Abstract
Three insertion elements were previously found in a family of germ line-limited mobile elements, the Tlr elements, in the ciliate Tetrahymena. Each of the insertions contains an open reading frame (ORF). Sequence analysis of the deduced proteins encoded by the elements suggests that they are homing endonucleases. The genes are designated TIE1-1, TIE2-1, and TIE3-1 for Tetrahymena insertion-homing endonuclease. The endonuclease motif occupies the amino terminal half of each TIE protein. The C-terminal regions of the proteins are similar to the APETELA2 DNA binding domain of plant transcription factors. The TIE1 and TIE3 elements belong to families of repeated sequences in the germ line micronuclear genome. Comparison of the genes and the deduced proteins they encode suggests that there are at least two distinct families of homing endonuclease genes, each of which appears to be preferentially associated with a specific region of the Tlr elements. The TIE1 and TIE3 elements and their cognates undergo programmed elimination from the developing somatic macronucleus of Tetrahymena. The possible role of homing endonuclease-like genes in the DNA breakage step in developmentally programmed DNA elimination in Tetrahymena is discussed.
Homing endonucleases are encoded by insertion elements. These elements are most often introns or inteins but can also be freestanding elements (14). The endonucleases make a double- or single-stranded cut in the DNA of a homologous allele lacking the element. The break in the DNA initiates the transposition of the element into the empty allele, a process known as homing (5). Transposition of the element occurs via a gene conversion event that is completed by the DNA repair systems of the host organism.
Homing endonucleases are site specific. Their recognition sites, at 12 to 40 bp, are much longer than those of most restriction enzymes. This limits the range of potential homing sites. The homing endonucleases are also distinct from restriction enzymes in that they can tolerate changes of a few base pairs in the recognition site. This provides flexibility such that the element can sometimes home into a site that has been slightly modified by mutation.
Homing endonucleases have been classified into four groups, based on the structures of the catalytic domains (5). The most widespread and well studied of these enzymes have one or more copies of the LAGLIDADG motif. The structures of several of these enzymes, bound to their target sites, have been solved (9, 17, 20, 26). They bind as dimers or pseudodimers to their substrates, and in a few cases the catalytic mechanisms of DNA cleavage are understood in detail. The second group of homing endonucleases are characterized by the motif GIY(X10-11)YIG. These are monomeric enzymes that cleave the DNA at a distance from the intron insertion site. The third class, the His-Cys box enzymes, are characterized by conservation of histidine and cysteine residues over a stretch of 100 amino acids. The least understood class of enzymes are the HNH (or in some cases, HNN) endonucleases. The catalytic sites of these enzymes contain histidine and asparagine residues with characteristic spacing over a range of 30 to 33 residues.
Homing endonucleases are widespread in nature. They have been described in all of the biological kingdoms and are found in nuclear genomes, in the genomes of organelles, and in various bacteriophages and eukaryotic viruses (14). Homing endonucleases are thought to be so successful because they are usually associated with elements such as introns and inteins. Thus, they are generally removed before a functional protein is produced and, therefore, do not reduce the fitness of the host organism. We describe here three putative homing endonuclease genes in Tetrahymena thermophila, TIE1-1, TIE2-1, and TIE3-1 (for Tetrahymena insertion-homing endonuclease). The genes encode deduced proteins with HNH endonuclease domains and APETELA2 (AP2) DNA binding domains. To the best of our knowledge, this is the first description of homing endonuclease genes in a ciliate.
The first three elements containing homing endonuclease genes were discovered as insertions in a family of germ line-limited elements. Tetrahymena have two nuclei: a germ line micronucleus and a transcriptionally active somatic macronucleus. During sexual reproduction the parental macronucleus is degraded, and a new macronucleus develops from a mitotic product of the zygotic micronucleus. The development of the new macronucleus involves extensive DNA rearrangement, resulting in the deletion of ∼6,000 specific DNA elements from the macronuclear genome (32). The deleted elements are termed internal eliminated sequences (IES). Most, but not all, of the IES are repeated in the micronuclear genome (33), and several of the characterized IES in Tetrahymena and other ciliates resemble mobile genetic elements of various types (1, 4, 8, 12, 21). One family of IES that has been well characterized in Tetrahymena is the Tlr (for Tetrahymena long repeat) element family. This is a family of about 30 putative mobile genetic elements residing in the micronuclear genome, all of which are eliminated during development of the macronuclear genome. Tlr family members have a highly conserved inner core of about 22 kb, containing 15 open reading frames (ORFs), and long, complex, terminal inverted repeats (29). We show here that there are at least two distinct families of TIE elements in Tetrahymena, each of which is preferentially associated with a specific site within the Tlr elements. Like the Tlr elements that contain them, the TIE elements are germ line-limited.
MATERIALS AND METHODS
DNA isolation and sequencing.
The isolation and sequencing of micronuclear genomic clones was described previously (29). Micronuclear and macronuclear DNA for Southern blots were isolated from strain CU428 according to the method of Gorovsky et al. (15).
Colony screening.
For duplicate colony screening, SURE cells (Stratagene) were transformed with an aliquot of the pMBR plasmid library (24) and colony lifts were done as described previously (29). The filters were placed face up on a Luria-Bertani agar plate containing 10 μg of chloramphenicol per ml, and a second filter was placed on top. The filter sandwich was incubated at 37°C overnight and autoclaved for 2 min to lyse the cells and denature the DNA. The duplicate filters were separated, and the DNA was UV cross-linked to the filter.
Probes and primers.
