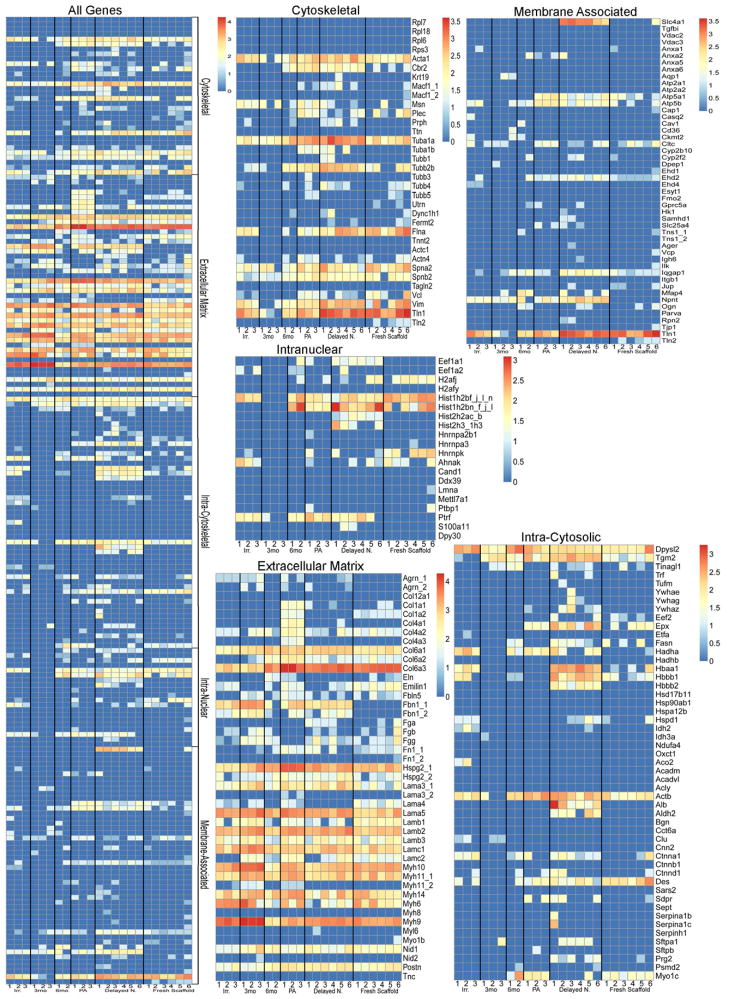
Proteins presented in Table 1 and the heat maps in Figure 3 were thus assigned to one of five groups; ECM, cytoskeletal, intracellular cytosolic, intracellular nuclear, and membrane associated, and ranked by the average number of unique peptides identified from the samples to investigate possible trends in the residual proteins. Heat maps were generated with each positively identified protein and its corresponding number of unique peptide hits. Values were log normal transformed to allow for more clear visual delineation on the heat maps. Most strikingly, strong trends emerged visually on the heat maps which were corroborated by statistical analyses (Figure 3, Table 1). The delayed necropsy lungs contained statistically significant increases in a large number of cellular associated proteins (non-ECM) as compared to freshly de-cellularized lungs. Notably, several proteins associated with erythrocytes including hemoglobins A and B and the erythrocyte membrane protein Slc4a were markedly increased in the delayed necropsy as compared to fresh de-cellularized lungs. Peracetic acid-treated lungs contained a significantly increased number of ECM components over the controls. Scaffolds which had been stored in PBS for 3 months at 4 °C contained several ECM components, such as aggrecan, fibrillin, laminins, and myosins, which were statistically increased over freshly de-cellularized scaffolds. There were no ECM proteins which achieved statistically significant increases in scaffolds which had been stored for 6 months. There were no statistically significant differences between the 3 months vs. 6 months or irradiated vs. peracetic acid-treated group comparisons. Protein identification information is provided in Supplemental Table 1.
Table 1.
Statistical analysis of the mass spectrometry data from the different sterilization and storage conditions for de-cellularized mouse lungs.
Values represent p-values from two group comparisons of raw unique peptide hits using the non-parametric test (pre-planned, selected, pairwise comparison of treatment group versus young decellularized lung controls). Protein group assignments were generated from UniProt entries of positively identified proteins (proteins containing two or more unique peptide hits). Grayed boxes with italicized text indicate the treatment group (labeled in the top row) reached statistical significance (p-values less than 0.05) with higher amounts of unique peptide hits than the control group (young de-cellularized lungs). White boxes with black, bolded text indicates the control group contained significantly more unique peptide hits.
