Abstract
The Notch pathway is an evolutionary conserved signaling cascade that has an essential role in melanoblast and melanocyte stem cell homeostasis. Notch signaling is emerging as a key player in melanoma, the most deadly form of skin cancer. In melanoma, Notch1 is inappropriately reactivated and contributes to melanoma tumorigenicity. Here, we propose a novel mechanism by which Notch1 promotes the disease. We found that Notch1 directly regulates the transcription of neuregulin1 (NRG1) by binding to its promoter region. NRG1 is the ligand for ERBB3 and 4, members of the Epidermal Growth Factor family of receptors that are involved in the genesis and progression of a number of cancers. Notch1 and NRG1 expression are associated in melanoma and inhibition of NRG1 signaling leads to melanoma cell growth inhibition and tumor growth delay. Mechanistically, these effects are associated with the inhibition of the PI3Kinase/Akt signaling pathway and with the accumulation of p27Kip1. On the other end, addition of recombinant NRG1 can partially restore melanoma cell growth that is inhibited by Notch1 ablation. Taken together, our findings underline a new, previously undescribed autocrine signaling loop between Notch1 and NRG1 that controls melanoma growth and provide experimental evidence that the targeting of Notch and ERBB signaling may represent a novel potential therapeutic approach in melanoma
Introduction
Melanoma is the deadliest form of skin cancer accounting for only 4% of all skin cancer cases but for 80% of all skin cancer related deaths (1). Melanoma has become the most common form of cancer for young adults between 25 and 29 years old (2). Because melanomas are highly resistant to the majority of standard therapies, the 5-year survival rate for patients with advanced disease is less than 15% which underscore a clear need for novel more effective therapies. The identification of the signaling pathways that are altered in melanoma provides opportunities for the development of such treatments.
Recent evidence suggests that normally dormant embryonic stem cell pathways may be reactivated in melanoma and contribute to melanoma tumorigenicity (3). The Notch pathway is an evolutionary conserved signaling cascade that has an essential role in embryonic development and cell renewal in the adult by participating in the maintenance of stem cell pluripotency in a variety of tissues (4-6). Remarkably, Notch signaling has been linked to the genesis and/or progression of tumors that originate in the same tissues (7-14). In the melanocytic lineage, elevated Notch1 signaling plays a key role in melanoblast and melanocyte stem cell homeostasis (15). While Notch1 expression is normally decreased in mature melanocytes, melanomas regain expression and activity of Notch1 (16, 17). Notch1 has been shown to be required for melanoma formation (16) and to be able to transform primary human melanocytes and to confer metastatic properties to primary melanoma cells when over-expressed (17-19). Together, these findings suggest that Notch1 is able to control multiple aspects of melanoma growth and progression. The mechanisms however are still not well understood.
Neuregulins and the ERBB receptor signaling cascade also play key roles during embryogenesis (20) and are emerging as new players in melanoblast and melanocyte homeostasis. NRG1 promotes melanoblast proliferation and survival and delays melanoblast maturation into melanocytes (21). In addition, ERBB3 has been found elevated in malignant melanoma and associated with reduced patient survival (22) whereas activating mutations in ERBB4 have been observed in 20% of melanomas (23).
The question of whether NRG1 signaling plays a role in melanoma growth and progression remains unanswered. Furthermore, although both Notch1 and NRG1 signaling are involved in melanocyte lineage homeostasis, it is not known whether cross-talk between these two pathways exists and whether it is a factor in melanoma development. Such cross-talk, for example, is observed in Schwann cell precursors where Notch1 promotes Schwann cell generation in vivo by maintaining high levels of ERBB2 (24, 25); and in normal and cancer stem cells of the breast where ERBB2 and Notch1 control their reciprocal levels of expression leading to an enrichment in HER2/Notch expressing stem cells (25, 26). Such enrichment in the cancer stem cell population results in increased migration, invasion and in vivo tumorigenesis.
In this study we show that Notch1 and NRG1 expression is associated in melanoma and that NRG1 is a previously unidentified, direct transcriptional target of Notch1. Furthermore, we show that NRG1 strongly affects melanoma growth both in vitro and in vivo through the modulation of the PI3Kinase/Akt signaling pathway and of the cell cycle regulator p27Kip1. Taken together, these findings provide compelling evidence of the existence of a Notch1-NRG1 autocrine signaling loop regulating melanoma growth.
Results
Notch1 and NRG1 are associated in melanoma cells
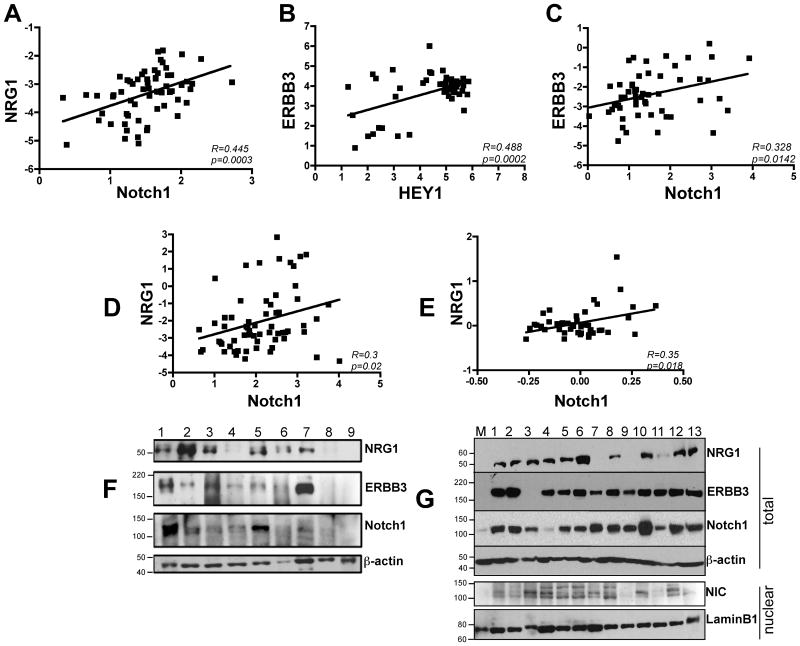
Given the fact that both Notch1 and NRG1 have been found to play a role in melanocyte precursor cell homeostasis we wanted to determine whether a correlation between Notch1 and NRG1 signaling in melanoma existed. We analyzed a series of publicly available data sets in the Oncomine database (27), two obtained from human melanoma tissue specimens (28, 29) and two from melanoma cell lines (30, 31). In the Talantov data set we observed a significant correlation between NRG1 and Notch1 (R=0.445, P=0.0003) (Fig. 1A) and between ERBB3, the receptor for NRG1, and HEY1, a direct transcriptional target of Notch1 (R=0.488, P=0.0002) (Fig. 1B). In the Riker data set (Fig. 1C), a significant correlation between ERBB3 and Notch1 was found (R=0.328, P=0.0142). Finally, NRG1 and Notch1 expression correlated significantly in the cell line data sets as well (Fig. 1D, E – R=0.3, P=0.02 and R=0.35, P=0.018 respectively). As a confirmation of the data obtained from the correlation analysis, we assessed the expression levels of Notch1 and NRG1 signaling elements in nine human samples obtained from resected melanoma metastases. Seven out of nine samples (77%) concurrently expressed NRG1, ERBB3 and Notch1 at various levels (Fig. 1F). Similarly, analysis of a number of human melanoma cell lines revealed that ten out of thirteen cell lines (77%) expressed high levels of NRG1 as compared to control melanocytes (Fig. 1G) and that twelve out of thirteen cell lines were also expressing high amounts of ERBB3 and Notch1 when compared to melanocytes. Interestinlgy, the pattern of expression of active Notch1 (NIC in figure 1G) more closely correlated with the expression pattern of NRG1 in the cells analyzed. Together these data support the existence of a correlation between Notch1 and NRG1 signaling. Furthermore, the fact that control melanocytes (M) showed no or low level of expression of either protein suggests that the NRG1 and Notch1 pathways may be reactivated as a result of malignant transformation.
