Abstract
Two of the unique events that occur in meiosis are high levels of genetic recombination and the reductional division. Our previous work demonstrated that the REC102, REC104, REC114, and RAD50 genes, required to initiate meiotic recombination in Saccharomyces cerevisiae, are needed for the proper timing of the first meiotic (MI) division. If these genes are absent, the MI division actually begins at an earlier time. This paper demonstrates that the meiotic recombination genes MER2/REC107, SPO11, and MRE2 and the synaptonemal complex genes HOP1 and RED1 are also required for the normal delay of the MI division. A rec103/ski8 mutant starts the MI division at the same time as in wild-type cells. Our data indicate no obvious correlation between the timing of premeiotic S phase and the timing of the first division in Rec− mutants. Cells with rec102 or rec104 mutations form MI spindles before wild-type cells, suggesting that the initiation signal acts prior to spindle formation. Neither RAD9 nor RAD24 is needed to transduce the signal, which delays the first division. The timing of the MI division in RAD24 mutants indicates that the pachytene checkpoint is not active in Rec+ cells and suggests that the coordination between recombination and the MI division in wild-type cells may occur primarily due to the initiation signal. Finally, at least one of the targets of the recombination initiation signal is the NDT80 gene, a transcriptional regulator of middle meiotic gene expression required for the first division.
Chromosomes passing through meiosis undergo a specific sequence of events that does not occur in mitotic cells. These events include an S phase that may well be unique to meiosis, high levels of genetic recombination, formation of the synaptonemal complex (SC), a reductional division, and an equational division that is not immediately preceded by an S phase (27, 33, 41). It has become clear that these meiotic events are coordinated, not only through a sophisticated temporal regulation of gene expression (5, 23, 36), but also by the existence of meiotic checkpoints that assess whether events have occurred properly (e.g., reference 42). A checkpoint in prophase I has been studied in detail and has been referred to variously as the pachytene checkpoint and the recombination checkpoint (18, 29).
At least two types of defects can be recognized by the pachytene checkpoint: defects caused by mutations in some late recombination genes such as dmc1 (i.e., “late” because they act after the formation of double strand breaks [DSBs]) and defects caused by mutations in some SC genes such as zip1. In such mutant strains, cells arrest in late prophase and do not undergo the first meiotic (MI) division. The ability to establish the checkpoint depends on the presence of part of the mitotic DNA damage checkpoint system; RAD24 and RAD17 are required for the arrest (29, 42). In contrast, RAD9 (which is required for the mitotic checkpoint) does not play a role in the pachytene checkpoint. Evidence suggests that the long single-strand tails that exist in dmc1 mutants (3) are recognized by the pachytene checkpoint. It has been suggested that the pachytene checkpoint might act in normal Rec+ cells to coordinate recombination and the MI division because exonuclease digestion after DSB formation normally results in single-stranded DNA (ssDNA) during recombination (29). Of course, in Rec+ cells, the ssDNA tails are shorter and not persistent (49), consistent with the idea that the normal ssDNA could cause a delay rather than an arrest of the first division.
In earlier work, we found that the genes that act in the initiation of meiotic recombination to form DSBs also play a role in coordinating the timing between recombination and the first division (15, 21). Ten genes are required to form DSBs: REC102, REC103/SKI8, REC104, MER2/REC107, REC114, MEI4, SPO11, RAD50, XRS2, and MRE11 (25). There are two genes (MRE2 and MER1) required to splice the intron present in MER2/REC107; mutations in these two genes confer phenotypes similar to those of initiation mutants. All of these genes have been classified as early recombination functions, because they are required to make DSBs and are rescued by the presence of a spo13 mutation (e.g., see reference 30). The SPO11 gene has been proposed to code for the protein that is directly involved with making DSBs; support for this hypothesis takes two forms. First, SPO11 has homology to type II topoisomerases involved in making DSBs as they pass one strand of DNA through the other (2). Second, Spo11p is found covalently attached to the ends of DSBs in certain mutants, which can make breaks but not process them (26). However, it is absolutely clear that the other nine genes are all required to make breaks. No breaks are observed when any of the genes are deleted (25), even if the Spo11p is tethered to the DNA by the binding domain of another protein (38).
The presence of all of the recombination initiation genes results in the normal timing of the first division. Our previous work discovered that the MI division begins at earlier times in cells lacking some of the recombination initiation genes (15, 21). It was initially tempting to attribute the delay of the MI division in Rec+ wild-type (WT) cells to the formation of DSBs, since even a single DSB in a mitotic cell can result in cell arrest (1, 12). However, mei4 mutants started the first division at the same time as Rec+ cells, yet no DSBs were observed in such cells (15, 21). This suggested that neither DSB formation nor recombination, per se, was the signal that resulted in the normal delay of the first division observed in Rec+ cells. We discovered that rec104 and rec114 cells displayed an early MI division starting about an hour before WT cells. Mutants containing either rad50 or rec102 exhibited a very early first division, starting about 2 h earlier than WT cells. Recent work by Kee and Keeney (24) indicates that spo11 mutants also start the MI division earlier than WT cells. When we constructed double mutants containing a “very early” mutation and an “early” mutation (e.g., rec102 rec104), the very early mutation was always epistatic to the early mutation. Because the recombination initiation mutations confer an earlier first division, it is important to realize that the delayed timing of MI observed in WT cells is a normal feature of meiosis.
Another group of genes is involved early in meiotic recombination; this group includes the genes coding for proteins involved in the formation of the SC. Two genes we examine in this paper are RED1 and HOP1. These two genes play a role in the formation of axial elements and are components of the mature SC (20, 47). Unlike mutations in the 10 recombination initiation genes required for DSBs, null mutations in hop1 or red1 do not completely abolish meiotic recombination or DSBs. Typically about 10% of the normal meiotic levels of recombination can be detected in red1 or hop1 mutants (19, 32, 40).
In this paper, we investigate what other recombination initiation functions are required for the normal signal to delay the first division and if components of the SC are required. Recently, Kleckner and colleagues (4) have published data that indicate that spo11 has a shorter premeiotic S phase than Rec+ cells; they suggested that this might be the reason that spo11 cells enter the MI division earlier. We address this proposal by examining the S phase in several recombination initiation mutants. Experiments are also presented that determine whether the initiation signal acts before or after MI spindle formation. Data are presented on the role of RAD24 and RAD9 in the initiation signal, and the results bear on the role of the pachytene checkpoint in WT Rec+ cells. Finally, experiments are presented that ask about the target of the recombination initiation signal that delays the MI division.
MATERIALS AND METHODS
Yeast strains and mutations.
All strains used in this paper are isogenic and are derivatives of the homothallic diploid K65-3D (15), ultimately derived from S288C. Our strains display classical kinetics of meiosis (11): slower than SK strains and faster than BR strains. K65-3D is homozygous for the following mutations: HO, lys2-1, tyr1-1, his7-2, can1r, ura3-13, ade5, met13-d, trp5-2, leu1-12, and ade2-1. All of the recombination and checkpoint mutations used were null deletion mutations. Some mutations have been described previously: red1Δ::URA3 (50), hop1Δ::URA3 (19), mei4Δ::URA3 (15), rec104-Δ1 (16), rec102Δ-2::LYS2 (15), and rad50Δ::URA3 (21). All strains containing the G418R gene were precise deletions of the coding region and were obtained from the Research Genetics deletion collection. The deletions are described by the Saccharomyces Genome Deletion Project (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). All deletion and deletion/insertion mutations were tested by both genetic analysis and Southern blottting.
