Abstract
We have isolated a Kluyveromyces lactis mutant unable to grow on all respiratory carbon sources with the exception of lactate. Functional complementation of this mutant led to the isolation of KlSDH1, the gene encoding the flavoprotein subunit of the succinate dehydrogenase (SDH) complex, which is essential for the aerobic utilization of carbon sources. Despite the high sequence conservation of the SDH genes in Saccharomyces cerevisiae and K. lactis, they do not have the same relevance in the metabolism of the two yeasts. In fact, unlike SDH1, KlSDH1 was highly expressed under both fermentative and nonfermentative conditions. In addition to this, but in contrast with S. cerevisiae, K. lactis strains lacking KlSDH1 were still able to grow in the presence of lactate. In these mutants, oxygen consumption was one-eighth that of the wild type in the presence of lactate and was normal with glucose and ethanol, indicating that the respiratory chain was fully functional. Northern analysis suggested that alternative pathway(s), which involves pyruvate decarboxylase and the glyoxylate cycle, could overcome the absence of SDH and allow (i) lactate utilization and (ii) the accumulation of succinate instead of ethanol during growth on glucose.
Succinate dehydrogenase (SDH) is a component of complex II of the respiratory chain that catalyses the oxidation of succinate to fumarate in the Krebs cycle and feeds electrons to the ubiquinone pool. The complex, which is highly conserved through evolution, is located in the inner mitochondrial membrane and consists of two catalytic and two structural subunits, all encoded by nuclear genes (38). In Saccharomyces cerevisiae, the four genes (SDH1 to SDH4) coding for SDH have been isolated and characterized (26, 27, 45, 47). The flavoprotein subunit (11, 42) responsible for the oxidation of succinate to fumarate is encoded by two paralogous genes, SDH1 and SDH1b, although only SDH1 is necessary for growth on respiratory carbon sources (11). SDH2 codes for the iron-protein subunit (31) that contains three different iron-sulfur centers (22) and, together with the protein Sdh1p, constitutes the catalytic core of the SDH complex, which conveys electrons from the covalently attached flavin adenine dinucleotide (FAD) of Sdh1p first to the iron-sulfur centers and then to ubiquinone. SDH3 and SDH4 code for two small hydrophobic peptides, which anchor the complex to the inner mitochondrial membrane (10, 15). In humans, the mutations in the SDH genes have been associated to several mitochondrial-related pathologies suggesting, beside the enzymatic activity of the complex in the Krebs cycle, its involvement in superoxide handling (39, 43).
In S. cerevisiae, the expression of the SDH genes is repressed by glucose and derepressed on respiratory carbon sources (31, 45), and the loss of SDH functions results in the inability of cells to grow on any respiratory carbon sources (12, 47).
In this paper we report the isolation of the KlSDH1 gene (EMBL accession number AJ555233) encoding the Kluyveromyces lactis flavoprotein subunit of the SDH complex. We show that, despite the general sequence conservation between K. lactis and S. cerevisiae genes, their regulation appears to be different, probably reflecting the predominant respiratory and fermentative nature, respectively, of these species (18, 51). The genes are expressed on both fermentable and nonfermentable carbon sources, and their deletion does not lead to a loss of the respiratory function.
MATERIALS AND METHODS
Strains, media and culture conditions.
The strains used in this work are reported in Table 1. Yeast cultures were grown overnight under aerated conditions on an orbital shaker at 28°C in YP medium (1% Difco yeast extract, 2% Difco Bacto-peptone) or in minimal medium (6.7 g of Difco yeast nitrogen base per liter), supplemented with different carbon sources at the concentrations specified in the text. Solid media were supplemented with 2% Bacto agar (Difco). Curve growth was performed by inoculating about 106 cells per ml of culture medium, and at time intervals aliquots of the cultures were taken, suitably diluted, and counted in a Thoma chamber to determine cell concentration (cells/milliliter). Minimal media were supplemented with the required auxotrophies at a final concentration of 10 μg/ml.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| K. lactis strains | ||
| MS14-1A | MATα, ade, trip1, ura3, sdh | This work |
| MW278-20C/1 | MATα, ade2, leu2, ura3, lac4-8 | 17 |
| MW179-1D | MATα, metA1, ade2, trip1, ura3, leu2, lac4-8 | 17 |
| MW179-1D/Klsdh1Δ | MW179-1D klsdh1::kanMX4URA3 | This work |
| CBS2359/152 | MATa, metA1 | 50 |
| GG1993a | MATa, Klpda1::Tn5BLE, ura3-49 | 54 |
| S. cerevisiae strains | ||
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Euroscarf |
| BY4741sdh1Δ | BY4741 sdh1::kanMX4 | Euroscarf |
| BY4741sdh1bΔ | BY4741 sdh1b::kanMX4 | Euroscarf |
Isogenic derivative of CBS2359
Escherichia coli strain DH5α was used for the propagation of plasmid DNA. Plasmid-carrying bacteria were grown at 37°C on LB medium (0.5% yeast extract, 1% tryptone, 0.5% NaCl) supplemented with 100 μg of ampicillin per ml.
Ethanol, glucose, and succinate concentrations in culture supernatants were determined by using commercial kits from Boehringer-Mannheim.
KlSDHI disrupting cassette.
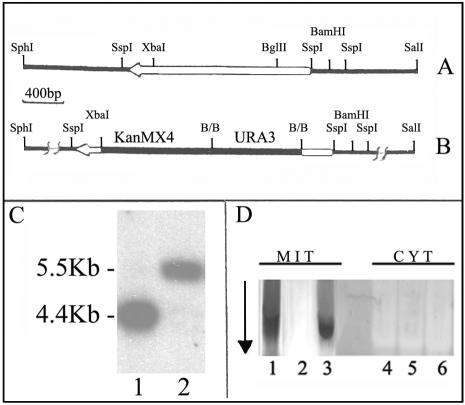
The plasmid p3AS, containing the complementing fragment (about 4.6 kbp, plus 0.2 kbp at the SphI site) in the Kep6 multicopy vector (5), was used for the construction of the disrupting cassette (see Fig. 3 for transformations). About 80% of the KlSDH1 open reading frame (XbaI-BglII fragment of 1.5 kbp) was replaced with the genes kanMX4 (49) and URA3 of S. cerevisiae. The wild-type MW179-1D strain was transformed with the linearized cassette to uracil prototroph and resistance to geneticin (G418). Positive clones were replica plated on minimal media containing glycerol to identify those impaired in growth. These clones were further analyzed by Southern blotting to verify the correct integration of the cassette into the KlSDH1 locus.
FIG. 3.
