Abstract
The complete nucleotide sequence of Sucra jujuba nucleopolyhedrovirus (SujuNPV) was determined by 454 pyrosequencing. The SujuNPV genome was 135,952 bp in length with an A+T content of 61.34%. It contained 131 putative open reading frames (ORFs) covering 87.9% of the genome. Among these ORFs, 37 were conserved in all baculovirus genomes that have been completely sequenced, 24 were conserved in lepidopteran baculoviruses, 65 were found in other baculoviruses, and 5 were unique to the SujuNPV genome. Seven homologous regions (hrs) were identified in the SujuNPV genome. SujuNPV contained several genes that were duplicated or copied multiple times: two copies of helicase, DNA binding protein gene (dbp), p26 and cg30, three copies of the inhibitor of the apoptosis gene (iap), and four copies of the baculovirus repeated ORF (bro). Phylogenetic analysis suggested that SujuNPV belongs to a subclade of group II alphabaculovirus, which differs from other baculoviruses in that all nine members of this subclade contain a second copy of dbp.
Introduction
Baculoviruses are rod-shaped, insect-specific viruses with double-stranded, circular DNA 80–180 kb genomes [1]. Baculoviruses have been widely used as bio-pesticides to control insect pests in agriculture and forestry [2], as vectors for protein expression, and as potential vectors for gene therapy [3], [4]. The family Baculoviridae used to be grouped into two genera: Nucleopolyhedroviruses (NPVs) and Granuloviruses (GVs), dependent upon differing morphologies of occlusion bodies (OBs) [5]. More recently, a new classification has subdivided the Baculoviridae into four genera, based on phylogeny and host specificities: Alphabaculovirus (lepidopteran-specific NPVs), Betabaculovirus (lepidopteran-specific GVs), Gammabaculovirus (hymenopteran-specific NPVs) and Deltabaculovirus (dipteran-specific NPVs) [6]. Alphavaculoviruses can be further gathered into group I and group II based on phylogenetic analyses, The NPVs are also characterized as single nucleocapsid NPVs (SNPVs) and multiple-nucleocapsid NPVs (MNPVs) according to the number of nucleocapsids per virion. To date, 62 baculovirus reference genomes are available in the National Centre for Biotechnology Information (NCBI) database; 42 of them are alphabaculoviruses, 15 betabaculoviruses, three gammabaculoviruses, one deltabaculovirus and one unclassified baculovirus.
Sucra jujuba Chu (Lepidopteral: Geometridae) is an important pest of jujube, and it is widespread in the jujube-growing regions of China. The larvae feed on the young leaves and buds of jujube, apple, pear and mulberry. In 2009, 1250 square hectometers of mulberry became infested with Sucra jujuba Chu in China [7]. Sucra jujuba NPV (SujuNPV) is a SNPV, which was first isolated from naturally diseased Sucra jujuba larvae in the early 1980s [8]. The virus is highly infectious to Sucra jujuba with an LC50 of 3.5×105 PIBs/mL in the third instar larvae [9]. It appears to be specific to Sucra jujuba as bioassay studies showed that it did not infect Antheraea pernyi, Arge captiva, Bombyx mori, Culcula panterinaria, Euproctis flava, Leucoma salicis, Lymantria dispar, Macaria elongaria, Phthonandria atrilineata, Plusia agnate or Semiothisa cineraria [8], [10], [11].
In the present study, the genome of SujuNPV is completely sequenced and annotated, and compared with those of the other representative baculoviruses. Results indicate that SujuNPV is a novel species belonging to a unique subclade of group II alphabaculoviruses, which contain a second copy of the DNA binding protein gene (dbp).
Materials and Methods
DNA extraction of the viral genome
The SujuNPV were purified from the dead Sucra jujube preserved in “Chinese general virus collection center” (CGVCC) with collection Number IVCAS 1.0048, which was originally isolated from Shandong Province, China, in 1983 [12]. The ODVs were purified as previously reported [13]. To extract DNA, the ODVs were incubated with four times volume 1 M DAS (5 M Nacl, 5 M NaCO3 and 0.5 M EDTA (pH8), mixed in the ratio of 3∶3∶0.6) at 37°C for 30 min. Then, the same volume of 1 M Tris (pH 7.4) was added followed by centrifugation at 10,000 rpm (5 min) to obtain the viral DNA.
