Abstract
The transcription factor Rim101p of Candida albicans has been shown to play a major role in pH-dependent gene regulation. Rim101p is involved in cell wall biosynthesis, since it regulates PHR1 and PHR2, two almost functionally redundant cell wall glycosidases important for adaptation to either neutral or acidic habitats within the human host. To identify additional cell wall components regulated by Rim101p, we performed transcriptional profiling with a cell wall-specific DNA microarray. We showed that Rim101p contributes to the activation of known hypha-specific genes such as HWP1 and RBT1 but is also required for repression of the previously uncharacterized potential cell wall genes RBR1, RBR2, and RBR3. Further characterization of RBR1 revealed that it encodes a small glycosylphosphatidyl inositol protein that is expressed under acidic conditions predominantly at low temperature. Deletion of the gene resulted in a filamentation defect at low pH. Most interestingly, NRG1, a transcriptional repressor of hyphal growth in C. albicans, was required for RBR1 expression. The apparently activating effect of NRG1 observed in this study has not been described before. In addition, we showed that expression of NRG1 is not only temperature but also pH dependent.
Candida albicans is the most frequent causative agent of candidiasis, which is among the most important nosocomial infections of humans. A key feature of C. albicans is its ability to grow in diverse microenvironments of the human body. Examples are the skin and oral and gut mucosae as well as the vaginal mucosa. Each of these niches imposes diverse stresses on the fungus, including nutrient limitation, temperature shifts, and change of pH. A complex network of signaling pathways mediates adaptation of C. albicans to these diverse environmental conditions (24). These signaling pathways converge on transcriptional activators such as Efg1p (39, 40) and Nrg1p, which is postulated to reconstitute a DNA-binding repressor complex (30).
The transcription factor Rim101p is crucial for the regulation of genes in response to external pH (33). Little is known about the processes involved in the pH-specific alteration of cell wall composition in C. albicans. The transcription of the two cell wall genes PHR1 and PRA1 has been shown to be activated in response to neutral pH in a Rim101p-dependent manner (8, 33, 36, 37), whereas Phr2p, encoding a Rim101p-dependent glycosidase with functions equivalent to Phr1p, is part of the cell wall in an acidic environment (10, 29).
RIM101 is the orthologue of Aspergillus nidulans PacC, the best studied of the above-mentioned pH-dependent transcription factors (16), and Saccharomyces cerevisiae RIM101 (23, 41). It contains a conserved DNA-binding Zn2+ finger region and is expressed as a preprotein. Rim101p activation at neutral pH depends on members of a proteolytic cascade, RIM8 and RIM20 (8). C-terminally truncated forms of the transcription factor lead to pH-independent constitutive activation or repression of Rim101p target genes (8, 13). For PHR1, it has recently been shown that direct binding of processed Rim101p to two consensus sites in the promoter results in transcription of the gene (32). On the other hand, PHR2 is repressed by RIM101 at neutral pH and becomes constitutively activated in a rim101Δ mutant (8, 33), although the promoter of PHR2 also contains two Rim101p consensus sites.
Similarly, PacC directly activates ipnA, an alkaline-expressed gene of the pH-dependent penicillin synthesis pathway and represses the acid-expressed gabA, which is subject to carbon catabolite and nitrogen metabolite repression and pH response in Aspergillus nidulans (14, 15, 42). S. cerevisiae RIM101 also has activating and repressing functions, since it plays a major activating role in meiosis and sporulation (23), but a variety of genes have also been shown to be repressed by transcriptional profiling. S. cerevisiae RIM101 indirectly regulates ion tolerance by repression of NRG1, a known repressor of transcription in S. cerevisiae and C. albicans (20, 21). In C. albicans, Nrg1p is known to interact with the transcription factors Tup1p and Mig1p to repress different subsets of hypha-expressed genes (30).
To investigate the RIM101-mediated pH-dependent changes in the cell wall of C. albicans, we performed transcriptional profiling experiments. We used a cell wall-specific DNA microarray comprising 117 open reading frames related to cell wall synthesis (39). Our results indicate both activating and repressing functions of RIM101. One of the genes repressed by RIM101, RBR1, was characterized in more detail, revealing a novel pH- and temperature-regulated gene from the cell wall of C. albicans required for proper hyphal development. Furthermore, our results indicate that NRG1 plays a mayor role in activating RBR1 and that NRG1 expression is regulated in a pH-dependent manner mediated by RIM101. These results correlate with induction of the hypha-specific gene HWP1. Moreover, RIM101 contributes to activation of RBT1, a hyphal gene repressed by Tup1p independent of Nrg1p (5). These results link pH regulation in C. albicans to both Nrg1p-dependent and Nrg1p-independent morphogenetic pathways.
MATERIALS AND METHODS
C. albicans strains.
The C. albicans strains used in this study are listed in Table 1. DNA microarray experiments were carried out with three different strains. The wild-type strain (SC5314), the homozygous rim101Δ deletion strain (CAF3-X ura3::imm434/ura3::imm434 rim101::hisG/rim101::hisG::URA3) (13) and the RIM101-1426 strain (CAF3-16-2 ura3::imm434/ura3::imm434 RIM101/RIM101-[pBSK+-RIM101-1426-URA3]n > 2), which overexpresses dominant active Rim101-1426p (13).
TABLE 1.
C. albicans strains used in this study
| Strain | Former name | Genotypea | Reference |
|---|---|---|---|
| Wild type | SC5314 | Clinical isolate | 19 |
| CAI4 | CAI4 | ura3::imm434/ura3::imm434 | 17 |
| rim101Δ | CAF3-X | ura3::imm434/ura3::imm434 rim101::hisG/rim101::hisG::URA3 | 13 |
| RIM101-1426 | CAF3-16-2 | ura3::imm434/ura3::imm434 RIM101/RIM101-(pBSK+-RIM101-1426-URA3)n > 2 | 13 |
| efg1Δ | HLC52 | ura3::imm434/ura3::imm434 efg1::hisG/efg1::hisG::URA3 | 25 |
| nrg1Δ | MMC3 | ura3::imm434/ura3::imm434 nrg1::hisG/nrg1::hisG::URA3 | 31 |
| nrg1-pMET3-NRG1 | MMC5 | ura3::imm434/ura3::imm434 nrg1::hisG/nrg1::hisG RP10::pMET3-NRG1-URA3 | 31 |
| tup1Δ | BCA2-9 | ura3::imm434/ura3::imm434 tup1::hisG/tup1::hisG::URA3 | 3 |
| rbr1Δ | HL12-2 and HL14-1 | ura3::imm434/ura3::imm434 rbr1::FRT/rbr1::FRT RP10::URA3 | This studyb |
| rbr1Δ-RBR1 | HL13-6 | ura3::imm434/ura3::imm434 rbr1::FRT/rbr1::FRT RP10::RBR1-URA3 | This study |
FRT denotes the FLP recognition sites remaining after excision of the URA3-FRT flipper cassette (28).