Media, growth, and sporulation conditions for sporulation experiments.
The media, growth, and sporulation conditions have been described previously (15). For each experiment, all cultures were grown in the same batch of medium and were treated identically. In every experiment, we examined sporulation of one culture of a WT strain (K65-3D), one culture of a rec104 strain, and two cultures of the mutant being examined. The WT and rec104 strains were included as normal and early timing controls, respectively. All timing experiments were repeated at least twice. Time points were taken every half hour through the period when the MI division initiates, and at least 1,000 cells were counted for each point. We note, as previously reported (15), that the exact timing of events in sporulation can vary slightly from one experiment to another, but the relative timing never varies (e.g., rec104 mutants always begin the MI division earlier than REC104 cells).
Microscopy.
Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and cells were examined with a fluorescent microscope as described previously (15), except that cells were suspended in mounting medium by using 65% glycerol. For time points of ≤3 h, at least 400 cells were counted; for all subsequent time points, at least 1,000 cells were counted. Mononucleate cells include those that have not yet undergone the first division. Binucleate cells consist of cells with two distinguishable DAPI-staining nuclei. (Binucleate cells that represent events in mitotic cells are easily distinguishable because the two nuclei are located at the bud/cell junction; such cells aren't counted in the data. The number of mitotic cells with two nuclei falls to <0.5% by 2 h in sporulation medium.) The final degree of sporulation for WT K65-3D cells in these experiments ranged from 65 to 75% mature asci. Meiosis-specific Rec− mutants displayed sporulation frequencies ranging from about 20 to 30%. Sporulation frequencies of rad50 and rec103/ski8 mutants ranged from 5 to 15%.
Tubulin staining used standard protocols (17, 39). Briefly, cells obtained from time points were immediately fixed in 4% formaldehyde for 24 h, and about 107 cells from each time point were permeabilized by incubation in 60-μg/μl zymolase, 2% glusulase, and 0.8% β-mercaptoethanol for 40 min at 37°C in 40 mM phosphate buffer with 1.2 M sorbitol and 0.5 mM MgCl2. Cells were then incubated in 75% ethanol for 5 min followed by an hour of incubation at room temperature with a 1:500 dilution of YOL1/34 antitubulin rat monoclonal antibody (no. MCA78S; Serotec) in a mixture of 0.1 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid; pH 6.9], 5 mM EGTA, 5 mM MgCl2, and 2% bovine serum albumin. The secondary antibody used was Alexa 488-conjugated goat anti-rat immunoglobulin G (no. A-11006; Molecular Probes) at a 1:1,000 dilution. Cells were resuspended in a DAPI solution consisting of 1-μg/ml DAPI in sodium bicarbonate buffer (pH 9.0) with 65% glycerol. Stained cells were examined with a Leitz Laborlux 12 microscope equipped with fluorescence filters and a Pixera Pro 150ES digital camera. Viewfinder 3.0, Studio 3.0, and Adobe Photoshop 7 software were used to obtain the images in Fig. 5. For time points of <4 h, at least 500 cells were counted; for time points of ≥4 h, at least 1,000 cells were counted. Both short and long MI spindles were counted as soon as they became detectable. Mitotic spindles and MII spindles were easily distinguished from MI spindles by their morphology.
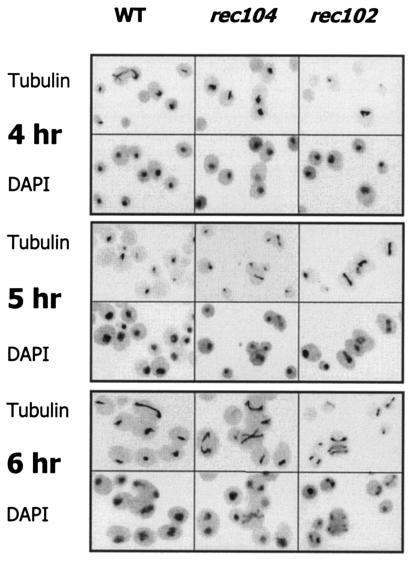
FIG. 5.
Meiosis I spindles in WT cells and Rec− mutants. Fields illustrating the presence of MI spindles are shown at illustrative time points in WT, rec104, and rec102 cells. For each time point, the top figure is a picture of the immunofluoresence observed due to antitubulin antibody. The bottom figure is a picture of the nucleus as observed by DAPI fluorescence.
Northern analysis.
Cell pellets obtained from meiotic time point experiments were frozen at −75°C until RNAs from these cells were isolated as previously described (31). Northern (RNA) blot analysis was performed as described previously (6). DNA probes were made by PCR and encompassed the coding regions and approximately 100 bp upstream and downstream. Probes were labeled with [α-32P]dATP by using the Invitrogen Life Technologies Random Primers DNA labeling system. Northern blots were analyzed with a Molecular Dynamics PhosphorImager (model 445SI) and ImageQuant software.
Analysis of premeiotic DNA synthesis by flow cytometry.
For the experiments analyzing premeiotic DNA synthesis, cells were grown and sporulated as described above with the following differences: Diploid cells were grown to saturation in YPD (1% yeast extract, 2% peptone, 2% glucose), diluted in YPA (1% yeast extract, 2% peptone, 1% potassium acetate), and grown to 4 × 107 to 6 × 107 cells/ml to reduce the number of mitotic cells that were 4C upon entering sporulation medium. Cells were then spun down, washed, and resuspended in sporulation medium as described above. Aliquots (0.5 ml) of cells were collected at each time point, pelleted, resuspended in 70% ethanol, and stored at 4°C. Prior to staining, cells were treated as follows. (i) The cells were twice pelleted and resuspended in 50 mM sodium citrate (pH 7.5) at room temperature for at least 30 min. (ii) They were then pelleted and resuspended in 50 mM sodium citrate containing 0.25-μg/ml RNase A and (iii) incubated at 37°C for 16 to 20 h. (iv) Proteinase K was added to a concentration of 20 μg/ml. (v) Finally, these samples were incubated at 50°C for 3 h. Propidium iodide was then added to a final concentration of 5 μg/ml. Cells were stored at 4°C in the dark. On the day of fluorescence-activated cell sorting analysis, cells were sonicated for 20 s at 40% power with an Artek 150 Sonic Dismembrator. Samples were analyzed on a Becton Dickinson FACSCalibur using CellQuest software. For each sample, 105 events were collected. Strains were sporulated and analyzed in three independent experiments to determine the average values, except for the rec114 strain, for which experiments were done twice.