Figure 1. Notch and NRG1 signaling components correlate in melanoma.
Correlation analysis of NRG1 and Notch1 (A); ERBB3 and HEY1 (B); and ERBB3 and Notch1 (C) of publicly available data sets of human melanoma samples. Data in A and B show a significant correlation between NRG1-Notch1 (R=0.445, p=0.0003) and ERBB3-HEY1 (R=0.488, p=0.0002) among both preneoplastic lesions (nevi, n=18) and primary melanomas (n=45) (Talantov et al (28)). Data in C show a significant correlation between ERBB3 and Notch1 (R=0.328, p=0.0142) in both primary (n=16) and metastatic melanomas (n=40) (Riker et al (29)). D-E) A significant correlation between NRG1 and Notch1 is observed in human melanoma cell lines (D, n=63, R=0.3, p=0.02 (30); E, n=45, R=0.35, p=0.018 (31). F) Western blot analysis of NRG1, ERBB3 and Notch1-TM in nine human metastatic melanoma samples. G) Upper panel: western blot analysis of NRG1, ERBB3 and Notch1-TM expression in thirteen melanoma cell lines compared with normal melanocytes (M). The melanocytes used are a mixed population of cells derived from at least five different foreskins. The bottom panel shows Notch1-NIC expression in nuclear lysates. β-actin or lamin-B1 were used a loading controls for either total or nuclear protein lysates.
Notch1 affects NRG1 expression through binding to the NRG1 promoter
The findings presented above suggest that Notch1 and NRG1 may be co-activated. We next sought to determine whether a molecular link between the two proteins existed in melanoma. By using a melanoma cell line with elevated NRG1 and Notch1 expression (WM266-4, cell line # 12, Fig. 1G upper panel), we inhibited Notch1 by using two specific shRNA sequences against human Notch1. The knock down was determined by quantitative real time PCR which showed a 65% and a 70% inhibition of Notch1 expression achieved by shN1-1 and shN1-2 respectively (Fig. 2A). Importantly, the downregulation of Notch1 was accompanied by a corresponding inhibition of NRG1 mRNA (Fig. 2A and suppl. Fig. 1 where results are shown for K457 melanoma cells -cell line #10 in Fig. 1G). NRG1 protein was also reduced following inhibition of Notch1 expression by the shN1-2 (Fig. 2B). Vice versa, expression of the active intracellular domain of Notch1 (NIC) in cells with low or no Notch1 (V2387, cell line # 4), resulted in increased NRG1 transcript and protein (Fig. 2C, D). Together these data suggest that NRG1 is a transcriptional target of Notch1.
Figure 2. Notch1 affects NRG1 expression.
A). Quantitative real time PCR on WM266-4 cells (cell line # 12 in Fig. 1G) expressing two shRNA sequences against Notch1. Notch1 and NRG1 mRNA levels were normalized with β-actin mRNA. B). Representative western blot for Notch1-TM, NRG1 and β-actin in WM266-4 cells expressing the shNotch1-2. C). V2387 (cell line #4 in figure 1G) expressing active Notch1 (NIC). Both Notch1 and NRG1 mRNA levels are reported after normalization with β-actin. D). Western blot analysis for Notch1-NIC and NRG1 in V2387 cells expressing an empty lentiviral vector (pLM) or the active Notch1-NIC (pLM-NIC). β-actin was used as a loading control. The data shown are representative of at least three independent experiments.
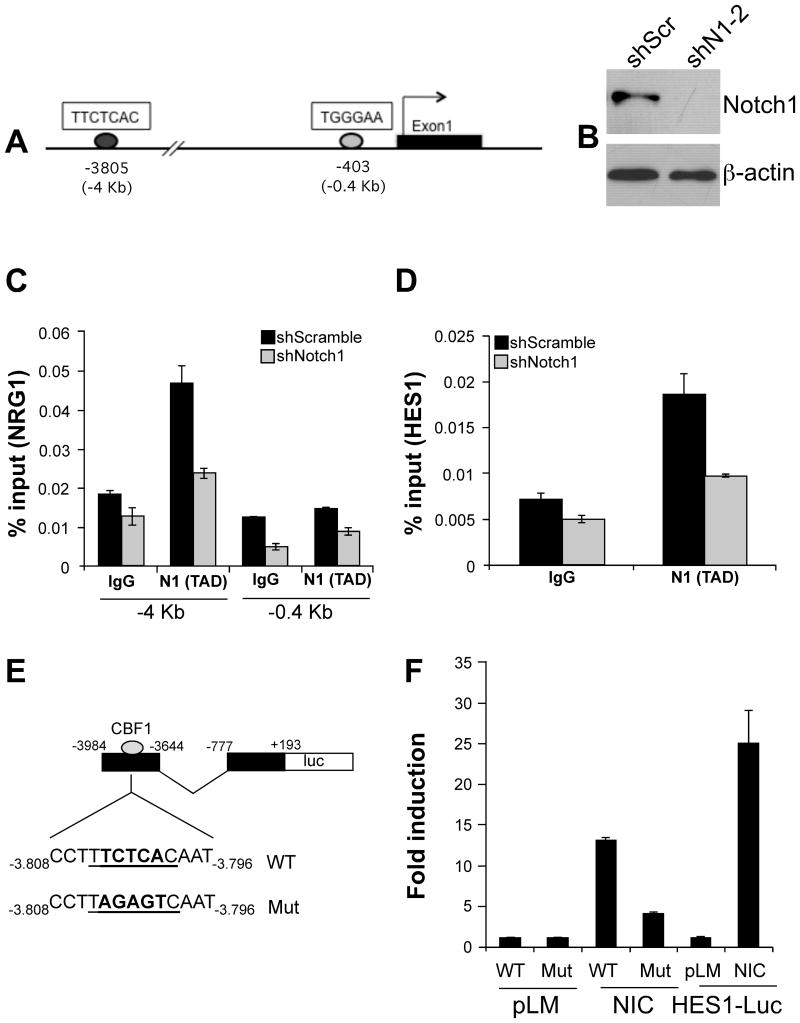
Active Notch proteins act as transcriptional co-activators by participating in the transcription of genes in a complex with the DNA binding protein CSL. To determine if NRG1 is a direct Notch target, we searched for cis elements within the NRG1 promoter that might be occupied by Notch1. Three putative CSL/Notch-binding sequences (TGGGAA - (33)) were identified within a 5 kb sequence upstream the NRG1 transcription start site either on the sense or antisense DNA strand: one at -3805, one at -1700 and one at -403 (Fig. 3A shows the sequences at -3805 and -403). The sites were then tested for Notch1 occupancy in K457 cells with a ChIP assay. To this end, K457 cells were infected with either shScramble (control) or shNotch1-2, fixed with formaldehyde, and lysed for analysis of Notch1-DNA complexes. Effective Notch1 knock down was assessed by western blotting and showed a significant inhibition of the protein (Fig. 3B). PCR amplification of Notch1-associated DNA demonstrated enrichment of the −3805 site in the K457 shScramble cells compared to the K457 shNotch1 cells. In contrast, amplification of the -403 sites was unchanged by the level of Notch1 expression (Fig. 3C). Of the three CSL/Notch-binding elements, only the -3805 complexes with Notch1 as shown in supplementary Fig. 2. Binding of Notch1 to the HES1 promoter was used as positive control and showed similar results to the ChIP performed on the NRG1 promoter (Fig. 3D).