Sequencing and sequence analysis of the SujuNPV genome
The SujuNPV genome sequence was determined by 454 pyrosequencing. A total of 92,684 reads were obtained and assembled into 10 contigs using GS De Novo Assembler software, covering 97.8% of the whole genome with a sequencing depth of 225x. The remaining gaps were filled using PCR and Sanger sequencing.
Briefly, the genome was broken randomly into small fragments of about 600–900 bp by nebulization and adapters were added to construct a genomic library. Subsequently, the library was amplified by emPCR before sequencing. The SujuNPV genome was assembled using a GS De Novo Assembler providing 454 programs. Additional verifications were performed for gaps and ambiguous sequences using sequence-specific primers. The hypothetical ORFs of the SujuNPV genome were predicted by fgenesV0 (http://www.softberry.com/berry.phtml) [14], adopting the criteria of a size of at least 50 aa with a minimal overlap with other ORFs. Predicted aa sequences were compared with homologues of typical baculoviruses of the four genera, including AcMNPV (NC_001623), HearNPV-G4 (NC_002654), CpGV (NC_002816), NeleNPV (NC_005906) and CuniNPV (NC_003084), and similarities were obtained by DNAStar software with default parameters.
Gene parity plots were generated in order to analyze the gene order of SujuNPV relative to three other closely related baculoviruses (ApciNPV, EcobNPV and OrleNPV) and the five representative viruses mentioned above.
Consensus promoter motifs were searched for in the upstream 150 bp region from the start codon of each ORF based on the characterization of baculovirus' promoters, that’s a TATA box linked with a CAKT motif 20–40 bp downstream and a DTAAG box.
Phylogenetic analysis
Phylogenetic analysis of baculoviruses was performed using the concatenated aa sequence of 37 core genes [15] from 62 baculovirus reference genomes (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10442, data update until Jan.5th, 2014). The sequences were aligned by ClustalW with default parameters of MEGA5. And the maximum likelihood (ML) phylogenetic tree was reconstructed according to the previous report [16] with 1000 bootstrap values. The phylogenetic trees of dbp, helicase and p26 were constructed based on the same parameters.
Prediction of secondary structure
The secondary structures of DNA sequences were predicted by the Mfold Web Server using default parameters [17].
Results and Discussion
Characteristics of the SujuNPV genome sequence
The full SujuNPV genome [GeneBank: KJ676450] was 135,952 bp in length with an A+T content of 61.34%. Following convention, the adenine coding for the start methionine of the polyhedrin gene (ph) was chosen as the zero point of the SujuNPV genome and ph was designated as the first ORF. Overall, 131 putative ORFs were detected in the SujuNPV genome with the criteria of a length of at least 50 amino acids (aas) and a minimal overlap with adjacent ORFs. The total ORFs covered 89.2% of the whole genome, distributed with 60 ORFs in a forward orientation and 71 ORFs in a reverse orientation. In addition, seven homologous regions (hrs) were identified in SujuNPV (Fig. 1).
Figure 1. Circular map of the SujuNPV genome.
The arrows inside or outside the circle indicate the orientation of putative ORFs. Arrows, red represent the core genes, blue represent Lepidoptera baculovirus conserved genes, grey represent genes common to baculoviruses, open are genes unique to SujuNPV, and yellow rectangles indicate hrs. The collinear region conserved in Lepidoptera baculoviruses is also shown.
BLAST comparisons of the 131 protein sequences of the SujuNPV, deduced from the homologous sequences of other baculoviruses, revealed that SujuNPV has 37 core genes (shown in red in Fig. 1) and 24 other genes conserved in lepidopteran baculoviruses (shown in blue in Fig. 1). It also contains 65 additional genes commonly found in various baculoviruses (shown in grey in Fig. 1) and five unique genes (shown as open arrows in Fig. 1). Consensus promoter motifs were searched for in the upstream 150 bp region of the start codon of each ORF. Amongst all 131 ORFs identified in the SujuNPV genome, 24 ORFs possessed the early promoter motif (a TATA box linked with a CAKT motif 20–40 bp downstream), whereas 61 ORFs had the late promoter motif DTAAG and 10 ORFs contained both the early and late promoter motifs (Table 1). No obvious baculoviral promoter motifs were detected for the remaining 36 ORFs.