Once events were collected, cell cycle analysis was performed with FlowJo software. The Dean-Jett-Fox model (7, 10, 13), incorporated into the cell cycle analysis option of FlowJo, was used to determine the fraction of cells in the 2C, S, and 4C phases at any particular time. Once the fraction of cells in S phase was determined, the method of Cha et al. (4) was employed to determine the length of S phase. Using the length of S phase, we calculated a cumulative curve (4) to determine the time at which 50% of cells had entered S phase. We also calculated the time when 50% of cells had entered S phase by plotting the fraction of 2C cells versus time and determining when 50% had left the 2C peak. This calculation assumes that cells no longer in 2C have entered S phase. This provides an independent measurement of S-phase entry that does not depend on the calculation of S-phase length to create a cumulative curve.
RESULTS
The recombination initiation signal for the delay of the first division.
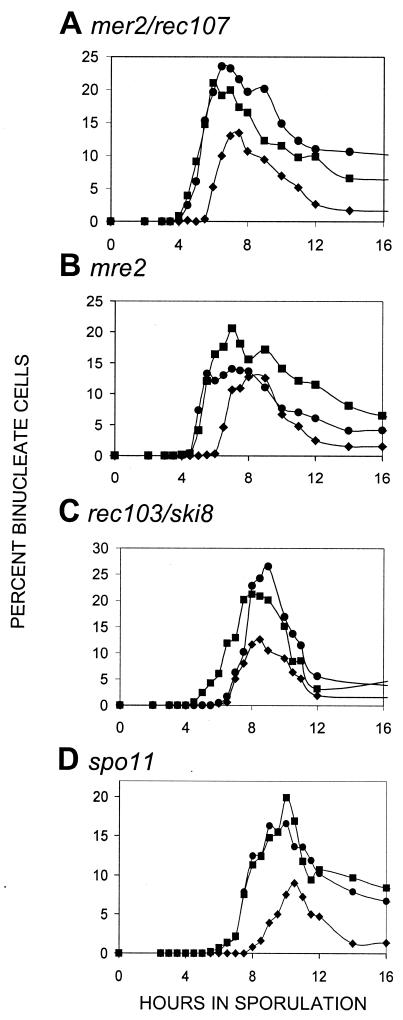
Throughout this paper, we define cells that have segregated their chromosomes into two separated and distinguishable nuclei as having undergone the MI division. Recombination initiation genes previously shown to be required for the normal delay of the first division include REC102, REC104, REC114, and RAD50. Kee and Keeney (24) recently demonstrated that spo11 mutants also begin the first division earlier. The data in Fig. 1 indicate that MER2/REC107 and MRE2 are also required for the WT delay; mutants in these genes have an earlier reductional division. The spo11 mutant in our strain background acted in the same manner as observed by Kee and Keeney in the SK1 background (24); our independent data in a different strain background support their conclusion that normal timing of the first division requires SPO11. A strain lacking REC103/SKI8 began the MI division at the normal time; REC103/SKI8 is apparently not needed for the recombination initiation signal to the first division.
FIG. 1.
Timing of the first division in mutants affecting recombination initiation. In all of the graphs, the first division of WT cells is shown as solid diamonds (♦) and is a control for normal timing. The control for early timing of the first division in all panels is a rec104 strain denoted by solid squares (▪). In all of the graphs, the mutant examined is shown as solid circles (•); for each mutant, at least two independent cultures were examined for each experiment and the data point shown is the mean. Another independent experiment was done for each mutant; in each case the repeat gave the same result (data not shown). The type of mutant examined is shown above each panel.
The timing of the first division in cells lacking SPO11 or MER2/REC107 was indistinguishable from the that of the early division that occurs in the rec104 control. We note that the timing of the first division in mre2 cells, which lack an mRNA processing function required for MER2/REC107 (35), also occurred at an earlier time that was indistinguishable from that of rec104 cells (See Discussion).
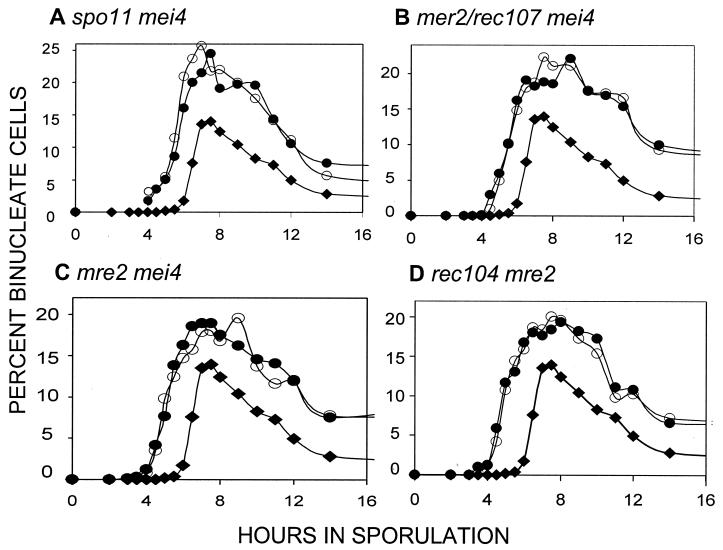
Double mutant analysis of meiotic recombination initiation mutants.
In our previous work (15, 21), the recombination initiation mutants displayed patterns of epistasis consistent with their acting in a linear pathway with respect to timing of the first division. To determine whether the mutations examined in Fig. 1 above could also be placed in the same formal pathway, we examined a number of double mutants. As expected from our earlier results, spo11 was epistatic to mei4 (Fig. 2A); the double mutant displayed the early division phenotype. Likewise, the mer2/rec107 and the mre2 mutations were epistatic to mei4 (Fig. 2B and C). The double rec104 mre2 mutant had timing indistinguishable from that of either mutant alone (Fig. 2D). This observation suggests that the rec104 and mre2 mutations affect the same process.
FIG. 2.
Timing of the first division in double mutants. The type of double mutant examined is shown above each panel. In each panel, WT cells are denoted by solid diamonds (♦). Two independent cultures of each double mutant are shown and denoted by open and solid circles (○, •). (A to D) All double mutant cultures and the WT control were done at the same time; the WT curves are identical in each panel.
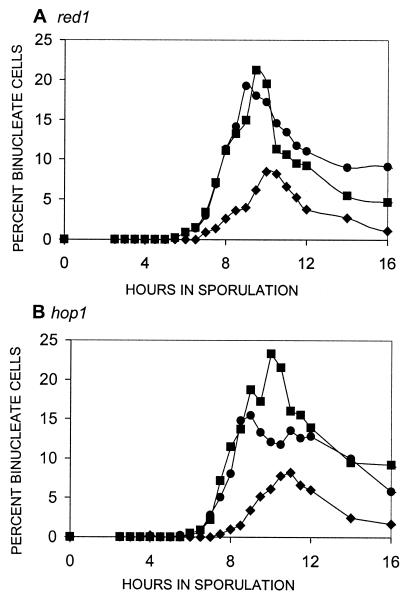
Are components of the SC required for the initiation signal?