Figure 3. NRG1 is a direct target of Notch1.
A) Consensus binding sequences for CSL/Notch, the hexamers TGGGAA and TTCTCAC, identified within the human NRG1 promoter at positions -3805 and -403. B) Western blot analysis on K457 cells showing Notch1 protein levels in control (shScramble) and shNotch1-2 expressing cells. β-actin serves as a loading control. C) ChIP analysis showing association of Notch1 to the CSL hexamer at position -3805Kb. The site at -0.403Kb shows absence of binding. D) The HES1 promoter was used as a positive control for CSL/Notch binding activity. The data are representative of four experimental repeats for the binding to NRG1 and of two repeats for the binding to HES1. E) schematic representation of the CBF1 reporter construct containing a wild type (WT) or mutated (Mut) CBF1 sequence at the -3.8 site. F) Luciferase reporter assay in WM266-4 cells uninduced (pLM) or induced by a construct expressing active Notch1 (NIC). An HES1-Luc reporter plasmid was used as a positive control of the treatment. A significant reduction of luciferase activity was observed in the mutant CBF1-Luc (P<0.05, Student's T test). The reporter assay was repeated twice and results were very similar in both cases.
To conclusively confirm that Notch1 binds and transcribes NRG1, we created a series of reporter constructs encompassing a minimal promoter region of NRG1 (-777 to +193) and a sequence comprising the −3805 binding site (-3984 to -3644) in which the -3805 site was either left unchanged or mutated in the core region to prevent the binding of CBF1 (Fig. 3E). The reporter assay showed that transfection of an active Notch1-NIC construct into WM266-4 cells, resulted in a thirteen fold induction of luciferase activity when compared to uninduced (pLM only) control (Fig. 3F), while the mutation in the -3805 site significantly reduced the reporter activity (P<0.05, Student T test). A reporter construct containing the HES1 promoter was used as positive control to assess the efficiency of the Notch1-NIC construct. Together, these data underscore a direct role for Notch1 in the modulation of NRG1 expression.
NRG1 signaling is required for melanoma cell growth
The data presented thus far support a direct effect of Notch1 on NRG1 activation in melanoma. To test whether NRG1 plays a role in melanoma, NRG1 was inhibited in melanoma cells and in vitro growth assessed. To control for off-target effects, two distinct shRNA sequences against NRG1 were employed. As shown in figure 4A, the shRNAs inhibited NRG1 mRNA accumulation by 75% and 90% respectively and resulted in a correlated decrease in NRG1 protein (Fig. 4C upper panel). More importantly, NRG1 knock down resulted in 50% and over 90% inhibition of melanoma cell growth in WM266-4 cells expressing either shNRG1-3 or -5 (Fig. 4B). Similar results where observed in additional cell lines (SKMEL2, SKMEL24 and K457 – suppl. Fig 3A-E) underlying a general effect of NRG1 on the growth of melanoma cells.
Figure 4. NRG1 affects melanoma cell growth.
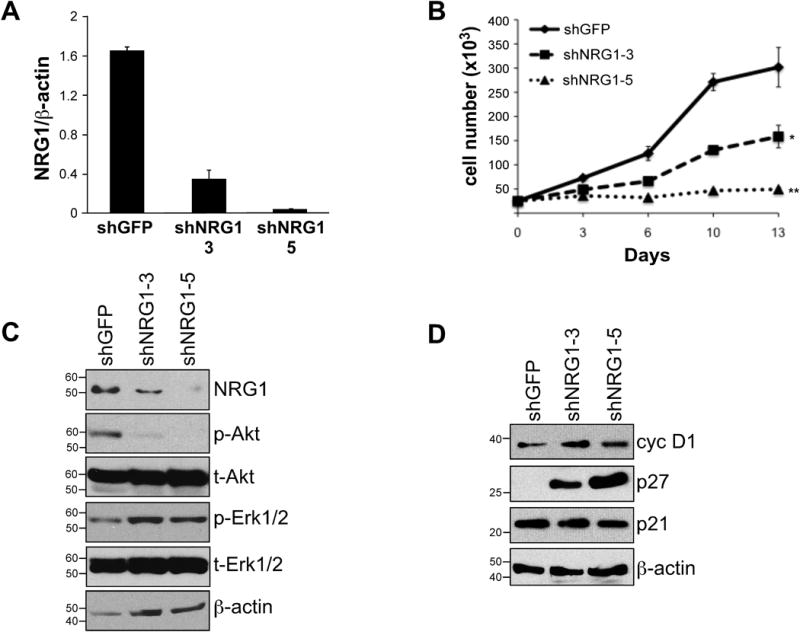
A) qRT-PCR showing NRG1 mRNA levels in WM266-4 cells transduced with a control shRNA (shGFP) or shRNAs against NRG1 (shNRG1-3 and -5). B) In vitro growth of cells expressing the two different shRNAs against NRG1 (shNRG1-3 and -5). C) Western blot analysis on NRG1 protein and downstreasm effectors Akt and ERK1/2 (both phosphorylated (p-) and total (t-) proteins). β-actin was used as a loading control. D) Western blot of the cell cycle regulators cyclin D1, p27Kip1 and p21Cip1 after NRG1 knock down by the shNRG1-3 and -5 in WM266-4 cells. β-actin serves as a loading control. Data are representative of three independent experimets.
The NRG1 signaling pathway has been shown to trigger the activation of major survival and growth signaling cascades such as the PI3kinase and MAPKinase pathways (32-34). Interestingly, we found that knock down of NRG1 affected the level of phosphorylation of Akt while leaving the MAPkinase signaling cascade, measured as phosphorylation of ERK1/2, unchanged (fig. 4C). Akt triggers a signaling network that positively regulates cell cycle progression by increasing cyclin D1, by inhibiting p21Cip1 (35) and by reducing p27Kip1 levels (36-39). Although the knock down of NRG1 by both shRNA sequences did not alter p21Cip1 or cyclin D1 expression, it resulted in a significant accumulation of p27Kip1 that was inversely correlated with the levels of inhibition of NRG1 (Fig. 4D).
Since NRG1 is known to exert its activity through binding to the ERBB3 and ERBB4 tyrosine kinase receptors (40), we next asked whether blocking receptor signaling might result in cell growth inhibition as is the case in cells ablated of NRG1. Indeed, we observed slower growth when cells were treated with an anti ERBB3 antibody that competes for NRG1 binding with the endogenous receptor (41) (Fig. 5A). Interestingly, as was the case following NRG1 knock down, the inhibition of receptor activity resulted in the inhibition of PI3kinase/Akt signaling and in an increase in p27Kip1 protein (Fig. 5B). These data suggest that NRG1 promotes melanoma cell growth in part through the engagement of the ERBB3 receptor and the activation of the PI3Kinase/Akt signaling cascade.
Figure 5. Inhibition of NRG1 binding to its receptor decreases cell growth.