(A) Restriction map of the KlSDH1 locus. (B) KlSDH1 (kanMX4 URA3) deletion cassette. Open bar, open reading frame; black bar, gene promoter and terminator. (C) Southern analysis of the wild-type MW179-1D (1) and Κsdh1Δ isogenic strain (2). Genomic DNAs were digested with SalI and SphI and probed with a PCR-amplified fragment (about 3.6 kbp) containing, besides the coding region, 1.2 kbp of promoter and 0.4 kbp of terminator. (D) Mitochondrial (MIT) and cytoplasmic (CYT) extracts were prepared from glucose-grown cultures of MW179-1D (lanes 1 and 4), Κlsdh1Δ (lanes 2 and 5), and the Κlsdh1Δ strain complemented with a centromeric plasmid harboring KlSDH1 (lanes 3 and 6). The proteins were separated on native gel and stained for SDH activity as described in Materials and Methods. The arrow indicates the direction of the protein migration.
SDH assay on electrophoresis gels.
Cell extracts for the SDH staining assay were prepared in the following way. Cultures were grown to the early stationary phase in 20 ml of YP or 100 ml of minimal medium containing 2% glucose. Cells were collected, washed with 0.6 M sorbitol, and resuspended in 300 μl of TE-sorb (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.6 M sorbitol), and cold glass beads were added at about two-thirds the final solution volume (diameter, 0.5 mm; B. Braun Melsungen AG). Cell extracts were prepared by vortexing samples in micro tubes for 3 to 4 min in a refrigerator. Cell debris was pelleted for 5 min at 4,000 rpm in a bench top centrifuge (Sigma 1-26) and discarded. Supernatants were collected and centrifuged for another 30 min at 15,000 to 20,000 rpm. The cytoplasmic supernatant was kept and used as a control for an SDH native staining assay, and the mitochondrial pellet was washed once with 0.5 μl of TE-Sorb and centrifuged for another 30 min at 15,000 to 20,000 rpm. The supernatant was discarded, and the final pellet was resuspended in 15 μl of TE-triton (10 mM TRIS-HCl [pH 7.5], 1 mM EDTA, 0.2% Triton X-100) plus 15 μl of 4× loading buffer (0.1 M Tris-HCl [pH 6.8], 50% glycerol, 0.02 M β-mercaptoethanol, 0.008% bromphenol blue) and dispensed mechanically by repeated pipetting. A total of 25 μl of the mixture was loaded on 5% polyacrylamide native gel (Amresco) prepared as previously described (33, 34, 44). Supernatant (25 μl) was used as a cytoplasmic SDH staining control. The gel was run in a refrigerator in a mini-apparatus (Hoefer Scientific Instruments), at 20 mA constant current until the dye reached the bottom of the gel (about 1 h). The SDH staining mixture for one gel (5 ml) contains the following: 15 μl of phenazine methosulfate (catalogue no. P-9625; Sigma), equivalent to a concentration of 40 mg/ml in distilled water; 30 μl of nitro blue tetrazolium (product no. N-6876; Sigma), equivalent to a concentration of 50 mg/ml in distilled water; 30 μl of 18% Na succinate (pH 7.0); and up to 5 ml of distilled water. SDH staining was performed by soaking the gel with the 5-ml mixture until the band of activity appeared (about 2 h).
Quantitative assays of SDH and d-lactate ferricytochrome c oxidoreductase (D-LCR) activities were performed according to Lodi and Ferrero (28).
Cell respiration.
A total of 100 ml of cells grown on YP medium containing glucose was harvested by using a bench top centrifuge, washed twice with cold distilled water, and suspended in 0.1% KCl. Before oxygen consumption measurements, cells were pelleted and suspended in a volume of cold water twice the pellet volume. The respiratory rate was measured at 30°C by using a Clark-type electrode in a reaction vessel with 3 ml of air-saturated respiration buffer (0.1 M phthalate-KOH, pH 5.0) containing glucose (10 mM), lactate (10 mM), or ethanol (10 mM) and starting the reaction with 20 mg of cells (wet weight), according to the method of Ferrero et al. (16). The respiratory rate was expressed as microliters of O2 consumed per hour per milligram of dry mass (QO2). Dry mass was determined by weighing 1 ml of cell suspension oven dried overnight at 90°C. The respiration rate values represent the average of three independent measurements.
General methods.
DNA manipulation, plasmid engineering, and other techniques were performed according to standard procedures. Yeast transformation was performed by electroporation with a Bio-Rad Gene-Pulser apparatus following the method described by Becker and Guarente (4). Nucleotide sequencing was performed by MWG-BIOTECH AG from the complementing yeast plasmid. This plasmid (Kep6) (5) contained about 4.6 kbp of K. lactis genomic DNA (see Fig. 3A). Total RNA preparation has been previously described (44). Probes for KlSDH1 KlPDCA, KlACS1, KlACS2, KlICL1, and KlMLS1 were amplified from the K. lactis genome by PCR using the following oligonucleotide primers: KlSDH1 forward, GTCGACTTTCCCGACGGTGAG; KlSDH1 reverse, GCTTTTCTTTCCATGCACAACAAACG; KlPDCA forward, GCAAGTCGAAGTTCAAACCAT; KlPDCA reverse, GTTCTTAGCGTTGGATGCAGC; KlACS1 forward, TCTAACGCTTCAGCTGCCGC; KlACS1 reverse, CACGGAATCGATCAAGTGCT; KlACS2 forward, TAACGCGCAGGAAGCAAGGT; KlACS2 reverse, TGAATGGATCGTCACTGTGGAA; KlICL1 forward, ATGGTCTCCGTTAAGGCTTC; KlICL1 reverse, GAACTGATCCTCAGTGACAC; KlMLS1 forward, CCTCAATTCTCTCCAAGCAC; and KlMLS1 reverse, GCTGTGTGTAAGTCGTCATC.
The DNA sequence of KlMLS1 was obtained from the alignments of S. cerevisiae amino acid sequences with the corresponding K. lactis hypothetical ortholog peptides.
Protein concentration was determined by the method of Bradford (9).
RESULTS
Isolation of the KlSDH1.