Probes for Southern hybridization and colony hybridization were made by PCR amplification with plasmid DNA as the substrate. The primers for TIE1-1 amplification from pMBR8E1 (GenBank accession no. AF451862) were 5′-TAGGATGAGATAGATGATTTAGAAG-3′ and 5′-CTCCTTTGTAAATTGATGAAGTTG-3′. A TIE3-1 probe was amplified from pMBR8 (GenBank accession no. AF451865) with primers 5′-ATTTGGATTAGGAGATTTGGGTTG-3′ and 5′-TTCGTTATATTTTTAGGCTGCTTC-3′. The probe for the Tlr region at the TIE1-1 insertion site was amplified from pMBR4C1 (GenBank accession no. AF451860) with primers 5′-CATTGGTATTCTGGTTAGTTCAGCGT-3′ and 5′-CCACTGTACAACCAAGATATT-3′. Probes were labeled by random priming (Roche Molecular), and the hybridization of Southern blots and colony filters were performed as described previously (29).
Sequence analysis.
Sequence data for additional TIE family members was obtained from the Institute for Genomic Research website at http://tigrBLAST.tigr.org/er-blast/index.cgi?project=ttg. The ratio of base pair changes between different members of a TIE family that produced synonymous versus nonsynonymous base changes was assessed by using the SNAP program at http://www.hiv.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html.
To assess the degree of amino acid conservation between putative proteins encoded by members of the TIE1 and TIE3 families, amino acid substitutions were categorized as identical, similar, or dissimilar. Similarity was defined according to the structure-genetic matrix scoring system (11), where amino acid pairs are assigned a number from 0 to 6 based on the structural similarity and likelihood of interchanges. A score of 6 indicates identity. For the purposes of this analysis (see Tables 4 and 5), similarity was defined as a structure-genetic matrix score of 4 or 5.
TABLE 4.
Nucleotide and amino acid substitutions among the TIE3 open reading frames
| Degree of amino acid substitution | No. of nucleotide changes by position
|
||
|---|---|---|---|
| First | Second | Third | |
| Identical | 3 | 1 | 21 |
| Similar | 8 | 2 | 6 |
| Dissimilar | 10 | 11 | 5 |
| Total | 21 | 14 | 32 |
TABLE 5.
Nucleotide and amino acid substitutions among the TIE1 open reading frames
| Nature of amino acid substitution | No. of nucleotide changes by position
|
||
|---|---|---|---|
| First | Second | Third | |
| Identical | 6 | 1 | 32 |
| Similar | 22 | 13 | 19 |
| Dissimilar | 13 | 23 | 7 |
| Total | 41 | 37 | 58 |
Sequence reads from clones in the TIGR database containing empty TIE1 insertion sites were identified by a BLASTn search with bp 4501 to 4800 of clone pMBR4C1 (GenBank accession no. AF451860). Sequences with the left end of TIE1 elements inserted at the homologous site in the Tlr elements were found by searching with bp 1 to 300 of pMBR8E1 (GenBank accession no. AF451862). A BLASTn search with the TIE1-1 ORF (bp 520 to 1254 of clone pMBR8E1) produced matches to clones with flanking sequences on the right end of the TIE1 elements. Clones with empty TIE3 sites were identified in a BLASTn search by using Tlr Int B bp 6868 to 7364 (GenBank accession no. AF232243) as the query sequence. Clones covering the left and right ends of TIE3 elements were identified by using bp 3938 to 4337 and 4137 to 4706 of clone pMBR8, respectively, as the query sequences. DNA sequences at TIE insertions sites were aligned by using the MultAlin program with the DNA-5-0 symbol comparison table (6).
RESULTS
Insertion elements encode HNH endonucleases.
In a chromosome walk through the Tlr family, clones of the various family members were found to be colinear and 90 to 97% conserved at the nucleotide level, with the notable exception of three apparent insertion elements. One of the insertions was ∼1,200 bp in length and the other two were ∼600 bp (29) (Fig. 1).
FIG. 1.
Insertion elements in the Tlr clones. A physical map of the Tlr elements, drawn as a composite of sequences from nine genomic clones. Clones containing the TIE element insertions and homologous clones lacking the TIE elements are shown below the map of the Tlr family. The Tlr Int clones and the pMBR clones were described previously (13, 29).
ORF analysis revealed a single ORF in each of the three insertion elements. The ORFs encode deduced proteins of 244, 181, and 189 amino acids, respectively. CLUSTAL W (27) alignment indicated that the three conceptual proteins are ∼31% identical and 60% similar to each other over the most conserved stretch of 169 amino acid residues.
The conceptual proteins encoded by the three insertion elements are all more similar to each other than to any other proteins in the nonredundant database, with expect values ranging from 7e-28 to 2e-21. For each insertion element, the next most significant alignments generated in a BLASTp search were with bacteriophage homing endonucleases, where the highest expect values ranged from e-13 to e-11. The genes encoding the putative Tetrahymena endonucleases were designated TIE1-1, TIE2-1, and TIE3-1, for Tetrahymena insertion-homing endonuclease.
The endonuclease domains of the TIE proteins reside in the N-terminal regions of the proteins. A match of Tie1-1p to the HNH domain was detected in the conserved domain database routine of the BLASTp search. Figure 2 shows the alignment of the three Tetrahymena proteins, Tie1-1p, Tie2-1p, and Tie3-1p with seven endonucleases listed in the Pfam (protein family database) analysis (2) of the HNH domain. Amino acids that are similar in at least five of the first eight proteins are shown in color. (Tie2-1p and Tie3-1p were not included in this first assessment of similarity, so as not to bias the analysis in favor of the Tetrahymena proteins). The most common amino acid at each position is shown in red, and similar amino acids are in blue. A consensus domain derived from the alignment is shown above the amino acid alignment. The region designated as the HNH domain in the Pfam analysis is underlined. Thus, the N-terminal ∼60% of each Tie protein apparently contains the HNH homing endonuclease domain. Three additional regions of striking similarity, designated motifs I, II, and III, become apparent after alignment of the proteins with two or three gaps at conserved positions.
FIG. 2.