Table 1. SujuNPV Genome Annotation.
| ORF | name | motif | start | end | length (aa) | str. | ORF position | amino acid identity (%) | ||||||||
| AcMNPV | HearNPV | CpGV | NeleNPV | CuniNPV | AcMNPV | HearNPV | CpGV | NeleNPV | CuniNPV | |||||||
| 1 | polyhedrin | E | 1 | 741 | 246 | + | 8 | 1 | 1 | 1 | 86.5 | 88.2 | 53.7 | 45.1 | ||
| 2 | orf1629 | L | 774 | 2474 | 566 | − | 9 | 2 | 2 | 14.9 | 19.6 | 17.2 | ||||
| 3 | pk-1 | 2467 | 3270 | 267 | + | 10 | 3 | 3 | 32.6 | 44.6 | 30.3 | |||||
| 4 | hoar | E | 3313 | 5259 | 506 | − | 4 | 15 | ||||||||
| 5 | orf5 | 5663 | 6724 | 353 | + | |||||||||||
| 6 | pif-5* | L | 6962 | 8059 | 365 | + | 148 | 15 | 18 | 23 | 102 | 53.4 | 53.7 | 45.1 | 37.5 | 19.7 |
| 7 | bro-1 | L | 8122 | 8466 | 114 | + | 59 | 23.7 | ||||||||
| 8 | cg30-2 | 8570 | 9388 | 272 | + | 88 | 77 | 15.5 | 12.1 | |||||||
| 9 | p10 | L | 9436 | 9696 | 86 | − | 137 | 21 | 22 | 27.4 | 35.7 | 31 | ||||
| 10 | p26-1 | E,L | 9747 | 10604 | 285 | − | 136 | 22 | 34.6 | 43.8 | ||||||
| hr1 | 10666 | 11269 | ||||||||||||||
| 11 | ac29 | 11438 | 11656 | 72 | + | 29 | 23 | 25.4 | 40.3 | |||||||
| 12 | lef-6 | L | 11762 | 12508 | 248 | − | 28 | 24 | 80 | 19.7 | 26.2 | 16.8 | ||||
| 13 | dbp-2 | 12532 | 13464 | 310 | − | 25 | 25 | 81 | 14 | 18.1 | 23.9 | 14.1 | 14.8 | |||
| hr2 | 13546 | 14517 | ||||||||||||||
| 14 | orf14 | 13638 | 13889 | 83 | + | |||||||||||
| 15 | p74* | L | 14521 | 16479 | 652 | − | 138 | 20 | 60 | 47 | 74 | 57.8 | 54.4 | 39.4 | 38.9 | 32.8 |
| hr3 | 16580 | 17170 | ||||||||||||||
| 16 | me-53 | E | 17219 | 18349 | 376 | − | 139 | 16–17 | 143 | 16.8 | 22.6 | 17.2 | ||||
| 17 | ie-0 | E,L | 18698 | 19495 | 265 | + | 141 | 8 | 24.5 | 27.5 | ||||||
| 18 | p49* | L | 19595 | 21028 | 477 | + | 142 | 9 | 15 | 60 | 30 | 44.4 | 55.8 | 26.3 | 19.2 | 6.3 |
| 19 | odv-e18* | L | 21058 | 21324 | 88 | + | 143 | 10 | 14 | 62 | 31 | 61.3 | 51.9 | 38.1 | 18.8 | 7.9 |
| 20 | odv-ec27* | L | 21428 | 22306 | 292 | + | 144 | 11 | 97 | 63 | 32 | 45.9 | 52.1 | 22.2 | 21 | 15.3 |
| 21 | chtb | L | 22336 | 22614 | 92 | + | 145 | 12 | 9 | 64 | 45.5 | 44.6 | 31.5 | 27.2 | ||
| 22 | ep23 | L | 22639 | 23262 | 207 | − | 146 | 13 | 8 | 29.4 | 25.6 | 19.3 | ||||
| 23 | ie-1 | E | 23325 | 25439 | 704 | + | 147 | 14 | 7 | 24.9 | 27.2 | 11.7 | ||||
| 24 | ac34 | L | 25586 | 26134 | 182 | − | 34 | 27 | 23.6 | 50 | ||||||
| 25 | orf25 | E | 26224 | 26802 | 192 | − | ||||||||||