In Saccharomyces cerevisiae, the axial elements (future lateral elements) of the synaptonemal complex appear to form at a time in recombination when the initiation functions are acting (41). We considered two possibilities for the role of Red1p and Hop1p in the initiation signal. First, if they were part of a recombination initiation complex that signaled for the normal delay of the first division, hop1 and red1 mutants should have an earlier MI division. Alternatively, both red1 and hop1 mutants display about 10% of the WT level of meiotic recombination (19, 32, 40); this level represents about a 20- to 30-fold induction over the mitotic background. If this level of recombination were sufficient to signal the initiation of recombination, hop1 and red1 strains should have normal timing. The data in Fig. 3 indicate that in both hop1 and red1 strains, the first division starts at an earlier time indistinguishable from that of the rec104 control. This indicates that these SC proteins do play a role in the normal signal for delay.
FIG. 3.
Timing of the first division in SC mutants. In all of the graphs, the first division of WT cells is shown as solid diamonds (♦) and is a control for normal timing. The control for early timing of the first division in all panels is a rec104 strain denoted by solid squares (▪). In all of the graphs, the mutant examined is shown as solid circles (•). For each mutant, at least two independent cultures were examined for each experiment and the data point shown is the mean. Another independent experiment was done for each mutant. In each case, the repeat gave the same result (data not shown). The mutant examined is shown above each panel.
Premeiotic DNA synthesis in meiotic recombination initiation mutants.
Recently, Cha et al. (4) examined premeiotic S phase in several meiotic mutants. Of the mutants examined, two were deficient in meiotic recombination initiation (spo11Δ and rec102Δ). The authors reported that the length of premeiotic S phase is shorter in spo11Δ cells (59 min) than in WT cells (77 min) or rec102Δ cells (72 min). However, the times at which 50% of the WT, rec102Δ, or spo11Δ cells entered premeiotic S phase were indistinguishable. The shorter S-phase length of spo11Δ mutants suggests a possible reason for the earlier MI division, although the normal length of S in rec102Δ cells (4) is not consistent with that idea.
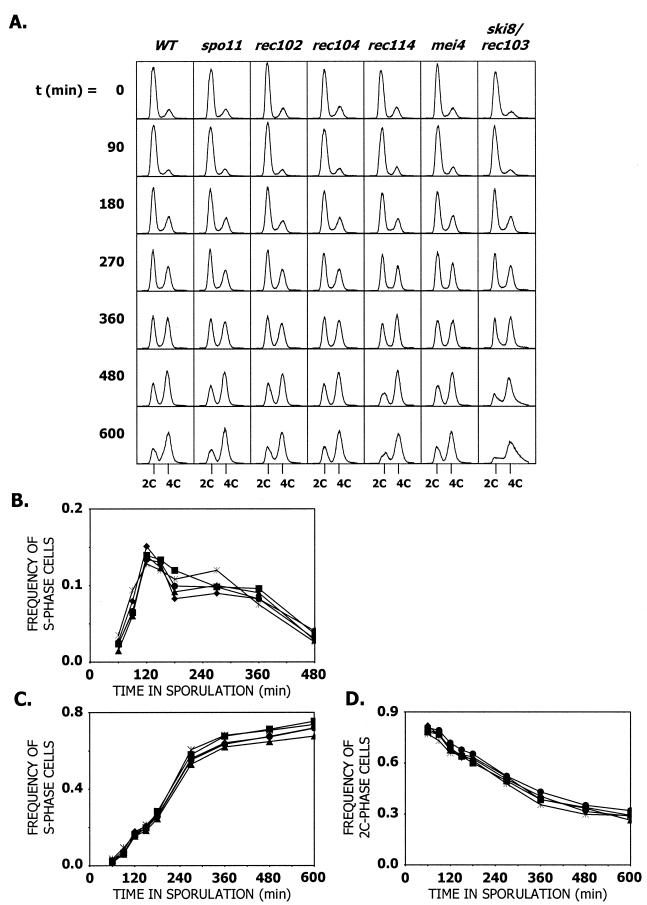
To test this possibility, premeiotic DNA synthesis was examined in the WT and spo11, rec102, rec104, rec114, mei4, and rec103/ski8 gene deletion mutants (Fig. 4). Using the methods of Cha et al. (4), curves were plotted for each culture (Fig. 4B), and the length of the S phase was calculated (Table 1). The average of three independent experiments shows that all of the mutants exhibit S-phase lengths of 55 to 59 min (Table 1). None of these values is significantly different from the S-phase length of WT cells or from each other (Table 1). The calculated S-phase lengths for WT cells and spo11, rec102, rec104, and rec114 mutants (all of which have an early MI division) are indistinguishable. We did not calculate S-phase length for mei4 or rec103/ski8 cells, since only one experiment was done for each. However, we do note that there are no obvious differences in the histograms (Fig. 4A) between the WT up to the 600-min time point with the mei4 mutant or up to the 360-min time point with the ski8/rec103 mutant. Cumulative curves of cells entering S phase were calculated by the methods described by Cha et al. (4) (Fig. 4C). In the WT and all mutants examined, 50% of the cells enter premeiotic S phase at about 213 min after introduction into sporulation medium. There were no significant differences between the Rec− mutants and the WT cells. Finally, we determined a time of entry into S phase, using a method independent of the calculation of S-phase life span (Fig. 4D). This method shows that, in WT cells and all mutants examined, 50% of the cells enter premeiotic S phase at about 240 min after introduction into sporulation medium. There is no significant difference between any of the mutants examined and WT, suggesting that differences in S-phase length do not explain why Rec− cells start the MI division earlier.
FIG. 4.
Analysis of premeiotic DNA synthesis by flow cytometry. (A) Representative histograms showing DNA content in cells proceeding through meiosis. The relevant genotype of each strain is designated above, minutes in sporulation are designated beside and 2C and 4C DNA contents are designated below histograms. (B) Noncumulative curve of cells proceeding through premeiotic S phase. (C) Cumulative curve of cells proceeding through premeiotic S phase. The curve was calculated by the method of Cha et al. (4). (D) Cumulative curve of cells exiting 2C phase (entering S phase). The curve was calculated by using the cell cycle analysis tool in the FlowJo software package. WT, ♦; spo11, ▴; rec102, •; rec104, ▪; rec114, *. For all graphs, the average values for three independent experiments are shown for all strains except the rec114 strain (which had only two independent experiments). The average sporulation frequencies for the experiments were as follows: WT = 74%, spo11 = 25%, rec102 = 31%, rec104 = 20%, and rec114 = 25%.
TABLE 1.
Premeiotic S-phase length and time of entry into S phase of meiotic recombination initiation mutantsa
| Strain type | S-phase length (min)b | Time (min) of entry into S phasec | Time (min) of exit from 2C phased |
|---|---|---|---|
| WT | 55.9 ± 1.3 | 214 ± 4 | 234 ± 14 |
| spo11 | 58.0 ± 5.0 | 212 ± 7 | 258 ± 18 |
| rec102 | 58.1 ± 3.2 | 215 ± 6 | 249 ± 25 |
| rec104 | 58.5 ± 4.5 | 213 ± 6 | 232 ± 28 |
| rec114 | 56.9 ± 0.9 | 213 ± 9 | 236 ± 8 |
All values are the average of three independent sporulation experiments, except for the rec114 strain (two independent experiments). None of the values is significantly different from WT, nor are they significantly different from each other (as examined by Student's t test).