| Gene | Protein Name | Cold | 3mos | 6mos | Irrad | Para | IPI |
|---|---|---|---|---|---|---|---|
| Cytoskeletal | |||||||
|
| |||||||
| Acta1 | Actin, alpha skeletal muscle | 0.026 | 0.060 | 1.000 | 0.155 | 0.262 | IPI00110827.1 |
| Cbr2 | Carbonyl reductase [NADPH] 2 | 0.002 | 0.321 | 0.214 | 0.321 | 0.012 | IPI00128642.1 |
| Flna | Isoform 1 of Filamin-A | 0.550 | 0.012 | 0.036 | 0.107 | 0.071 | IPI00131138.10 |
| Msn | Moesin | 1.000 | 0.286 | 1.000 | 0.083 | 0.024 | IPI00110588.4 |
| Tuba1a | Tubulin alpha-1A chain | 0.002 | 0.131 | 0.429 | 0.274 | 0.036 | IPI00110753.1 |
| Tuba1b | Tubulin alpha-1B chain | 0.182 | 1.000 | 1.000 | 1.000 | 0.012 | IPI00117348.4 |
| Tubb2b | Tubulin beta-2B chain | 0.009 | 0.202 | 0.536 | 0.345 | 0.131 | IPI00109061.1 |
|
| |||||||
| Extracellular Matrix Proteins | |||||||
|
| |||||||
| Agrn_1 | Isoform 2 of Agrin | 0.455 | 0.012 | 1.000 | 0.012 | 0.083 | IPI00378698.6 |
| Col1a1 | Isoform 1 of Collagen alpha-1(I) chain | 1.000 | 0.500 | 1.000 | 0.500 | 0.012 | IPI00329872.1 |
| Col1a2 | Collagen alpha-2(I) chain | 0.002 | 0.012 | 0.036 | 0.012 | 0.012 | IPI00222188.4 |
| Col4a1 | Collagen alpha-1(IV) chain | 1.000 | 1.000 | 1.000 | 1.000 | 0.012 | IPI00109588.4 |
| Col4a2 | Collagen alpha-2(IV) chain | 0.517 | 0.702 | 0.714 | 0.214 | 0.024 | IPI00338452.3 |
| Col4a3 | Collagen alpha-3(IV) chain | 1.000 | 1.000 | 1.000 | 1.000 | 0.012 | IPI00137938.7 |
| Col6a1 | Collagen alpha-1(VI) chain | 0.004 | 0.155 | 0.071 | 0.060 | 0.024 | IPI00339885.2 |
| Col6a2 | Collagen alpha-2(VI) chain | 0.732 | 0.060 | 0.071 | 0.012 | 0.012 | IPI00621027.2 |
| Col6a3 | collagen alpha-3(VI) chain | 0.734 | 0.012 | 0.143 | 0.012 | 0.060 | IPI00830749.3 |
| Emilin1 | Emilin-1 | 0.006 | 0.012 | 0.036 | 0.107 | 0.214 | IPI00115516.1 |
| Fbn1_1 | Fibrillin-1 | 0.002 | 0.012 | 0.250 | 0.012 | 0.012 | IPI00122438.1 |
| Fbn1_2 | Fibrillin -1 | 0.015 | 0.012 | 0.250 | 0.012 | 0.083 | IPI00338565.2 |
| Fn1_1 | Fibronectin | 0.818 | 0.024 | 0.321 | 0.012 | 0.095 | IPI00113539.2 |
| Hspg2_1 | Basement membrane-specific heparan sulfate proteoglycan core protein | 0.074 | 0.798 | 0.179 | 0.012 | 0.012 | IPI00113824.1 |
| Lama3_1 | Isoform B of Laminin subunit alpha-3 | 0.039 | 0.024 | 0.893 | 0.869 | 0.036 | IPI00125058.2 |
| Lama4 | Laminin subunit alpha-4 | 0.002 | 0.143 | 0.036 | 0.107 | 0.250 | IPI00223446.5 |
| Lama5 | Laminin subunit alpha-5 | 0.054 | 0.024 | 0.393 | 0.095 | 0.012 | IPI00116913.3 |
| Lamb1 | laminin subunit beta-1 | 0.004 | 0.750 | 0.036 | 0.036 | 0.048 | IPI00338785.3 |
| Lamb2 | laminin subunit beta-2 precursor | 0.011 | 0.012 | 0.321 | 0.500 | 0.012 | IPI00109612.2 |
| Lamc1 | Laminin subunit gamma-1 | 0.199 | 0.012 | 0.679 | 0.488 | 0.262 | IPI00400016.1 |
| Lamc2 | laminin subunit gamma-2 | 0.792 | 0.202 | 0.071 | 0.012 | 0.036 | IPI00117115.3 |
| Myh10 | Myosin, heavy polypeptide 10, non-muscle | 0.006 | 0.012 | 0.036 | 0.726 | 0.857 | IPI00338604.5 |
| Myh11_2 | myosin-11 isoform 1 | 1.000 | 0.012 | 1.000 | 0.333 | 0.012 | IPI00938530.1 |
| Myh6 | Myosin-6 | 0.006 | 0.321 | 0.071 | 0.060 | 0.845 | IPI00129404.1 |
| Myh9 | Myosin-9 | 0.121 | 0.012 | 0.036 | 0.012 | 0.369 | IPI00123181.4 |
| Nid1 | Nidogen-1 | 1.000 | 0.214 | 0.250 | 0.024 | 0.071 | IPI00111793.1 |
|
| |||||||
| Intracellular cytoplasmic | |||||||
|
| |||||||
| Actb | Actin, cytoplasmic 1 | 0.024 | 0.060 | 0.250 | 0.512 | 0.024 | IPI00110850.1 |
| Alb | Serum albumin | 0.002 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00131695.3 |
| Aldh2 | Aldehyde dehydrogenase,mitochondrial | 0.015 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00111218.1 |
| Des | Desmin | 0.043 | 0.012 | 0.071 | 0.012 | 0.131 | IPI00130102.4 |
| Epx | Eosinophil peroxidase | 0.019 | 0.167 | 0.357 | 0.167 | 0.476 | IPI00113854.1 |
| Hadha | Trifunctional enzyme subunit alpha, mitochondrial | 0.061 | 1.000 | 0.036 | 0.012 | 0.012 | IPI00223092.5 |