Alignment of the partial sequence of the deduced proteins encoded by TIE1-1, TIE2-1, and TIE3-1 with the amino acid sequences of seven viral homing endonucleases, two proteins containing the AP2 domains, and the AP2 consensus sequence. Red, identical residues in the first eight proteins; blue, conserved residues; green, the AP2 DNA binding domain consensus sequence and identical sequences in the last five proteins. A consensus sequence for the N-terminal region is above the sequence alignment in red. Uppercase letters designate amino acids that are invariant among these proteins. The HNH domain as designated by the Pfam program is underlined. Motifs I, II, and III are additional motifs that are conserved among the TIE elements and the bacteriophage endonucleases. In motif II amino acids that may be either positively or negatively charged are indicated (±). The sequences of the termini of several of the proteins, including the N termini of Tie1-1p and Tie2-1p were omitted from the figure where they do not align with consensus domains. Accession numbers: Bacillus subtilis SPO1, P34081; Lactococcus lactis phage r1t, AAB18716; B. subtilis, CAB13895; phage SP82, AAA56884; lactococcal bacteriophage bIL170, AAC27218; Bacteroides thetaiotaomicron VPI-5482, NP_810939; bacteriophage phi-E, U04813; TIE1-1, AF451862; TIE2-1 and TIE3-1, AF451865; Atriplex hortensis AP2 domain protein, AAF76898; A. thaliana AP2 domain protein, NP_177931.
AP2 DNA binding domains.
The Pfam analysis in the same BLASTp search identified a second conserved domain in the Tie proteins. The C-terminal region of all three putative proteins showed matches to AP2 domains (expect values of 3e-08 to 8e-06). AP2 domains of ∼60 amino acids are the DNA binding domains found in hundreds of plant transcription factors (22, 23). The AP2 consensus domain from the SMART program (25) is shown in green below the proteins aligned in Fig. 2. Two of the AP2 domain-containing proteins identified in the BLASTp search, one from Arabidopsis thaliana and one from Atriplex hortensis, were aligned with the consensus domain and with the Tetrahymena proteins. Residues found in at least three of the sequences are shown in green and similar amino acids are in blue. The C-terminal regions of all three Tie proteins have significant similarity to the AP2 consensus domain and to the AP2 domain-containing proteins.
It was surprising to find putative AP2 domains in the Tetrahymena endonucleases because this domain was previously thought to be limited to the plant kingdom (28). One possibility is that the elements encoding these endonucleases entered Tetrahymena by lateral transmission from plants. The GC content and codon usage of the TIE genes were examined as a measure of how recently these genes might have invaded Tetrahymena. First, the TIE genes have a relatively high AT content characteristic of the overall Tetrahymena genome (AT content of 75%). For example, the TIE1-1 gene has a GC content of 22%. In contrast, the Bacteriodes thetaiotamicron VPI5482 gene (the closest match among the homing endonuclease genes) has a GC content of 49%, and the AP2 domain gene from Atriplex hortensis has a GC content of 48%. This suggests that the introduction of the homing endonuclease genes into Tetrahymena was not a recent event.
A more definitive argument can be made from an analysis of codon usage. Several ciliates have unusual codon usage. In Tetrahymena, the canonical stop codons TAA and TAG encode glutamine (16, 18). In TIE1-1, TIE2-1 and TIE3-1, 14 of 17, 12 of 14, and 8 of 9 glutamine codons, respectively, are TAA or TAG. Thus, the TIE genes have resided in the Tetrahymena genome for sufficient time to allow for adaptation to Tetrahymena codon usage.
The alignments in Fig. 2 suggest that the catalytic and DNA binding functions of the Tetrahymena endonucleases are present in separate domains. This has been demonstrated to be the case for I-TevI, a homing endonuclease encoded by the phage T4 td intron. In I-TevI, the catalytic and DNA binding domains are separated by a flexible, protease-sensitive hinge (7). Eight of the 26 amino acid residues in this hinge region are basic (K or R). In the proteins encoded by the TIE genes, the regions designated as the catalytic (HNH) and DNA binding (AP2) domains in each protein are separated by 8 to 9 amino acid residues, 3 to 4 of which are basic. Although I-TevI belongs to a different family of homing endonucleases, in which the catalytic site is characterized by a GIY-YIG motif, it is possible that the Tetrahymena endonucleases may share a common architecture with I-TevI, in which distinct catalytic and DNA binding domains are separated by a basic region.
TIE sequences repeated in the micronucleus are germ line-limited.
TIE1-1, TIE2-1, and TIE3-1 were identified as insertions in two of the cloned fragments of the Tlr elements of Tetrahymena (29). That is, some Tlr elements contain TIE elements and the homologous site in other members of the Tlr family are empty (Fig. 1). Southern blot analysis was done in order to determine whether TIE elements were repeated in the Tetrahymena genome. Figure 3A shows hybridization of the TIE1-1 probe to micronuclear and macronuclear DNA digested with HindIII or XbaI. Multiple fragments were observed in the lanes containing micronuclear DNA. Since there are no HindIII or XbaI sites in the TIE1-1 gene, at least some of these fragments are likely to represent different members of a family of TIE1-1 sequences in the genome.
FIG. 3.
TIE elements are repeated in the micronuclear genome and are germ line limited. Southern blots of genomic DNA probed with the TIE elements TIE1-1 (A) and TIE3-1 (B). Open circles indicate the expected positions of bands if the TIE3 probe had cross-hybridized to TIE2-1 sequences. The sizes of the hybridizing fragments were determined relative to Ho-Lo DNA marker (Minnesota Molecular). Mi, micronuclear DNA; Ma, macronuclear DNA; p8E1, plasmid pMBR8E1 DNA; p8, plasmid pMBR8 DNA; H, HindIII; E, EcoRV; X, XbaI.
There was considerable variation in the intensity of the bands in the Southern blot. One possible explanation for this observation is that sequence differences between the different family members resulted in differences in the strength of the hybridization signal. Another possibility is that some bands contain fragments from two or more family members. Restriction sites are somewhat conserved between members of the Tlr family (29). If the various members of the TIE1 family are located within Tlr elements and the restriction sites in the surrounding Tlr DNA are conserved, then fragments from two or more TIE1 family members would comigrate in the Southern blot. Thus, the data suggest that TIE1 sequences are repeated in the micronuclear genome, but the copy number cannot be determined from this experiment.