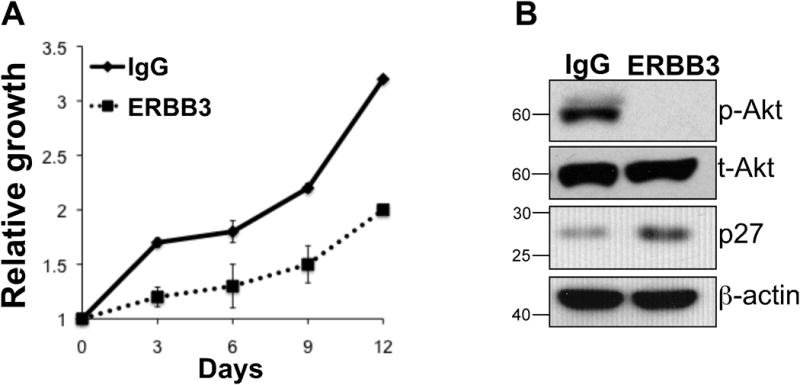
A) Cell growth of WM266-4 in the presence of an anti ERBB3 antibody (ERBB3) or a control IgG. The ERBB3 antibody prevents NRG1 binding to the receptor. B) Western blot analysis of the cells treated with anti ERBB3 or IgG showing the effect of NRG1 binding inhibition on phospho-Akt and p27Kip1. The growth in the presence of anti ERBB3 antibody was repeated twice and data in A are the average between the two independent repeats.
NRG1 supports the tumorigenic potential of melanoma cells
To assess the role of NRG1 as a promoter of the tumorigenic potential of melanoma cells, WM266-4 cells expressing shRNAs against NRG1 were either seeded in soft agar or implanted in vivo in immunodeficient mice. Efficient NRG1 knock down was assessed by qRT-PCR and again showed a significant inhibition of NRG1 mRNA (Fig. 6A). Reduced NRG1 expression resulted in a decreased ability of the cells to grow in an anchorage independent manner (Fig. 6B) and significantly delayed their growth in vivo (Fig. 6C). The growth delay observed was more pronounced in tumors expressing the shNRG1-5, which is more effective at inhibiting NRG1 expression. In a similar manner, the growth of K457 cells in vivo was inhibited by the knock down of NRG1 (Suppl. Fig. 3F). In addition, the tumor growth delay was associated with an accumulation of p27Kip1 within the tumors to a degree that correlates with NRG1 inhibition: shNRG1-5 shows the highest p27Kip1 accumulation both in the cytoplasm and the nucleus (Fig. 6D). This observation is in line with other findings where p27Kip1 accumulation results in even distribution or the protein between the cytosol and the nucleus (36, 37). Notch1 levels were not affected by the NRG1 knock down as shown by immunohistochemical staining using an anti Notch1 antibody that recognizes the transmembrane form of Notch1 (Notch1-TM) (Fig. 6D, lower panel). Overall, we conclude that the NRG1 signaling pathway plays a critical role in the growth and tumorigenic potential of melanoma cells.
Figure 6. NRG1 affects the tumorigenic potential of melanoma cells and their ability to grow in vivo.
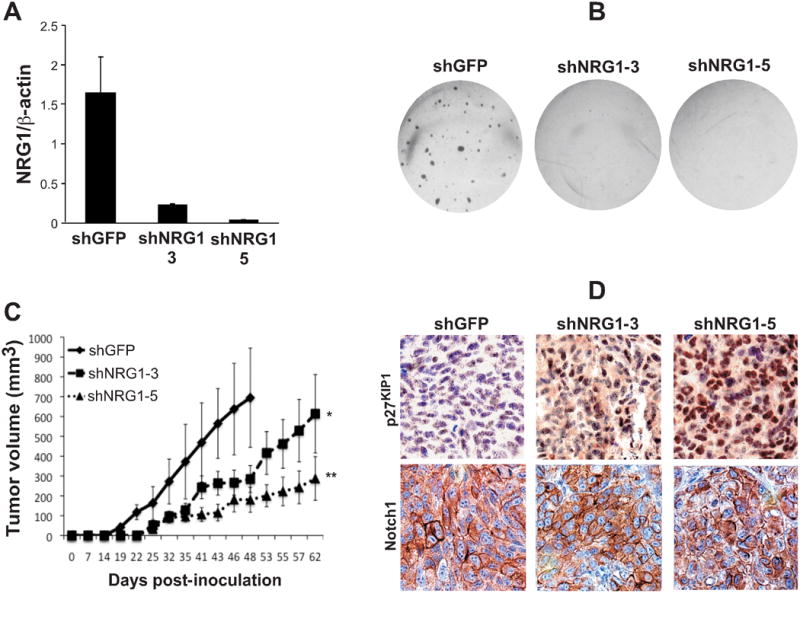
A) qRT-PCR showing the levels of inhibition of NRG1 after transduction with the specific NRG1 shRNA sequences -3 and -5 in WM266-4 cells. B) Soft agar assay performed on WM266-4 cells expressing an shRNA control (shGFP), or the shNRG1-3 and -5. C) in vivo growth of the cells in B. D) Representative immunohistochemistry showing p27Kip1 and Notch1 levels in tumors originated from the assay in C and collected at 48 (shGFP) or 62 (shNRG1-3 and -5) days post-inoculation. The antibody against Notch1 is raised against Notch1-TM. The soft agar assay was repeated four times and the data in B is a representation of one of the experimental repeats. The in vivo growth was done twice.
NRG1 is required for Notch1-mediated melanoma cell growth
Previous work by us and others have shown that Notch1 is required for the efficient growth of a number of cancer cells including melanoma (16), breast (42) and leukemia (43). Based on the data presented above, we hypothesized that Notch1 may modulate melanoma cell growth in part by regulating NRG1 expression. To test this hypothesis, cells expressing a control shRNA (shGFP) or a specific shNotch1 (shN1-2) were subjected to treatment with recombinant NRG1 for four days and the relative growth between control and treated groups was subsequently analyzed. While Notch1 knock down resulted in a 70% growth inhibition compared to control cells, treatment with recombinant NRG1 led to a significant rescue of the growth capacity of cells depleted of Notch1 (Fig. 7A). Interestingly, Notch1 inhibition resulted in the inhibition of Akt phosphorylation that was reconstituted to the levels of controls (shGFP) by recombinant NRG1 treatment (Fig. 7B). Furthermore, while Notch1 inhibition was followed by p27Kip1 accumulation, addition of NRG1 to the cells reduced the levels of p27Kip1 to the levels observed in control cells. These data suggest that NRG1 is sufficient, at least in part, to restore the cell growth capacities that are hindered by Notch1 inhibition.
Figure 7. NRG1 is required for Notch1-mediated melanoma cell growth.
