Abstract
Background and Objectives
The Colton blood group antigens are carried by the AQP1 water channel. AQP1−/− individuals, also known as Colton-null since they express no Colton antigens, do not suffer any apparent clinical consequence but may develop a clinically significant alloantibody (anti-CO3) induced by transfusion or pregnancy. Identification and transfusion support of Colton-null patients are highly challenging, not only due to the extreme rarity of this phenotype, the lack of appropriate reagents in most laboratories, as well as the possibility of confusing its with the recently described CO:-1,-2,3,-4 phenotype where AQP1 is present. This study investigated a new Colton-null case and evaluated three commercially available anti-AQP1s to identify Colton-null red blood cell samples.
Methods
The Colton-null phenotype was investigated by standard serological techniques, AQP1 sequencing, immunoblot and flow cytometry analyses.
Results
We identified and characterized the Colton-null phenotype in a Gypsy woman who developed an anti-CO3 during her first pregnancy. After developing a simple and robust method to sequence AQP1, we showed that she was apparently homozygous for a new AQP1 null allele, AQP1 601delG, whose product is not expressed in her red blood cells. We also established the Colton specificity of three commercially available anti-AQP1s in immunoblot and/or flow cytometry analyses.
Conclusion
This Gypsy woman represents the sixth Colton-null case characterized at the serological, genetic and biochemical levels. The validation here of new reagents and methods should facilitate the identification of Colton-null individuals.
Keywords: Blood groups, Colton system, AQP1, null mutation
Introduction
The Colton blood group system (ISBT Number 015) currently comprises four antigens: CO1 (aka Coa, present in 99.8% of the population [1]), the antithetical antigen CO2 (aka Cob, present in 8.5% of the population [1]), and two high-prevalence antigens, CO3 and CO4 (described hereafter in detail). The Colton antigens are carried by the water channel aquaporin 1 (AQP1), which is highly expressed in the membrane of red blood cells (RBCs) and also carries the ABH antigens (see Figure 1D). Expression of the CO2 antigen results from a single nucleotide polymorphism (SNP) in the AQP1 gene (NM_198098:c.134C>T; rs28362692) causing an alanine to valine substitution at amino acid position 45 (NP_932766:p.Ala45Val) in the first extracellular loop of AQP1 (see Figure 1D). Of note, the rs28362692 SNP is systematically analyzed on current blood group genotyping platforms (e.g. BioArray HEA BeadChip™ and Progenika BLOODchip®). The CO3 antigen, whose exact nature remains unknown, has been found expressed in all tested individuals except those who have inherited two null alleles of AQP1 [2]. In fact, these individuals express no Colton antigens and are commonly called Colton-null (Conull). The recently described CO4 antigen [3] is also a high-prevalence Colton antigen, which is not only absent in Conull individuals (CO:-1,-2,-3,-4) but also in CO:-1,-2,3,-4 individuals, who do express AQP1 on their RBCs despite their CO:-1,-2 phenotype. The expression of the CO4 antigen seems to depend on glutamine 47 since the CO:-1,-2,3,-4 phenotype has been found in two unrelated individuals who were apparently homozygous for the allele AQP1 140G encoding a functional water channel AQP1 with a glutamine to arginine substitution at amino acid position 47 [3].
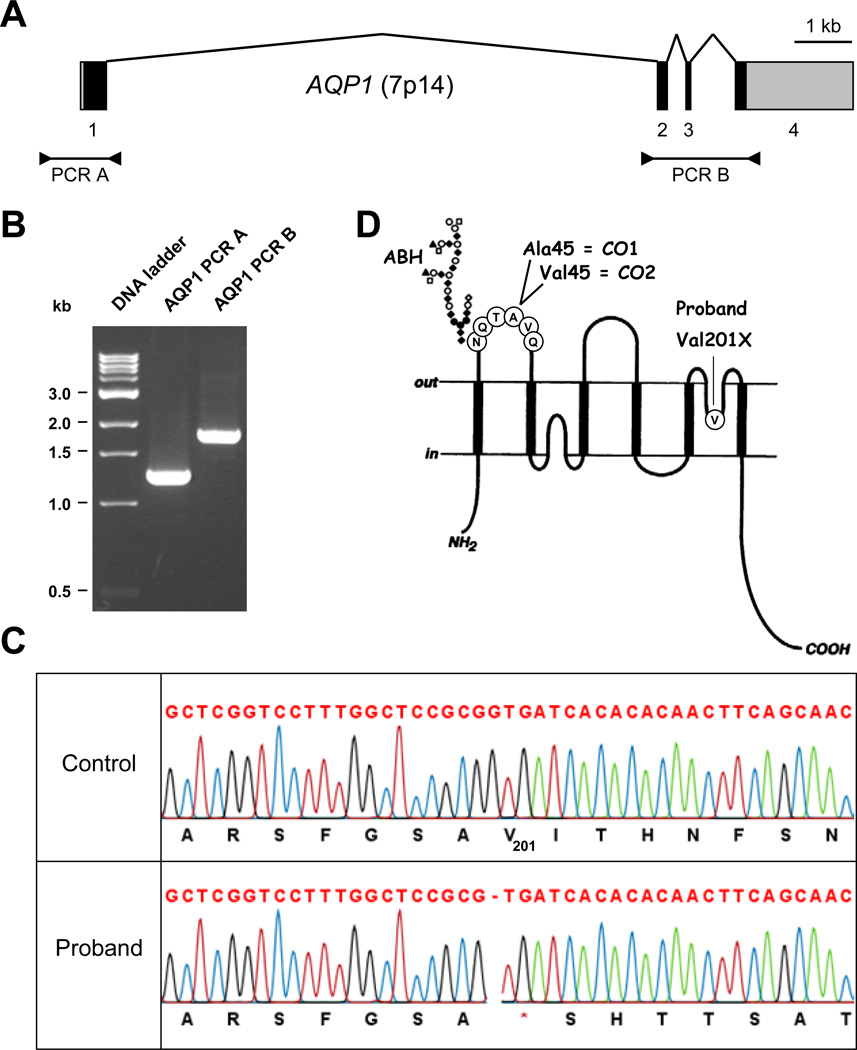
Figure 1. Identification of an AQP1 mutation in the proband.