Mutant strains described in this study were constructed three times independently.
RBR1 deletion mutants were constructed by sequential homologous recombination of CAI4 with a URA3 flipper cassette (28). One inner and one outer pair of sequences flanking the RBR1 coding sequence were amplified by PCR. The outer pair of flanking regions (FR1 and FR2) were used for deletion of the first allele; the inner flanking regions (FR3 and FR4) were used for deletion of the second RBR1 allele. The following primers were used to amplify the outer and inner flanking regions by divergent PCR in a Peltier Thermal Cycler 200 (MJ Research) within 30 cycles at 55°C: FR1_for, 5′-AAGGGCCCCACAAAATAAAAGCAGCAGGAA, and FR1_rev, 5′-CCGCTCGAGTTCCAACTTTAATCCCGCAC (product length 457 bp); FR2_for, 5′-ATAAGAATGCGGCCGCTTGCCACCAGTCAAATTCAA, and FR2_rev, 5′-CGAGCTCCCGAAATGCCACCATAGTTT (product length 527 bp); FR3_for, 5′-AAGGGCCCGTGCGGGATTAAAGTTGGAA, and FR3_rev, 5′-CCGCTCGAGTTGTTGTTGTAAGCGAAGCC (product length 563 bp); FR4_for, 5′-ATAAGAATGCGGCCGCTGAATGAGAATGAGGGGGAC, and FR4_rev, 5′-CGAGCTCTTGAATTTGACTGGTGGCAA (product length 565 bp). Embedded in the primer sequences were unique cleavage sites (italics) for directed ligation of the flanking regions into plasmid pSFU1 (28).
FR1 and FR3 were ligated into the vector after ApaI and XhoI digestion, FR2 and FR4 were integrated via NotI and SacI, resulting in plasmids pSFUR1-2 for the outer and pSFUR1-4 for the inner deletion constructs. For reversion of homozygous rbr1Δ strains, the coding sequence and promoter region of RBR1 were cloned into the integrative C. albicans expression vector pCaExp (6) with primers 2736f-1_FR1_for (AAGGGCCCCACAAAATAAAAGCAGCAGGAA) and RBR1_RVT_rev (CCGCTCGAGCCGAAATGCCACCATAGTTT). This vector carries the URA3 gene under its native promoter and was designed for integration into the RP10 locus. We transformed three independent rbr1Δ mutant strains each with pCaExp and pCaExp-RBR1. Transformants expressing URA3 were selected on synthetic complete medium lacking uridine (SC-uri). Single colonies were picked and cultivated for 6 h in SC-uri at 30°C to confirm recombination of the RBR1 locus by Southern blotting. To monitor mRNA abundance by Northern blot experiments, strains were grown at 30°C in alpha minimal essential medium (α-MEM) buffered to pH 4.5.
Media and growth conditions.
All media used in this study contained final concentrations of 100 mM HEPES and 0.1 mM uridine. Desired pHs were adjusted with either 1 M NaOH to pH 7.4 or 1 M HCl to pH 4.5 before filter sterilization. For DNA microarray experiments, cells from overnight cultures grown at 30°C in YPD (Difco) at pH 7.4 and pH 4.5 were pelleted, decanted, and resuspended in the residual medium. Warmed medium of equal pH was inoculated with the cell suspension to an optical density at 600 nm of 0.05. The rim101Δ mutant and wild-type reference were grown at pH 4.5; to compare the RIM101-1426 mutant and wild-type reference strain, both strains were cultivated at pH 7.4. To avoid undesirable effects due to significant changes in pH, it was monitored at the end of each experiment. Fluctuation was not more than 0.2 pH units. When Northern blot experiments were conducted to confirm the DNA microarray data, the strains were cultivated under identical conditions.
Northern blot experiments and phenotypic studies were carried out in YPD, α-MEM (Gibco) and α-MEM without cysteine and methionine containing 2% glucose, and tissue culture medium M-199 (Gibco) with 0.1 mM uridine and 100 mM HEPES. Strains were grown at 25, 29, 30, and 37°C at acidic and neutral pHs as indicated in the Results. Solid medium contained either 2% agar or 0.3% agar for soft agar plates. Heated 4% agar was added to warmed, autoclaved, and filter-sterilized medium up to the final concentration.
Composition of the C. albicans cell wall DNA microarray.
The cell wall-specific DNA microarray was recently designed and described (39). This array consists of 117 different probes for known genes and not-yet-characterized open reading frames of C. albicans; 65 of these genes were designated homologues of cell wall-localized genes in S. cerevisiae. Open reading frames in this work are labeled following the assembly 6 names of the Stanford C. albicans database (http://www-sequence.stanford.edu/group/candida). In addition, the array comprises genes coding for proteins that are already known to be localized to the cell wall of C. albicans. Probes for actin, ACT1 (X16377), were included as controls.
DNA microarray preparation, isolation of RNA, labeling of cDNA, and hybridization to DNA microarrays.
DNA microarray preparation, isolation of RNA, labeling of cDNA, and hybridization to the DNA microarrays were done as previously described (39) with the following modifications: PCR was performed in PCR buffer (100 mM Tris-HCl [pH 8.8], 600 mM KCl, 15 mM MgCl2), containing 1 M betaine (Sigma) and 0.3 μM cresol red (Sigma). After analysis of the PCR products by agarose gel electrophoresis, the amount of liquid was reduced by evaporation overnight at 8°C to achieve a final betaine concentration of at least 2 M before spotting of the probes on polylysine-coated glass slides (12).
Cells for isolation of RNA were cultivated for 6 h, optical density was checked, and the cultures were harvested by brief centrifugation at 1,700 × g. Cell pellets were immediately frozen in liquid nitrogen, and the pH of the supernatant was checked. From the frozen cell pellets, total RNA was isolated as described before (39).
Fluorescently labeled cDNA for microarray hybridization was obtained by reverse transcription of 30 μg of RNA with Superscript II (Invitrogen) with either indocarbocyanine-dUTP or indodicarbocyanine-dUTP (Amersham) and hybridized to the DNA microarrays as described before (39).
Analysis of DNA microarray data.