| Hbaa1 | Putative uncharacterized protein | 0.002 | 0.393 | 0.607 | 0.119 | 0.393 | IPI00110658.1 |
| Hbbb1 | LOC100503164 Beta-globin | 0.002 | 0.083 | 0.643 | 0.429 | 0.083 | IPI00762198.2 |
| Hbbb2 | Hemoglobin subunit beta-2 | 0.002 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00316491.4 |
| Idh2 | Isocitrate dehydrogenase [NADP], mitochondrial | 1.000 | 1.000 | 1.000 | 0.012 | 1.000 | IPI00318614.9 |
| Myo1c | Isoform 2 of Myosin-Ic | 0.013 | 0.012 | 0.250 | 0.012 | 0.036 | IPI00467172.2 |
| Prg2 | bone marrow proteoglycan | 0.028 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00126365.1 |
| Tgm2 | Protein-glutamine gamma- glutamyltransferase 2 | 0.002 | 0.024 | 0.679 | 0.024 | 0.060 | IPI00126861.3 |
|
| |||||||
| Intracellular nuclear | |||||||
|
| |||||||
| Eef1a1 | Elongation factor 1-alpha 1 | 0.002 | 1.000 | 0.250 | 1.000 | 0.333 | IPI00307837.6 |
| Eef1a2 | Elongation factor 1-alpha 2 | 1.000 | 1.000 | 0.036 | 0.083 | 1.000 | IPI00119667.1 |
| Hist1h2bf_j_l_n | Histone H4 | 0.006 | 0.012 | 0.286 | 0.060 | 0.012 | IPI00407339.7 |
| Hist2h2ac_b | Histone H2A type 2-C | 0.002 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00272033.3 |
| Hnrnpk | Isoform 2 of Heterogeneous nuclear ribonucleoprotein K | 0.030 | 0.060 | 0.107 | 0.083 | 0.131 | IPI00224575.1 |
| Ptrf | Polymerase I and transcript release factor | 0.015 | 0.333 | 0.036 | 0.012 | 0.012 | IPI00117689.1 |
|
| |||||||
| Membrane | |||||||
|
| |||||||
| Atp5a1 | ATP synthase subunit alpha, mitochondrial | 0.255 | 0.071 | 0.179 | 0.071 | 0.036 | IPI00130280.1 |
| Atp5b | ATP synthase subunit beta, mitochondrial | 0.017 | 0.881 | 0.429 | 0.524 | 0.012 | IPI00468481.2 |
| Ehd2 | EH domain-containing protein 2 | 0.006 | 0.012 | 1.000 | 0.452 | 0.119 | IPI00402968.1 |
| Iqgap1 | Ras GTPase-activating-like protein IQGAP1 | 0.006 | 0.488 | 0.071 | 0.024 | 0.429 | IPI00467447.3 |
| Mfap4 | Isoform 1 of Microfibril-associated glycoprotein 4 | 0.182 | 0.333 | 0.036 | 1.000 | 1.000 | IPI00133751.1 |
| Npnt | Isoform 4 of Nephronectin | 0.002 | 0.012 | 0.250 | 0.012 | 0.012 | IPI00124689.1 |
| Slc4a1 | Isoform Erythrocyte of Band 3 anion transport protein | 0.002 | 1.000 | 1.000 | 1.000 | 1.000 | IPI00120761.3 |
| Tln1 | Talin-1 | 0.907 | 0.012 | 0.107 | 0.131 | 0.095 | IPI00465786.3 |
italics treatment group significantly higher
gray rll significantly higher
Figure 3. Heat map displays of mass spectrometric assessments of residual proteins demonstrate both similarities and significant differences between the experimental conditions.
The compiled heat map and specific heat maps for each of the designated protein categories is depicted. The color scheme represents log order transform values indicating the relative abundance of each protein compared to presence in freshly de-cellularized lungs.
Growth of MSCs and C10 cells in de-cellularized lungs
Similar initial localization and distribution (day 1) of both C10s and MSCs throughout the lungs were observed with the different storage and sterilization conditions as that seen in freshly de-cellularized scaffolds (Figures 4,5). Notably, de-cellularized lungs that had been atelectatic or deranged appearing prior to inoculation (3 month storage, irradiated, peracetic acid, and to a limited, degree the 6 month storage condition) demonstrated some restoration of normal appearing architecture, likely reflecting an inflation effect during cell inoculation as demonstrated in Figure 1D. As previously observed [5,14,15], many of the MSCs that initially lodged in parenchymal lung regions developed a spindle-shaped elongated appearance over time (Figure 4). This appeared consistent in the various lung treatment conditions. Also as previously observed, C10 cells develop an elongated phenotype over time as they grow along alveolar septa [5]. This observation was also consistent between the different lung treatment conditions.