| 26 | ubiquitin | L | 26988 | 27254 | 88 | + | 35 | 28 | 54 | 74 | 72.3 | 68.2 | ||||
| 27 | 39k | 27529 | 28428 | 299 | − | 36 | 31 | 57 | 33.1 | 35.5 | 5.8 | |||||
| 28 | lef-11 | 28430 | 28786 | 118 | − | 37 | 32 | 58 | 33 | 30.5 | 27.1 | |||||
| 29 | bv-e31 | L | 28711 | 29433 | 240 | − | 38 | 33 | 69 | 53.7 | 48.3 | 39.5 | ||||
| 30 | dbp-1 | E,L | 29783 | 30712 | 309 | + | 25 | 25 | 81 | 24.9 | 34 | 12.4 | ||||
| 31 | p47* | 30827 | 32011 | 394 | − | 40 | 35 | 68 | 46 | 73 | 53.6 | 50.8 | 45.2 | 25.2 | 14 | |
| 32 | lef-12 | E | 32272 | 33114 | 280 | − | 41 | 36 | 30.9 | 29.6 | ||||||
| 33 | lef-8* | 33321 | 36038 | 905 | − | 50 | 38 | 131 | 78 | 26 | 63 | 68.9 | 49.1 | 30.5 | 18.3 | |
| 34 | orf34 | 33504 | 34118 | 204 | + | 34 | orf34 | |||||||||
| 35 | djbp | 36078 | 37316 | 412 | + | 51 | 39 | 15.4 | 20.1 | |||||||
| hr4 | 37323 | 37964 | ||||||||||||||
| 36 | iap-1 | L | 38038 | 38844 | 268 | − | 27 | 103 | 17 | 11 | 17 | 24.3 | 23.1 | 24.3 | 12.7 | 3.1 |
| 37 | ac52 | 39221 | 39895 | 224 | − | 52 | 42 | 19.5 | 30.6 | |||||||
| 38 | ac53 | L | 39855 | 40328 | 157 | + | 53 | 43 | 134 | 77 | 28 | 46 | 52.2 | 17.3 | 12.1 | 8.9 |
| 39 | orf39 | L | 40332 | 41459 | 375 | − | 44 | 17.3 | ||||||||
| 40 | orf40 | L | 41473 | 41709 | 78 | − | ||||||||||
| 41 | vp1054 | L | 41760 | 42869 | 369 | + | 54 | 47 | 138 | 83 | 8 | 39.7 | 47.9 | 30.1 | 18.8 | 18.5 |
| 42 | ac55 | 43039 | 43242 | 67 | + | 55 | 48 | 35.8 | 56.7 | |||||||
| 43 | ac56 | L | 43184 | 43534 | 116 | + | 56 | 49 | 11.9 | 32.8 | ||||||
| 44 | ac57 | E,L | 43717 | 44208 | 163 | + | 57 | 50 | 35.4 | 38.7 | ||||||
| 45 | chaB | E,L | 44279 | 44800 | 173 | − | 59 | 51 | 37.7 | 34.4 | ||||||
| 46 | chaB | L | 44876 | 45145 | 89 | − | 60 | 52 | 41.4 | 39.8 | ||||||
| 47 | bro-2 | L | 45281 | 45697 | 138 | − | 60 | 27.5 | ||||||||
| 48 | fp/25k | L | 45905 | 46549 | 214 | − | 61 | 53 | 118 | 51.9 | 56.1 | 28 | ||||
| 49 | lef-9 | 46687 | 48216 | 509 | + | 62 | 55 | 117 | 37 | 59 | 66.2 | 66.6 | 52.3 | 34 | 18.9 | |
| 50 | dna ligase | 48547 | 50361 | 604 | + | 120 | 23.5 | |||||||||
| 51 | bro-3 | 50436 | 51428 | 330 | − | 2 | 105 | 36 | 13.3 | |||||||
| 52 | gp37 | E | 51534 | 52391 | 285 | − | 64 | 58 | 13 | 45.3 | 55.6 | 41.4 | ||||
| 53 | orf53 | 52471 | 53133 | 220 | + | |||||||||||
| 54 | chitinase | L | 53319 | 55037 | 572 | − | 126 | 41 | 10 | 68.8 | 63.7 | 57.3 | ||||