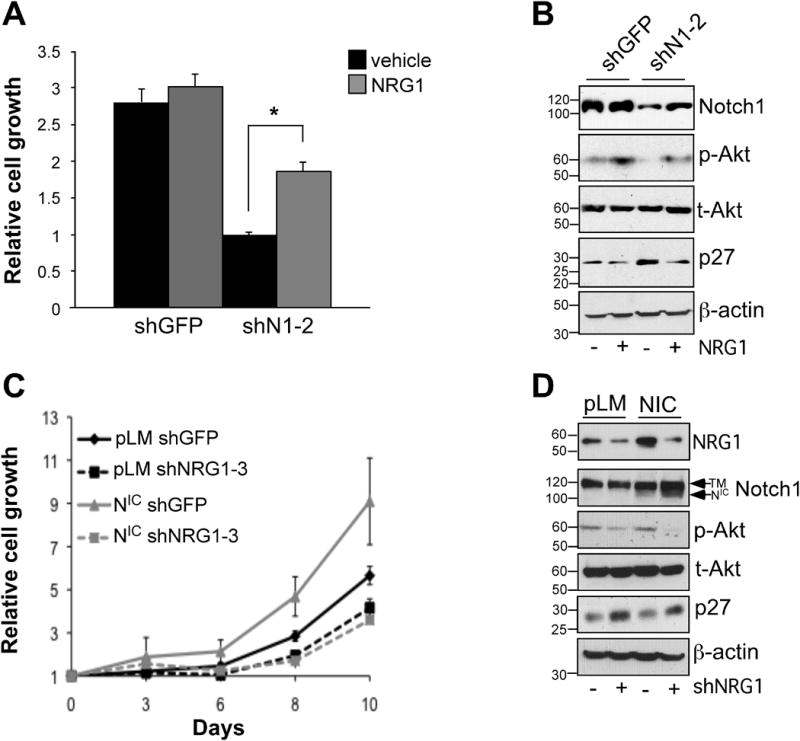
A) Relative growth of WM266-4 cells after Notch1 knock down (shN1-2) at four days post treatment with recombinant NRG1 or vehicle control. The growth recovery of shN1-2 cells treated with recombinant NRG1 is statistically significant with respect to vehicle treated shN1-2 cells (p<0.001, Student's t test). B) Western blot analysis on WM266-4 cells for Notch1-TM , phospho- and total-Akt and p27Kip1. β-actin was used as loading control. C) Growth assay on V2387 cells expressing a control vector (pLM) or active Notch1 (NIC) and further infected with either a shRNA control (shGFP) or shNRG1-3. D) Western blot analysis on V2387 cells decribed in C for Notch1-TM , phospho- and total-Akt and p27Kip1. β-actin was used as loading control. The blot for NRG1-CRD was performed on cell supernatants and loading corrected to the number of cells present at the time of media collection in the various treatment groups. Data are representative of at least three independent experiments.
To further determine the requirement of NRG1 in Notch1-mediated melanoma cell growth, V2387 cells, that express lower endogenous levels of Notch1, were infected with a lentiviral construct expressing active Notch1 (Notch1-NIC) and subsequently with a specific shRNA against NRG1 (shNRG1-3). We found that, while forced expression of Notch1-NIC accelerated melanoma cell growth, inhibition of NRG1 expression abolished the growth promoting function of Notch1-NIC (Fig. 7C). Mechanistically, the growth inhibitory effect was associated with a decrease in phosphorylation of Akt followed by accumulation of p27KIP1 (Fig. 7D). Taken together, these data suggest that the growth phenotype associated with Notch1 is in part dependent on NRG1 expression and the modulation of a Notch1-NRG1-Akt-p27Kip1 axis.
The fact that NRG1 only in part rescued cell growth following Notch1 knock down, suggests that additional Notch1 dependent functions are involved in the regulation of melanoma cell proliferation. We therefore speculated that blockade of both Notch and ERBB signaling may enhance melanoma growth inhibition. Indeed, preliminary data show that by combining a γ-secretase inhibitor (GSI) with lapatinib, an EGFR/ERBB2 inhibitor, we obtain additive cell growth inhibition in a number of melanoma cells (Suppl. Fig. 4). Although preliminary and warrant of more detailed mechanistic studies, these data suggest that targeting both Notch and ERBB signaling may be a novel approach in the treatment of melanoma.
Discussion
Here we demonstrate that Notch1 and NRG1 signaling are associated in melanoma and that NRG1 constitutes a new direct Notch1 transcription target. We present evidence that NRG1 signaling is required for effective melanoma cell growth through the modulation of Akt and p27Kip1. Furthermore, we show that the NRG1 pathway is critical for the maintenance of the malignant phenotype. Finally, we show that the growth phenotype associated with Notch1 is in part attributable to the expression of NRG1 and the modulation of a Notch1-NRG1-Akt-p27Kip1 axis. Together these data provide strong indications that Notch1 lies upstream of NRG1 highlighting the existence of an autocrine signal-transducing loop involving Notch1 and NRG1 in the majority of the tumor cell lines studied herein.
Notch1 and NRG1 signaling are essential in embryogenesis and play a crucial role in the maintenance of melanoblasts and melanocyte stem cells (15, 21, 44). Interestingly, while Notch1, NRG1 and ERBB3 were significantly associated in a number of melanoma data sets and highly expressed in most of the melanoma samples and cell lines analyzed, they were barely detectable or absent in melanocytes suggesting that the expression of Notch1 and NRG1 signaling components might be reactivated during malignant transformation. This observation is intriguing particularly in view of other reports in which the aberrant re-expression of embryonic signaling pathways, such as Notch4 and Nodal, was shown to regulate melanoma cell plasticity and aggressiveness (3). Our data would suggest that a similar mechanism might be in place for Notch1 and NRG1 signaling. Indeed, previous work demonstrated that inhibition of Notch1 in melanoma cells decreased their growth in vitro and in vivo (16, 17). The data presented here highlight a new potential mechanism of Notch1 dependent melanoma growth in the identification of NRG1 as a Notch1 target. Our data show that downregulation of NRG1 in melanoma cells strongly affects their growth capacity both in vitro and in vivo and their anchorage independent growth. Furthermore, the growth defects observed appear to be dependent on the level of NRG1 remaining in the cells. In fact, in the presence of the strongest knock down achieved with shNRG1-5, cell growth was inhibited by 85-90% in a number of cell lines and tumor volumes at forty-seven (Fig. 6C) or fifty-three (suppl. Fig. 3F) days post inoculation were still, roughly, 70 to 90% smaller than control tumors (200 mm3 vs 700 mm3 and 100 mm3 vs 900 mm3, respectively).
Whether a Notch1-NRG1 autocrine signaling loop is involved in melanoma metastasis is not known and it will be addressed in future studies. However, evidence from other tumor types would suggest that it is a likely possibility. For example, a Notch/ERBB2 cross-talk was observed in cancer stem cells of the breast and shown to contribute to cancer cell invasion and tumorigenesis (25, 26).
NRG1 signals through the engagement of ERBBB3 and ERBB4, members of the epidermal growth factor family of tyrosine kinase receptors, and promotes the activation of major survival and growth signaling cascades such as PI3Kinase and MAPKinase (40). Although no change in Erk1/2 phosphorylation was observed, we found a strong reduction of Akt phosphorylation after NRG1 knock down. Furthermore, we also observed the accumulation of the cell cycle inhibitor p27Kip1 following NRG1 knock down that was associated with the levels of NRG1 inhibition achieved with the two shRNA sequences employed. Based on these results we hypothesize that the effects of NRG1 on melanoma cells may be dependent on the engagement of the ERBB receptors. In fact, not only is NRG1 known to be the ligand for ERBB3 and ERBB4, both receptors have also been implicated in melanoma. Recently, ERBB3 has been found activated in both melanomas and melanoma cell lines (21, 45) and its expression levels associated with poor patient outcome (22); ERBB4 activating mutations have been observed in up to 20% of melanomas (23). Interestingly, when we prevented NRG1 binding to ERBB3 by using an anti ERBB3 antibody, we observed inhibition of melanoma cell growth that was associated with the inhibition of the PI3Kinase/Akt signaling pathway and with an increase in p27Kip1. Taken together, these results strongly support a role for the NRG1 pathway in the modulation of melanoma growth and tumorigenic potential and suggest a potential autocrine signaling pathway. In this pathway, NRG1 exerts its growth promoting activity through signaling via the ERBB receptors, activation of the PI3Kinase/Akt signaling cascade that culminates with the suppression of the cell cycle inhibitor p27Kip1 (38, 39).