DNA microarrays were scanned by epifluorescence microscopy with a GMS 418 array scanner (Genetic Microsystems) and the ImaGene software version 3.0 (BioDiscovery, Los Angeles, Calif.) (39). Data presented are means of duplicates from at least three different experiments. DNA microarray data were largely confirmed by Northern blot analysis under identical conditions (see above, Strains and growth conditions).
Northern hybridization.
For Northern hybridization, 15 μg of total RNA was separated by denaturing gel electrophoresis in 1% agarose-morpholinepropanesulfonic acid (MOPS) buffer (0.02 M MOPS, 8 mM sodium acetate, 1 mM EDTA) and 2.2 M formaldehyde. RNA was blotted onto Hybond-N (Amersham) nylon membranes with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) following standard protocols (35) and UV cross-linked on wet blots twice at 120 mJ in a Stratalinker (Stratagene). Blots were either prehybridized for 1 h at 65°C or frozen at −80°C before further processing. PCR-amplified probes (for primers, see Table 2) were purified and subsequently labeled with [α-32P]dCTP by Klenow reaction with random primers (Stratagene). Hybridization with single probes was performed for at least 6 h, followed by three washing steps in 1× SSC-0.1% sodium dodecyl sulfate (SDS). Blots were exposed to a phosphor screen (Molecular Dynamics) for 24 to 72 h. Screens were scanned with a PhosphorImager (Molecular Dynamics), and individual bands were quantified with ImageQuant 5.2 (Molecular Dynamics). Signals were either quantified after normalization to actin mRNA levels or related to ethidium bromide-stained 18s and 28s rRNA bands, as indicated in the Results.
TABLE 2.
Synthetic oligonucleotide primers used for Northern hybridization
| Gene | Primer | Sequence |
|---|---|---|
| ACT1 | act1for | TTTCCAACTGGGACGATA |
| act1rev | TCTTGGACAAATGGTTGG | |
| ALS1 | als1for | ACTGGTTGCTACTACACA |
| als1rev | GGACAATAATGTGATCAA | |
| ALS5 | 2848f + 3bfor | ACTGCCGATGGTGTTAAA |
| 2848f + 3brev | TGGTGCCACAAAAAGAGT | |
| HWP1 | 2426for | CTAAACCAGCTGCTCCAAAAT |
| 2436rev | GTTGTTACCAGCACCTTCAAA | |
| NRG1 | NRG1for | ATGCTTTATCAACAATCATATCC |
| NRG1rev | CTATACTAGGCTCTTGGTG | |
| PHR1 | phr1for | TTGCAATGTCCAGGAACT |
| phr1rev | CTGCCACTAGCACTAGCC | |
| PHR2 | phr2for | GAATCTGCCTCCTCCATT |
| phr2rev | AATGAAGCAGAACCACCA | |
| RBR1 | forRBR1prox | CTACCAGCACCAGCGACAG |
| revRBR1 down | CTGTTGGTGTGGGTTGTGAG | |
| RBR2 | RBR2proxfor | AACAGGGGCAACAAGTTCAC |
| RBR2downrev | AGTTTGGATTCGACTGTGGG | |
| RBR3 | 1477for | CCCTGCCTTCCTTAACATCGT |
| 1477rev | TAAAAATTTTGCGATGTGGAA | |
| RBT1 | 2768f + 2for | ACTGCCGATGGTGTTAAAT |
| 2768f + 2rev | TGGTGCCACAAAAAGAGTT | |
| RIM101 | RIM101/1-23 | ATGGGTAACAGTCCCCATTCCTC |
| RIM101/1048-1027 | GGCTTCAATGGGACATGGACTC | |
| TUP1 | CaTUP1for | ATGTATCCCCAACGCACC |
| CaTUP1rev | TTATTTTTTGGTCCATTTCC |
Isolation of chromosomal DNA and Southern hybridization.
Chromosomal DNA of C. albicans strains was isolated as described previously (27), digested with EcoRI and PstI, and separated on 0.8% agarose. Blotting onto nylon membranes in 20× SSC and hybridization with an [α-32P]dCTP randomly labeled gene-specific probe was done by standard protocols (35). Membranes were exposed on a phosphor screen (Molecular Dynamics) for 12 h. They were scanned and visualized as described for Northern hybridization.
RESULTS
DNA microarray analysis to identify novel RIM101-regulated cell wall genes in C. albicans.
The cell wall of C. albicans protects the fungus from its environment and is crucial for host-pathogen interactions. Thus, the cell wall has to be adapted and remodeled constantly to cope with various environmental conditions. The pH-dependent transcription factor Rim101p is known to regulate PHR1 and PHR2, two glycosidases involved in cell wall biosynthesis in C. albicans (29). To identify additional RIM101-regulated cell wall genes, we performed transcriptional profiling with a cell wall-specific DNA microarray. The array has been described previously (39) and consists of known C. albicans cell wall genes and 65 open reading frames (ORFs) with significant homology to already-characterized cell wall proteins in S. cerevisiae. Microarray experiments were carried out with a rim101Δ mutant and strain RIM101-1426, overexpressing C-terminally truncated, dominant active Rim101-1426p (Table 1). These mutant strains were compared to wild-type cells grown in 30°C YPD at pH 7.4 and pH 4.5, respectively. In order to focus on gene regulation induced by pH changes, all strains were grown under conditions favoring blastospore morphology (see Materials and Methods). Thus, none of the strains showed filamentous growth when the cells were harvested for RNA isolation after 6 h of cultivation (data not shown).
RIM101 activates and represses cell wall genes of C. albicans.
Our results indicate that RIM101 is required for both activation and repression of transcription. In this focused approach, looking at cell wall genes only, we predominantly found RIM101-repressed genes. In total, comparing the rim101Δ mutant and the RIM101-1426 mutant with the wild type, 32 genes out of 117 were differentially regulated by RIM101. All of these genes showed at least a 2.5-fold change in expression rate in three independent experiments. RIM101 was involved in repression of 23 potential cell wall genes, whereas nine genes turned out to be RIM101 activated under the conditions tested. Table 3 gives an overview of genes regulated by RIM101, as revealed by transcriptional profiling.
TABLE 3.