Plasmid DNAs pMBR8E1 and pMBR8 (Fig. 1) were blotted to the filter as positive and negative controls for TIE1 hybridization. Major fragments of 6 kb (HindIII) and 5.4 kb (XbaI) were present in the lanes containing pMBR8E1 DNA, as expected since this Tlr clone contains the TIE1 element. (The faint band in lane 7 is the size expected for cross-hybridization of the probe to vector sequences on the plasmid.) In contrast, the TIE1-1 probe did not cross-hybridize to the TIE2 or TIE3 sequences in pMBR8 (lanes 8 and 9). Thus, the TIE2-1 and TIE3-1 genes were not represented among the fragments detected in lanes 1 and 3 containing micronuclear DNA hybridized with the TIE1-1 probe.
Similar results were obtained with a probe derived from TIE3-1 (Fig. 3B). The probe hybridized to multiple fragments in the micronuclear DNA. Fragments of the expected sizes were detected in lanes containing plasmid DNA digested with EcoRV, XbaI, and HindIII, corresponding to the fragments of pMBR8 known to include TIE3 sequences. In contrast, no bands were detected at the mobility expected for pMBR8 restriction fragments containing TIE2, nor was there any hybridization to TIE1-1 sequences in lanes 4 to 6, containing pMBR8E1, even after overexposure of the blot (data not shown).
No hybridization of the TIE1-1 or TIE3-1 probes was detected in macronuclear DNA, despite the fact that ethidium staining of the gels showed there was an equal amount, or slightly more, DNA in these lanes than in the micronuclear DNA lanes (data not shown). Thus, all copies of the TIE1 and TIE3 families are eliminated from the genome during development of the macronucleus. This finding is consistent with a model in which the TIE elements are exclusively associated with Tlr elements. According to this model, TIE elements might be deleted along with the Tlr elements containing them. Alternatively, they might be deleted as independent IES.
TIE1 elements are preferentially associated with Tlr elements.
Colony screening was performed in order to determine whether the family of sequences that hybridize to TIE1-1 are generally found within Tlr elements, or whether they are present in other regions of the genome as well. Duplicate colony lifts were made of cells transformed with plasmid clones from a library of micronuclear DNA (24). One copy of each filter was hybridized with a probe specific for TIE1-1, and the duplicate filter was hybridized with a probe for the surrounding Tlr sequences (Fig. 4). A total of 108 colonies from 10 different filters hybridized only to the Tlr probe. An additional 74 colonies that hybridized to the Tlr probe also hybridized to the TIE1-1 probe. There were no colonies that contained TIE1 sequences, but not Tlr sequences. The data suggest that the multiple copies of TIE1 sequences in the Tetrahymena genome are preferentially associated with the homologous region of the Tlr elements.
FIG. 4.
Probes for screening duplicate colony lifts. Thin line, micronuclear DNA in plasmids pMBR8E1 (the plasmid with the TIE1-1 insertion) and pMBR4C1 (a plasmid covering the same region of the Tlr elements but lacking the TIE1-1 insertion); heavy line, TIE1-1 insertion; angled arrow, TIE1-1 ORF; open box, TIE1-1 probe; gray boxes, Tlr probe obtained by PCR amplification of pMBR4C1 and the corresponding regions on the pMBR8E1 plasmid.
Sequence diversity of the TIE element genes and homing endonucleases.
TIE1-1, TIE2-1, and TIE3-1 are significantly different from one another in nucleotide sequence. Approximately 500 bp of the most highly conserved nucleotide sequence from the three elements were compared by the LALIGN program (19). These included the sequences encoding the proteins from the tryptophan codon at the beginning of motif I to the ends of the ORFs encoded by TIE1 and TIE2, omitting the sequences encoding the extended N terminus of TIE1-1 and the glutamic acid-rich tail in the TIE3 ORF (Fig. 2). (Hereinafter this will be referred to as the conserved region of the endonuclease genes or proteins.) Even in this conserved region, there was only ∼60% nucleotide identity between the various TIE elements (Table 1). This was consistent with the observation that these elements did not cross-hybridize with one another on Southern blots under conditions of normal stringency (Fig. 3).
TABLE 1.
Percent nucleotide identity between conserved regions of the TIE element open reading frames
| TIE element | TIE1-2 | TIE1-3 | TIE2-1 | TIE3-1 | TIE3-2 | TIE3-3 |
|---|---|---|---|---|---|---|
| TIE1-1 | 88 | 87 | 62 | 61 | 63 | 63 |
| TIE1-2 | 87 | 60 | 60 | 61 | 63 | |
| TIE2-1 | 57 | 57 | 56 | |||
| TIE3-1 | 96 | 87 |
The blot shown in Fig. 3A suggests that TIE1-1 belongs to a family of homing endonuclease genes that are more similar to each other than they are to the genes encoded by TIE2 and TIE3 elements. Similarly, TIE3-1 apparently belongs to a family of genes that are more similar to each other than they are to TIE1-1 or TIE2-1 (Fig. 3B). After the colony screening was completed, a preliminary version of the Tetrahymena genome project became available. Although the genome project was designed to obtain only macronuclear DNA sequences, a number of micronucleus-limited sequences were also present in the database. To identify additional TIE elements, BLASTn searches of the Tetrahymena genome database were performed by using the conserved regions of TIE1-1, TIE2-1, and TIE3-1 as the query sequences.