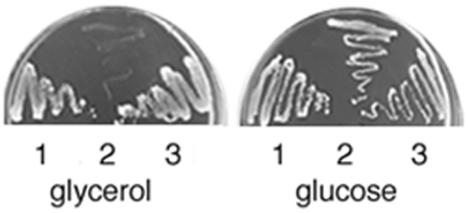
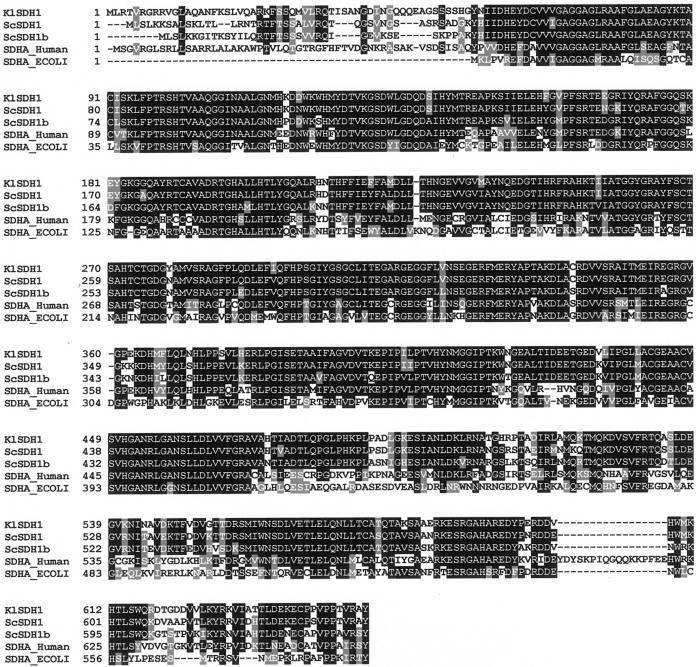
During a screening of K. lactis mutants affected in the respiratory metabolism, we isolated MS14-1A, a mutant that failed to grow on minimal media containing respiratory carbon sources such as glycerol (Fig. 1), acetate, ethanol, pyruvate and succinate (data not shown), but it was still able to grow in the same medium containing lactate (see Fig. 6). We performed the genetic analysis of this strain by crossing it with the wild-type strain CBS2359/152. The presence of a single mutation in the MS14-1A strain was demonstrated by the meiotic 2+:2− segregation of the growth phenotype on minimal medium containing glycerol. In a functional complementation experiment, MS14-1A was transformed with a genomic library in the Kep6 multicopy vector (5). Out of 14,000 Ura+ transformants analyzed, two showed a growth rate comparable to that of the wild-type reference strains (Fig. 1). Restriction analysis of the plasmids recovered from the two clones showed identical DNA inserts of 6.1 kbp. By deletion, we reduced the complementing region to a fragment of about 4.6 kbp. Sequence analysis showed the presence in this fragment of an open reading frame of 651 codons located between the SspI sites (see Fig. 3). A deduced amino acid sequence comparison revealed a significant degree of identity with the genes coding for the flavoprotein subunit of the SDH complex from various organisms (7, 11, 24, 42, 52). As shown in Fig. 2, a multiple protein alignment showed high sequence conservation along the entire protein between the Klsdh1p and other Sdh1 proteins. The K. lactis protein was 52% identical to that of E. coli, 62% identical to the Homo sapiens protein, and 84 and 79% identical to the S. cerevisiae Sdh1 and Sdh1b proteins, respectively. There was a higher degree of identity if we exclude the first 56 amino acids, which represent the mitochondrial targeting sequence that is absent in the E. coli protein (11, 40). We therefore called the K. lactis gene KlSDH1.
FIG. 1.
Growth was compared between strains MW179-1D (1), MS14-1A (2), and MS14-1A (3) complemented with the multicopy plasmid containing the KlSDH1 gene. The plates are minimal media containing glycerol or glucose as carbon sources.
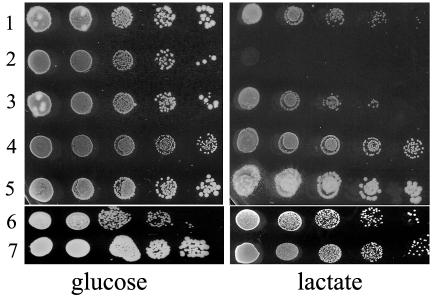
FIG. 6.
Growth test of the S. cerevisiae wild type BY4741 (1), BY4741sdh1Δ (2), and BY4741sdh1bΔ (3) strains and K. lactis MW179-1DKlsdh1Δ (4), MW179-1D (5), GG1993 (Klpda1Δ) (6), and CBS2359/152 (7). The plates are minimal media containing glucose or lactate. GG1993 is an isogenic derivative of CBS2359 (50). The initial cell concentration was 2 × 107 to 4 × 107 with a 10-fold dilution.
FIG. 2.
Sequence alignments of the flavoprotein subunit of the SDH complex from K. lactis (KlSDH1), S. cerevisiae (ScSDH1 and ScSDH1b), H. sapiens (SDHA_HUMAN), and E. coli (SDHA_ECOLI). The alignments have been performed with MultiAlin (14). Identical amino acids are shaded in black, and isofunctional ones are shaded in gray.
Construction of the KlSDH1 null mutant.
In S. cerevisiae, strains lacking SDH1 are unable to grow on nonfermentable carbon sources while SDH1b is slightly expressed and dispensable (13, 47). To study the effect of the deletion of KlSDH1 in K. lactis cells, we constructed a Klsdh1 disrupted mutant. Southern analysis (Fig. 3C) confirmed the correct integration of the cassette at the KlSDH1 locus. In fact, the wild-type 4.4-kbp fragment (lane 1) was replaced, as expected, by a 5.5-kbp fragment in the mutant (lane 2). The single hybridization band observed by Southern blotting and the analysis of the entire genome sequences confirmed that K. lactis has a single SDH1 gene (Yvan Zivanovic [Université de Paris-Sud], personal communication). A growth test on minimal media containing various carbon sources confirmed that the deleted strain, like the MS14-1A mutant, was unable to grow on all respiratory carbon sources tested, with the exception of lactate.
We also checked for the presence of SDH activity in both the wild type and in the deletion strains. Mitochondrial and cytoplasmic extracts were prepared from glucose-grown cultures, electrophoresed on native gels, and stained for SDH activity. As shown in Fig. 3D, the wild-type strain showed a single band of activity (lane 1) that was missing both in the deletion strain (lane 2) and in the MS14-1A mutant strain (data not shown). The activity could be detected again when the deletion strain was transformed with the KlSDH1 gene on a centromeric plasmid (lane 3). No SDH activity was detected in the cytoplasmic fractions of any strains (lanes 4, 5, and 6). The same result was obtained in lactate-grown cells. A quantitative spectrophotometric determination (28) in the mitochondrial extracts revealed an SDH activity value of 60 (expressed as nanomoles of substrate utilized per minute per milligram of protein) for the wild-type strain while no activity was detected in the Klsdh1Δ mutant. As a control, the extracts tested for D-LCR (28) activity showed high levels in both the wild type and the Klsdh1Δ mutant (857 and 1,099 nmol of substrate per min per mg of protein, respectively).
KlSDH1 is not repressed by glucose.
The SDH complex of S. cerevisiae has been extensively studied. Like many other genes of the Krebs cycle, the respiratory chain, and gluconeogenesis, SDH1 and SDH2 are regulated at the transcription level by carbon sources (19, 31, 41, 45).