S-phase length calculated by the method of Cha et al. (4).
Time in which 50% of active cells have entered S phase calculated by the method of Cha et al. (4) (Fig. 4C).
Time in which 50% of active cells have exited 2C phase (and therefore entered S phase) as calculated with the cell cycle analysis tool in the FlowJo software package (Fig. 4D).
Meiosis I spindle formation in recombination initiation mutants.
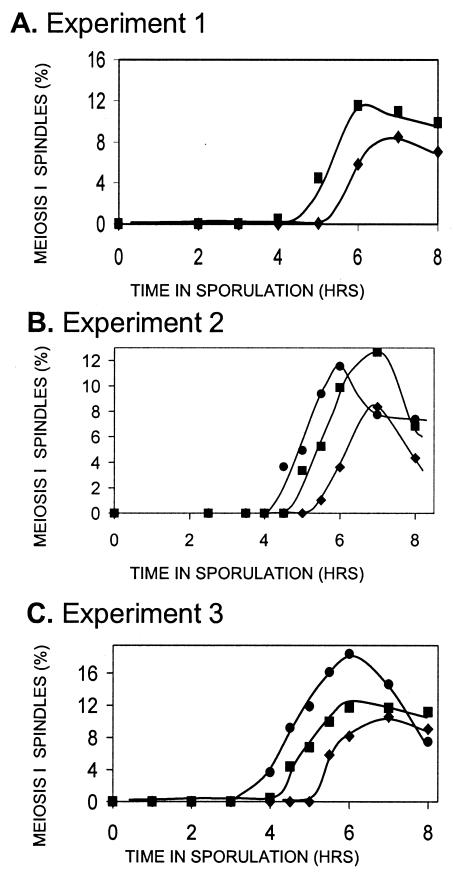
Shonn et al. (46) have argued that the reason chromsomes separate at an earlier time in Rec− initiation mutants is that, without chiasmata to provide tension, chromosomes are pulled apart as soon as the spindle is formed in Rec− mutants; division in WT Rec+ cells with chiasmata is delayed by the spindle checkpoint system. This idea predicts that MI spindles form at the same time in initiation mutants and WT cells, but that chromosome segregation is delayed in WT cells. To test this, we measured the fraction of cells containing MI spindles versus time in sporulation for WT, rec102, and rec104 cells by using fluorescence microscopy with antitubulin antibodies (Fig. 5 and 6). The data clearly show that MI spindles form earliest in rec102, then in rec104, and then in WT cells. The fact that spindles form at an earlier time in Rec− cells is not easily reconciled with the hypothesis of Shonn et al. (46) (see Discussion).
FIG. 6.
The timing of MI spindle formation in WT cells and Rec− mutants. Three independent experiments were performed on different days to determine the time of MI spindle formation in WT cells and rec104 and rec102 mutant strains. In each experiment, DAPI staining verified that the timing of the first division was earlier in the Rec− mutants than in the WT cells (data not shown).
Do known meiotic checkpoint functions transduce the recombination initiation delay?
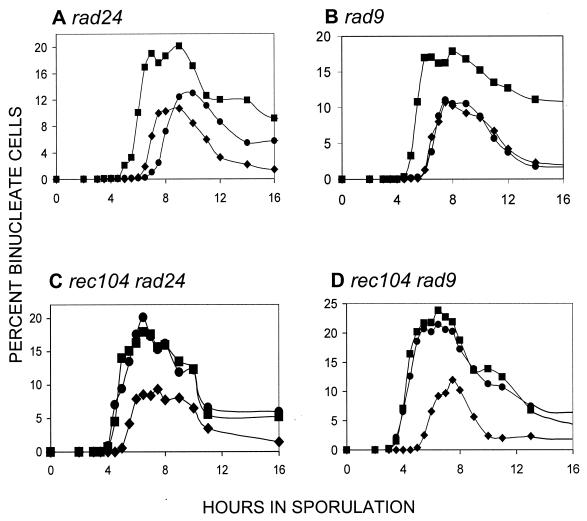
The pachytene checkpoint (e.g., as defined by dmc1 or zip1 mutants) results in arrest before the first division (3, 42). Mutations in RAD24 eliminate the arrest; for example, rad24 dmc1 cells proceed through the first (and second) division (29, 42). We asked if RAD24 was required for the recombination initiation signal that normally delays the first division: if it were, rad24 mutants should display an earlier first division. The data in Fig. 7 indicate that RAD24 does not play a role in the process. In fact, exactly as observed by Shinohara et al. (45), we find that rad24 mutants begin the first division at a later time. The timing of rad24 makes the role of pachytene checkpoint in WT cells problematic (see Discussion). Although both RAD9 and RAD24 are needed for proper checkpoint activity in the DNA damage pathway in mitosis (8), the RAD9 gene does not play a role in the meiotic pachytene checkpoint (29). The RAD9 gene is also not required for the recombination initiation delay signal (Fig. 7B); the timing of the first division is indistinguishable from that in WT cells. The rec104 mutation is epistatic to both rad24 (Fig. 7C) and rad9 (Fig. 7D); we observe no synergistic effects in the double mutant. The data indicate that neither RAD9 nor RAD24 is required to transduce the initiation signal that delays the MI division.
FIG. 7.
Timing of the first division in checkpoint mutants. In all of the graphs, the first division of WT cells is shown as solid diamonds (♦). The control for early timing of the first division in all panels is a rec104 strain denoted by solid squares (▪). In panels A and B, the mutant in question is shown by solid circles (•); in panels C and D, the solid circles refer to the double mutant. The type of mutant examined is shown above each panel. For each mutant, at least two independent cultures were examined for each experiment, and the data point shown represents the mean. Another independent experiment was done for each mutant; in each case, the repeat gave the same result (data not shown).
The target of the signal for recombination initiation.
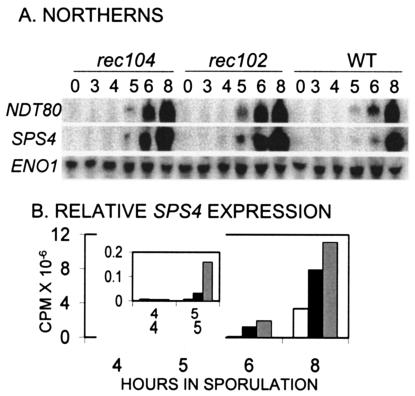
A candidate for the target of the recombination initiation signal that delays the first division is the NDT80 gene. This gene codes for a positive regulator of the middle meiotic genes, and ndt80 mutants arrest before the first division (5, 18, 51). We therefore examined the expression of NDT80 in the WT and rec104 and rec102 mutants (Fig. 8). Expression of the SPS4 gene was used as a reporter of total NDT80 activity, since evidence indicates that Ndt80p is also posttransciptionally regulated (36, 51). NDT80 transcription begins earlier in both rec102 and rec104 mutants than in WT cells, although our current data do not distinguish between the two Rec− mutants. SPS4 transcription, however, begins earlier and reaches higher levels in rec102 than in rec104 (e.g., compare SPS4 expression at 5 and 6 h of sporulation in Fig. 8A and B), consistent with the earlier start of the MI division. Both Rec− mutants clearly express SPS4 prior to the time it is expressed in WT cells. The data indicate that NDT80 is a target of the recombination initiation signal.