C. albicans cell wall genes activated and repressed by RIM101
| Category | Gene (accession no.)/ORF | Fold reduction
|
Mean devia- tiona | |
|---|---|---|---|---|
| RIM101-1426/ wild type (pH 4.5) | rim101Δ/ wild type (pH 7.4) | |||
| Activated genes | PHR1 (AF247190) | 32.1 | 19.6 | |
| HWP1 (U64206)/ECE2 (AF001978) | 25.8 | 20.0 | ||
| ALS1 (L25902) | 12.8 | 8.8 | ||
| orf6.7834 | 8.0 | 5.8 | ||
| SKN1 (D88491) | 5.6 | 1.1 | ||
| ALS5 (AF068866) | 4.2 | 2.7 | ||
| orf6.1982 | 3.4 | 1.1 | ||
| RBT1 (AF254142) | 3.1 | 0.7 | ||
| orf6.1230/orf6.1231/ orf6.1232 | 2.9 | 0.8 | ||
| Repressed genes | PHR2 (AF011386) | 15.2 | 2.9 | |
| RBR1 (orf6.6747) | 15.2 | 10.9 | ||
| PHO genes (orf6.4212/ orf6.1652/orf6.1832) | 9.2 | 2.3 | ||
| PHO11 (orf6.1832) | 6.7 | 1.4 | ||
| RBR2 (orf6.6744) | 5.2 | 1.3 | ||
| RBR3 (orf6.1159) | 4.8 | 2.1 | ||
| CHS1 (X52420) | 4.5 | 1.9 | ||
| orf6.4590 | 4.5 | 2.0 | ||
| INT1 (U35070) | 4.1 | 1.6 | ||
| orf6.3954 | 3.6 | 0.9 | ||
| orf6.4388 | 3.6 | 0.6 | ||
| orf6.3476 | 3.4 | 0.7 | ||
| orf6.2804 | 3.4 | 0.7 | ||
| orf6.4736/orf6.4737 | 3.1 | 1.0 | ||
| orf6.799 | 3.0 | 0.7 | ||
| orf6.7718 | 3.0 | 1.6 | ||
| orf6.8118 | 2.9 | 0.5 | ||
| orf6.474/orf6.475 | 2.7 | 0.5 | ||
| orf6.6972 | 2.7 | 0.5 | ||
| ALS4 (AF024586, AF024584) | 2.7 | 0.8 | ||
| KRE6 (D88490) | 2.6 | 0.4 | ||
| orf6.5422/orf6.5423/ orf6.537/orf6.538 | 2.6 | 0.7 | ||
| orf6.7977 | 2.6 | 0.6 | ||
| orf6.4073/orf6.4765 | 2.5 | 0.7 | ||
The cited mean deviations were derived from least three experimental repeats in duplicate.
Genes activated by Rim101p.
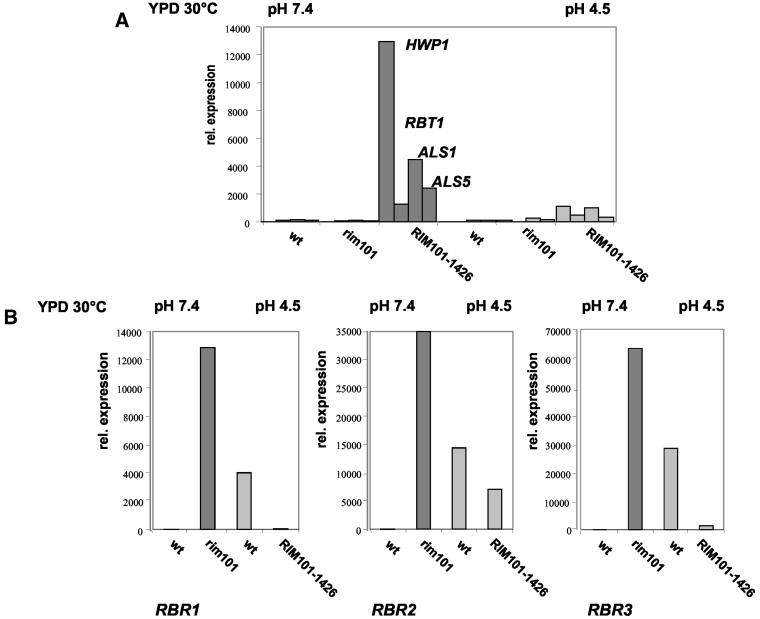
Transcriptional profiling revealed cell wall genes activated by RIM101. Consistent with existing data, PHR1 and SKN1, another putative cell wall glycosidase gene, were strongly upregulated in the RIM101-1426 strain compared to the wild-type strain (32) (Table 3). Notably, HWP1 and RBT1 were revealed to be induced in a RIM101-dependent way in blastospores (YPD, 30°C, pH 7.5). Overexpression of Rim101-1426p under these conditions strongly induced transcription of HWP1, RBT1, ALS1, and ALS5 at pH 7.4. At pH 4.5, this effect was much weaker, indicating that additional pH-dependent factors may be relevant for activation of these cell wall genes by Rim101p (Fig. 1a). This finding was unexpected, because Hwp1p is a well-characterized hyphal wall protein (38). Rbt1p was defined as a cell wall protein repressed by Tup1p, a transcription factor negatively regulating hyphal growth (2), and expression of both genes has been shown to depend on Efg1p (4, 24).
FIG. 1.
Rim101p activates and represses cell wall genes of C. albicans. (a) Activation of hyphal genes by RIM101. (b) Repression of RBR1, RBR2, and RBR3 by RIM101. RNA for Northern blotting was prepared from the wild-type strain SC5314 and the RIM101-1426 and rim101Δ mutants grown for 6 h in YPD with 100 mM HEPES at 30°C. Media were adjusted to pH 7.4 for SC5314 and the rim101Δ mutant (dark bars) or to pH 4.5 for SC5314 and the RIM101-1426 mutant (light bars). Quantification of mRNA abundance was done by normalizing hybridization signals to the ACT1 signal of SC5314 at pH 7.4.
Genes repressed by Rim101p.
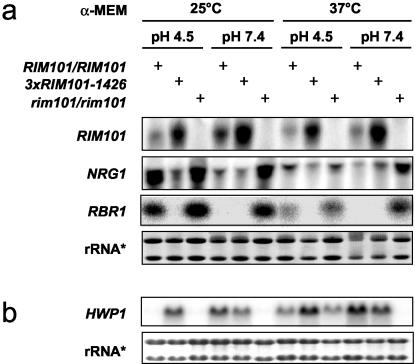
As expected, PHR2 was strongly repressed in the presence of active Rim101p under all conditions tested (8, 33) (Table 3). Additionally, PHO11 was repressed by RIM101, as recently revealed by transcription analysis of C. albicans by others (32). This result also parallels the situation in S. cerevisiae (21) and in A. nidulans (44). In addition, several as yet uncharacterized ORFs were strongly repressed. Three of the ORFs repressed by RIM101 were examined further and named RBR1 (orf6.6747), RBR2 (orf6.6744), and RBR3 (orf6.1159). To confirm our microarray data and to quantify transcription of these genes, we subsequently performed Northern blot analysis (Fig. 1b). RBR1, RBR2, and RBR3 were expressed under acid conditions, and no significant expression was detected in the wild type at neutral pH. Furthermore, the RBR genes were repressed by dominant active RIM101-1426 at acidic pH and strongly upregulated in rim101Δ mutant strains under all conditions (Fig. 1b and Fig. 4).