The sequence that was the first hit with TIE1-1 was 100% identical to the query sequence. It is likely that this was a clone of the TIE1-1 element. The next fifteen hits had a high degree of similarity to TIE1-1 (expect values of >e-40). Six of these were analyzed in detail. Five of the six were located in Tlr elements, as judged by the criterion that they contained at least 100 bp of sequence with high similarity to pMBR4C1. pMBR4C1 is a partial clone of a Tlr family member that overlaps the region of the Tlr clone pMBR8E1 containing the TIE1-1 insertion (Fig. 1 and 5). The sixth also appeared to be located in the same region of the Tlr element, but the clone did not extend far enough to confirm the similarity to pMBR4C1. These data are consistent with the colony screening in suggesting that most or all members of the TIE1 family are inserted at homologous positions within the Tlr elements. The nucleotide sequences in the ORFs of the five confirmed TIE1 clones were 87 to 89% identical to TIE1-1, and 84 to 90% identical to each other (Table 1).
FIG. 5.
Alignment of the TIE sequence reads from the Tetrahymena genome project. (A) Alignment of plasmid pMBR4C1 with plasmid pMBR8E1 (29), containing the TIE1-1 insertion and four reads from clones sequenced as part of the Tetrahymena genome project. Box, TIE1-1 insertion element; angled arrows, TIE1 ORFs, with the initiation codon represented by a vertical bar and the stop codon indicated by the arrowhead; black boxes above the clone, regions of homology with pMBR8E1; gray boxes below the clone, homology with pMBR4C1. (B) Alignment of plasmid Tlr Int C (13) with the homologous region of plasmid pMBR8 (29), containing the TIE3-1 insertion and with four reads from clones in the Tetrahymena genome project. For the TIE3-1 insertion element and TIE3 ORFs, the format corresponds to that of panel A. Black boxes, homology to pMBR8; gray boxes, homology to Tlr Int C. For both panels, letters to the right are the plasmid name or the identification number for the read in the Tetrahymena genome project database.
The first five hits in the search with the conserved region of TIE3-1 as a query sequence contained sequences encoding ORFs, or partial ORFs, that were 84 to 96% identical in nucleotide sequence to TIE3-1. These were identified as coming from the homologous site in the Tlr elements on the basis of having at least 100 bp of homology with the pMBR8 clone in a region outside the TIE3-1 element, or with the overlapping Tlr clone Tlr Int C (Fig. 1 and 5). Besides the similarity in nucleotide sequence, the clones were similar to each other in the structure of the deduced proteins they encoded. Three of the clones included the region encoding the C terminus of the proteins, and the conceptual proteins encoded by all three had C-terminal glutamate repeats, similar to those in the deduced TIE3-1 protein (Fig. 2). The consensus sequence for the C termini of Tie3-1p and the three additional members of the TIE3 family was E5-12D(Q/E)E3 (Table 2).
TABLE 2.
3′ Termini of Tie3p deduced proteins
| Protein | Sequence |
|---|---|
| TIE3-1 | FAKINFPREDYQE9 DQE3 |
| TIE3-2 | FAKINFPREDYQE5 DQE3 |
| TIE3-3 | FAKLNFPIEDYQE12DQE3 |
| TIE3-6 | YAKLNFPREDYQE5 DEE3 |
The search using TIE2-1 as a query produced only one sequence that appeared to be homologous to TIE2-1. The partial ORF of TIE2-2 (346 bp) was 83% identical in nucleotide sequence to the homologous region of TIE2-1. The expect values for additional hits in the search with TIE2-1 dropped dramatically from e-48 to e-26. Six of the next seven hits were to members of the TIE1 family, and the seventh did not have sufficient flanking sequence to determine whether it was associated with Tlr elements.
In summary, the nucleotide sequences of various members of the TIE1 and TIE3 families are identical in 87% or more of the nucleotides. In contrast, nucleotide identities between members of different families are on the order of only 60 to 62%. (Selected data are shown in Table 1.) This finding supports the hypothesis that there are at least two distinct families of homing endonuclease genes in Tetrahymena.
The concept of distinct families of TIE elements is supported at the protein level. Sequences containing family members with the complete ORF, or most of the ORF, were chosen for comparison (Table 3). In order to provide comparable alignments between and among families, all proteins were compared in the conserved region of ∼169 amino acids. Different proteins within the Tie1p family were identical in ∼77% of the amino acids, and proteins within the Tie3p family were identical in ∼85% or more of the amino acid sequence, while proteins in different families were only identical in ∼47% of the amino acids.
TABLE 3.
Percent amino acid identity between homologous regions of the TIE element open reading frames
| TIE element | Tie1-2 | Tie1-3 | Tie3-1 | Tie3-2 | Tie3-3 |
|---|---|---|---|---|---|
| Tie1-1 | 78 | 78 | 47 | 47 | 46 |
| Tie1-2 | 76 | 48 | 48 | 48 | |
| Tie3-1 | 99 | 85 | |||
| Tie3-2 | 85 |
The computational analysis was consistent with the Southern hybridization and colony hybridization data. All three data sets support a model of at least two, perhaps three, or even more distinct TIE families in the Tetrahymena genome. TIE1 and TIE3 elements are preferentially associated with Tlr elements, and various members of each TIE family are inserted at homologous positions within some, but not all, members of the Tlr family.
The 5′ region of the TIE1 elements.
TIE1-1, at 1,203 bp in length, is considerably larger that TIE2-1 (622 bp) and TIE3-1 (618 bp). This difference in length is due to the fact that TIE1-1 has a much longer region 5′ to the TIE1-1 ORF. Conceptual translation of the region 5′ to the TIE1-1 ORF on the complementary strand produces a polypeptide fragment of a homing endonuclease gene with similarity to amino acids 57 to 167 of the Tie1p element. The region of similarity covers most of the HNN domain, including motif I (Fig. 2) but ending two amino acids after the first asparagine.
Computer analysis was performed to determine whether this gene fragment was an unusual feature of the TIE1-1 element, or if it was a common feature of all insertion elements at the TIE1 site in Tlr elements.