(A) Diagram showing the structure of AQP1 and the two fragments that were PCR-amplified for sequencing; exons are depicted as boxes (coding sequences are in black and untranslated regions in gray) and introns as broken lines; PCR products are depicted below.
(B) PCR products of the two AQP1 fragments used for sequencing and analyzed using a 1.5% agarose gel; PCR A contains the AQP1 proximal promoter and exon 1, while PCR B contains the rest of the AQP1 coding sequence (exons 2 to 4).
(C) Detail of AQP1 sequencing from the proband (lower traces) and a control (upper traces) showing the apparently homozygous mutation c.601delG, p.Val201X in the proband.
(D) Diagram showing the membrane topology of AQP1 and the localization of ABH, CO1 and CO2 antigens (adapted from [8]).
The Conull phenotype is extremely rare worldwide and its genetic basis has been published for only five unrelated cases, who were all apparently homozygous for private mutations in AQP1 [2, 4, 5]. Despite the postulated essential role of AQP1, the Conull individuals described so far exhibit no clinical consequences. Nevertheless, AQP1 deficiency may result in an impaired response to physiological stress. Indeed, King and colleagues showed a decreased ability to maximally concentrate urine after more than 20 hours of water deprivation, as well as a decreased pulmonary vascular permeability after serial i.v. injections of 1 liter of saline solution, in two Conull individuals [6, 7]. More importantly, AQP1 deficiency may result in the development of a clinically significant alloantibody (anti-CO3) induced by blood transfusion or pregnancy [1]. In fact, the Conull phenotype is usually unveiled by the identification of an anti-CO3, when this phenotype is not identified during a targeted study in the family of a Conull proband. However, identifying anti-CO3 may be difficult since reference Conull RBCs are missing in most laboratories. Moreover, some of the RBCs labeled “Conull” due to their CO:-1,-2 phenotype have not been fully investigated and may actually correspond to CO:-1,-2,3,-4 RBCs. Altogether, these circumstances make the transfusion support of Conull patients highly challenging.
While the Conull phenotype has apparently no clinical consequences, it is worth mentioning that the lack of Colton blood group antigens has been observed in two blood disorders. The RBCs of patients with congenital dyserythropoietic anemia type IV (CDAIV) are completely deficient in AQP1 [8, 9] due to a dominant mutation in the erythroid transcription factor KLF1 [9], which governs the expression of AQP1 in the erythroid lineage. Of note, the RBCs of CDAIV patients are also deficient in CD44 [9, 10]. Some patients with monosomy 7 in acute myeloid leukemia (AML) or myelodisplastic syndrome (MDS) also lack the Colton antigens [11, 12]. However, in these cases the causative genetic mechanism is still unknown and most likely independent of the location of AQP1 on chromosome 7.
We report here a new case of the Conull phenotype unveiled by an anti-CO3 induced by a first pregnancy. We have characterized this new case at the genetic and biochemical levels. Furthermore, while we show the complete AQP1 deficiency in the proband’s RBCs, we have also validated several commercially available AQP1 antibodies in order to facilitate future identifications of this extremely rare blood group phenotype.
Patient, materials and methods
Proband
The proband was an 18-year-old woman from a Gypsy community settled near Marseilles, France. The proband self-declared that she was primipara. Her extended RBC phenotype was: ABO:-1,-2,-3; RH:1,2,-3,-4,5; KEL:-1,2,-3,4; FY:1,2; JK:1,2; MNS:1,-2,-3,4; P1PK:1; LU:-1,2; GE:2; YT:1; VEL:1. The proband was not followed for possible AML or pre-leukemia; she had a normal blood count and apparently suffered no diseases. The work carried out is consistent with the consent given by the proband and was performed within the ethics guidelines of the maternity hospital, the regional blood center, the French National Reference Center for Blood Groups, and the French National Institute of Blood Transfusion.