FIG. 4.
(a) Expression of NRG1 and RBR1 is RIM101 and temperature dependent. (b) pH-dependent repression of NRG1 correlates to Rim101p-dependent expression levels of HWP1. Wild-type SC5314 and RIM101-1426, and rim101Δ mutants were grown for 6 h in α-MEM with 100 mM HEPES buffered to pH 4.5 and 7.4 at 25 and 37°C to isolate RNA for Northern blot analysis. rRNA*, ethidium bromide-stained loading control.
Following in silico examination, all three RIM101-repressed genes carried an approximately 20-amino-acid N-terminal signal sequence for translocation into the endoplasmic reticulum and a C-terminal hydrophobic transmembrane domain (http://www.cbs.dtu.dk/services/SignalP). All of the novel RBR protein sequences also enclose a hydrophobic N-terminal domain (http://129.194.185.165/dgpi/DGPI_demo_en.html). The hydrophobic tail of RBR3 was predicted to be too short for location in the plasma membrane, most likely because orf6.1159 was annotated as a C-terminally incomplete ORF in the Stanford C. albicans database. The predicted domains are characteristic of glycosylphosphatidyl inositide (GPI)-anchored cell wall proteins, and they are followed by an upstream omega site sequence defining the cleavage site for attachment of the GPI anchor and further determinants for location in either the cell wall or the plasma membrane (7, 11, 18). Rbr1p (111 amino acids) and Rbr2p (168 amino acids) have potential cleavage sites at positions 81 and 143, respectively; Rbr3p (560 amino acids) shows a potential cleavage site at position 540. In addition, the protein sequence, excluding C- and N-terminal hydrophobic sequences, exhibits several sites with a high O-β-glycosylation potential typical of fungal cell wall proteins (http://www.cbs.dtu.dk/services/YinOYang).
Searching the pro- and eukaryotic genome databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST), we found no significant homology of the RBR genes with genes in S. cerevisiae or any other organisms. A protein query for nearly exact matches (BLASTP 2.2.6) at NCBI revealed 40% identity of Rbr3p with Hyr1p of C. albicans, a GPI-anchored nonessential cell wall protein abundant in hyphae (1).
rbr1Δ mutant strain shows a pH-dependent filamentation defect.
Since RBR1 showed the strongest expression among the novel RIM101-repressed genes, we characterized this gene further. The open reading frame (orf6.6747) encoding RBR1 is 336 bp long and predicted to encode a GPI-anchored protein of only 111 amino acids in length. For functional characterization of the potential cell wall protein Rbr1p in C. albicans, we constructed three independent rbr1Δ mutant strains by transformation and homologous recombination of CAI4 with the URA3 flipper cassette (28). Strains were routinely first selected for transformants on SC-uri and in a second step after induction of the FLP recombinase for ura3 mutant colonies. Homozygous rbr1Δ mutants were reverted either with the URA3 gene only or with URA3 and RBR1 under its native promoter (Table 1).
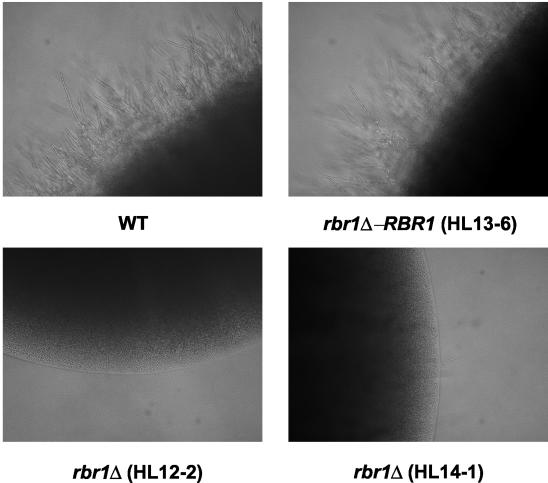
Southern and Northern blot analyses confirmed the genotype and the absence of RBR1 transcription in the homozygous rbr1Δ mutant strains. Heterozygous and homozygous rbr1Δ mutants showed no growth defects under any conditions used. Since RBR1 was expressed prominently at low pH (Fig. 1b), we expected potential alterations in growth, resistance to cell wall stress, or filamentation defects especially under these conditions. We examined growth and filament induction of the rbr1Δ mutants in the presence of hydrogen peroxide, Calcofluor White, and 0.3 M NaCl on different solid media at 25, 30, and 37°C. No distinct phenotype was observed on media with 2% agar, but we were able to detect a filamentation defect of rbr1Δ strains on soft agar containing 0.3% agar. On M-199 soft agar buffered to pH 4.5, rbr1Δ did not show filamentous growth within 3 days of incubation (Fig. 2), while the wild-type and the RBR1 revertant strains readily induced filamentation after 24 h at 30°C. Under the same conditions at pH 7.4, the rbr1Δ strain did induce filaments after 24 h (data not shown).
FIG. 2.
pH-dependent filamentation defect of rbr1Δ mutants on M-199 soft agar. The independent rbr1Δ mutants HL12-2 and HL14-1 are shown in the lower panels; HL13-6 is derived from HL12-2. Strains were precultured in SC-uri for 24 h and pelleted by brief centrifugation. Then 5 μl of concentrated cell suspension was spotted onto the agar surface. Plates were incubated for 72 h at 30°C.
This defect in filament induction implied that RBR1 was especially needed in the cell wall of C. albicans in an acidic environment, consistent with its expression profile (Fig. 1b). After 5 days on acidic M-199 soft agar at 30°C, possibly as an unrelated starvation response, rbr1Δ mutant strains also started to induce hyphae, growing out of the colonies from the center to the edge and downwards into the agar (data not shown). When tested in liquid media, no differences in growth or filamentation rate were detected between the wild-type and rbr1Δ mutant strains (data not shown), indicating that surface-mediated effects may be responsible for the observed phenotype.
NRG1 activates expression of RBR1.