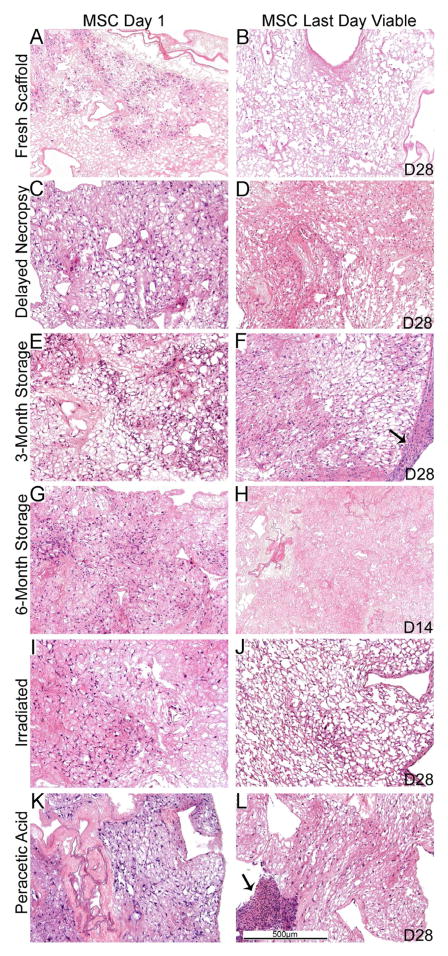
Figure 4. MSC cell growth is similar in the different sterilized and stored de-cellularized lungs except for the 6 month storage condition.
Representative H and E images of de-cellularized lungs treated under different storage and sterilization techniques and intratracheally inoculated with MSCs are shown. Images demonstrate characteristic re-cellularization one day after inoculation for all conditions (left). Robust re-cellularization was maintained for 28 days in culture (right) for all conditions except for de-cellularized lungs stored for 6 months in which no viable cells were observed after 7–14 days in culture. a = airways, bv = blood vessels. Arrows indicate areas where the MSCs have migrated out of the de-cellularized lung slices and formed agglomerates on plastic of the tissue culture dish. N = 3 for each condition (N = 2 for the 6 month storage condition). Original magnification 100X.
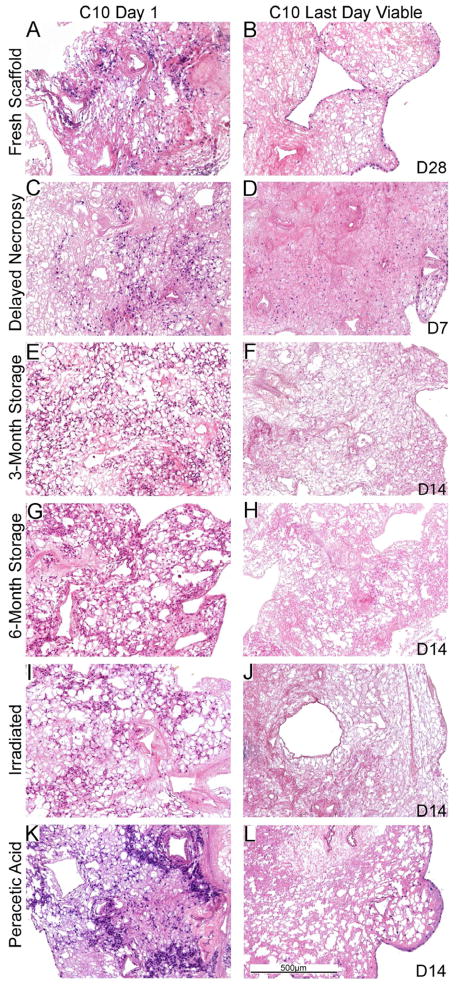
Figure 5. C10 cell growth varies in the different sterilized and stored de-cellularized lungs.
Representative H and E images of various de-cellularized lungs treated under different storage and sterilization techniques intratracheally inoculated with C10 cells. Images show characteristic re-cellularization one day after inoculation (left) in addition to the time point at which the last viable cells were observed (right). The last day cells were observed is indicated on the figure (e.g. D14 is day 14). a = airways, bv = blood vessels. N = 3 for each condition (N = 2 for the 6 month storage condition). Original magnification 100X.
The length of time in which the cells remained viable in the scaffolds was variable, dependent upon both the cell type and the treatment of the de-cellularized scaffold (Figures 4, 5). With the exception of the 6-month storage condition, in which no viable cells were observed after 7 to 14 days in culture, MSCs survived robustly through 28 days of culture (Figure 4, Panels B, D, F, H, J, L). The cells were localized throughout the tissue and retained their characteristic spindle-shaped elongated phenotype. This was despite a gradual re-development of atelectasis in the lung slices over the 28 day culture period compared to the initial lung inflation achieved with the cell inoculations. We had previously found robust Ki67 staining and minimal caspase-3 staining of MSCs at both 1 and 28 days when inoculated into freshly de-cellularized lungs [5,14]. In the current studies, Ki67 staining demonstrated actively proliferating cells throughout the lung slices for the different storage and sterilization conditions both at day 1 and day 28 after inoculation (Figure 6). Minimal early apoptosis was observed by caspase-3 staining at days 1 or 28 for all conditions except the 6 month storage in which increased caspase-3 staining was observed at 7 days(Figure 6) and also at 14 days (data not shown). In some conditions, aggregates of MSCs were observed on the periphery of the slices after 28 days in culture (Figure 4, Panels F, L). These likely represent MSCs that had migrated out of the slices to grow on the underlying tissue culture plastic.