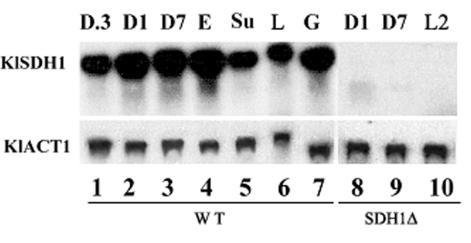
To investigate the regulation of KlSDH1, we performed a Northern analysis in three different reference strains of K. lactis, namely CBS2359/152, MW278-20C/1, and MW179-1D. Results showed that, unlike S. cerevisiae, the KlSDH1 transcript was always present in all strains grown on both fermentable and nonfermentable carbon sources (Fig. 4). In particular, the amount of the KlSDH1 mRNA from ethanol and glycerol cultures was very similar to that of cells grown in 1 and 7% glucose, indicating that the gene was not sensitive to carbon catabolite repression. These results are in agreement with the general observation that respiratory genes of K. lactis escape transcriptional repression during growth on glucose (26, 35, 36, 53). No KlSDH1 mRNA could be detected in the Klsdh1Δ strain grown on different concentrations of glucose or lactate (lanes 8, 9, and 10).
FIG. 4.
Transcription analysis of KlSDH1 in strain MW179-1D (lanes 1 to 7) and the Κlsdh1Δ (lanes 8 to 10) mutant grown in rich medium (YP) containing 0.3 (D.3), 1 (D1) or 7% (D7) glucose, 2% ethanol (E), 1% succinate (Su), 2% lactate (L), or 2% glycerol (G). Total RNAs were probed with the KlSDH1 1.5-kbp BglII-XbaI fragment (represented in Fig. 3A). Expression of the actin gene (KlACT1) was included as a relatively constant reference.
Growth of the Klsdh1Δ mutant on glucose.
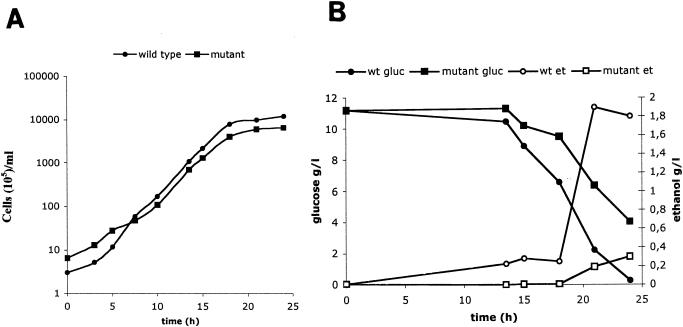
We also examined the growth of the mutant on glucose. The wild-type and Klsdh1Δ strains were grown on 50 ml of YP medium containing 1% glucose, and at time intervals, cell numbers, glucose consumption, and ethanol production were determined. Results showed that the Klsdh1Δ mutant had a reduced growth rate both in lag and stationary phases, while the growth curves of the two strains were similar during the exponential phase (Fig. 5A). Despite the small difference between the two strains in biomass yield, the levels of glucose consumption and ethanol production in the mutant cultures were significantly reduced. In fact, after 25 h of growth, about 40% of the glucose was still present in the medium while, in the wild-type cultures, the sugar was completely exhausted (Fig. 5B). Moreover, alcohol production in the mutant cultures was strongly delayed and only after 25 h reached a concentration of about 0.3g/liter, which is about one-seventh the amount produced by the wild type (about 2g/liter). These data clearly indicated that the growth of the Klsdh1Δ mutant was essentially due to the respiration of glucose and not to increased fermentative activity. Interestingly, dry biomass measurements of the same cell concentrations (for instance, 3 × 108 cells/ml) showed that the mutant weighs on average 15% less than the wild type. This difference could not alone account for the big difference in ethanol production (see below).
FIG. 5.
Growth curves of the MW179-1D (wild type or wt) and the Κlsdh1Δ (mutant) strain on YP medium containing 1% glucose (A). Glucose (gluc) and ethanol (et) concentrations were determined at time intervals on the culture supernatant (B). Each value in the figures represents the average of three independent determinations. In no case was the variation higher than 15%.
KlSDH1 is dispensable for growth on lactate.
In both S. cerevisiae and K. lactis, dl-lactate is oxidized to pyruvate by the mitochondrial enzymes d-lcr and l-lactate ferricytochrome c oxidoreductase (L-LCR), encoded by the genes DLD1 (KlDLD1) and CYB2 (KlCYB2), respectively, which use cytochrome c as the electron acceptor of the reaction (see Fig. 8) (1, 23, 28, 29). In S. cerevisiae, pyruvate derived from lactate is channeled into the Krebs cycle, and the disruption of the SDH1 gene blocks the capability of cells to utilize all respiratory carbon sources including lactate (Fig. 6, line 2) (27). In contrast, in K. lactis, the Klsdh1Δ strain, as well as the MS14-1A sdh mutant originally isolated, failed to grow on respiratory carbon sources with the exception of lactate (Fig. 6, line 4), indicating that lactate utilization in this yeast can be achieved through an alternative pathway bypassing SDH. Therefore, we analyzed the respiratory capability of the Klsdh1Δ strain. Cells grown on YP-glucose medium until the late exponential phase were starved for 24 h previous to the determination. Oxygen consumption was measured in the presence of glucose, ethanol, or lactate. As shown in Table 2, we found that the respiration rate in the presence of glucose was similar or even higher in the mutant compared to the level in the wild type. Moreover, high respiratory activity was also observed in the presence of ethanol, a substrate that the mutant cannot use for growth. On lactate, the mutant showed a rate of respiration that was significant, although reduced to one-eighth of that of the wild type (3.6 instead of 31 μl of oxygen consumed per hour per mg of dry cells). The addition of glucose to the lactate system restored high respiration levels (57.6 μl of oxygen). The addition of antimycin A blocked oxygen consumption completely, consistent with the observation that neither the wild type nor the deletion strain was able to grow on lactate medium containing this respiratory chain inhibitor. The oxygen consumption observed on lactate indicated that the SDH-alternative lactate catabolism of Klsdh1Δ cells is mediated by the antimycin A-sensitive respiratory chain. Interestingly, as observed in the Klsdh1Δ mutant, we found that the K. lactis strain devoid of KlPDA1 (54), the gene encoding the E1α subunit of the mitochondrial pyruvate dehydrogenase complex, was able to grow on lactate (Fig. 6, line 6). This suggested that in both mutants, pyruvate might be dissimilated through the pyruvate decarboxylase (PDC) pathway that bypasses the pyruvate dehydrogenase complex.