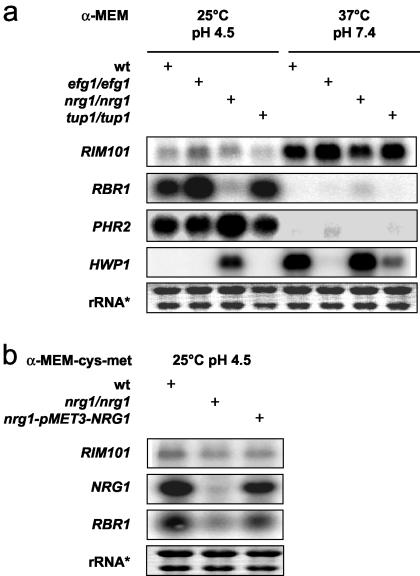
Since Rbr1p seemed to be involved in the dimorphic switch, we asked if, in addition to Rim101p, other transcriptional regulators of morphogenesis in C. albicans were involved in RBR1 regulation. Therefore, we examined expression of RBR1 in the efg1Δ, nrg1Δ, and tup1Δ mutant strains at pH 4.5 at 25°C and pH 7.4 at 37°C in hypha-inducing medium (Fig. 3a). Notably, we found expression of RBR1 to be strongly reduced in the nrg1Δ mutant at pH 4.5, while wild-type transcript levels were detected in the efg1Δ and tup1Δ mutants. At neutral pH, we did not detect significant RBR1 expression in either the wild type or the mutant strains tested.
FIG. 3.
(a) RBR1 expression is activated by NRG1. Transcript levels of RBR1, PHR2, and HWP1 in the wild type (wt) and efg1Δ, nrg1Δ, and tup1Δ mutant strains were determined by Northern blotting. RNA derived from strains grown in α-MEM at pH 4.5 (lanes 1 to 4) and pH 7.4 (lanes 5 to 8) for 4 h under noninducing and hypha-inducing temperatures of 25 and 37°C, respectively. (b) Reversion of the nrg1Δ mutant strain with pMET3-NRG1 restores RBR1 expression. The wild type and nrg1Δ, and nrg1-pMET3-NRG1 mutants were grown for 4 h in α-MEM at pH 4.5 without cysteine and without methionine. rRNA*, ethidium bromide-stained loading control.
The nrg1Δ mutant strain MMC4 was reverted with NRG1 under the control of the MET3 promoter. In this revertant strain, transcription of NRG1 and RBR1 was restored in α-MEM without cysteine and methionine (Fig. 3b). These data implied that Nrg1p directly or indirectly activated RBR1 transcription, while Efg1p and Tup1p were not involved in RBR1 regulation under the conditions tested. In parallel, we looked at the influence of Efg1p, Nrg1p, and Tup1p on PHR2 expression (Fig. 3a). PHR2, like RBR1, is acid expressed (29), but we did not find Nrg1p to be required for activation of PHR2. These results demonstrated that although both RBR1 and PHR2 are acid-induced genes, they are activated by different mechanisms. Furthermore, these data showed that RIM101 transcription did not depend on the presence of the transcription factor Efg1p, Nrg1p, or Tup1p.
Expression of NRG1 is repressed by RIM101 and negatively related to temperature.
Northern blot analysis revealed that the cell wall gene RBR1 was inversely regulated by the two transcription factors Rim101p and Nrg1p (Fig. 1b and 3). Furthermore, expression of RIM101 was independent of NRG1 (Fig. 3). We additionally performed experiments to examine whether expression of NRG1 depended on RIM101. Figure 4a demonstrates that both NRG1 and RBR1 were expressed in a pH-dependent manner, directly correlated to the activity of RIM101. In the wild type, NRG1 and RBR1 were strongly expressed at pH 4.5, while very low transcript levels were detected at pH 7.4. The presence of dominant active Rim101p significantly reduced expression of NRG1 and RBR1 at acidic pH, whereas deletion of RIM101 led to activation of NRG1 and RBR1 at neutral pH under all conditions tested.
Searching the promoter regions of NRG1 and RBR1 for Rim101p binding sites (CCAAGAAAA) (32), we found consensus sequences for Rim101p binding at positions −1302 bp for NRG1 and −1400 bp for RBR1. To prove that Rim101p binds directly to these consensus sequences, further studies will be necessary.
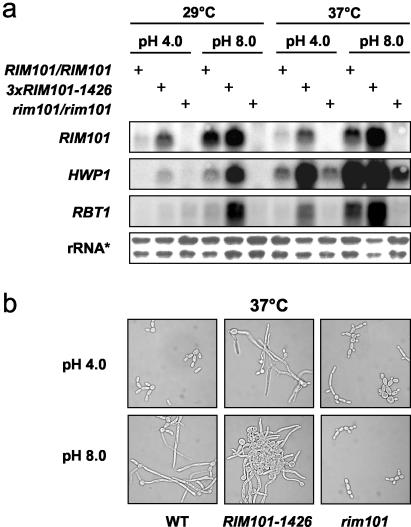
In addition, the Northern blot experiments revealed that expression of NRG1 and RBR1 was negatively correlated to temperature (Fig. 4a). Compared to the wild type at pH 4.5, 25°C, expression of NRG1 and RBR1 was strongly reduced at 37°C and pH 4.5. This result indicated a temperature-dependent factor restricting NRG1 expression in addition to the Rim101p-dependent and therefore pH-modulated repression of RBR1 and NRG1. Since Nrg1p is known to repress a variety of hypha-specific genes, we moreover did explore Rim101p-mediated induction of HWP1 under hypha-inducing conditions. HWP1 transcription was induced at 25 and 37°C, when Rim101p-mediated repression of Nrg1p was observed (Fig. 4b). The same effect was also found for both HWP1 and RBT1 in M-199 at 29 and 37°C (Fig. 5a). This Northern blot analysis additionally showed that temperature had a major effect on the expression of HWP1 and RBT1, which was also detected for the expression of NRG1. The corresponding phenotype of the wild-type and RIM101 mutant strains in M-199 (37°C) is shown in Fig. 5b. The RIM101-1426 mutant exhibited filamentous growth at acidic pH, as expected, but we also microscopically observed flocculation of C. albicans in response to RIM101-1426 overexpression at neutral pH.
FIG. 5.
Expression of HWP1 and RBT1 is activated by RIM101 and correlated to the RIM101-1426 overexpression phenotype. (a) RIM101-dependent expression of HWP1 and RBT1 in wild-type SC5314 and the RIM101-1426 and rim101Δ mutants was determined by Northern blotting. RNA was derived from cells grown in M-199 with 100 mM HEPES at pH 4.0 and 8.0 for 4 h. The pH and temperature values are indicated. rRNA*, ethidium bromide-stained loading control. (b) Phenotypes of the strains grown for Northern blot analysis after 4 h in M-199 at 37°C and pH 4.0 (upper panel) or pH 8.0 (lower panel).