The collection of sequence reads from the TIGR database was searched with 167 bp of Tlr DNA flanking the TIE1-1 insertion site. This search produced five distinguishable sequences that did not contain TIE elements and five sequences with TIE insertions containing the region homologous to the 5′ end of the TIE1-1 element. No examples were found of a TIE element lacking the 5′ region of TIE1-1. Thus, all elements inserted into Tlr DNA at the TIE1-1 site contain the extended region 5′ to the TIE1-1 ORF.
The five sequences of the 5′ region were 93 to 95% identical to each other at the nucleotide level; thus, they represent a family of closely related sequences. However, the conceptual peptide fragments encoded by these sequences are distinct from the deduced proteins encoded by the ORFs in the TIE elements. In fact, the closest match in a BLASTp search using the peptide fragment as the query sequence was to the phage SP82 endonuclease, with an expect value of 8e-08. The second match was to the TIE1-1 ORF, with an expect value of only 3e-06. The conceptual peptide fragments encoded by the 5′ end of the TIE1 element has 33% identity with 57% similarity to the corresponding region of Tie1-1p and 35% identity with 59% similarity to the corresponding region of Tie2-1p.
Tlr sequences at the site of TIE insertions.
Elements encoding homing endonucleases are most often introns or inteins but sometimes exist as free-standing elements (14). The Tlr sequences surrounding the TIE elements were examined to determine the nature of the Tlr sequences at the point of TIE element insertion. Clones containing empty TIE insertion sites and clones covering the left and right ends of the TIE1 and TIE3 families of elements were identified in BLASTn searches of the sequence reads from the TIGR database. Homologous regions from the clones were aligned by using the MultAlin program (see Materials and Methods for details).
TIE1 elements are inserted into intergenic Tlr DNA, 839 bp downstream of the ORF encoding 7Rp and 57 bp upstream of the ORF encoding 8Rp (Fig. 1). The DNA in this region has a low GC content of 15.8%, characteristic of intergenic DNA (30), and the TIE1 insertions lack consensus RNA splice sites at the termini and thus do not appear to be introns. The five elements depicted in Fig. 5 were found using the TIE1-1 ORF as the query sequence. Thus, there was no bias in the search for the orientation of the element. Nonetheless, all five insertions are in the same orientation, with the TIE1 ORFs on the opposite strand from the 7Rp and 8Rp ORFs (Fig. 5).
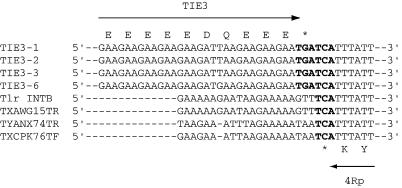
Similar to the TIE1 insertions, all of the TIE3 insertions were in the same orientation in the Tlr element, with the TIE3 ORFs on the opposite strand from the 4Rp and 5Rp ORFs of the Tlr element (Fig. 1 and 5). The 3′ ends of four TIE3 elements are aligned with four sequences containing empty TIE3 sites in Fig. 6. The GAA repeats that encode the polyglutamic acid repeats at the C termini of the TIE3 deduced proteins extend into the Tlr element. That is, the DNA encoding the last six amino acids of the TIE3 elements and the TIE3 stop codons are located within the Tlr element. The stop codons of the TIE3 ORFs abut the stop codon of the 4Rp ORF on the opposite strand (Fig. 6). Although the five empty site sequences are A rich in this region, they do not have pronounced GAA repeats, and none of them have TGA at the position of the TIE3 stop codon.
FIG. 6.
The TIE3 ORFs extend into the Tlr elements. The sequences of four clones containing the 3′ end of the TIE3 elements were aligned with the corresponding sequence from Tlr Int B and three additional empty site clones from the TIGR database. The single letter consensus code for the Tie3p deduced proteins is above the alignment, and the last two amino acids of the deduced protein Tlr4Rp are below. Stop codons are in bold letters.
Functionality of the TIE genes.
The data presented here show that the TIE ORFs encode deduced proteins with similarity to homing endonucleases. If these ORFs encode polypeptides, they might be expected to show evidence of evolutionary pressure to maintain a functional protein. First, TIE element families were examined to determine whether the reading frames were open. The first family examined was the TIE3 element family. TIE3-1, TIE3-2, and TIE3-3 all have complete ORFs.
Chi-square (χ2) analysis was done to determine whether there was a bias among the three ORFs in the position of the nucleotide changes within the codon. Since nucleotide changes in the third position of the codon are most likely to produce identical amino acids, a preponderance of nucleotide changes in the third position suggests that selective pressure has acted on the genes to maintain functional proteins. A consensus sequence was determined for the three TIE3 genes and the positions of departures from the consensus sequence were noted (Table 4). Chi-square analysis suggested that nucleotide changes were nonrandom with respect to position within the codons (P < 0.025). This is evidence that at some point the TIE3 ORFs were under selective pressure to encode functional proteins.
It appears that the TIE genes encode chimeric proteins with an N-terminal HNH homing endonuclease domain and a C-terminal AP2-like DNA binding domain. SNAP analysis was done in order to determine the positions of the synonymous versus nonsynonymous codon changes along the length of the protein. The analysis showed a sharp increase in the relative number of nonsynonymous codons in the C-terminal region of the protein. Thus, it appears that the homing endonuclease domain was under more stringent selection than the AP2 domain.
A similar analysis was done for the TIE1 family (Table 5). In this case, only the TIE1-1 element contained a complete ORF. The reading frames of the other four elements shown in Fig. 4 were open but incomplete. Nucleotide sequences that were present in TIE1-1, TIE1-2, and TIE1-3 ORFs were compared by χ2 analysis. Codons encoding motif I and most of motif II were omitted from the analysis because they were not present in one or more of the sequences. The analysis did not support the hypothesis that there was a statistically significant bias in the position of nucleotide changes within the codons (0.1 > P > 0.05). However, SNAP analysis of the TIE1 ORFs was consistent with the results from the TIE3 ORFs in that nonsynonymous codons accumulated much more rapidly in the C-terminal region of the protein (data not shown). Thus, it may be that the χ2 analysis of the TIE1 elements did not provide statistically significant evidence for the conservation of the proteins because the N termini of the deduced TIE1 proteins, which are likely to contain the most conserved region, were not available for analysis.