FIG. 8.
Expression and activity of NDT80 in Rec− mutants undergoing an early first division. (A) The genotype of the strain examined is shown at the top. Numbers below the genotypes indicate the time in sporulation that RNA was isolated. The probes used for the Northern blots are shown to the left. (B) The amount of SPS4 expression (corrected for loading by ENO1) is shown versus time. The white bar represents expression in WT cells, the black bar represents expression in rec104 cells, and the gray bar represents expression in rec102 cells.
DISCUSSION
For complex biological processes to occur properly, cells must ensure that events happen at the right time and in the correct order. During meiosis, chromosomes undergo premeiotic DNA synthesis, recombination and synapsis, the reductional division, the equational division, and packaging into gametes or spores. Given the complexity of these events, it is not surprising either that there is communication between them or that a number of checkpoint systems exist in S. cerevisiae that arrest cells in the progression through meiosis if the preceding step has occurred improperly. For example, a checkpoint for proper premeiotic DNA synthesis (48) arrests cells prior to recombination. One of the best-studied checkpoints occurs in pachytene (42), where defects in some of the steps of recombination and synapsis (e.g., dmc1, zip1, and hop2) result in arrest. This checkpoint is mediated by components of the mitotic DNA damage checkpoint system and requires RAD24, RAD17, DDC1, etc. Unlike the mitotic DNA damage checkpoint, the pachytene checkpoint does not require RAD9. The data suggest that at least part of the signal detected by the checkpoint in dmc1 mutants is the accumulation of large amounts of ssDNA (29). Since recombination and the MI division need to be coordinated in normal WT cells, it has been suggested that the transient amount of ssDNA present in WT cells might set off the checkpoint system, causing a transient delay for the first division until recombination is completed.
The components of the initiation signal for delay of the first division.
Because mutations completely deficient in the initiation of meiotic recombination go through the meiotic divisions, it had been thought that cells did not monitor initiation (41). Our previous work demonstrated that meiotic cells do monitor recombination initiation; they do so by assessing the presence of some of the meiotic recombination gene products required for initiation (15, 21). In the absence of REC102, REC104, REC114, and RAD50 the first meiotic division actually begins at an earlier time. The presence of the proteins encoded by these genes results in a signal leading to the delay of the MI division in WT cells. The SPO11 gene was also shown to be required for this normal MI delay (24). The work presented here confirms that the Spo11p is required for the MI delay and shows that the MER2/REC107 gene product is required as well. The MRE2 gene is needed for meiosis-specific splicing of MER2/REC107 and MER3 (a late recombination gene) (34, 35); it does not play a direct role in recombination initiation. A very low level of DSBs was detectable in an mre2 mutant at the his4::LEU2 locus (see Fig. 5 in reference 34). The mre2 mutant undergoes MI at a time indistinguishable from the rec104 mutant control. We presume that the early MI division in the mre2 mutant is due to the reduction in the amount of the MER2/REC107 gene product. The data also indicate that rec103/ski8 mutants initiate the first division at the same time as a WT cell; like MEI4, REC103/SKI8 is not needed for the signal to delay. Since the MI division starts at the same time as WT cells in both rec103/ski8 and mei4 mutants, recombination is not required for the normal delay.
HOP1 and RED1 are also required for the MI division delay signal. These two components of the axial elements and the tripartite SC are required for full levels of recombination, although null mutants still display a large induction of recombination over the mitotic background level, reaching about 10% of the meiotic level mutants (19, 32, 40). Different levels of DSBs have been detected in red1 and hop1 mutants, although all experiments agree that the amounts of DSBs in red1 and hop1 mutants are reduced (32, 44, 52). Estimates of the level of DSBs in these mutants range from 5 to ∼50% of normal levels: this range may reflect both strain differences and/or locus differences. It may also reflect the difference of monitoring DSBs by using a rad50S mutation versus a com1/sae2 mutation. The MI division in red1 and hop1 mutants occurs at an early time indistinguishable from that of rec104. The normal timing of the MI division in rec103/ski8 and mei4 mutants indicates that DSBs are not necessary for the recombination initiation signal. The levels of DSBs in hop1 and red1 strains, along with the early division observed in those strains, strongly suggest that moderately high levels of meiotic DSBs are not sufficient either. At this time, the signal for recombination initiation that results in the normal delay of the first division consists of Rec102p, Rec104p, Rec107p, Rec114p, Spo11p, Rad50p, Hop1p, and Red1p.
Mechanisms for the initiation signal that delays the MI division.
We address three possible mechanisms for the earlier start of the MI division in initiation mutants. The first hypothesis is that WT cells have a longer S phase because of the presence of recombination initiation proteins and that this results in a normal delay of the MI division. Cha et al. (4) showed that premeiotic S phase is shorter in spo11 mutants than in WT cells (4). This is consistent with the view that the shorter S phase results in the earlier timing of the MI division observed in spo11 mutants. The same authors, however, also showed that rec102 mutants had normal timing of premeiotic S phase (4). This is something of a paradox, since rec102 mutants clearly begin the MI division earlier than WT cells. We observe no significant differences in the timing of S phase (measured in three different ways) between WT cells and rec102, rec104, rec114, and spo11 mutants (Table 1). We note that the exact time at which 50% of cells have entered S phase is slightly different depending, which of the two methods is used, but the conclusion that the mutants are the same as the WT remains the same. If the reason that the MI division starts earlier in recombination initiation mutants is that the premeiotic S phase is shorter, then it would seem that all Rec− mutants displaying an earlier MI division should have a shorter S phase. We do not see it in our examination of four different mutants; Cha et al. (4) saw it in spo11 mutants but not in rec102 mutants. At the moment, we can only attribute the difference in the spo11 phenotypes to strain differences. However, since rec102 has normal S-phase timing in both strain backgrounds, even this explanation seems less than appealing. We conclude that neither the time of entry into S phase, the exit from 2C DNA content, nor the length of S phase is easily correlated with the time the MI division starts. We argue that alterations in S-phase length do not appear to explain the earlier MI division in recombination initiation mutants.
A second mechanism for the normal delay of MI caused by the presence of the recombination initiation functions was proposed by Shonn et al.(46). They suggested that the absence of chiasmata in spo11 mutants (and presumably in all initiation mutants) resulted in immediate separation of chromosomes, since there would be no tension on the MI spindle. In WT Rec+ cells, the presence of chiasmata and the consequent tension would result in a delay of chromosome separation and hence the normal delay of the MI division. While appealing, we discuss four observations indicating that this hypothesis doesn't seem to explain our observations. First, this view suggests that spindles should form at the same time in Rec− initiation mutants and in WT cells, but the chromosomes would be separated earlier in the mutants due to lack of chiasmata. Our data indicate that this is not the case. The MI spindle forms very early in rec102 mutants, early in rec104 mutants, and at the normal time in WT cells. This indicates that the delaying signal caused by recombination initiation functions acts prior to MI spindle formation. Second, both rec103 and mei4 mutants lack recombination but start the MI division at the same time as WT cells. Third, we have shown that NDT80 expression and activity are delayed in WT cells compared to the initiation mutants, which display earlier divisions. Since NDT80 is required for spindle formation for the first division, it is difficult to understand how the spindle checkpoint could monitor events where the spindle hasn't formed yet. Fourth, Shonn, et al. (46) showed that a spo11 mad2 strain started the MI division at the same time as a spo11 strain. Although the timing of a WT strain was not shown in the same experiment, we presume that the spo11 mutant began MI earlier than a WT strain would have. We conclude that loss of the spindle checkpoint did not affect the early start of the MI division.