Blood samples
RBC samples from the Conull subject SAR (proband 2 in [2]), the Conull subject CHA (SAR’s sister) and blood donors typed CO:1,-2, CO:1,2 and CO:-1,2 were from the frozen blood collection of the National Reference Center for Blood Groups (Paris, France). Reference anti-CO2 (serum GAM), anti-CO3 (serum SAR) and anti-CO4 (serum TOR) were also from the frozen antibody collection of the National Reference Center for Blood Groups (Paris, France).
Serological testing
Antibody identification and RBC typing were performed by indirect antiglobulin gel-test (ID-Micro Typing System Coombs Anti-IgG, DiaMed) with in-house panels of untreated, trypsin- or papain-treated RBCs according to the manufacturer’s instructions.
AQP1 sequencing
Blood genomic DNA was isolated using the Wizard Genomic DNA Purification Kit (Promega). The primers used in this study are described in Table 1. PCR fragment A (Figure 1A) was amplified and sequenced with primers AQP1-35 and AQP1-31, while PCR fragment B (Figure 1A) was amplified with primers AQP1-11 and AQP1-9, and sequenced with primers AQP1-12 and AQP1-7 (of note, AQP1-9 can’t be used for sequencing given the frequent deletion/insertion variation rs72463314). PCR amplification was performed in 20 µL reactions containing 100 ng of genomic DNA, 1X Advantage GC-Melt Buffer (Clontech), 4 pmoles of each primer (Eurofins MWG Operon), 5 nmoles of each dNTP (GE Healthcare) and 1U of Advantage GC Genomic LA Polymerase Mix (Clontech), in a 2720 Thermal Cycler (Applied Biosystems) with the following program: 2 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 57 °C, 2 min at 72 °C, and finally 5 min at 72 °C. PCR products were quality-checked by 1.5% agarose gel electrophoresis in 1X TBE buffer then sequenced with ABI BigDye terminator chemistry after ExoSAP treatment (GATC Biotech). Sequencing data analysis was performed using DNA Workbench software (CLC bio). Mutations were screened by unidirectional sequencing and confirmed by bidirectional sequencing.
Table 1.
Primers used in this study
| Name | Sequence | Locationa | Directionb | Positiona |
|---|---|---|---|---|
| AQP1-7 | TCTCCTACCTGCCTCCATC | Intron 3 | Sense | 16469–16487 |
| AQP1-9 | CTTGGGGAAGTGACTTTGG | Exon 4 | Antisense | 16877–16895 |
| AQP1-11 | GCCTGATTTCCACTACCTGC | Intron 1 | Sense | 15036–15055 |
| AQP1-12 | GCCGTGTAGGCTTGCCTG | Intron 1 | Sense | 15054–15071 |
| AQP1-31 | CCTCCAGCAACCTCTTGTC | Intron 1 | Antisense | 5547–5565 |
| AQP1-35 | GTCGAGAAGTTTGGGAGC | Promoter | Sense | 4275–4292 |
Location and position of the primers based on NCBI AQP1 genomic Reference Sequence NG_007475.1.
Direction of the primers compared to the AQP1 transcript.
AQP1 antibodies
The polyclonal goat AQP1 antibody (anti-AQP1(T-13), Santa Cruz Biotechnology) was produced and affinity-purified against a proprietary peptide (AQP1(T-13)P) which officially maps between amino acid (aa) 100 and 150 of AQP1 but whose mass spectrometry analysis indicates that it corresponds to aa 121 to 133 of AQP1 (details upon request). The polyclonal rabbit AQP1 antibody (AB2219, Millipore) was produced and affinity purified against a 19 aa peptide from the C-terminus of AQP1. The monoclonal mouse AQP1 antibody (1/A5F6, originally provided by Prof. P. Nemeth and later purchased from Thermo Scientific and then from AbD Serotec) was produced against a 20 aa peptide corresponding to the C-terminus of AQP1 [13].
Immunoblot analysis
RBC membranes were prepared at 0–4 °C by hypotonic lysis with 5P8 buffer (5 mM Na2HPO4 pH8.0 and 350 µM EDTA pH8.0) supplemented with 1 mM AEBSF, and solubilized with an equal volume of 4X LDS Sample Buffer (Invitrogen). Equal amounts of RBC membrane lysates were resolved by Tris-Glycine 12 % SDS-PAGE and transferred to PolyScreen PVDF Transfer Membrane (Perkin Elmer) by submarine transfer. The Mark12 Unstained Standard (Invitrogen) was used as a molecular weights reference. For immunoblot analysis with the goat AQP1 antibody anti-AQP1(T-13), membranes were blocked in 1X Animal-Free Blocker (Vector Laboratories) overnight at 4 °C and then incubated for 2 h at 21 °C in the same buffer with anti-AQP1(T-13) (33 ng/mL) which was detected with a donkey anti-goat IgG-HRP (1:5,000; Santa Cruz Biotechnology). These membranes were re-probed with the mouse monoclonal anti-GAPDH 6C5 (50 ng/mL; Millipore). For immunoblot analysis with the rabbit AQP1 antibody AB2219, membranes were blocked in 1X Blocking Buffer (Sigma) overnight at 4 °C and then incubated for 2 h at 21 °C in the same buffer with AB2219 (1 ng/mL) which was detected with an anti-rabbit IgG(H+L) horseradish-peroxidase-linked goat antibody (1:5,000; P.A.R.I.S Biotech). These membranes were re-probed with the mouse monoclonal anti-dematin 18 (2 ng/mL; Santa Cruz Biotechnology). For immunoblot analysis with the mouse AQP1 antibody 1/A5F6, membranes were blocked in 1X Blocking Buffer (Sigma) overnight at 4 °C and then incubated for 2 h at 21 °C in the same buffer with 1/A5F6 (166 ng/mL) which was revealed with an anti-mouse IgG(H+L) horseradish-peroxidase-linked goat antibody (1:5,000; P.A.R.I.S Biotech). These membranes were re-probed with a rabbit serum raised against p55 (1:100,000; [14]).