DISCUSSION
Three insertion elements (TIE1-3 elements) were found within the members of the germ line-limited Tlr family of putative mobile genetic elements in the ciliate Tetrahymena. BLASTp analysis suggests that they encode chimeric proteins. The N-terminal portion of each gene has similarity to homing endonucleases encoded by bacteriophages, and the C-terminal regions contain putative AP2 DNA binding domains.
For both the TIE1 and TIE3 families, all of the elements examined had ORFs. The TIE3 elements displayed a bias in the position of nucleotide changes within the codons that suggested the ORFs were at some point under selective pressure to encode functional proteins. Although the analysis of the ORFs from TIE1 elements did not show a statistically significant bias in the position of modified codons, the region that was likely to show the highest level of amino acid conservation was not included in the analysis. It should be noted that these TIE elements were extracted from the database based on their similarity in nucleotide sequence. Once an element has become nonfunctional, it would be expected to diverge more rapidly than active elements that are under functional constraints. There was no bias toward functionality in TIE1-1, TIE2-1, and TIE3-1 since they were discovered as insertions in the Tlr elements. However, if the query sequences happened to be active genes, the search would be biased toward functional elements. Nonetheless, the data suggest that at some point these elements were under selection to produce functional proteins, thus supporting the hypothesis that the ORFs encoded proteins that were active at some point. Whether these are currently active homing elements is unknown.
An analysis of the sequence reads from the Tetrahymena genome project, at both the nucleotide and predicted protein levels, supports the hypothesis that the TIE1 elements and the TIE3 elements represent different families. Besides the differences in sequence in the conserved region of the ORFs, the Tie1p proteins are characterized by an extended amino terminus, and the Tie3p family is distinguished by C termini containing variable lengths of polyglutamate. The data suggest that the two families may have been founded by independent invasion events. Although the TIE2 elements appear to represent a third family, the available data are too limited to confirm that hypothesis.
The TIE1 elements all contain a partial ORF of another HNH homing endonuclease gene in the region 5′ to the TIE1 ORFs, on the opposite strand. The sequences containing the partial ORF are highly conserved in different TIE1 elements, but the conceptual peptides are significantly different from the corresponding region encoded by the TIE1 ORFs. This may suggest that a progenitor TIE1 element homed into a different homing endonuclease gene. This composite element may have subsequently transposed as a unit or may have been inactivated so far as independent transposition goes.
The TIE elements were found within a larger family of germ line-limited elements, the Tlr elements. Colony hybridization of the TIE1 elements and computational analysis of TIE1 and TIE3 sequences were consistent and suggested that the multiple TIE elements within a family were all located at homologous positions within the Tlr elements in the micronuclear genome. Estimates from Southern analysis are that the Tlr elements are repeated ∼30-fold in the micronuclear genome. Colony hybridization suggested that ∼30 to 40% of the Tlr elements had a TIE1 element at a position similar to that of TIE1-1 in pMBR8E1. This could have arisen in at least three ways. First, there were multiple invasions of the Tetrahymena genome by TIE1 elements, all of which inserted at homologous positions in the Tlr elements. This seems unlikely, particularly since the TIE1 sites are all occupied by complex elements that seem to have resulted from the insertion of one TIE element into another. Second, the TIE1 elements may be able to home into repeated targets at nonallelic loci. Third, the invasion of the first TIE1 element occurred soon after the appearance of the Tlr element, such that transposition of the Tlr elements resulted in a concomitant proliferation of the TIE1 elements within them.
The TIE3 elements contain variable numbers of GAA repeats at the 3′ ends, which encode polyglutamate tails at the C terminus of the deduced endonucleases. In principle, these might be generated by polymerase slippage during insertion of the element or by unequal crossing over during the homologous recombination that presumably facilitates homing of the TIE elements. In any case, the extension of the GAA repeats into the Tlr flanking sequences might represent an interesting mechanism through which homing endonucleases could evolve new functions. For example, if the TIE3 element were to home into a coding region, the result might be the generation of a fusion protein with a new function. Such a model might explain how the TIE HNH domains became associated with AP2 domains.
The endonuclease activity of elements containing homing endonucleases is expected to promote a high frequency of the lateral transmission of elements containing these genes (14). The lateral transmission of genes from plants to Tetrahymena might be expected, since Tetrahymena inhabit fresh water ponds and since decaying plant matter is a major food source for the ciliate in the wild. Codon usage analysis indicates that, if the endonucleases were transferred from plants to Tetrahymena, the time elapsed since the supposed invasion of the TIE elements must have been sufficient to allow for the mutations necessary to bring them into conformance to the Tetrahymena genetic code.
In some cases, homing endonucleases have evolved into proteins with functions that contribute to the host biology. The most striking example of this is the HO gene that initiates mating type switching in yeast (14). Fan and Yao have suggested that an enzyme involved in developmentally programmed chromosome breakage in Tetrahymena may be evolutionarily related to homing endonucleases (10). During development of the new macronucleus in Tetrahymena, the five germ line chromosomes are broken at ∼50 to 200 specific sites to produce the macronuclear chromosomes. A 15-bp chromosome breakage sequence (CBS) at these sites (34) has been shown to be necessary and sufficient for chromosome breakage (31). CBSs have two features in common with the target sites of homing endonucleases. First, their length is reminiscent of the long homing endonuclease recognition sites, which range from 12 to 40 bp (3). Second, the enzymes that produce chromosome breakage are similar to homing endonucleases in that they can tolerate a small degree of variation in the CBS (10). In fact, about 40% of the naturally occurring CBSs vary by a nucleotide or two from the canonical CBS sequence (E. Hamilton, S. Williamson, S. Dunn, D. Cassidy-Hanley, and E. Orias, personal communication). It is unlikely that the homing endonucleases encoded by the TIE elements are the same as the enzyme that recognizes the CBS, because homing does not occur at the CBS cleavage sites. However, codon usage in the TIE genes suggests that they have been present in the Tlr elements for a substantial period of time, perhaps long enough to allow for the evolution of a cognate protein to assume the function of chromosomal breakage.