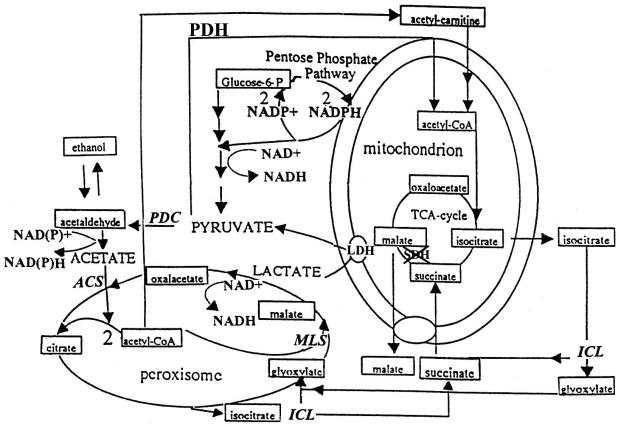
FIG. 8.
Schematic drawing of the hypothetical metabolic routes (glycolysis, Krebs, and glyoxylate cycles), modified from Tabak et al. (46), involved in succinate accumulation in the Klsdh1Δ mutant. The major sources of NAD(P)H production during growth on glucose and lactate are indicated. The pentose phosphate shunt has been included because in K. lactis this pathway is probably constitutively operative. The NADPH produced in this pathway can be reoxidized directly via mitochondrial membrane dehydrogenase; ACS, PDC, MLS, and ICL indicate the genes coding for the following activities, respectively: acetyl-coA synthetase, pyruvate decarboxylase, malate synthase, and isocitrate lyase. LDH indicates the mitochondrial activities responsible for lactate oxidation encoded by the KlCYB2 and KlDLD1 genes. PDH, pyruvate dehydrogenase complex. To indicate the block in the SDH route, SDH has been crossed out (×).
TABLE 2.
Respiration rates of strains grown on YP medium containing glucose
| Growth in the presence of: | O2 consumption (μl of O2 consumed/h/mg of dry mass)a
|
|
|---|---|---|
| Wild typeb | KlsdhΔ | |
| Glucose | 45 | 56 |
| Lactate | 31 | 3.6 (57.6)c |
| Ethanol | 90 | 68.4 |
Strains were grown on YP medium containing glucose until late exponential phase, starved for 24 h, and then shifted on glucose, lactate, and ethanol. All values represent the average of three independent experiments. In no case was the variation greater than 15%.
Strain MW179-1D.
The value in parentheses represents oxygen consumption after the addition of glucose to the sample.
Expression of pyruvate-related genes in the Klsdh1Δ mutant.
The Klsdh1Δ strain was further analyzed to explain the growth on lactate and the reduced fermentative capabilities. We performed a Northern analysis of the genes involved in pyruvate utilization to look at the metabolic route of its dissimilation. Therefore, we analyzed the expression levels of the genes for pyruvate decarboxylase (KlPDCA) (6) and acetyl-coenzyme A (acetyl-CoA) synthetase (KlACS1 and KlACS2) (55), and two genes of the glyoxylate cycle, namely isocitrate lyase (KlICL1) (32) and malate synthase (KlMLS1) (see Fig. 8 for a representation of the reactions catalyzed by these activities). Wild-type and Klsdh1Δ mutant strains were grown overnight on YP medium containing 1% glucose or 2% lactate, and total RNAs were prepared from these cultures. In K. lactis the regulation of the two genes for acetyl-CoA synthetase has been previously described: KlACS1 is expressed at low level on glucose or ethanol and induced on acetate or lactate, while KlACS2 is preferentially expressed on glucose and ethanol (30, 55). Such regulation of the ACS genes is present also in our wild-type strain grown on glucose (Fig. 7 lane 1) and lactate (lane 3). Interestingly, KlACS1 and KlACS2 in the mutant strain were expressed at higher levels compared to the wild type when the mutant strain was grown in lactate (lane 4) and glucose (lane 2), respectively. Compared to expression in the wild-type strain (lanes 1 and 3), increased expression of these genes in the mutant (lanes 2 and 4) was also observed for KlMLS1 and KlICL1 on both glucose and lactate. While the low level of the KlPDCA transcript observed in the Klsdh1Δ mutant on glucose (lane 2) was unexpected since this gene is normally induced by this carbon source (6), the level of expression is in agreement with the reduced fermentative capabilities of the mutant. The high level of expression of KlPDCA on lactate in both strains (lanes 3 and 4) confirmed that this gene could play a role in lactate utilization.
FIG. 7.
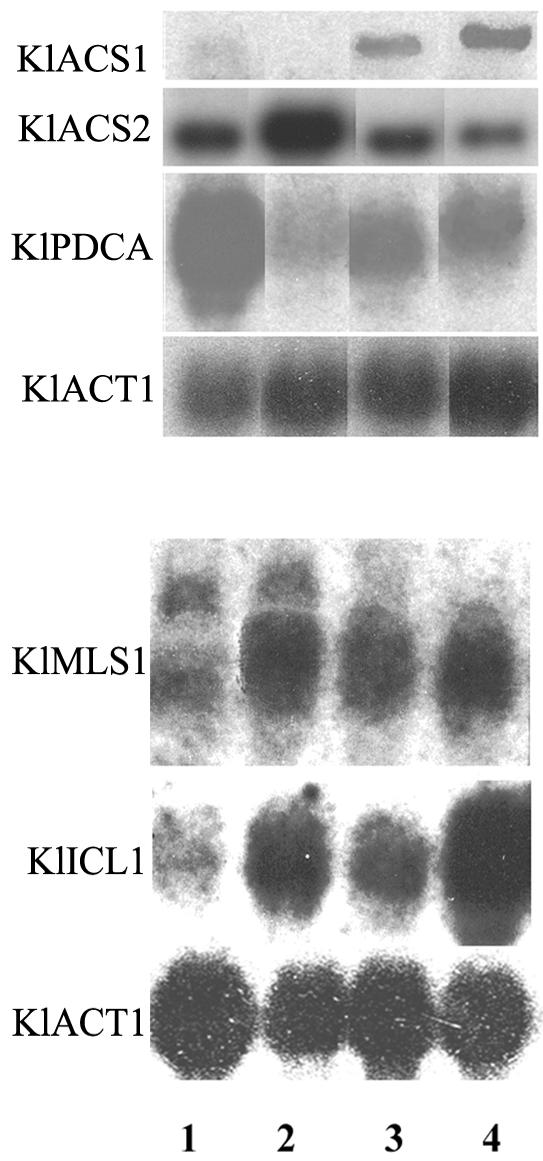
Transcription analysis of KlACS1, KlACS2, KlPDCA, KlMLS1, and KlICL1 genes in wild-type MW179-1D (lanes 1 and 3) and the Klsdh1Δ mutant (lanes 2 and 4) grown in YP medium containing 1% glucose (lanes 1 and 2) or YP medium containing 2% lactate (lanes 3 and 4). Transcription of the actin gene (KlACT1) was included as a relatively constant reference.
Succinate determination in the Klsdh1Δ mutant.