Flow cytometry analysis
For intracellular staining of AQP1, RBCs were extensively washed with D-PBS (Invitrogen), fixed 15 min at 21 °C in fixation solution (1 % formaldehyde, 0.025 % glutaraldehyde in D-PBS), washed twice with D-PBS, permeabilized 15 min at 21 °C in permeabilization solution (1 % n-Octyl-β-D-glucopyranoside in D-PBS), washed twice with D-PBS, blocked 30 min at 21 °C in blocking solution (1 % bovine serum albumin, 20 % normal goat serum in D-PBS) and incubated overnight at 4 °C with the mouse monoclonal AQP1 antibody 1/A5F6 (5 µg/mL; AdD Serotec) or the isotype control MOPC-31C (5 µg/mL; BD Pharmingen) in D-PBS supplemented with 1 % bovine serum albumin and 2 % normal goat serum. For extracellular staining of CD44, RBCs were extensively washed with D-PBS, and incubated for 1 h at 21 °C with the mouse monoclonal CD44 antibody 515 (1 µg/mL; BD Pharmingen) or the isotype control MOPC-31C (1 µg/mL; BD Pharmingen) in D-PBS supplemented with 0.15 % bovine serum albumin. Staining of AQP1 and CD44 was revealed with goat F(ab’)2 anti-mouse IgG(H+L)-PE (1:200; Beckman Coulter) and analyzed with a FACSCanto II flow cytometer (BD Bioscience) equipped with FACSDiva software (v. 6.1.2) (BD Bioscience). Ten thousand RBCs, gated on forward scatter (FSC) vs. side scatter (SSC), were collected for each sample. Data were further analyzed with FlowJo software (v. 7.2.5) (TreeStar).
Results
Identification of an anti-CO3 in a primipara Gypsy woman
An alloantibody against a high prevalence RBC antigen was detected in a Gypsy woman of Spanish origin at the 34th week of her first pregnancy. Her serum reacted with all tested RBCs (more than 30 RBCs including one sample of the following rare phenotypes: VEL:-1, YT:-1, 2, LU:-1,2, MNS:-3,-4,-5 and RH:-1,-2,-3,-4,-5) but not her own RBCs. Of note, her serum exhibited a stronger reactivity with papain-treated (4+), trypsin-treated (3+) than non-treated RBCs (2+). Furthermore, the reactivity of her serum (titer 8, score 19 by indirect antiglobulin gel test on non-treated RBCs) was not altered by DTT treatment suggesting that her alloantibody was an IgG isotype.
The proband’s RBCs reacted with sera containing antibodies against the following high-prevalence antigens: DI2, DO4, IN2, KEL7, GE2, LU2, MNS5, YT1, SC1, VEL1, Lan and Jra. In contrast, her RBCs did not react with two anti-CO1. Further investigations indicated her RBCs also did not react with anti-CO2, anti-CO3 and anti-CO4, suggesting that the proband was a new Conull subject and had developed an anti-CO3. Her serum did react with five CO:-1,2 RBCs, as well as CO:1,-2 RBCs, but not with RBCs of two previously characterized Conull subjects (SAR and CHA), which confirmed that her alloantibody was anti-CO3. The potential existence of common underlying alloantibodies was ruled out by differential warm allogeneic adsorption of the proband’s serum on three selected donor RBCs.
The proband had an uneventful pregnancy and delivered a nonhydropic male child at full term. The newborn’s RBCs had a positive direct anti-IgG test (3+) but a negative direct anti-C3d test. At one day of age, the newborn had a normal blood count (e.g. Hb 191 g/L) and was allowed to go home five days later.