To further support the notion that NRG1 may be a key mediator in Notch1 dependent melanoma growth, we show that inhibition of Notch1 results in a reduction in the activity of the PI3Kinase/Akt signaling pathway and increased accumulation of p27Kip1. Interestingly, treatment with recombinant NRG1 partially restored the cell growth capabilities, increased Akt phosphorylation and counteracted the accumulation of p27Kip1. On the other hand, forced expression of active Notch1 (Notch1-NIC) accelerated cell growth but such growth promoting capabilities were ablated by the inhibition of NRG1. This behavior was associated with the deregulation of Akt and p27KIP1. Together, these data suggest that Notch1 promotes melanoma cell growth in part by driving the expression of NRG1, which, in turn, maintains an active PI3Kinase/Akt signaling cascade and keeps in check the levels of the cell cycle inhibitor p27Kip1. The fact that NRG1 did not completely restore the cell growth capacities of cells deprived of Notch1, suggests that Notch1 may control melanoma growth via additional pathways. For example, Notch1 has been recently shown to affect lung adenocarcinoma cell survival by directly regulating the expression of IGF-1R (Insulin-like Growth Factor 1 Receptor) (46).
Interestingly, an antagonistic relationship between Notch and ERBB signaling has been observed. For example, recent work (47) has demonstrated an antagonistic relationship between Notch1 and ERBB2 in breast cancer cells such that inhibition of ERBB2 activity by trastuzumab led to the activation of Notch1 signaling and resulted in acquired resistance to the drug. Dual inactivation of ERBB2 and Noch1 by trastuzumab and a GSI reversed the resistant phenotype. Similarly, Dong et al (48) showed that only the combined inhibition of EGFR and γ-secretase by gefitinib and a GSI promoted apoptosis of basal-like breast cancer cells. These results are quite in contrast with our findings where Notch and NRG1 are part of a positive autocrine signaling loop that promotes melanoma growth, and suggests that the interaction between Notch and ERBB signaling could regulate different cell fates depending on the components of the ERBB pathway involved and the context of the tissue in which such interaction occurs.
Recent advances in melanoma research have led to the successful development of therapies targeted against activated BRAF, thus far the most frequently mutated gene in melanoma. However, the benefits achieved with these experimental approaches are still limited to patients bearing BRAF mutations and by the emergence over time of resistant tumors. Melanomas rely on the activity of multiple signaling pathways for their growth and survival. The study presented here identifies a new mechanism of Notch1 dependent melanoma growth and provides experimental evidence that Notch1 and NRG1, two evolutionary conserved signaling cascades, are reactivated in melanoma and functionally linked. Furthermore, this work suggests that the inhibition of Notch and ERBB signaling may represent a novel potential therapeutic approach in melanoma. The therapeutic potential of this study is particularly attractive since inhibitors for both Notch signaling and the ERBB pathway already exist and are either being tested in clinical trials or are being used for the treatment of other cancers. Indeed, in preliminary studies we observed that the combination of a GSI with lapatinib, an anti EGFR/ERBB2 compound, additively inhibits the growth of a number of melanoma cell lines (suppl. Fig. 4). Furthermore, the treatment was equally effective on either mutated or wild type BRAF cells. Given the fact that only about 50% of melanoma patients harbor BRAF mutations and are therefore eligible for anti BRAF therapy, these data suggests that the use of a combined anti Notch-ERBB treatment may be beneficial to a broader cohort of melanoma patients. In conclusion, while these initial results require further validation and a more in depth mechanistic analysis, they support the potential use of Notch and ERBB inhibitors in the treatment of melanoma.
Materials and methods
Cells and tissue specimens
Primary and metastatic melanoma cells were in part purchased from ATCC and in part were a gift of Dr. Marianne Broome Powell (Stanford University) (Bedogni et al., 2008). The use of these cells was approved by the Case Cancer IRB. The cell lines used in this study are, in the order they appear in the blot in Fig. 1G: immortalized human melanocytes; A375.52; C32TG; WM3211; V2387; WM35; WM115; UACC2521;SKMEL2; D1594; K457; UACC3291; WM266-4; SKMEL24. Functional assays were performed on the cell lines WM266-4, K457 and V2387, SKMEL2 and SKMEL24. These are human melanoma cells derived from metastases of the skin (WM266-4 and K457, SKMEL2) and lymph node (V2387, SKMEL24) and are either BRAF mutants (WM266-4, K457, V2387) or wild type (SKMEL2 and 24). All cells were maintained in DMEM supplemented with 10% FCS, 1% glutamine, and 1% penicillin-streptomycin. Immortalized human melanocytes were a gift of Dr. Robert Weinberg (Harvard University) and were maintained in DMEM supplemented with 5% FCS, 1% glutamine, and 1% penicillin-streptomycin (49).
De-identified melanoma tissues were redundant diagnostic specimens obtained at surgery for clinically indicated removal at metastatic sites. The collection of such specimens followed the guidelines of the IRB-approved Human Biospecimen Resource Core in the Cleveland Clinic Department of Anatomic Pathology (IRB protocol n° 06-050).
shRNAs and expression plasmids
shRNAs against human Notch1 (TRCN0000003359 and TRCN0000003362) and human NRG1 (TRCN0000058303 and TRCN0000058305) were purchased from Open Biosystems. The Notch1-NIC lentiviral expression vector was constructed by inserting the cDNA sequence corresponding to human Notch1-NIC (base pairs 5278-7668 of full length human Notch1) into the pLM-CMV-Ha-puro-PL3 lentiviral plasmid (50), between XbaI-XhoI. Viral particles were produced in 293-FT cells using the Fugene6 reagent (Roche) as per manufacturer's instructions. The packaging plasmid used were pMD2.G and psPAX2 that were purchased from Addgene.
Luciferase assays
The NRG1-reporter plasmid was constructed by inserting the -3.8 CBF1 core region (contained between the sequence -3.808 and -3.796 upstream of the NRG1 transcription stat site) fused with a minimal promoter region of the NRG1 promoter (-777-+ 193), into the pGL3-basic luciferase plasmid. The HES1-reporter construct was a gift of R. Kageyama (Kyoto University, Kyoto, Japan). WM266-4 cells (5 × 104/well, in 24-well plates) were transfected using the Fugene6 reagent (Roche) per the manufacturer's instructions. After 36–48 hours, cells were lysed in 100 μl lysis buffer (Promega). A Renilla luciferase reporter plasmid driven by a CMV promoter was co-transfected with NRG1 and HES1 reporter constructs at a 1:5 ratio to assess transfection efficiency. Activities of Firefly and Renilla were assessed by the dual-luciferase Assay system (Promega) and light production was measured for 10 s in a Monolight 2010 Luminometer (Analytical Luminescence Laboratory).
Western blot analysis
Cells (2×106 cells/dish, 100-mm dishes) were plated in DMEM plus 10% FCS, allowed to adhere, and then they were either collected after 24 hours post seeding or serum starved for 16 hours prior to NRG1 stimulation for 4 days. Total protein was extracted with urea lysis buffer (9 M urea; 75 mM Tris-HCl, pH 7.5; and 100 mM 2-ME), and 40–50 μg per sample were separated by 8-10% SDS-PAGE and transferred onto nitrocellulose membranes. Nuclear proteins were extracted as follows: cells were disrupted with a Dounce homogenizer in a hypotonic buffer (20 mM HEPES, pH 7; 10 mM KCl; 1 mM MgCl2; 0.1% Triton X-100; 20% glycerol; 2 mM PMSF; 5 μg/ml aprotinin; and 5 μg/ml leupeptin) and centrifuged at 850 g to obtain a pellet of nuclei. Nuclear lysates were prepared in urea buffer, and 40–50 μg were loaded onto 8% SDS-PAGE gels. Tumor samples were homogenized in RIPA buffer (50 mM Tris-Cl, pH 7.4; 150 mM NaCl; 1% NP-40; 0.25% Na-deoxycholate; 1 mM PMSF; 1 μg/ml aprotinin; 1 μg/ml leupeptin; and 1 mM Na-ortovanadate), and 50 μg were loaded on an 8% gel. Membranes were probed with the following antibodies: anti-neuregulin1 CRD (NeuroMab, UC Davis); anti–phospho-Akt (Ser473), anti-phospho-Erk1/2, anti-total Akt, anti-total Erk1/2 and anti-Notch1-NIC (Cell Signaling Technologies); anti–p27Kip1 and anti-Notch1-TM (Santa Cruz Biotechnologies). Bands were detected using Super Signal Detection Reagent (Thermo Scientific). Loading was normalized with anti–β-actin for total lysates or LaminB1 for nuclear lysates (Santa Cruz Biotechnology).