| 55 | v-cath | L | 55144 | 56136 | 330 | + | 127 | 56 | 11 | 66.6 | 47 | 42.4 | ||||
| 56 | p26-2 | E | 56200 | 56919 | 239 | − | 136 | 22 | 17.2 | 15.9 | ||||||
| 57 | helicase-2 | 57035 | 58390 | 451 | − | 126 | 27.5 | |||||||||
| 58 | ac150 | L | 58435 | 58752 | 105 | − | 150 | 12 | 79 | 25.3 | 21.7 | 25.7 | ||||
| 59 | iap-2 | E | 58756 | 59682 | 308 | − | 71 | 62 | 17 | 28.9 | 35.2 | 16 | ||||
| 60 | pif-6 | 59711 | 60088 | 125 | − | 68 | 64 | 114 | 38 | 58 | 35.2 | 44 | 24 | 21.6 | 16 | |
| 61 | lef-3 | 60087 | 61340 | 417 | + | 67 | 65 | 113 | 17.9 | 23.7 | 4.5 | |||||
| 62 | desmoplakin | L | 61434 | 64043 | 869 | − | 66 | 66 | 112 | 21 | 92 | 17.6 | 17.1 | 12.3 | 11.7 | 10.7 |
| 63 | dna polymerase | 64042 | 67248 | 1068 | + | 65 | 67 | 111 | 20 | 91 | 44.5 | 51.6 | 30.9 | 24.3 | 16.5 | |
| 64 | ac75 | L | 67287 | 67679 | 130 | − | 75 | 69 | 108 | 20 | 34.6 | 10.8 | ||||
| 65 | ac76 | L | 67774 | 68031 | 85 | − | 76 | 70 | 107 | 41.7 | 65.9 | 35.7 | ||||
| 66 | vlf-1 | L | 68185 | 69354 | 389 | − | 77 | 71 | 106 | 42 | 18 | 67.5 | 69.4 | 28.6 | 25.1 | 18.2 |
| 67 | ac78 | L | 69378 | 69710 | 110 | − | 78 | 72 | 105 | 43 | 34 | 33 | 45.5 | 13.6 | 21.3 | 16.7 |
| 68 | gp41 | L | 69758 | 70963 | 401 | − | 80 | 73 | 104 | 44 | 33 | 43.1 | 53.4 | 28.4 | 26.3 | 11.5 |
| 69 | ac81 | 70932 | 71621 | 229 | − | 81 | 74 | 103 | 45 | 106 | 48.9 | 52.4 | 41.9 | 33.1 | 16.1 | |
| 70 | tlp-20 | L | 71515 | 72237 | 240 | − | 82 | 75 | 102 | 26.7 | 33.8 | 11.6 | ||||
| 71 | vp91 | L | 72206 | 74731 | 841 | + | 83 | 76 | 101 | 82 | 35 | 37.6 | 42.8 | 21.8 | 23.4 | 22.3 |
| 72 | cg30 | E | 74792 | 75739 | 315 | − | 88 | 77 | 19.7 | 18.7 | ||||||
| 73 | vp39 | L | 75861 | 76835 | 324 | − | 89 | 78 | 96 | 88 | 24 | 36.4 | 42 | 22.5 | 19 | 14.5 |
| 74 | lef-4 | 76834 | 78264 | 476 | + | 90 | 79 | 95 | 59 | 96 | 45.7 | 45.1 | 30.3 | 25 | 13.7 | |
| 75 | orf75 | L | 78301 | 78726 | 141 | − | 77 | 9.9 | ||||||||
| 76 | p33 | E,L | 78822 | 79580 | 252 | − | 92 | 80 | 93 | 16 | 14 | 50.4 | 57.9 | 35.1 | 19.4 | 19 |
| 77 | p18 | L | 79579 | 80055 | 158 | + | 93 | 81 | 92 | 17 | 13 | 52.5 | 62 | 31 | 17.1 | 4.8 |
| 78 | odv-e25 | L | 80057 | 80728 | 223 | + | 94 | 82 | 91 | 18 | 15 | 39 | 59.2 | 48.8 | 13 | 11.2 |
| 79 | helicase-1 | L | 80777 | 84505 | 1242 | − | 95 | 84 | 90 | 58 | 89 | 41 | 46.9 | 22.5 | 17.4 | 11.1 |