Identification of a new AQP1 allele encoding AQP1 Val201X
In order to identify the mutation responsible for the Conull phenotype of the proband, we sequenced the coding regions, the exon-intron boundaries and the proximal promoter of the AQP1 gene, which spans approximately 14 kb at 7p14 (Figure 1A). For this purpose, two gDNA fragments were amplified by PCR (Figure 1B) and subsequently sequenced by a Sanger method. The proband’s AQP1 gene harbored a single nucleotide deletion in exon 3 (c.601delG) leading to an immediate stop codon (p.Val201X) (Figure 1C). No other difference was found in the proband’s AQP1 gene when compared with the reference genomic sequence NG_007475, which corresponds to the AQP1 allele CO*01 encoding the CO1 antigen. The proband was apparently homozygous for this new AQP1 allele, AQP1 601delG, consistent with the likely consanguinity of her parents. We could not confirm this hypothesis because her parents and siblings refused to give blood samples.
Confirmation by immunoblot analysis that no AQP1 is expressed on the proband’s RBCs
The allele AQP1 601delG potentially encodes an AQP1 protein with a 68 amino acid C-terminal truncation (AQP1 Val201X). We wanted to determine whether this truncated protein was expressed or not in the RBC membrane. For this purpose, we looked for an AQP1 antibody able to recognize AQP1 Val201X. To our knowledge, the only commercially available AQP1 antibody directed against an epitope present in AQP1 Val201X was a goat polyclonal antibody that had been produced against a peptide corresponding to amino acids 121 to 133 of APQ1 (anti-AQP1(T-13), see Materials for details). We used this goat antibody to probe blots of RBC membrane lysates prepared from the proband and control donors, as well as a previously characterized Conull subject (SAR, who was apparently homozygous for the AQP1 mutation c.308_309insT, p.Gly104fsX24; first reported by Preston and colleagues [2] and reanalysed in this study). As shown in Figure 2A, this goat antibody strongly detected a polypeptide from control RBC membrane lysates at approximately 28 kDa, which corresponds to the theoretical molecular mass of AQP1 (lanes 1 and 4) but nothing from the Conull subject SAR (lane 2) or the proband (lane 3). Not only does this result establish the AQP1 specificity of this goat antibody in immunoblot analysis, but it also demonstrates that AQP1 Val201X is not expressed in the RBC membrane (AQP1 Val201X has a theoretical molecular mass of 21 kDa). Thus, AQP1 601delG is a new AQP1 null allele and the Conull phenotype of the proband was ultimately established.
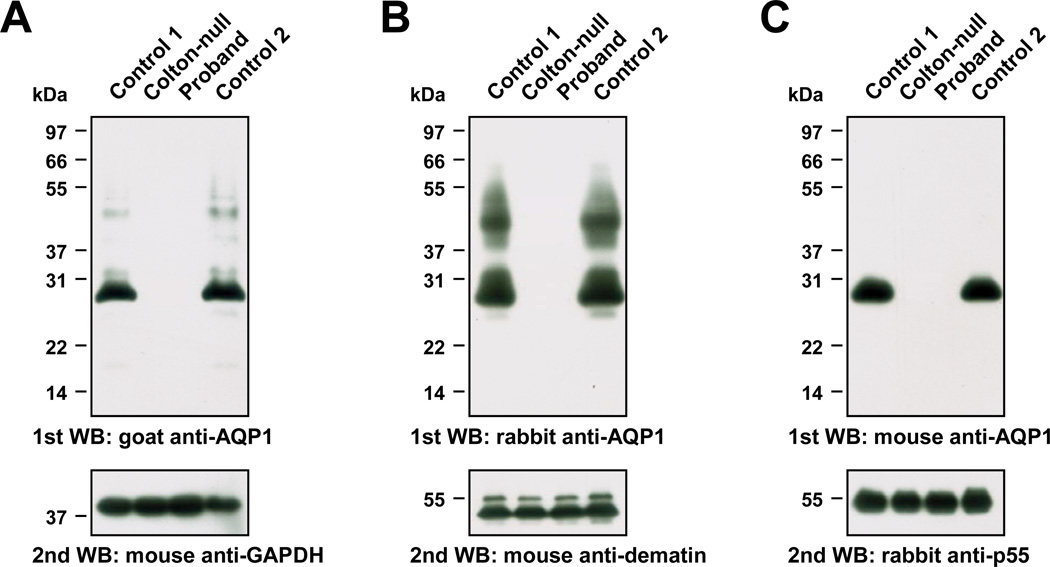
Figure 2. Immunoblot analysis of AQP1 from RBC membrane lysates prepared from the proband.
Equal amounts of membrane lysates prepared from the RBCs of two control donors (lanes 1 and 4), a previously characterized Conull subject (SAR, lane 2) and the proband (lane 3) were resolved by Tris-Glycine 12% SDS-PAGE electrophoresis. Samples were not reduced or heat-denatured prior to electrophoresis, and were transferred to PVDF membrane for immunoblot analysis.
(A) PVDF membrane probed with goat anti-AQP1 (upper panel) and re-probed with mouse anti-GAPDH (lower panel).
(B) PVDF membrane probed with rabbit anti-AQP1 (upper panel; the smear migrating between 37 and 55 kDa corresponds to glycosylated AQP1) and re-probed with mouse anti-dematin (lower panel; erythroid dematin is expressed as two isoforms produced by alternative splicing).