In conclusion, our data demonstrated that active RIM101 was able to repress NRG1, as has been shown for NRG1 in S. cerevisiae (20). We also found that Nrg1p, a repressor of hyphal genes, showed an activating function towards the acid-expressed cell wall gene RBR1 in C. albicans. Moreover, our data led to the hypothesis that repression of NRG1 by RIM101 is responsible for pH-dependent induction of the cell wall protein Hwp1p and that Rim101p in parallel activates Rbt1p.
DISCUSSION
Repression and activation of cell wall genes by RIM101.
The pH-dependent transcription factor Rim101p is known to act as both a transcriptional repressor and activator. In accordance with this, our transcriptional profiling experiments to detect novel downstream targets of Rim101p in C. albicans revealed a set of cell wall genes that are either repressed or induced by Rim101p in response to changes in the environmental pH (Table 1).
RIM101 is responsible for pH-dependent activation of hyphal genes.
In our experimental approach, we focused on pH-dependent cell wall gene regulation by Rim101p and attempted to avoid activation of the developmental program inducing hyphae by growing all strains as blastospores (see Materials and Methods). Nevertheless, our microarray data reveal significant activation of the hypha-specific genes HWP1, RBT1, ALS1, and ALS5 by RIM101 in a pH-dependent way (Table 3 and Fig. 1a). RIM101 has been shown to contribute to filamentation of C. albicans on a phenotypic level and on a molecular level to the regulation of HWP1 (8, 9, 13). Our studies show that Rim101p also contributes to the regulation of additional hypha-specific cell wall genes (Fig. 1a and 5a). These results apparently correlate with the RIM101-1426 phenotype, which is characterized by filamentous growth at acidic pH and an excess adhesion phenotype at neutral pH and 37°C (Fig. 5b).
At acidic pH (pH 4.0), activation of both RBT1 and HWP1 by RIM101-1426 was significant but not as strong as at pH 8.0 (Fig. 5a and 1a). In addition, we found that the level of RIM101-mediated activation depended on the temperature. Higher temperature in general results in stronger activation of HWP1 and RBT1. Obviously, additional factors besides RIM101 contribute to the pH regulation as well as the temperature-mediated induction of hyphal genes. A pH-responsive pathway defined by Mds3p that could participate in inducing the pH-related effects observed has recently been identified (9).
EFG1 is required for expression of Hwp1p, Rbt1p, and Als1p (4, 22). Repression of HWP1 has been shown to depend on both Nrg1p and Tup1p, while RBT1 seems to be mainly controlled by Tup1p (2, 4). Our experiments revealed that expression of RBT1 depended on active Rim101p (Fig. 5a) and was strongly induced at neutral pH and higher temperatures in the wild type. Under these conditions, equal levels of TUP1 mRNA were detected in the wild-type and rim101Δ stains (data not shown), and expression of RIM101 was independent of Tup1p (Fig. 3a). These results suggest that RIM101-dependent activation of RBT1 is not mediated by repression of TUP1 but that Rim101p, in addition to Efg1p, functions as an activator of RBT1 in C. albicans.
Our data furthermore showed that Rim101p expression was independent of Efg1p (Fig. 3a). Thus, Rim101p and Efg1p seem to independently converge on the regulation of several hypha-specific genes and precisely modulate gene expression in order to adapt to distinct external conditions. These findings are in agreement with a proposed regulation of the hypha-specific genes such as HWP1, RBT1, and HYR1 by multiple transcription factors with DNA-binding capacity (22). Similar complex regulation mechanisms for the expression of cell wall proteins were also observed for Flo11p, a protein necessary for pseudohyphal differentiation in S. cerevisiae (26, 34).
Repression by RIM101.
The vast majority of the genes identified in our experiments are repressed by Rim101p (Table 1). In S. cerevisiae, RIM101 has also been shown to have significant repressing functions in gene regulation (20). Recently, genomewide transcriptional profiling experiments identified 13 genes that are repressed by RIM101 (32). Two of these genes (PHR2 and PHO11) were also identified in the course of our experiments focused on cell wall genes. The three most strongly repressed and so far uncharacterized ORFs identified in our screen were named RBR1, RBR2, and RBR3 (repressed by Rim101p) and encode a set of GPI-anchored proteins with so far unknown function. All three genes are expressed under acidic conditions and repressed by RIM101 at neutral pH. Similar to the regulation of PHR2 (13), deletion of RIM101 resulted in constitutive expression of the RBR genes, and dominant active RIM101-1426 strongly repressed these genes at every pH tested (Fig. 1b and 4a). Therefore, RIM101 can be considered a repressor of RBR1, RBR2, and RBR3. An in silico promoter analysis of RBR1 revealed a consensus sequence for Rim101p binding at position −1400 bp, while no established binding site for the transcription factor was found 2 kb upstream of the other two Rim101p-repressed genes.
Functional characterization of RBR1.
To functionally characterize RBR1, we deleted the gene in C. albicans and investigated the phenotype of the resulting strain under various conditions, including cell wall stress and osmotic stress and different hypha-inducing media. Deletion of RBR1 results in a filamentation defect on M-199 soft agar buffered to pH 4.5 (Fig. 2), indicating a function in hyphal development under acidic conditions. The presence of other acid-expressed genes such as RBR2 and RBR3 may have compensated for a more severe phenotype.
The phenotype observed is in accordance with the expression pattern of RBR1, which displays strong transcription at acidic pH. Our results show that Nrg1p is required for RBR1 expression. NRG1 transcription parallels the temperature- and pH-related regulation of RBR1 (Fig. 4a). This is surprising because so far Nrg1p has only been described as acting as a transcriptional repressor, most likely by recruiting Tup1p to the promoters of target genes, e.g., HWP1 (31). The transcription factors Efg1p and Tup1p show no effect on RBR1 expression (Fig. 3a). Although this is the first time that an apparently activating role for Nrg1p in C. albicans has been reported, data resulting from transcriptional profiling experiments indicate this activating function of Nrg1p for other genes as well (30). In contrast to RBR1, activation of PHR2 under acidic conditions did not require NRG1 under the conditions tested, and thus different regulation mechanisms are present for these two acid-expressed genes. Since GPI proteins, as shown for Hwp1p and Als1p, are involved in tissue adhesion by and virulence of C. albicans, these proteins might be targets for new antifungal substances (43).