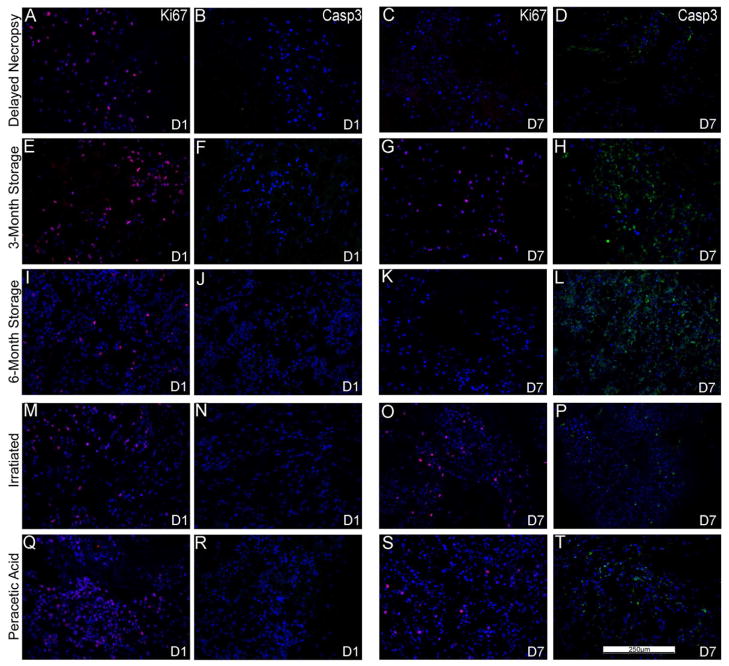
Figure 6. MSCs inoculated into the de-cellularized lungs demonstrate similar sustained proliferation by Ki67 staining and minimal apoptosis by caspase-3 staining.
Representative photomicrographs of characteristic Ki67 (red) or caspase-3 (green) are shown at 1 day post-inoculation as well as at the last viable time point at which cells were observed (7 days for 6 month storage, 28 days for the other conditions). A marked increase in caspase-3 staining was observed at the last time point in which viable cells were observed in the 3 and 6 month storage conditions. For both panels, caspase-3 immunofluorescence is depicted in green and Ki67 immunofluorescence is depicted in red. DAPI nuclear staining is depicted in blue. a = airways, bv = blood vessels. N = 3 for each condition (N = 2 for the 6 month storage condition). Original magnification 200X.
We had previously observed sustained growth and spreading of intratracheally inoculated C10 cells along alveolar walls following either 1 or 14–28 days in culture [5]. In parallel, robust Ki67 and minimal caspase-3 staining was observed [5]. In contrast, C10 cells inoculated into the different sterilization and storage conditions were largely non-viable or absent at different time points ranging between 7 and 14 days in culture (Figure 5). At the last viable time point for each condition (Figure 5, Panels B, D, F, H, J, L), the C10 cells were largely localized on the periphery of the tissue or lining the major airways. Ki-67 and caspase-3 staining demonstrated active proliferation and minimal early apoptosis 1 day after seeding but a significant increase in apoptosis at or before the last viable time point at which cells were observed for each storage and sterilization condition (Figure 7).
Figure 7. C10 inoculated into various storage and sterilized de-cellularized lungs demonstrate diminished proliferation by Ki67 staining and increased apoptosis by caspase-3 staining.
Representative photomicrographs of characteristic Ki67 (red) or caspase-3 (green) are shown at 1 day post-inoculation (left) as well as at time points in which caspase-3 staining was increased at or prior to the time when viable cells were last observed (e.g. D7 is day 7). For both panels, caspase-3 immunofluorescence is depicted in green and Ki67 immunofluorescence is depicted in red. DAPI nuclear staining is depicted in blue. a = airways, bv = blood vessels. N = 3 for each condition (N = 2 for the 6 month storage condition). Original magnification 200X.
Discussion
Use of de-cellularized whole lung scaffolds for ex vivo generation of functional lung tissue is a rapidly growing area that may provide a viable option for clinical lung transplantation [4–6,14–16,20–25]. As demonstrated in other tissue types such as skin, muscle, bladder, and others, successful use of biologic scaffolds has already entered clinical practice [1–4]. However, the complicated 3-dimensional structure-functional biology of the lung makes this a more formidable challenge. A number of recent reports, including those from our laboratory, have begun to explore lung de-cellularization, re-cellularization, and implantation in rodent and primate models [5,6,14–16,20–24]. While these reports show the promise of this approach, a number of questions remain. For example, there is no consensus on the optimal means of producing clinically relevant de-cellularized lungs and different detergent and physical approaches have been utilized. Comparison studies demonstrate significant differences between the structure and protein content and also the mechanical properties of de-cellularized lungs produced using the different approaches [5,6,24]. However, whether these differences will significantly affect subsequent re-cellularization and also the potential immunogenicity of the de-cellularized scaffolds remains unclear. Recent data suggests that initial binding and short term growth of stromal and epithelial cells inoculated into mouse lungs de-cellularized using different detergent-based approaches is similar [5]. However, more data on longer term growth and also on growth of other cell types to be inoculated into the de-cellularized lungs, including vascular endothelial cells, is necessary.