(C) PVDF membrane probed with mouse anti-AQP1 (upper panel) and re-probed with rabbit anti-p55 (lower panel).
Having RBC membranes from two unrelated Conull subjects (the proband and SAR), which constituted a unique opportunity to validate the specificity of any AQP1 antibody, we decided to test two other commercially available AQP1 antibodies, one rabbit polyclonal (AB2219) and one mouse monoclonal (1/A5F6). Both antibodies were raised against a peptide in the intracellular C-terminus of AQP1 (see Materials for details), as most commercially available AQP1 antibodies. While each of these antibodies showed an unambiguous AQP1 specificity in immunoblot analysis (Figure 2B and 2C), only the rabbit polyclonal was able to recognize glycosylated AQP1, which migrates as a smear between 37 and 55 kDa [2]. This was unexpected since both AQP1 antibodies recognized an epitope in the intracellular C-terminus of AQP1, and this difference of reactivity between the mouse and rabbit AQP1 antibodies is currently unexplained. Of note, the goat AQP1 antibody barely detected glycosylated AQP1 (Figure 2A).
Flow cytometry assay to determine the expression of AQP1 in RBCs
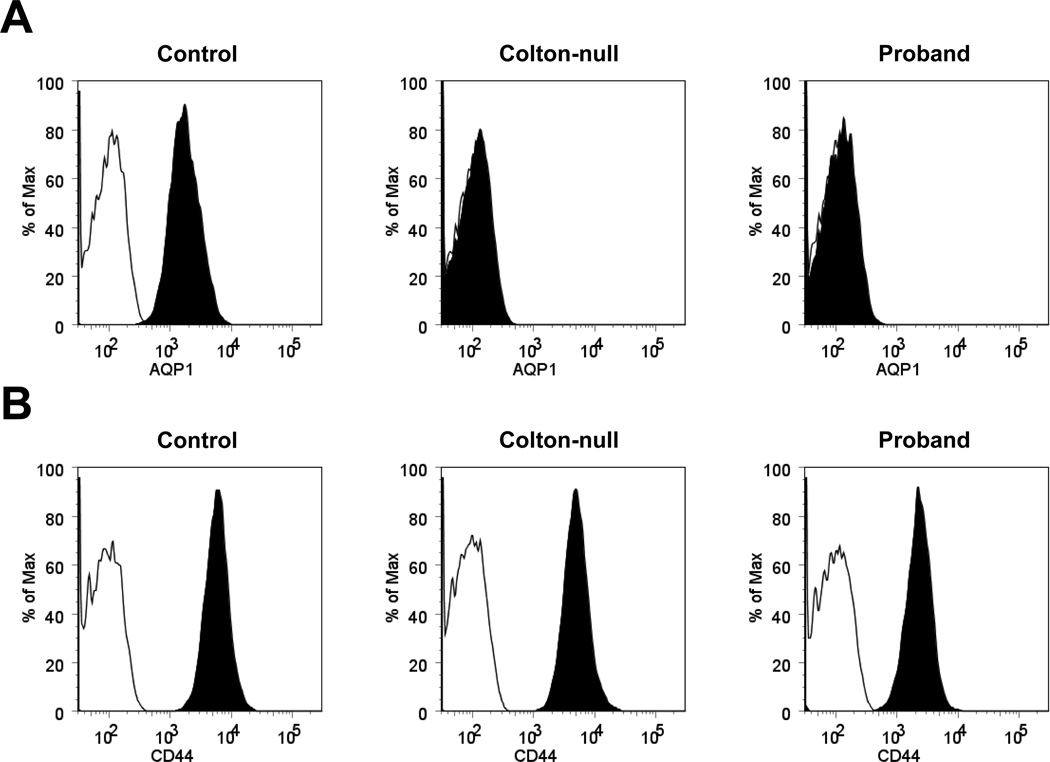
Finally, we decided to develop a flow cytometry assay for AQP1 expression in RBCs in order to facilitate the detection of Conull subjects. Since no AQP1 antibody allowed the extracellular labeling of AQP1, we performed its intracellular labeling in fixed and permeabilized RBCs by using the mouse monoclonal AQP1 antibody 1/A5F6. As shown in Figure 3A, the 1/A5F6 antibody exhibited the same AQP1 specificity in flow cytometry analysis, i.e. producing no signal in Conull samples, as in immunoblot analysis (Fig. 2C). This result demonstrated that AQP1 expression in RBCs can be tested by flow cytometry, which is a technique more rapid and accessible than immunoblot analysis in immunohematology laboratories. Actually, we used flow cytometry to confirm that the AQP1-deficient RBCs of the two Conull subjects studied here (the proband and SAR) were not also deficient in CD44 (Figure 3B) in contrast with the RBCs of patients with CDAIV [9, 10].
Figure 3. Flow cytometry analysis of AQP1 and CD44 in the proband’s RBCs.
Intracellular staining of AQP1 (A) and extracellular staining of CD44 (B) in the RBCs of a control donor (left overlays), a previously characterized Conull subject (SAR, middle overlays) and the proband (right overlays) analyzed by flow cytometry; black histograms correspond to the staining with anti-AQP1 or anti-CD44, and white histograms to the isotype control.