RIM101 regulates the transcriptional repressor NRG1.
Our studies revealed coregulation of RBR1 by Rim101p and Nrg1p. Further investigations to identify cross talk between Rim101p-dependent pH signaling and NRG1 showed that active Rim101p represses the transcription of NRG1 (Fig. 4a), while deletion of NRG1 had no significant influence on RIM101 transcripts (Fig. 3a and b). This indicates that Rim101p acts as a repressor of NRG1, as has been reported for S. cerevisiae (20). Although our data suggest that Rim101p functions similar to S. cerevisiae Rim101p, ectopic expression of Rim101p in an S. cerevisiae rim101Δ mutant did not complement its invasive growth defects (data not shown). A recent study on Rim101p binding specificity also involved transcriptional profiling of wild-type and rim101Δ strains (32). The authors indicate that transcriptional profiling did not reveal activation of NRG1 or Nrg1p-repressed genes in the rim101Δ mutant compared to the wild type (M-199, 28°C, and pH 7.4). The Northern blot experiments shown in Fig. 5a were performed under similar conditions (M-199, 29°C, and pH 8.0) and display a relatively weak induction of the NRG1-repressed gene HWP1 in the wild-type cells. This might explain why significant downregulation of NRG1-repressed genes versus wild-type expression could not be detected in DNA microarray experiments under the conditions chosen. We observed that the effect of Rim101p on NRG1 was most prominent at lower temperatures in α-MEM (Fig. 4a).
Our results suggest that Rim101p not only acts as a transcriptional activator of PHR1 but also significantly controls gene expression through downregulation of the transcriptional repressor Nrg1p (Fig. 4a). This leads to induction of the hypha-specific gene HWP1, as has been shown before (4, 5). Furthermore, the expression of RBT1 strongly correlates with Rim101p activity (Fig. 5a). High levels of Nrg1p at acidic pH suppress filamentous growth under these conditions (31). Thus, derepression of NRG1 by deletion of RIM101 corresponds to the filamentation defect described for the rim101Δ mutant at neutral pH (8), while enhanced adhesion and induction of hyphae in the RIM101-1426 strain (13) reflect downregulation of NRG1. How Nrg1p in concert with Rim101p directly or indirectly mediates regulation of RBR1 expression is still unclear and will be the subject of further research.
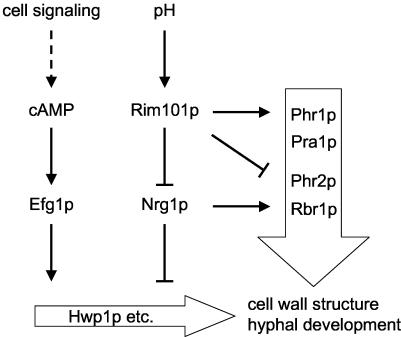
Our results document that Rim101p is woven into the complex network of signaling pathways required for morphogenesis (Fig. 6), giving C. albicans the ability to integrate pH signals with other environmental signals to react specifically to external stresses.
FIG. 6.
Model of Rim101p cross talk with the NRG1 pathway. Integration of external pH signals into other signaling pathways is mediated by RIM101-dependent repression of Nrg1p. The presence of active Rim101p at neutral pH has three consequences: first, direct activation of pH-specific cell wall genes (PHR1, PRA1), second, direct or indirect repression of acid-expressed cell wall genes (PHR2, RBR1) and third, enhanced expression of hypha-specific genes by repression of NRG1. C. albicans is thus able to integrate pH signals via Rim101p and Nrg1p with other signaling pathways required, e.g., for morphogenesis to react specifically to changes in environmental conditions.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG grant RU608/2-1,2). Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
We thank Alistair Brown, Gerald Fink, and Alexander Johnson for providing C. albicans mutant strains. Joachim Morschhäuser and Peter Sudbery are acknowledged for the gift of plasmids. Sequence data for C. albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida.
REFERENCES
- 1.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 7.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende, and F. M. Klis. 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 8.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bernardis, F., F. A. Muhlschlegel, A. Cassone, and W. A. Fonzi. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 12.Diehl, F., S. Grahlmann, M. Beier, and J. D. Hoheisel. 2001. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 29:E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Barkani, A., O. Kurzai, W. A. Fonzi, A. Ramon, A. Porta, M. Frosch, and F. A. Muhlschlegel. 2000. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol. Cell. Biol. 20:4635-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espeso, E. A., and H. N. Arst, Jr. 2000. On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol. Cell. Biol. 20:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espeso, E. A., and M. A. Penalva. 1996. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J. Biol. Chem. 271:28825-28830. [DOI] [PubMed] [Google Scholar]
- 16.Espeso, E. A., J. Tilburn, L. Sanchez-Pulido, C. V. Brown, A. Valencia, H. N. Arst, Jr., and M. A. Penalva. 1997. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 274:466-480. [DOI] [PubMed] [Google Scholar]
- 17.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frieman, M. B., and B. P. Cormack. 2003. The omega-site sequence of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol. Microbiol. 50:883-896. [DOI] [PubMed] [Google Scholar]
- 19.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 20.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 22.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 23.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 25.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 26.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millon, L., A. Manteaux, G. Reboux, C. Drobacheff, M. Monod, T. Barale, and Y. Michel-Briand. 1994. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J. Clin. Microbiol. 32:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morschhauser, J., S. Michel, and P. Staib. 1999. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 32:547-556. [DOI] [PubMed] [Google Scholar]
- 29.Muhlschlegel, F. A., and W. A. Fonzi. 1997. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17:5960-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 31.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramon, A. M., and W. A. Fonzi. 2003. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell 2:718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sentandreu, M., M. V. Elorza, R. Sentandreu, and W. A. Fonzi. 1998. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J. Bacteriol. 180:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 40.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su, S. S., and A. P. Mitchell. 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21:3789-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukahara, K., K. Hata, K. Nakamoto, K. Sagane, N. A. Watanabe, J. Kuromitsu, J. Kai, M. Tsuchiya, F. Ohba, Y. Jigami, K. Yoshimatsu, and T. Nagasu. 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol. Microbiol. 48:1029-1042. [DOI] [PubMed] [Google Scholar]
- 44.van den Hombergh, J. P., A. P. MacCabe, P. J. van de Vondervoort, and J. Visser. 1996. Regulation of acid phosphatases in an Aspergillus niger pacC disruption strain. Mol. Gen. Genet. 251:542-550. [DOI] [PubMed] [Google Scholar]