| 80 | pif-4 | 84459 | 84980 | 173 | + | 96 | 85 | 89 | 57 | 90 | 50.3 | 57.8 | 35.4 | 26.2 | 25.4 | |
| 81 | 38k | L | 85007 | 85927 | 306 | − | 98 | 86 | 88 | 56 | 87 | 42.8 | 50.3 | 39.5 | 27.2 | 24.8 |
| 82 | lef-5 | L | 85811 | 86662 | 283 | + | 99 | 87 | 87 | 55 | 88 | 46.8 | 54.4 | 38 | 27.2 | 7.9 |
| 83 | p6.9 | 86739 | 86999 | 86 | + | 100 | 88 | 86 | 28 | 23 | 7.3 | 8.1 | 4.1 | 8.1 | 13.8 | |
| 84 | p40 | E,L | 87012 | 88154 | 380 | − | 101 | 89 | 85 | 29 | 22 | 40.2 | 41.2 | 17.6 | 13.7 | 7.6 |
| 85 | p12 | L | 88194 | 88553 | 119 | − | 102 | 90 | 84 | 21.8 | 20.2 | 13.8 | ||||
| 86 | p48/p45 | E,L | 88540 | 89727 | 395 | − | 103 | 91 | 83 | 31 | 55 | 42.4 | 47.5 | 31.9 | 15.6 | 6 |
| 87 | vp80 | 89820 | 92120 | 766 | + | 104 | 92 | 12 | 17.2 | |||||||
| 88 | ac110 | 92160 | 92334 | 57 | + | 110 | 93 | 53 | 28.6 | 40.4 | 25 | |||||
| 89 | odv-ec43 | L | 92343 | 93482 | 379 | + | 109 | 94 | 55 | 67 | 69 | 48.3 | 58.4 | 30.4 | 15.3 | 10.3 |
| 90 | ac108 | L | 93532 | 93789 | 85 | + | 108 | 95 | 29.4 | 41.2 | ||||||
| 91 | orf91 | 93807 | 94313 | 168 | − | |||||||||||
| 92 | endonuclease | L | 94403 | 94762 | 119 | + | 79 | 65 | 24 | 26.6 | ||||||
| 93 | ac112 | 94746 | 95765 | 339 | + | 112 | 31 | |||||||||
| hr5 | 95796 | 96554 | ||||||||||||||
| 94 | nrk1 | 96753 | 97811 | 352 | + | 33 | 16 | 26.4 | 29.6 | |||||||
| 95 | p43 | E | 97850 | 99010 | 386 | − | 39 | 18.2 | ||||||||
| 96 | iap-3 | 99009 | 99479 | 156 | + | 27 | 103 | 94 | 23.1 | 24.4 | 27.6 | |||||
| 97 | ac106 | E,L | 99518 | 100228 | 236 | − | 106 | 101 | 52 | 32 | 50.8 | 49.6 | 25.4 | 16.5 | ||
| 98 | parg | L | 100286 | 101974 | 562 | − | 100 | 16.7 | ||||||||
| 99 | orf99 | 102055 | 102522 | 155 | − | 99 | 17.8 | |||||||||
| 100 | pif-3 | L | 102512 | 103135 | 207 | − | 115 | 98 | 35 | 66 | 46 | 44.1 | 47.2 | 35.7 | 28.5 | 31.5 |
| 101 | orf101 | 103207 | 103581 | 124 | − | |||||||||||
| 102 | sod | L | 103683 | 104159 | 158 | + | 31 | 106 | 59 | 60.9 | 62.7 | 46.2 | ||||
| 103 | ac117 | 104202 | 104513 | 103 | + | 117 | 110 | 22.1 | 39.8 | |||||||
| 104 | calyx/pep | L | 104559 | 105578 | 339 | − | 131 | 120 | 22 | 50 | 22.6 | 35.7 | 12.1 | 12.1 | ||
| 105 | orf105 | 105634 | 106980 | 448 | + | |||||||||||
| 106 | orf106 | 107008 | 107532 | 174 | − | |||||||||||
| 107 | orf107 | L | 108086 | 108496 | 136 | + | 68 | 13.2 | ||||||||
| hr6 | 108513 | 109329 | ||||||||||||||