Discussion
We describe a new AQP1 null allele, AQP1 601delG, found in a Gypsy woman who had developed an anti-CO3 during her first pregnancy. The mutation c.601delG results in an immediate stop codon in the reading frame, thus mimicking a nonsense mutation (p.Val201X). We further show that the corresponding truncated water channel protein is not expressed at the RBC membrane, which is fully consistent with the Conull phenotype of this woman. The absence of AQP1 Val201X at the RBC membrane may result from its mislocalization, as previously observed for AQP1 P38L and AQP1 N192K [3] that are also encoded by AQP1 null alleles [2, 4]. Alternatively, and more likely, is the targeting of the mutant mRNA to the nonsense-mediated decay pathway since the “nonsense” mutation c.601delG occurs in the penultimate exon of AQP1 gene [15].
Few cases of the Conull phenotype and even less of anti-CO3 induced by pregnancy have been reported so that this report is important for the pregnancy follow-up in Conull women. The proband has developed an anti-CO3, albeit with a low titer, during her first pregnancy, which suggests that AQP1 is highly immunogenic. Consistent with its low titer, her anti-CO3 was not associated with hemolytic disease of the fetus and newborn (HDFN). However, severe HDFN may be expected in any future pregnancies of the proband [16, 17] who should be monitored accordingly.
This study also highlights the difficulty to identify anti-CO3 as well as the Conull phenotype. Even though we had access to previously-characterized anti-CO3 and Conull RBCs, which is not the case of most laboratories, the identification of anti-CO3 in the proband’s serum was reagent- and time-consuming. Therefore, we decided to validate new antibodies and methods that would dectect complete AQP1 deficiency of RBCs; the absence of AQP1 is indeed the ultimate proof of the Conull phenotype. Historically, AQP1 deficiency of RBCs was evaluated by measuring their water permeability with a stopped-flow spectrophotometer [2, 18]. However, this equipment is not available to most laboratories. We have validated three commercially available AQP1 antibodies to check the expression of AQP1 in RBC membrane by immunoblot analysis. Furthermore, we have developed a flow cytometry assay for AQP1 expression in RBCs with the mouse monoclonal antibody 1/A5F6. Altogether, these methods should greatly facilitate the identification of Conull samples.
We also provide a robust and simple method to sequence AQP1. While the Colton blood group is routinely analyzed on current blood group genotyping platforms, it should be kept in mind that only the SNP corresponding to the CO1/CO2 polymorphism is determined. This methodology would neither have allowed the identification of the new AQP1 null allele characterized here, nor the previously published AQP1 null allele with a single nucleotide mutation (i.e. c.308_309insT, p.Gly104fsX24 [2]; c.113C>T, p.Pro38Leu [2]; c.576C>A, p.Asn192Lys [4]; c.232delG, p.Ala78fsX42 [5]; c.112C>T, p.Pro38Ser [19]). Therefore, AQP1 sequencing is essential when there is a phenotype/genotype discrepancy in the Colton blood group system. Furthermore, we suggest to sequence AQP1 in all registered Conull individuals who have not been fully investigated since they may actually be CO:-1,-2,3,-4 [3] and lead to erroneous conclusions in immunohematological studies.
Acknowledgments
The authors thank Virginie Helias, Sandrine Genetet, Sylvain Bigot, Joëlle Nataf and Eliane Vera (National Institute of Blood Transfusion, Paris, France) for their help. This study was supported in part by the National Institute of Blood Transfusion (INTS), the National Institute for Health and Medical Research (INSERM) and Paris Diderot University (Paris 7). B.A.B. and K.A.S. were funded by the Vermont Genetics Network through NIH/NCRR grant P20 RR16462. C.S., C.L. and S.A.S. acquired and analyzed the data; T.P., B.A.B., I.D. and L.A. analyzed and interpreted the data; C.C. and P.N. provided essential materials; L.A. drafted the paper, which was revised critically by J.-P.C., T.P. and B.A.B. All authors approved the submitted version.
Footnotes
Competing interests
The authors have no competing financial interests.