The activation of the glyoxylate cycle, indicated by the high levels of the KlICL1 and KlMLS1 transcripts (Fig. 7, lanes 2 and 4), should follow the deletion of KlSDH1. If the activation of this cycle occurred, we would expect from one mole of glucose the production by isocitrate lyase of one mole of glyoxylate and one mole of succinate. Figure 8 shows a schematic representation of the metabolic routes of the Krebs and the glyoxylate cycles that determine succinate utilization and production. The inability of the Klsdh1Δ mutant to oxidize succinate would lead to succinate accumulation into the medium. To test this hypothesis, the wild type and Klsdh1Δ strain were grown on YP medium containing 1% glucose or 2% lactate, and the production of succinate was determined at 24 and 36 h. Indeed, the Klsdh1Δ mutant, when grown on glucose, accumulates succinate at concentrations of about 0.5g/liter at 24 h and 1.2 g/liter at 36 h. In contrast, the amount of succinate in the wild type was, under the same conditions, below the sensitivity of the system. No succinate was detected in either the wild-type or mutant strain during growth on lactate.
DISCUSSION
Glucose metabolism in the Klsdh1Δ mutant.
SDH is an important complex of the Krebs cycle that catalyzes the oxidation of succinate to fumarate and feeds electrons to the respiratory chain ubiquinone pool. SDH1 is the catalytic component of this complex. While S. cerevisiae has two paralogues of SDH1, KlSDH1 appears to be a unique gene in K. lactis. More important, the regulation of the expression of these genes is very different in the two yeasts. In fact, in S. cerevisiae SDH1 is derepressed at the transcriptional level in the presence of nonfermentable carbon sources, while in K. lactis, KlSDH1 is expressed even in glucose-grown cells. S. cerevisiae strains devoid of SDH1 were unable to grow on respiratory carbon sources, whereas a Klsdh1 null strain was still able to grow on lactate. Moreover, the Klsdh1Δ mutant showed decreased glucose consumption and severely reduced ethanol production. Accordingly, we found a reduced level of expression of the pyruvate decarboxylase gene (KlPDCA) during growth on glucose. Recently, it has been reported that KlICL1, the K. lactis gene coding for isocitrate lyase, a key enzyme of the glyoxylate cycle, is glucose repressed (32). On the contrary, in the Klsdh1Δ mutant we found not only specific induction of this gene but also increased expression of the malate synthase (KlMLS1) and acetyl-CoA synthetase (KlACS2) genes, which suggested a rerouting of the glucose flux towards the glyoxylate cycle. It follows that acetaldehyde, due to the inability of the Klsdh1Δ mutant to accumulate ethanol, is converted to acetate activated to acetyl-CoA that is finally channeled into the glyoxylate cycle. The use of this cycle in a strain blocked in SDH would lead to the accumulation of succinate, as demonstrated in the scheme of Fig. 8 that shows the metabolic pathways involved in the production and utilization of this organic acid. This hypothesis was confirmed in the Klsdh1Δ mutant by the accumulation of succinate (1.2g/liter at 36 h). Therefore, the accumulation of succinate instead of ethanol in K. lactis suggests a bond between the expression of the KlSDH1 gene on glucose and efficient fermentation. The production of succinate was previously observed to a lower extent under very different growth conditions (15% glucose) also on the S. cerevisiae sdh1Δ mutant (2). According to the authors of that study, in this yeast succinate accumulation was not dependent on the glyoxylate cycle but on the activation of the oxidative part of the Krebs cycle (2). Since this cycle is the main source of the precursors, in particular α-ketoglutarate and oxaloacetate, necessary for the synthesis of amino acids, nucleotides, heme, etc. (56), the mutation in SDH may block the supply of the intermediates necessary for the second part of the Krebs cycle. In K. lactis, unlike S. cerevisiae, anaplerotic pathways like the glyoxylate cycle are activated to bypass this block. The high respiration rate observed in the mutant not only on glucose but also on ethanol suggests that the respiratory chain is fully functional.
In fact, the activation of the glyoxylate cycle and the production of succinate instead of ethanol require the reoxidation of the NAD(P)H produced at the level of the glyceraldehyde 3-phosphate and acetaldehyde (Fig. 8), whereas the high oxygen consumption observed on ethanol could be associated to the reoxidation of the NAD(P)H produced by alcohol and aldehyde dehydrogenases. It has been reported that K. lactis, unlike S. cerevisiae, is able via mitochondria to oxidize directly not only NADH but also NADPH (3, 20, 21, 25, 37). In both cases, the redox potential may be directly transferred via mitochondrial membrane transdehydrogenases (3, 37) to the ubiquinone pool, thus avoiding the SDH block in the Krebs cycle (8).
How can the Klsdh1Δ mutant grow on lactate and not on other respiratory substrates?
K. lactis cells lacking SDH showed a high respiration rate on ethanol but failed to grow on this carbon source as well as on pyruvate and glycerol, substrates that in principle can be dissimilated through the PDC pathway. This fact indicates that respiration is a condition necessary but not sufficient for the utilization of nonfermentable carbon sources. One possible explanation is that, following the impairment of the Krebs cycle, the synthesis of ATP required for acetate activation becomes the limiting step for the utilization of these substrates. In the case of lactate, the fundamental step for the utilization of this substrate is its oxidation to pyruvate. This reaction catalyzed by the mitochondrial d-lcr and l-lcr use cytochrome c as the electron acceptor of the reaction. Reoxidation of cytochrome c at the level of complex IV may produce the proton force gradient for ATP synthesis necessary for pyruvate activation. Since we have found that the Klpda1Δ strain is able to grow on lactate (Fig. 6, line 6), lactate-derived pyruvate must be utilized through the pyruvate decarboxylase pathway. In fact, the high transcription levels of the KlPDCA, KlACS1, KlICL1, and KlMLS1 genes suggest activation of this pathway that leads to the glyoxylate cycle, as shown in the scheme of Fig. 8. Although this metabolic route is required in the K. lactis Klsdh1Δ mutant for growth on lactate, it is not clear how this mutant avoids succinate accumulation. We can hypothesize that, on lactate, the request of intermediates necessary for the synthesis of heme, gluconeogenesis, or other metabolites is so high that succinate accumulation is prevented. Alternatively, we could think that other activities, not expressed on glucose and specifically induced by lactate, avoid succinate accumulation. Further investigation will be necessary to unveil the metabolic route(s) beyond lactate utilization. The reduced oxygen consumption of the Klsdh1Δ mutant cells on lactate, compared to consumption in the wild type, could be due to the low levels of the reducing equivalents produced mainly in the conversion from acetaldehyde to acetate and from malate to oxaloacetate (Fig. 8). We also asked why S. cerevisiae sdh1 mutants that harbor mitochondrial D-LCR and L-LCR are unable to grow on lactate. One possible explanation is that respiration, inferred by many authors only by nongrowth on respiratory carbon sources, and/or the PDC pathways are not active in such mutants. In conclusion, the reported pathway for lactate utilization could be a peculiar trait of K. lactis, acquired in the milk-derived niches that are the natural habitats of this yeast (1, 48). We hypothesize that the evolution of this SDH-independent lactate pathway in K. lactis might have evolved from the coexistence in the same habitat of lactic acid-producing bacteria and microaerobic conditions. The idea that K. lactis could choose the modality of lactate utilization depending on oxygen availability is appealing, and we wonder if a similar mechanism may exist also in higher eukaryotes.