Other practical issues need to be considered for use of de-cellularized lung scaffolds. Biologic scaffolds such as bone, cartilage, and skin can be stored for prolonged periods of time prior to use, particularly if treated with an adjunct method such as low level irradiation or final rinse in peracetic acid for sterilization [9–13]. However, it is unclear whether these approaches or long term storage in general will be applicable for de-cellularized whole lungs. Recent data demonstrates that significant tissue breakdown can occur in de-cellularized tracheas stored for up to one year [8].
To assess this, we initially evaluated freshly de-cellularized lungs that were stored under refrigerated sterile conditions for up to 6 months. Most of the lungs remained sterile with only infrequent episodes of bacterial or fungal contamination. Histologic assessment of the stored lungs demonstrated development over time of lung atelectasis and loss of native architecture. While partially reversible with re-inflation in lungs stored for 3 months, these changes were particularly marked and mostly irreversible with inflation following 6 months storage. These results suggest that de-cellularized lungs should not be stored beyond 3 months if indeed that long. Irradiation, even at a dose under that commonly utilized for other biologic materials according to International Standard of Organizations (15–25 kGy) [11–13], produced significant distortion that was only partly responsive to subsequent lung re-inflation. Peracetic acid, a denaturing agent used both for sterilization and to rinse out residual detergents and other reagents utilized during tissue de-cellularization [9,10], had less effect on the resulting architecture.
There were significant differences in residual protein content, as assessed by mass spectrometry, between the different storage and sterilization procedures examined here. Compared to freshly de-cellularized lungs, the most striking differences observed were in the scaffolds following delayed necropsy and with use of peracetic acid sterilization of freshly de-cellularized lungs. The presence of erythrocyte components and other intracellular proteins in the delayed necropsy group suggests that autolysis of red blood cells and other cells present in the lungs occurred over time, despite cold storage, and that the proteins released from autolysed cells are not completely removed by the de-cellularization approach utilized. Additionally, peracetic acid can act as a protein denaturing agent has previously been utilized to solubilize ECM components for protein detection. Concordantly, freshly de-cellularized peracetic acid-treated lungs contained a significantly increased number of ECM components compared to non-treated lungs, most noticeably among the collagens detected (I, IV, VI). The increase in ECM components is most likely not indicative of an absolute increase in ECM components, but rather increased solubilization of ECM components which were then more readily detected using mass spectrometry. These results suggest that use of peracetic acid may not be optimal as a final sterilization approach for preservation of de-cellularized lungs. However, further investigation into the effects of peracetic acid treatment on the resulting de-cellularized lung scaffold and also the ramifications for re-cellularization are necessary. Similarly, scaffolds stored for 3 months had more detectable ECM proteins than controls, but these increases were absent in the scaffolds stored similarly for 6 months. Those proteins which were readily solubilized in the 3 month scaffolds were likely degraded beyond positive identification by the 6 month analysis.
Initial binding and subsequent survival and proliferation of a stromal cell line (MSCs) inoculated into the airways was comparable between the delayed necropsy, 3 month storage, peracetic acid, and irradiated de-cellularized lungs. This suggests that, despite some derangements in architecture and ECM protein content observed in some of these conditions, appropriate preservation of the ECM structures necessary for initial binding and subsequent cell growth and proliferation were preserved. The notable exception was in de-cellularized lungs stored for 6 months in which architecture was markedly abnormal and, although initial MSC binding and growth was preserved, sustained survival was diminished compared to the other conditions. Markers of early apoptosis were observed following relatively short periods of MSC culture in de-cellularized lungs stored for either 3 or 6 months, further suggesting that prolonged storage of de-cellularized lungs may not support sustained cell growth.
Despite good initial survival and proliferation, viability of a type 2 alveolar epithelial cell line (C10) was diminished over time in all of the experimental conditions compared to survival and proliferation of C10 cells inoculated into freshly de-cellularized lungs [5]. Markers of early apoptosis were also observed following relatively short time in culture, again particularly in C10 cells seeded into freshly de-cellularized lungs stored for 3 or 6 months. At present the reasons for the decreased survival of the C10 as opposed to the MSCs remains unclear. The C10 cells may be more stringent in their requirements for epitopes necessary for binding and subsequent proliferation remaining in the de-cellularized lung scaffolds than the perhaps more adaptable MSCs. Further investigation with a wider range of cell types will help clarify whether de-cellularized lungs produced, stored, or sterilized under different conditions are capable of supporting the growth of the range of cells necessary to produce functional lung tissue.