References
- 1.Daniels G. Colton blood group system; Human Blood Groups. Blackwell Science Ltd; 2002. pp. 398–403. [Google Scholar]
- 2.Preston GM, Smith BL, Zeidel ML, Moulds JJ, Agre P. Mutations in aquaporin-1 in phenotypically normal humans without functional CHIP water channels. Science. 1994;265:1585–1587. doi: 10.1126/science.7521540. [DOI] [PubMed] [Google Scholar]
- 3.Arnaud L, Helias V, Menanteau C, Peyrard T, Lucien N, Ripoche P, Lapegue R, Pham BN, Le Pennec PY, Moulds JJ, Cartron JP. A functional AQP1 allele producing a Co(a-b-) phenotype revises and extends the Colton blood group system. Transfusion. 2010;50:2106–2116. doi: 10.1111/j.1537-2995.2010.02687.x. [DOI] [PubMed] [Google Scholar]
- 4.Chrétien S, Cartron JP. A single mutation inside the NPA motif of aquaporin-1 found in a Colton-null phenotype. Blood. 1999;93:4021–4023. [PubMed] [Google Scholar]
- 5.Joshi SR, Wagner FF, Vasantha K, Panjwani SR, Flegel WA. An AQP1 null allele in an Indian woman with Co(a-b-) phenotype and high-titer anti-Co3 associated with mild HDN. Transfusion. 2001;41:1273–1278. doi: 10.1046/j.1537-2995.2001.41101273.x. [DOI] [PubMed] [Google Scholar]
- 6.King LS, Choi M, Fernandez PC, Cartron JP, Agre P. Defective urinary-concentrating ability due to a complete deficiency of aquaporin-1. N Engl J Med. 2001;345:175–179. doi: 10.1056/NEJM200107193450304. [DOI] [PubMed] [Google Scholar]
- 7.King LS, Nielsen S, Agre P, Brown RH. Decreased pulmonary vascular permeability in aquaporin-1-null humans. Proc Natl Acad Sci U S A. 2002;99:1059–1063. doi: 10.1073/pnas.022626499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agre P, Smith BL, Baumgarten R, Preston GM, Pressman E, Wilson P, Illum N, Anstee DJ, Lande MB, Zeidel ML. Human red cell Aquaporin CHIP. II. Expression during normal fetal development and in a novel form of congenital dyserythropoietic anemia. J Clin Invest. 1994;94:1050–1058. doi: 10.1172/JCI117419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, Prehu C, Foliguet B, Montout L, de Brevern AG, Francina A, Ripoche P, Fenneteau O, Da Costa L, Peyrard T, Coghlan G, Illum N, Birgens H, Tamary H, Iolascon A, Delaunay J, Tchernia G, Cartron JP. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am J Hum Genet. 2010;87:721–727. doi: 10.1016/j.ajhg.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons SF, Jones J, Anstee DJ, Judson PA, Gardner B, Wiener E, Poole J, Illum N, Wickramasinghe SN. A novel form of congenital dyserythropoietic anemia associated with deficiency of erythroid CD44 and a unique blood group phenotype [In(a-b-), Co(a-b-)] Blood. 1994;83:860–868. [PubMed] [Google Scholar]
- 11.De la Chapelle A, Vuopio P, Sanger R, Teesdale P. The Colton blood groups in monosomy 7 of the bone marrow. Birth Defects Orig Artic Ser. 1976;12:280–283. [PubMed] [Google Scholar]
- 12.Pasquali F, Bernasconi P, Casalone R, Fraccaro M, Bernasconi C, Lazzarino M, Morra E, Alessandrino EP, Marchi MA, Sanger R. Pathogenetic significance of "pure" monosomy 7 in myeloproliferative disorders. Analysis of 14 cases. Hum Genet. 1982;62:40–51. doi: 10.1007/BF00295602. [DOI] [PubMed] [Google Scholar]
- 13.Nagy G, Szekeres G, Kvell K, Berki T, Nemeth P. Development and characterisation of a monoclonal antibody family against aquaporin 1 (AQP1) and aquaporin 4 (AQP4) Pathol Oncol Res. 2002;8:115–124. doi: 10.1007/BF03033720. [DOI] [PubMed] [Google Scholar]
- 14.Arnaud L, Salachas F, Lucien N, Maisonobe T, Le Pennec PY, Babinet J, Cartron JP. Identification and characterization of a novel XK splice site mutation in a patient with McLeod syndrome. Transfusion. 2009;49:479–484. doi: 10.1111/j.1537-2995.2008.02003.x. [DOI] [PubMed] [Google Scholar]
- 15.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Lacey PA, Robinson J, Collins ML, Bailey DG, Evans CC, Moulds JJ, Daniels GL. Studies on the blood of a Co (a-b-) proposita and her family. Transfusion. 1987;27:268–271. doi: 10.1046/j.1537-2995.1987.27387235637.x. [DOI] [PubMed] [Google Scholar]
- 17.Savona-Ventura C, Grech ES, Zieba A. Anti-Co3 and severe hemolytic disease of the newborn. Obstet Gynecol. 1989;73:870–872. [PubMed] [Google Scholar]
- 18.Mathai JC, Mori S, Smith BL, Preston GM, Mohandas N, Collins M, van Zijl PC, Zeidel ML, Agre P. Functional analysis of aquaporin-1 deficient red cells. The Colton-null phenotype. J Biol Chem. 1996;271:1309–1313. doi: 10.1074/jbc.271.3.1309. [DOI] [PubMed] [Google Scholar]
- 19.Karpasitou K, Frison S, Longhi E, Drago F, Lopa R, Truglio F, Marini M, Bresciani S, Scalamogna M, Poli F. A silenced allele in the Colton blood group system. Vox Sang. 2010;99:158–162. doi: 10.1111/j.1423-0410.2010.01332.x. [DOI] [PubMed] [Google Scholar]