Acknowledgments
We thank Claudia Getuli for help in the genetic analysis, Wésolowski-Louvel (Universitè Claude Bernard Lyon1) for the K. lactis genomic bank and for the strains used in this work, and H. Y. Steensma (University of Leiden) for providing GG1993 and PM6-7A/Klpda1Δ null strains. We thank Daniela Uccelletti for helpful discussion and critical reading of the manuscript and M. Bolotin-Fukuhara (Universitè of Paris Sud) for the sequence of the K. lactis KlMLS1 genes.
This work was supported by the COFIN2002 grant (code 2002052349) from Ministero Istruzione Università e Ricerca (MIUR).
REFERENCES
- 1.Alberti, A., P. Goffrini, I. Ferrero, and T. Lodi. 2000. Cloning and characterization of the lactate-specific inducible gene KlCYB2, encoding the cytochrome b(2) of Kluyveromyces lactis. Yeast 16:657-665. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa, Y., T. Kuroyanagi, M. Shimosaka, H. Muratsudaki, K. Enomoto, R. Kodaira, and M. Okazaki. 1999. Effect of gene disruptions of the TCA cycle on production of succinic acid in Saccharomyces cerevisiae. J. Biosci. Bioeng. 87:28-36. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, B. M., K. M. Overkamp, A. J. van Maris, P. Kotter, M. A. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. [DOI] [PubMed] [Google Scholar]
- 4.Becker, D. M., and L. Guarente. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194:182-187. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi, M. M., C. Falcone, M. Wésolowski-Louvel, L. Frontali, and H. Fukuhara. 1987. Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1.6 μm circular plasmid pKD1. Curr. Genet. 12:185-192. [Google Scholar]
- 6.Bianchi, M. M., L. Tizzani, M. Destrulle, L. Frontali, and M. Wésolowski-Louvel. 1996. The “petite-negative” yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol. Microbiol. 19:27-36. [DOI] [PubMed] [Google Scholar]
- 7.Birch-Machin, M. A., L. Farnsworth, B. A. Ackrell, B. Cochran, S. Jackson, L. A. Bindoff, A. Aitken, A. G. Diamond, and D. M. Turnbull. 1992. The sequence of the flavoprotein subunit of bovine heart succinate dehydrogenase. J. Biol. Chem. 267:11553-11555. [PubMed] [Google Scholar]
- 8.Boumans, H., L. A. Grivell, and J. A. Berden. 1998. The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 273:4872-4877. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Bullis, B. L., and B. D. Lemire. 1994. Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J. Biol. Chem. 269:6543-6549. [PubMed] [Google Scholar]
- 11.Chapman, K. B., S. D. Solomon, and J. D. Boeke. 1992. SDH1, the gene encoding the succinate dehydrogenase flavoprotein subunit from Saccharomyces cerevisiae. Gene 118:131-136. [DOI] [PubMed] [Google Scholar]
- 12.Ciriacy, M. 1977. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 154:213-220. [DOI] [PubMed] [Google Scholar]
- 13.Colby, G., Y. Ishii, and A. Tzagoloff. 1998. Suppression of sdh1 mutations by the SDH1b gene of Saccharomyces cerevisiae. Yeast 14:1001-1006. [DOI] [PubMed] [Google Scholar]
- 14.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daignan-Fourier, B., M. Valens, B. D. Lemire, and M. Bolotin-Fukuhara. 1994. Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J. Biol. Chem. 269:15469-15472. [PubMed] [Google Scholar]
- 16.Ferrero, I., A. M. Viola, and A. Goffeau. 1981. Induction by glucose of an antimycin-insensitive, azide-sensitive respiration in the yeast Kluyveromyces lactis. Antonie Van Leeuwenhoek 47:11-24. [DOI] [PubMed] [Google Scholar]
- 17.Fiori, A., M. Saliola, P. Goffrini, and C. Falcone. 2000. Isolation and molecular characterization of KlCOX14, a gene of Kluyveromyces lactis encoding a protein necessary for the assembly of the cytochrome oxidase complex. Yeast 16:307-314. [DOI] [PubMed] [Google Scholar]
- 18.Flores, C. L., C. Rodriguez, T. Petit, and C. Gancedo. 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 24:507-529. [DOI] [PubMed] [Google Scholar]
- 19.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez Siso, M. I., M. A. Freire Picos, and M. E. Cerdan. 1996. Reoxidation of the NADPH produced by the pentose phosphate pathway is necessary for the utilization of glucose by Kluyveromyces lactis rag2 mutants. FEBS Lett. 387:7-10. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez Siso, M. I., M. A. Freire Picos, E. Ramil, M. Gonzalez Dominguez, A. Rodriguez Tarres, and M. E. Cerdan. 2000. Respirofermentative metabolism in Kluyveromyces lactis: insights and perspectives. Enzyme Microb. Technol. 26:699-705. [DOI] [PubMed] [Google Scholar]
- 22.Gould, S. J., S. Subramani, and I. E. Scheffler. 1989. Use of the DNA polymerase chain reaction for homology probing: isolation of partial cDNA or genomic clone encoding the iron-sulphur protein of succinate dehydrogenase from several species. Proc. Natl. Acad. Sci. USA 86:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guiard, B. 1985. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 4:3265-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirawake, H., H. Wang, T. Kuramochi, S. Kojima, and K. Kita. 1994. Human complex II (succinate-ubiquinone oxidoreductase): cDNA cloning of the flavoprotein (Fp) subunit of liver mitochondria. J. Biochem. (Tokyo) 116:221-227. [DOI] [PubMed] [Google Scholar]
- 25.Jacoby, J., C. P. Hollenberg, and J. J. Heinisch. 1993. Transaldolase mutants in the yeast Kluyveromyces lactis provide evidence that glucose can be metabolized through the pentose phosphate pathway. Mol. Microbiol. 10:867-876. [DOI] [PubMed] [Google Scholar]
- 26.Kiers, J., A. M. Zeeman, M. Luttik, C. Thiele, J. I. Castrillo, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1998. Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 14:459-469. [DOI] [PubMed] [Google Scholar]
- 27.Lemire, B. D., and B. D. Oyedotun. 2002. The mitochondrial succinate: ubiquinone reductase. Biochim. Biophys. Acta 1553:102-116. [DOI] [PubMed] [Google Scholar]