These results suggest that commonly utilized approaches for storage and sterilization of other de-cellularized tissues and other types of biologic scaffolds may not be applicable for de-cellularized lungs. In addition to gross alterations in lung architecture, changes in the protein content and distribution in residual scaffold may occur which can adversely affect survival and proliferation of certain inoculated cell types, particularly in combination with other physical factors such as gas diffusion into more central atelectatic regions of the de-cellularized lung scaffolds. Different cell types may react differently to the various storage and sterilization conditions, further complicating development of an optimal overall strategy for use of de-cellularized lungs. These studies do not address the impact of different storage and sterilization conditions on a wider range of cells that will be inoculated into the de-cellularized lung constructs including both mature pulmonary vascular endothelial cells as well as a range of stem and progenitor cells that might be utilized for re-cellularization. This will need to be done in a rigorous manner in future studies.
At present, our bias is that use of mass spectrometry to assess residual proteins remaining in de-cellularized lung scaffolds provides mixed messages. On one hand, as a high throughput screening approach, the range of proteins detected is far more accessible than that obtained with histologic or immunohistologic (IHC) assessments. As such, one can detect a range of minor proteins remaining in the scaffold that make up the complex ECM scaffold and that would not normally be probed by IHC or western analyses, as we have utilized in previous investigations of de-cellularized lung scaffolds [5,6,14]. This could provide critical information on whether necessary proteins remain and also whether undesirable proteins, ECM or otherwise, may remain. As such, this approach may be best utilized as a tool for comparing different de-cellularization approaches. On the other hand, the sensitivity and specificity of mass spectometry may be less than that afforded by IHC or western analyses using highly specific monoclonal antibodies directed towards relevant epitopes critical for binding and or growth and differentiation of inoculated cells. For example, the specificity of detecting a particular isoform depends on whether a peptide specific to the isoform was picked up and sequenced in the mass spectrometry run. As such, biologically relevant information provided by mass spectometry may be limited. A comparable proteomics approach has recently been utilized by Welham and colleagues to assess residual proteins remaining in decellularized human vocal fold mucosa [26]. In particular, this article validates our choice of grouping by cellular compartment rather than function.
Conclusions
In summary, the current results demonstrates that conditions of storage or of sterilization of de-cellularized lung scaffolds can significantly impact both the structure and residual protein content as well as the ability of different cell types to survive and proliferate following inoculation. While excellent progress is being made in developing techniques for utilizing cadaveric de-cellularized lungs as scaffolds for ex vivo generation of functional lung tissue suitable for clinical use, these and other parameters remain to be better defined for optimal use of de-cellularized lungs. Careful attention needs to be given to better understanding all of the conditions that will affect lung procurement, de-cellularization, sterilization, storage, re-cellularization, and eventual implantation.
Supplementary Material
Average raw unique peptide hits for all proteins positively identified in the different sterilization and storage conditions for de-cellularized mouse lungs. Protein group assignments were generated from UniProt entries of positively identified proteins (proteins containing two or more unique peptide hits). Summary rows indicate total number of replicates for each experimental group as well as the total number of individual proteins identified in each condition.
Representative images showing the lack of non-specific secondary antibody staining. Top images depict DAPI staining overlaid with autofluoresence. Bottom images are identical images with DAPI removed. Original magnification 200x.
Acknowledgments
The authors gratefully acknowledge the staffs of the Offices of Animal Care Management at the University of Vermont, Bruce Bunnell, Christine Finck, and Andrew Hoffman for critical reads of the manuscript and Tyler Bittner and Ian Johnson for valuable contributions to the experimental studies. Studies were supported by NIH ARRA RC4HL106625 (DJW), NHLBI R21HL094611 (DJW), UVM Lung Biology Training grant T32 HL076122 from the NHLBI, and UVM Environmental Pathology Training grant T32 ES007122 from the NIEHS. Facilities and equipment were supported by the UVM Lung Biology COBRE (NIH NCRR P20 RR-155557, the Vermont Cancer Center DNA Analysis facility (NIH P30 CA22435), and the Vermont Genetics Network through Grant Number 8P20GM103449 from the INBRE Program of the National Institute of General Medical Sciences (NIGMS) and the National Center for Research Resources (NCRR),.
Footnotes
Disclosure Statement
No competing financial interests exist
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicholas R. Bonenfant, Email: nicholas.bonenfant@med.uvm.edu.
Ying Wai Lam, Email: ylam@uvm.edu.
Bin Deng, Email: bdeng@uvm.edu.
Roberto Loi, Email: rloi@unica.it.