Real-time PCR analysis
cDNA was synthesized from total RNA, treated with DNaseI (Invitrogen), using SuperScript first-strand synthesis system for RT-PCR (Invitrogen). 1 μl cDNA was used for PCR amplification using SYBR Green PCR Master Mix (Biorad). The following primer sets were used to amplify specific target genes: human Notch1 forward 5′-GAGGCGTGGCAGACTATGC-3′, reverse 5′-CTTGTACTCCGTCAGCGTGA-3′; human NRG1 forward 5′-CAAAGAAGGCAGAGGCAAAG-3′, reverse 5′-AACTGGTTTCACACCGAAGG-3′; human β-actin forward 5′-CATGTACGTTGCTATCCAGGC-3′, reverse 5′-CTCCTTAATGTCACGCACGAT-3′. PCR amplification was done in a Biorad I-Cycler. β-actin was used to normalize mRNA. Relative quantification of mRNA expression levels was determined using the relative standard curve method according to the manufacturer's instructions (Biorad).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using ChIP assay kits (Upstate Biotechnology) following the manufacturer's recommendations. The following primer sequences were used: NRG1 promoter at -4kb: forward: 5′-CCCCCAAATTTTATGAGCTTC-3′; reverse: 5′ TTGTGAGAAAGGCATAAAAACAAA-3′; NRG1 promoter at -0.4kb: forward: 5′-AGGAATGAACCCCGAACTCT-3′; reverse: 5′ AGAGTCTCCGTGGTCAGGAA-3′; HES1 promoter: forward: 5′-CGTGTCTCCTCCTCCCATTG-3′; reverse: 5′-CCAGGACCAAGGAGAGAGGT-3′. The antibody employed for the immunoprecipitation was previously described (51).
Cell proliferation assays
WM266-4 cells (2×104) expressing shRNA control (shGFP) or shRNA against NRG1 (#3 and #5) were plated in triplicate in 24 well plates and were counted every 3–4 days for the duration of the experiment using an electronic particle counter (Beckman Coulter). For the ERBB3 antibody treatment, 3000 cells were seeded in 96 well plates in triplicate and 10 mg/ml ERBB3 antibody (Ab-5, Lab Vision-Thermo Scientific) was added in each well. Cell growth was assessed by a crystal violet assay as previously described (52). For the treatment with recombinant NRG1 (Cell signaling Technology), 5000 cells were seeded in 96 well plates in triplicate and treated with 5 ng/ml NRG1 for four days. At the end of the treatment, cells were fixed with 10% buffered formalin and subjected to the crystal violet assay. V2387 cells expressing an empty vector (pLM) or active Notch1 (NIC) in conjunction with shGFP and shNRG1-3 were seeded in a 96 well plate (3000 cells/well) in triplicate, fixed with formalin at the time points shown and subjected to the crystal violet assay. Treatment with the γ-secretase compounds RO4929097 (10 μM) and DBZ (10 μM) (dibenzazepine) and the EGFR/ERBB2 inhibitor lapatinib (20 μM) were done on WM266-4 (4000/well), K457 (2000/well) and SKMEL2 (4000/well) cells seeded in a 96 well plate. The drugs were added the following day, either alone or in combination, and four days later cell growth was assessed by the crystal violet assay.
Anchorage independent growth
Cells (105 cells/well, 24-well plates) were suspended in DMEM + 10% FBS containing 0.3% agar and spread onto a 0.6% agar layer. Photographs were taken after four weeks of incubation in agar.
In vivo tumor growth
Male athymic nude mice (nu/nu of 3–5 wk old) were supplied by the Athymic Animal Facility at Case Western Reserve University. All experimental protocols were approved by the Administrative Panel on Laboratory Animal Care of Case Western Reserve University. Cells (2 × 106) were injected subcutaneously in the dorsal flanks of mice for a total of 8 tumors per experimental group. Mice were sacrificed 48 (shGFP) and 62 days (ShNRG1) after injection. Tumors were measured, and tumor volume was calculated as (w2 × l) × 0.52, in which w and l represent width and length, respectively (53). Tumors were formalin fixed, and 5-μm sections were cut for staining.
Immunohistochemistry
Formalin-fixed tumor sections were incubated with anti p27Kip1 or anti Notch1-TM primary antibody (both from Santa Cruz Biotechnologies, Palo Alto, CA), followed by biotinylated secondary antibody and HRP-conjugated Streptavidin (Vector Laboratories). Hematoxylin was used as counterstain. Antigen unmasking was performed by treating sections with low pH unmasking solution (for p27KIP1) or high pH unmasking solution (for Notch1-TM) (Vector Laboratories) in a pressure cooker as per manufacturer's instructions.
Statistics
The significance of the correlation between NRG1-Notch1, ERBB3-HEY1 and ERBB3-Notch1was calculated by the Pearson correlation using Prism 4. A correlation was considered significant for p≤0.05. Statistical differences between groups of data generated by quantitative real time PCR, growth assays, chromatin-IP and reporter assay were determined by the Student's t test. In all cases a p value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported in part by grant # ACS-IRG-91-022 and grant # ACS-RSG-11-139-01-DDC from the American Cancer Society and by start up funds awarded by the National Institute of Health (award number: P30CA147877: New Faculty Recruitment to Enhance Melanoma Research) .
We wish to thank Drs. Scott Welford and David Samols for critical discussion; Dr. Marianne Broome Powell for making available many of the melanoma cell lines used in this work.