- 28.Lodi, T., and I. Ferrero. 1993. Isolation of the DLD1 gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol. Gen. Genet. 238:315-324. [DOI] [PubMed] [Google Scholar]
- 29.Lodi, T., D. O'Connor, P. Goffrini, and I. Ferrero. 1994. Carbon catabolite repression in Kluyveromyces lactis: isolation and characterization of the KIDLD gene encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol. Gen. Genet. 244:622-629. [DOI] [PubMed] [Google Scholar]
- 30.Lodi, T., M. Saliola, C. Donnini, and P. Goffrini. 2001. Three target genes for the transcriptional activator Cat8p of Kluyveromyces lactis: acetyl coenzyme A synthetase genes KlACS1 and KlACS2 and lactate permease gene KlJEN1. J. Bacteriol. 183:5257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombardo, A., K. Carine, and I. E. Scheffler. 1990. Cloning and characterization of the iron-sulfur subunit gene of succinate dehydrogenase from Saccharomyces cerevisiae. J. Biol. Chem. 265:10419-10423. [PubMed] [Google Scholar]
- 32.Lopez, M. L., B. Redruello, E. Valdes, F. Moreno, J. J. Heinisch, and R. Rodricio. 2004. Isocitrate lyase of the yeast Kluyveromyces lactis is subject to glucose repression but not to catabolite inactivation. Curr. Genet. 44:305-316. [DOI] [PubMed] [Google Scholar]
- 33.Lutstorf, U., and R. Megnet. 1968. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 126:933-944. [DOI] [PubMed] [Google Scholar]
- 34.Mazzoni, C., M. Saliola, and C. Falcone. 1992. Ethanol-induced and glucose-insensitive alcohol dehydrogenase in the yeast Kluyveromyces lactis. Mol. Microbiol. 6:2279-2286. [DOI] [PubMed] [Google Scholar]
- 35.Mulder, W., I. H. Scholten, and L. A. Grivell. 1995. Carbon catabolite regulation of transcription of nuclear genes coding for mitochondrial proteins in the yeast Kluyveromyces lactis. Curr. Genet. 28:267-273. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, C., M. Bolotin-Fukuhara, M. Wesolowski-Louvel, and H. Fukuhara. 1995. The respiratory system of Kluyveromyces lactis escapes from HAP2 control. Gene 152:113-115. [DOI] [PubMed] [Google Scholar]
- 37.Overkamp, K. M., B. M. Bakker, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 2002. Two mechanisms for oxidation of cytosolic NADPH by Kluyveromyces lactis mitochondria. Yeast 19:813-824. [DOI] [PubMed] [Google Scholar]
- 38.Oyedotun, K. S., and B. D. Lemire. 1999. The Saccharomyces cerevisiae succinate-ubiquinone reductase contains a stoichiometric amount of cytochrome b562. FEBS Lett. 442:203-207. [DOI] [PubMed] [Google Scholar]
- 39.Parfait, B., D. Chretien, A. Roetig, C. Marsac, A. Munnich, and P. Rustin. 2000. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum. Genet. 106:236-243. [DOI] [PubMed] [Google Scholar]
- 40.Pon, L., and G. Schatz. 1991. Biogenesis of yeast mitochondria, p. 333-406. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Prieto, S., B. J. de la Cruz, and I. E. Scheffler. 2000. Glucose-regulated turnover of mRNA and the influence of poly(A) tail length on half-life. J. Biol. Chem. 275:14155-14166. [DOI] [PubMed] [Google Scholar]
- 42.Robinson, K. M., and B. D. Lemire. 1992. Isolation and nucleotide sequence of the Saccharomyces cerevisiae gene for the succinate dehydrogenase flavoprotein subunit. J. Biol. Chem. 267:10101-10107. [PubMed] [Google Scholar]
- 43.Rustin, P., and A. Roetig. 2002. Inborn errors of complex II: unusual human mitochondrial diseases. Biochim. Biophys. Acta 1553:117-122. [DOI] [PubMed] [Google Scholar]
- 44.Saliola, M., and C. Falcone. 1995. Two mitochondrial alcohol dehydrogenase activities of Kluyveromyces lactis are differentially expressed during respiration and fermentation. Mol. Gen. Genet. 249:665-672. [DOI] [PubMed] [Google Scholar]
- 45.Scheffler, I. E. 1998. Molecular genetics of succinate: quinone oxidoreductase in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 60:267-315. [DOI] [PubMed] [Google Scholar]
- 46.Tabak, H. F., I. Braakman, and B. Distel. 1999. Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol. 9:447-453. [DOI] [PubMed] [Google Scholar]
- 47.Tzagoloff, A., and C. L. Dieckmann. 1990. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54:211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Walt, J. P., and E. Johannensen. 1984. Genus 13. Kluyveromyces van der Walt emend. van der Walt, p. 224-251. In N. J. W. Kreger-van Rij (ed.), The yeasts: a taxonomic study. Elsevier, New York, N.Y.
- 49.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 50.Wésolowski-Louvel, M., A. Algeri, P. Goffrini, and H. Fukuhara. 1982. Killer DNA plasmids of the yeast Kluyveromyces lactis: mutation affecting the killer character. Curr. Genet. 5:191-197. [DOI] [PubMed] [Google Scholar]
- 51.Wésolowski-Louvel, M., K. D. Breunig, and F. Fukuhara. 1996. Kluyveromyces lactis, p. 139-201. In K. Wolf (ed.), Nonconventional yeasts in biotechnology. Springer-Verlag, Heidelberg, Germany.
- 52.Wood, D., M. G. Darlison, R. J. Wilde, and J. R. Guest. 1984. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem. J. 222:519-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaror, I., F. Marcus, D. L. Moyer, J. Tung, and J. R. Shuster. 1993. Fructose-1, 6-bisphosphatase of the yeast Kluyveromyces lactis. Eur. J. Biochem. 212:193-199. [DOI] [PubMed] [Google Scholar]
- 54.Zeeman, A. M., M. A. Luttik, C. Thiele, J. P. van Dijken, J. T. Pronk, and H. Y. Steensma. 1998. Inactivation of the Kluyveromyces lactis KlPDA1 gene leads to loss of pyruvate dehydrogenase activity, impairs growth on glucose and triggers aerobic alcoholic fermentation. Microbiology 144:3437-3446. [DOI] [PubMed] [Google Scholar]
- 55.Zeeman, A. M., and H. Y. Steensma. 2003. The acetyl co-enzyme A synthetase genes of Kluyveromyces lactis. Yeast 20:13-23. [DOI] [PubMed] [Google Scholar]
- 56.Zhengchang, L., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]