We note that Rec− initiation mutants display another alteration of the MI division in addition to the earlier onset; the fraction of cells that are binucleate is higher and the number of binucleate cells persists longer. This is true even in a rec103/ski8 mutant, which starts MI at the normal time. Shonn et al. (46) showed that a spo11 mad2 strain had a lower level of binucleate cells than a spo11 strain. From the data presented, it is harder to determine if the mad2 mutation also reduced the persistence of binucleates. We propose that the spindle checkpoint does not affect the start of the MI division, but does affect the ability of Rec− cells to proceed through the division.
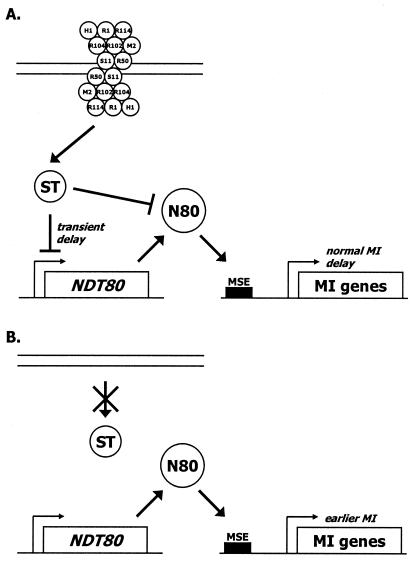
A third mechanism for the normal delay of the MI division would be the presence of a putative recombination initiation complex that is recognized as a signal that recombination has started and that the MI division should be delayed (Fig. 9). This complex would consist of (at least) Rec102p, Rec104p, Rec107p, Rec114p, Spo11p, Rad50p, Hop1p, and Red1p. (Rec103p and Mei4p might be part of the complex, but would not be recognized by the signal transduction system.) If any of the eight proteins are missing, no delay occurs. Seven of these proteins are specifically produced in meiosis; Rad50p is also expressed in mitotic cells. Considerable support exists for interactions among these eight proteins. Spo11p and Rec102p have been shown to interact by high-copy suppression of specific alleles, negative interactions of specific alleles, and coimmunoprecipitation (22, 24). Rec102p and Rec104p have been shown to interact by allele-specific suppression, high-copy suppression of specific alleles, and coimmunoprecipitation (22, 43). Hop1p and Red1p have been shown to interact by high-copy suppression, allele-specific suppression, coimmunoprecipitation, and colocalization on the SC (9, 14, 20, 47). In addition, some hop1 mutations can be suppressed by overexpression of REC104 (14). Certainly the hypothesis that a recombination initiation complex exists (25) and is recognized as a signal seems both plausible and testable. It is not completely clear why rad50 and rec102 mutants undergo the first division even earlier than the other Rec− mutants: perhaps the putative initiation complex is built in stages, and defects in different steps have greater or lesser effects upon timing (21). If this view of the differences in early timing has merit, it suggests that the Rad50p and Rec102p might be early steps in assembly. The model also predicts that there should be functions that transduce the signal to the target(s).
FIG. 9.
Model for the earlier reductional division observed in some meiotic recombination initiation mutants. (A) The recombination initiation proteins necessary for the normal transient delay of the start of the MI division are shown on double-stranded DNA as a complex. The proteins are symbolized as follows: Spo11p, S11; Rad50p, R50; Rec104p, R104; Rec102p, R102; Mer2/Rec107p, M2; Rec114p, R114; Hop1p, H1; and Red1p, R1. The organization of the proteins is not intended to reflect the actual complex. Other recombination proteins (e.g., the protein coded for by MEI4) may well be present in the putative initiation complex, but are not shown either because mutations in them do not affect the start of the division or because they have not yet been tested. The recombination initiation complex on the DNA is relayed through an as yet unknown signal transduction pathway (ST). This pathway inhibits transcription of NDT80 and (from the SPS4 expression patterns) Ndt80p (N80) activity. Since the activity of Ndt80p is delayed, middle meiotic genes (MMGs) containing middle sporulation elements (MSEs) that require Ndt80p for expression are also delayed. Among the MMGs that are affected are genes required for the MI division. The delay in Ndt80p activity is transient, because the complex is transient and at least part of it leaves the DNA as recombination proceeds. (B) In the absence of the appropriate recombination initiation proteins, the complete complex does not form on DNA. Since no signal is detected, there is no inhibition of NDT80 expression or activity. Thus, the MMGs are expressed earlier, and the MI division begins at an earlier time.
Transduction of the initiation signal.
Because components of the mitotic DNA damage checkpoint clearly play a role in arresting cells defective in some late recombination events, it seemed important to investigate whether they played a role in the MI division delay caused by the normal initiation signal. Because RAD9 was not involved in the pachytene checkpoint, we hypothesized that it might instead have a role in the normal delay of MI caused by the presence of the early recombination functions. The predicted phenotype of a mutation in a gene involved in transducing the signal is an earlier first division. The data obtained show that RAD9 is not required to transduce the signal; rad9 mutants begin the first division indistinguishably from WT cells. Likewise, deletion of RAD9 has no effect on the early division observed in a rec104 mutant.
We next examined the effect of a rad24 mutation on the timing of the first division. As with the prediction for rad9, rad24 mutants should have had an earlier MI division if the Rad24p was required to transduce the initiation signal. This was not the case; rad24 mutants actually go through the first division at a later time than WT cells. A similar result was recently reported by Shinohara et al. (45). From the timing of the rec104 rad24 double mutant, it's clear that rec104 is epistatic to rad24. This suggests that the delay in the first division observed in the rad24 single mutant is solely due to presence of recombination. Shinohara et al. (45) suggested that the rad24 observation calls into question the concept that the checkpoint acting in dmc1 cells is also active in DMC1 cells (as a delay). We had raised a similar concern (21). Since rad24 mutants do not exhibit an earlier first division, it would appear that the simplest conclusion is that the pachytene checkpoint is not active in WT cells. This view is also consistent with the normal start of the MI division observed in mei4 and rec103/ski8 mutants; since recombination doesn't initiate, the putative DMC1 delay signal is never reached. In order to completely rule out the DNA damage checkpoint pathway as involved in signal transduction, we must examine the double rad9 rad24 mutant. In the mitotic DNA damage checkpoint, both Rad9p and Rad24p have parallel and additive input into the subsequent steps of the pathway (8). Deletion of either one partially removes the ability to respond to DNA damage by arrest. The double mutant, however, is even more deficient in the checkpoint. It remains possible that both RAD9 and RAD24 must be removed in meiosis to eliminate transduction of the initiation signal and the consequent delay.