Acknowledgments
This work was supported by grant MCB-9974885 from the National Science Foundation. K.M.K. was supported in part by a Summer Faculty Fellowship from Marquette University. J.D.W. was supported in part by fellowships from the Arthur J. Schmitt Foundation, the Richard W. Jobling Trust Fund, the Scholl Foundation, and the Marquette University Graduate School. P.R.L and A.E.M were supported by supplemental REU grants from the National Science Foundation.
Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. The authors thank the anonymous reviewer for very helpful suggestions.
REFERENCES
- 1.Baird, S. E., G. M. Fino, S. L. Tausta, and L. A. Klobutcher. 1989. Micronuclear genome organization in Euplotes crassus: a transposon-like element is removed during macronuclear development. Mol. Cell. Biol. 9:3793-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belfort, M., and R. J. Roberts. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherry, J. M., and E. H. Blackburn. 1985. The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell 43:747-758. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier, B. S., and B. L. Stoddard. 2001. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29:3757-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbyshire, V., J. C. Kowalski, J. T. Dansereau, C. R. Hauer, and M. Belfort. 1997. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 265:494-506. [DOI] [PubMed] [Google Scholar]
- 8.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. H. Herrick. 1994. A proposed superfamily of transposase-related genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan, X., F. S. Gimble, and F. A. Quiocho. 1997. Crystal structure of PI-SciI, a homing endonuclease with protein splicing activity. Cell 89:555-564. [DOI] [PubMed] [Google Scholar]
- 10.Fan, Q., and M. C. Yao. 2000. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res. 28:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, D. F., M. S. Johnson, and R. F. Doolittle. 1985. Aligning amino acid sequences: comparison of commonly used methods. J. Mol. Evol. 21:112-125. [DOI] [PubMed] [Google Scholar]
- 12.Fillingham, J. S., T. A. Thing, N. Vythilingum, A. Keuroghlian, D. Bruno, G. B. Golding, and R. E. Pearlman. 2004. A non-LTR retrotransposon family is restricted to the germ-line micronucleus in the ciliated protozoan Tetrahymena thermophila. Eukaryot. Cell 3:157-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gershan, J. A., and K. M. Karrer. 2000. A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein. Nucleic Acids Res. 28:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimble, F. S. 2000. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 185:99-107. [DOI] [PubMed] [Google Scholar]
- 15.Gorovsky, M. A., M.-C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 16.Hanyu, N., Y. Kuchino, and N. Susumu. 1986. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAsGln. EMBO J. 5:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath, P. J., K. M. Stephens, R. J. Monnat, Jr., and B. L. Stoddard. 1997. The structure of I-CreI, a group I intron-encoded homing endonuclease. Nat. Struct. Biol. 4:468-476. [DOI] [PubMed] [Google Scholar]
- 18.Horowitz, S., and M. A. Gorovsky. 1985. An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. USA 82:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, X., and W. Miller. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337-381. [Google Scholar]
- 20.Ichiyanagi, K., Y. Ishino, M. Ariyoshi, K. Komori, and K. Morikawa. 2000. Crystal structure of an archael intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 300:889-901. [DOI] [PubMed] [Google Scholar]
- 21.Jahn, C. L., M. F. Krikau, and S. Shyman. 1989. Developmentally coordinated en masse excision of a highly repetitive element in E. crassus. Cell 59:1009-1018. [DOI] [PubMed] [Google Scholar]
- 22.Ohme-Takagi, M., and H. Shinshi. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamuro, J. K., B. Caster, R. Villarroel, M. Van Mantagu, and K. D. Jofuku. 1997. The AP2 domain of APETELA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94:7076-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers, M. B., and K. M. Karrer. 1989. Cloning of Tetrahymena genomic sequences whose message abundance is increased during conjugation. Dev. Biol. 131:261-268. [DOI] [PubMed] [Google Scholar]
- 25.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, G. H., J. Z. Dalgaard, M. Belfort, and P. Van Roey. 1999. Crystal structure of the thermostable archael intron-encoded endonuclease I-DmoI. J. Mol. Biol. 286:1123-1136. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weigel, D. 1995. The APETELA2 domain is related to a novel type of DNA binding domain. Plant Cell 7:388-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuitschick, J. D., J. A. Gershan, A. J. Lochowicz, S. Li, and K. M. Karrer. 2002. A novel family of mobile genetic elements is limited to the germ line genome in Tetrahymena thermophila. Nucleic Acids Res. 30:2524-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuitschick, J. D., and K. M. Karrer. 1999. Analysis of genomic G + C content, codon usage, initiator codon context and translation termination sites in Tetrahymena thermophila. J. Euk. Microbiol. 46:239-247. [DOI] [PubMed] [Google Scholar]
- 31.Yao, M.-C., C.-H. Yao, and B. Monks. 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63:763-772. [DOI] [PubMed] [Google Scholar]
- 32.Yao, M.-C., J. Choi, S. Yokoyama, C. Austerberry, and C.-H. Yao. 1984. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36:433-440. [DOI] [PubMed] [Google Scholar]
- 33.Yao, M.-C., and M. Gorovsky. 1974. Comparison of the sequences of macronuclear and micronuclear DNA of Tetrahymena pyriformis Chromosoma 48:1-18. [DOI] [PubMed] [Google Scholar]
- 34.Yao, M.-C., K. Zheng, and C.-H. Yao. 1987. A conserved nucleotide sequence at the sites of developmentally regulated chromosome breakage in Tetrahymena. Cell 48:779-788. [DOI] [PubMed] [Google Scholar]