References
- 1.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatia SK. Tissue engineering for clinical applications. Biotechnol J. 2010;5:1309–23. doi: 10.1002/biot.201000230. [DOI] [PubMed] [Google Scholar]
- 3.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Badylak SF, Weiss DJ, Caplan A, Macchiarini P. Engineered whole organs and complex tissues. Lancet. 2012;379:943–52. doi: 10.1016/S0140-6736(12)60073-7. [DOI] [PubMed] [Google Scholar]
- 5.Wallis JM, Borg ZD, Daly AB, Deng B, Ballif BA, Allen GB, et al. Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng Part C Methods. 2012;18:420–32. doi: 10.1089/ten.tec.2011.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen T, Roszell B, Zang F, Girard E, Matson A, Thrall R, et al. A rapid lung de-cellularization protocol supports embryonic stem cell differentiation in vitro and following implantation. Tissue Eng Part C Methods. 2012;18:632–46. doi: 10.1089/ten.tec.2011.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haykal S, Soleas JP, Salna M, Hofer SO, Waddell TK. Evaluation of the structural integrity and extracellular matrix components of tracheal allografts following cyclical decellularization techniques: comparison of three protocols. Tissue Eng Part C Methods. 2012;18:614–23. doi: 10.1089/ten.TEC.2011.0579. [DOI] [PubMed] [Google Scholar]
- 8.Baiguera S, Del Gaudio C, Jaus MO, Polizzi L, Gonfiotti A, Comin CE, et al. Long-term changes to in vitro preserved bioengineered human trachea and their implications for decellularized tissues. Biomaterials. 2012;33:3662–72. doi: 10.1016/j.biomaterials.2012.01.064. [DOI] [PubMed] [Google Scholar]
- 9.Hodde JP, Record RD, Tullius RS, Badylak SF. Retention of endothelial cell adherence to porcine-derived extracellular matrix after disinfection and sterilization. Tissue Eng. 2002;8:225–34. doi: 10.1089/107632702753724996. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan DC, Mirmalek-Sani SH, Deegan DB, Baptista PM, Aboushwareb T, Atala A, et al. Decellularization methods of porcine kidneys for whole organ engineering using a high-throughput system. Biomaterials. 2012;33:7756–64. doi: 10.1016/j.biomaterials.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Freytes DO, Stoner RM, Badylak SF. Uniaxial and biaxial properties of terminally sterilized porcine urinary bladder matrix scaffolds. J Biomed Mater Res B Appl Biomater. 2008;84:408–14. doi: 10.1002/jbm.b.30885. [DOI] [PubMed] [Google Scholar]
- 12.ISO. ISO Technical Committee TC 1982006. Sterilization of health care products -- Radiation -- Part 1: Requirements for development, validation and routine control of a sterilization process for medical. [Google Scholar]
- 13.ISO. ISO Technical Committee TC 1982011. Sterilization of health care products – Radiation – Part 2: Establishing the sterilization dose. [Google Scholar]
- 14.Daly AB, Wallis JM, Borg ZD, Bonvillain RW, Deng B, Ballif BA, et al. Initial binding and recellularization of decellularized mouse lung scaffolds with bone marrow-derived mesenchymal stromal cells. Tissue Eng Part A. 2012;18:1–16. doi: 10.1089/ten.tea.2011.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonvillain RW, Danchuk S, Sullivan DE, Betancourt AM, Semon JA, Eagle ME, et al. A Nonhuman Primate Model of Lung Regeneration: Detergent-Mediated Decellularization and Initial In Vitro Recellularization with Mesenchymal Stem Cells. Tissue Eng Part A. 2012;18:2437–52. doi: 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Development of a decellularized lung bioreactor system for bioengineering the lung: the matrix reloaded. Tissue Eng Part A. 2010;16:2581–91. doi: 10.1089/ten.tea.2009.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–41. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 18.Malkinson AM, Dwyer-Nield LD, Rice PL, Dinsdale D. Mouse lung epithelial cell lines--tools for the study of differentiation and the neoplastic phenotype. Toxicology. 1997;123:53–100. doi: 10.1016/s0300-483x(97)00108-x. [DOI] [PubMed] [Google Scholar]
- 19.Zar JH. Biostatistical analysis. Englewood Cliffs, N.J: Prentice-Hall; 1974. [Google Scholar]
- 20.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 21.Cortiella J, Niles J, Cantu A, Brettler A, Pham A, Vargas G, et al. Influence of acellular natural lung matrix on murine embryonic stem cell differentiation and tissue formation. Tissue Eng Part A. 2010;16:2565–80. doi: 10.1089/ten.tea.2009.0730. [DOI] [PubMed] [Google Scholar]
- 22.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song JJ, Kim SS, Liu Z, Madsen JC, Mathisen DJ, Vacanti JP, et al. Enhanced in vivo function of bioartificial lungs in rats. Ann Thorac Surg. 2011;92:998–1005. doi: 10.1016/j.athoracsur.2011.05.018. discussion-6. [DOI] [PubMed] [Google Scholar]
- 24.Petersen TH, Calle EA, Colehour MB, Niklason LE. Matrix composition and mechanics of decellularized lung scaffolds. Cells Tissues Organs. 2012;195:222–31. doi: 10.1159/000324896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panoskaltsis-Mortari A, Weiss DJ. Breathing new life into lung transplantation therapy. Mol Ther. 2010;18:1581–3. doi: 10.1038/mt.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welham NV, Chang Z, Smith LM, Frey BL. Proteomic analysis of a decellularized human vocal fold mucosa scaffold using 2D electrophoresis and high-resolution mass spectrometry. Biomaterials. 2013;34:669–676. doi: 10.1016/j.biomaterials.2012.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average raw unique peptide hits for all proteins positively identified in the different sterilization and storage conditions for de-cellularized mouse lungs. Protein group assignments were generated from UniProt entries of positively identified proteins (proteins containing two or more unique peptide hits). Summary rows indicate total number of replicates for each experimental group as well as the total number of individual proteins identified in each condition.
Representative images showing the lack of non-specific secondary antibody staining. Top images depict DAPI staining overlaid with autofluoresence. Bottom images are identical images with DAPI removed. Original magnification 200x.