Target of the initiation signal.
The discovery that the recombination initiation signal for MI division delay acts before the formation of spindles suggested that mutations in any target for the signal would block meiosis before division and before formation of MI spindles. The NDT80 gene is just such a candidate. Null mutations in NDT80 arrest in pachytene and although the spindle pole body duplicates, it does not separate and MI spindles do not form (53). The data in this paper indicate that Ndt80p is involved in the communication between recombination initiation functions and the MI division. The transcription of NDT80 is increased at earlier times in both rec102 and rec104 mutants. SPS4 is a middle meiotic gene regulated only by NDT80 and not by other meiotic transcriptional regulators, and (36, 37; J. Segall, personal communication) SPS4 transcription therefore measures active Ndt80p. SPS4 is clearly transcribed at earlier times in rec102 than in rec104, and earlier in rec104 than in WT cells. The control of NDT80 is complex and occurs at the level of transcription and posttransciptional events (36). We are currently investigating known regulators of NDT80 to determine which, if any, of them are involved in conveying the recombination initiation signal. The SUM1 repressor of NDT80 (28, 36) would seem one likely candidate.
Acknowledgments
This work was supported by an NSF grant MCB00-83816 to R.E.M. M.L.P. was supported during the summer of 2003 by an undergraduate fellowship from the College of Medicine, University of Iowa. L.C. was supported by a Howard Hughes summer research fellowship in 2002. B.S., D.R.H., and K.E.L. did their work while on a first year graduate student rotation and were supported by the Department of Biological Sciences during that period.
We sincerely thank Jodie Haring, John Harty, and Justin Fishbaugh for providing instruction about flow cytometry and time on the FACSCalibur.
REFERENCES
- 1.Bennett, C. B., A. L. Lewis, K. K. Baldwin, and M. A. Resnick. 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 90:5613-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 4.Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker, and N. Kleckner. 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14:493-503. [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 6.Cool, M., and R. E. Malone. 1992. Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol. Cell. Biol. 12:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean, P. N., and J. H. Jett. 1974. Mathematical analysis of DNA distributions derived from flow microfluorometry. J. Cell Biol. 60:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de los Santos, T., and N. M. Hollingsworth. 1999. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J. Biol. Chem. 274:1783-1790. [DOI] [PubMed] [Google Scholar]
- 10.Dien, B. S., M. S. Peterson, and F. Srienc. 1994. Cell-cycle analysis of Saccharomyces cerevisiae. Methods Cell Biol. 42:457-475. [DOI] [PubMed] [Google Scholar]
- 11.Esposito, R. E., and M. S. Esposito. 1974. Genetic recombination and commitment to meiosis in Saccharomyces. Proc. Natl. Acad. Sci. USA 71:3172-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairhead, C., and B. Dujon. 1993. Consequences of unique double-stranded breaks in yeast chromosomes: death or homozygosis. Mol. Gen. Genet. 240:170-178. [DOI] [PubMed] [Google Scholar]
- 13.Fox, M. H. 1980. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry 1:71-77. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, D. B., N. M. Hollingsworth, and B. Byers. 1994. Insertional mutations in the yeast HOP1 gene: evidence for multimeric assembly in meiosis. Genetics 136:449-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galbraith, A. M., S. A. Bullard, K. Jiao, J. J. Nau, and R. E. Malone. 1997. Recombination and the progression of meiosis in Saccharomyces cerevisiae. Genetics 146:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galbraith, A. M., and R. E. Malone. 1992. Characterization of REC104, a gene required for early meiotic recombination in the yeast Saccharomyces cerevisiae. Dev. Genet. 13:392-402. [DOI] [PubMed] [Google Scholar]
- 17.Hasek, J., and E. Streiblova. 1996. Fluorescence microscopy methods. Methods Mol. Biol. 53:391-405. [DOI] [PubMed] [Google Scholar]
- 18.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingsworth, N. M., and B. Byers. 1989. HOP1: a yeast meiotic pairing gene. Genetics 121:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingsworth, N. M., and L. Ponte. 1997. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147:33-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao, K., S. A. Bullard, L. Salem, and R. E. Malone. 1999. Coordination of the initiation of recombination and the reductional division in meiosis in Saccharomyces cerevisiae. Genetics 152:117-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao, K., L. Salem, and R. Malone. 2003. Support for a meiotic recombination initiation complex: interactions among Rec102p, Rec104p, and Spo11p. Mol. Cell. Biol. 23:5928-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 24.Kee, K., and S. Keeney. 2002. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 26.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 27.Kleckner, N. 1996. Meiosis: how could it work? Proc. Natl. Acad. Sci. USA 93:8167-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 30.Malone, R. E., S. Bullard, M. Hermiston, R. Rieger, M. Cool, and A. Galbraith. 1991. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone, R. E., D. L. Pittman, and J. J. Nau. 1997. Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol. Gen. Genet. 255:410-419. [DOI] [PubMed] [Google Scholar]
- 32.Mao-Draayer, Y., A. M. Galbraith, D. L. Pittman, M. Cool, and R. E. Malone. 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144:71-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martini, E., and S. Keeney. 2002. Sex and the single (double-strand) break. Mol. Cell 9:700-702. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa, T., and H. Ogawa. 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18:5714-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa, H., K. Johzuka, T. Nakagawa, S. H. Leem, and A. H. Hagihara. 1995. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv. Biophys. 31:67-76. [DOI] [PubMed] [Google Scholar]
- 36.Pak, J., and J. Segall. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pak, J., and J. Segall. 2002. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6430-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecina, A., K. N. Smith, C. Mezard, H. Murakami, K. Ohta, and A. Nicolas. 2002. Targeted stimulation of meiotic recombination. Cell 111:173-184. [DOI] [PubMed] [Google Scholar]
- 39.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 40.Rockmill, B., and G. S. Roeder. 1990. Meiosis in asynaptic yeast. Genetics 126:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roeder, G. S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600-2621. [DOI] [PubMed] [Google Scholar]
- 42.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 43.Salem, L., N. Walter, and R. Malone. 1999. Suppressor analysis of the Saccharomyces cerevisiae gene REC104 reveals a genetic interaction with REC102. Genetics 151:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwacha, A., and N. Kleckner. 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90:1123-1135. [DOI] [PubMed] [Google Scholar]
- 45.Shinohara, M., K. Sakai, T. Ogawa, and A. Shinohara. 2003. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164:855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shonn, M. A., R. McCarroll, and A. W. Murray. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289:300-303. [DOI] [PubMed] [Google Scholar]
- 47.Smith, A. V., and G. S. Roeder. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136:957-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart, D., and C. Wittenberg. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12:2698-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155-1161. [DOI] [PubMed] [Google Scholar]
- 50.Sym, M., and G. S. Roeder. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79:283-292. [DOI] [PubMed] [Google Scholar]
- 51.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woltering, D., B. Baumgartner, S. Bagchi, B. Larkin, J. Loidl, T. de los Santos, and N. M. Hollingsworth. 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 20:6646-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]