Key words
- NIC
Notch1 intracellular domain
- Notch1-TM
transmembrane Notch1
- NRG1
neuregulin1
- ERBB
Erythroblastic Leukemia Viral Oncogene Homolog or EGF (Epidermal Growth Factor) family of transmembrane Receptor Tyrosine Kinases
- p27Kip1
CDKN1B, cyclin-dependent kinase inhibitor 1B
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Literature Cited
- 1.Altekruse SF, K C, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK. SEER Cancer Statistics Review, 1975-2007. 2010 [Google Scholar]
- 2.Herzog Cynthia, MAP, MD, Bondy Melissa, PhD, Bleyer Archie, MD, Kirkwood John., MD Cancer Epidemiology in Older Adolescents and Young Adults. SEER AYA Monograph. 2007:53–63. [Google Scholar]
- 3.Hardy KM, Kirschmann DA, Seftor EA, Margaryan NV, Postovit LM, Strizzi L, et al. Regulation of the embryonic morphogen Nodal by Notch4 facilitates manifestation of the aggressive melanoma phenotype. Cancer Res. 2010 Dec 15;70(24):10340–50. doi: 10.1158/0008-5472.CAN-10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou K, Huang L, Zhou Z, Hu C, Liu W, Zhou J, et al. Wnt and Notch signaling pathways selectively regulating hematopoiesis. Ann Hematol. 2010 Aug;89(8):749–57. doi: 10.1007/s00277-010-0923-3. [DOI] [PubMed] [Google Scholar]
- 5.Brabletz S, Schmalhofer O, Brabletz T. Gastrointestinal stem cells in development and cancer. J Pathol. 2009 Jan;217(2):307–17. doi: 10.1002/path.2475. [DOI] [PubMed] [Google Scholar]
- 6.Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010 Mar 3;30(9):3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991 Aug 23;66(4):649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 8.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007 Sep 1;67(17):8051–7. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 9.Pahlman S, Stockhausen MT, Fredlund E, Axelson H. Notch signaling in neuroblastoma. Semin Cancer Biol. 2004 Oct;14(5):365–73. doi: 10.1016/j.semcancer.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology. 2001 Jul 20;286(1):23–30. doi: 10.1006/viro.2001.0867. [DOI] [PubMed] [Google Scholar]
- 11.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004 Oct 1;64(19):6854–7. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 12.Sjolund J, Johansson M, Manna S, Norin C, Pietras A, Beckman S, et al. Suppression of renal cell carcinoma growth by inhibition of Notch signaling in vitro and in vivo. J Clin Invest. 2008 Jan;118(1):217–28. doi: 10.1172/JCI32086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert CA, Daou MC, Moser RP, Ross AH. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. Sep 1;70(17):6870–9. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovinga KE, Shimizu F, Wang R, Panagiotakos G, Van Der Heijden M, Moayedpardazi H, et al. Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells. Jun;28(6):1019–29. doi: 10.1002/stem.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osawa M, Fisher DE. Notch and melanocytes: diverse outcomes from a single signal. J Invest Dermatol. 2008 Nov;128(11):2571–4. doi: 10.1038/jid.2008.289. [DOI] [PubMed] [Google Scholar]
- 16.Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest. 2008 Nov;118(11):3660–70. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009 Jul 1;69(13):5312–20. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating Ncadherin expression. Cancer Res. 2006;66:4182–90. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 19.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005 Nov;115(11):3166–76. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci. 2008 May;65(10):1566–84. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buac K, Xu M, Cronin J, Weeraratna AT, Hewitt SM, Pavan WJ. NRG1 / ERBB3 signaling in melanocyte development and melanoma: inhibition of differentiation and promotion of proliferation. Pigment Cell Melanoma Res. 2009 Dec;22(6):773–84. doi: 10.1111/j.1755-148X.2009.00616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reschke M, Mihic-Probst D, van der Horst EH, Knyazev P, Wild PJ, Hutterer M, et al. HER3 is a determinant for poor prognosis in melanoma. Clin Cancer Res. 2008 Aug 15;14(16):5188–97. doi: 10.1158/1078-0432.CCR-08-0186. [DOI] [PubMed] [Google Scholar]
- 23.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009 Oct;41(10):1127–32. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009 Jul;12(7):839–47. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnifico A, Albano L, Campaner S, Delia D, Castiglioni F, Gasparini P, et al. Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab. Clin Cancer Res. 2009 Mar 15;15(6):2010–21. doi: 10.1158/1078-0432.CCR-08-1327. [DOI] [PubMed] [Google Scholar]
- 26.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008 Oct 16;27(47):6120–30. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004 Jan-Feb;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005 Oct 15;11(20):7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 29.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007 Jun;20(3):216–21. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 31.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006 Aug;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 32.Calvo M, Zhu N, Grist J, Ma Z, Loeb JA, Bennett DL. Following nerve injury neuregulin-1 drives microglial proliferation and neuropathic pain via the MEK/ERK pathway. Glia. 2011 Apr;59(4):554–68. doi: 10.1002/glia.21124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ieguchi K, Fujita M, Ma Z, Davari P, Taniguchi Y, Sekiguchi K, et al. Direct binding of the EGF-like domain of neuregulin-1 to integrins ({alpha}v{beta}3 and {alpha}6{beta}4) is involved in neuregulin-1/ErbB signaling. J Biol Chem. 2010 Oct 8;285(41):31388–98. doi: 10.1074/jbc.M110.113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukazawa R, Miller TA, Kuramochi Y, Frantz S, Kim YD, Marchionni MA, et al. Neuregulin-1 protects ventricular myocytes from anthracycline-induced apoptosis via erbB4-dependent activation of PI3-kinase/Akt. J Mol Cell Cardiol. 2003 Dec;35(12):1473–9. doi: 10.1016/j.yjmcc.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003 Jul-Aug;2(4):339–45. [PubMed] [Google Scholar]
- 36.Shin I, Rotty J, Wu FY, Arteaga CL. Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin alpha during G1 and prevents nuclear re-entry. J Biol Chem. 2005 Feb 18;280(7):6055–63. doi: 10.1074/jbc.M412367200. [DOI] [PubMed] [Google Scholar]
- 37.Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002 Oct;8(10):1145–52. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 38.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009 Apr;11(4):420–32. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009 Apr;11(4):397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009 Jul;9(7):463–75. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Levkowitz G, Tzahar E, Karunagaran D, Lavi S, Ben-Baruch N, et al. An immunological approach reveals biological differences between the two NDF/heregulin receptors, ErbB-3 and ErbB-4. J Biol Chem. 1996 Mar 29;271(13):7620–9. [PubMed] [Google Scholar]
- 42.Rasul S, Balasubramanian R, Filipovic A, Slade MJ, Yague E, Coombes RC. Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells. Br J Cancer. 2009 Jun 16;100(12):1879–88. doi: 10.1038/sj.bjc.6605034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pancewicz J, Taylor JM, Datta A, Baydoun HH, Waldmann TA, Hermine O, et al. Notch signaling contributes to proliferation and tumor formation of human T-cell leukemia virus type 1-associated adult T-cell leukemia. Proc Natl Acad Sci U S A. 2010 Sep 21;107(38):16619–24. doi: 10.1073/pnas.1010722107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birchmeier C. ErbB receptors and the development of the nervous system. Exp Cell Res. 2009 Feb 15;315(4):611–8. doi: 10.1016/j.yexcr.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 45.Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, Muthusamy V, et al. Phospho-Proteomic Screen Identifies Potential Therapeutic Targets in Melanoma. Mol Cancer Res. 2011 Apr 26; doi: 10.1158/1541-7786.MCR-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eliasz S, Liang S, Chen Y, De Marco MA, Machek O, Skucha S, et al. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene. 2010 Apr 29;29(17):2488–98. doi: 10.1038/onc.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008 Aug 28;27(37):5019–32. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- 48.Dong Y, Li A, Wang J, Weber JD, Michel LS. Synthetic lethality through combined Notch-epidermal growth factor receptor pathway inhibition in basal-like breast cancer. Cancer Res. 2010 Jul 1;70(13):5465–74. doi: 10.1158/0008-5472.CAN-10-0173. [DOI] [PubMed] [Google Scholar]
- 49.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005 Oct;37(10):1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Razorenova OV, Agapova LS, Budanov AV, Ivanov AV, Strunina SM, Chumakov PM. Retroviral reporter systems for the assessment of activity of stress-induced signal transduction pathways controlled by p53, HIF-1 and HSF-1 transcription factors. Mol Biol (Mosk) 2005 Mar-Apr;39(2):286–93. [PMC free article] [PubMed] [Google Scholar]
- 51.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997 Mar 7;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 53.Arbiser JL, Moses MA, Fernandez CA, Ghiso N, Cao Y, Klauber N, et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc Natl Acad Sci U S A. 1997 Feb 4;94(3):861–6. doi: 10.1073/pnas.94.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.