| 108 | p24 | L | 109521 | 110312 | 263 | + | 129 | 118 | 71 | 37.9 | 52 | 24.1 | ||||
| 109 | orf109 | L | 110450 | 110818 | 122 | + | ||||||||||
| 110 | lef-2 | 110826 | 111515 | 229 | + | 6 | 117 | 41 | 54 | 25 | 36.2 | 36.7 | 18.7 | 15.4 | 11.6 | |
| 111 | pkip | L | 111630 | 112145 | 171 | + | 24 | 130 | 13 | 29.6 | ||||||
| 112 | orf112 | 112189 | 112524 | 111 | − | |||||||||||
| 113 | pif-2 | L | 112615 | 113769 | 384 | + | 22 | 132 | 48 | 52 | 38 | 60.2 | 68.1 | 50.3 | 44.3 | 46.4 |
| 114 | ac111 | 113827 | 114033 | 68 | − | 111 | 116 | 55.2 | 30.9 | |||||||
| hr7 | 114159 | 114766 | ||||||||||||||
| 115 | F | E,L | 114888 | 116945 | 685 | − | 23 | 133 | 31 | 104 | 14.2 | 37.8 | 24.5 | 15.3 | ||
| 116 | orf116 | 117225 | 120083 | 952 | + | 129 | 24.6 | |||||||||
| 117 | ac17 | 120196 | 120909 | 237 | − | 17 | 128 | 16.5 | 29.1 | |||||||
| 118 | orf118 | E,L | 120959 | 121600 | 213 | − | 127 | 21.4 | ||||||||
| 119 | egt | E | 121812 | 123353 | 513 | − | 15 | 126 | 141 | 45.1 | 51.7 | 35.5 | ||||
| 120 | orf120 | L | 123560 | 123916 | 118 | − | ||||||||||
| 121 | lef-1 | 124016 | 124717 | 233 | + | 14 | 124 | 74 | 65 | 45 | 35.6 | 42.1 | 33.9 | 28.4 | 21.5 | |
| 122 | 38.7k | 124783 | 126009 | 408 | + | 13 | 123 | 73 | 22.9 | 31.9 | 13.1 | |||||
| 123 | ac19 | L | 126065 | 126508 | 147 | − | 19 | 115 | 21.3 | 23.3 | ||||||
| 124 | orf124 | L | 126507 | 127745 | 412 | + | ||||||||||
| 125 | alk-exo | L | 127807 | 129066 | 419 | + | 133 | 114 | 125 | 33 | 54 | 36.3 | 40.3 | 32.4 | 24.4 | 21.5 |
| 126 | orf126 | L | 129111 | 129875 | 254 | − | ||||||||||
| 127 | fgf | 130158 | 131180 | 340 | + | 32 | 113 | 123 | 23.8 | 19.6 | 9.1 | |||||
| 128 | orf128 | 131201 | 131440 | 79 | − | 112 | ||||||||||
| 129 | pif-1 | L | 131447 | 133045 | 532 | − | 119 | 111 | 75 | 76 | 29 | 52.1 | 46.4 | 33.3 | 29.1 | 26 |
| 130 | bro-4 | 133123 | 133962 | 279 | − | 2 | 13.6 | |||||||||
| 131 | dna photolyase | E | 134303 | 135796 | 497 | + | ||||||||||
ORFs listed are those predicted in the SujuNPV genome and their homologues in the five representative genomes (AcMNPV, HearNPV-G4, CpNPV, NeleNPV and CuniNPV). The start gene is polyhedrin and core genes were marked with*. E and L indicate the Early and Late promoter motifs, respectively. ‘+’ and ‘−’ means the transcription direction; ‘+’ clockwise; ‘−’ anticlockwise.
Relationship with other baculoviruses
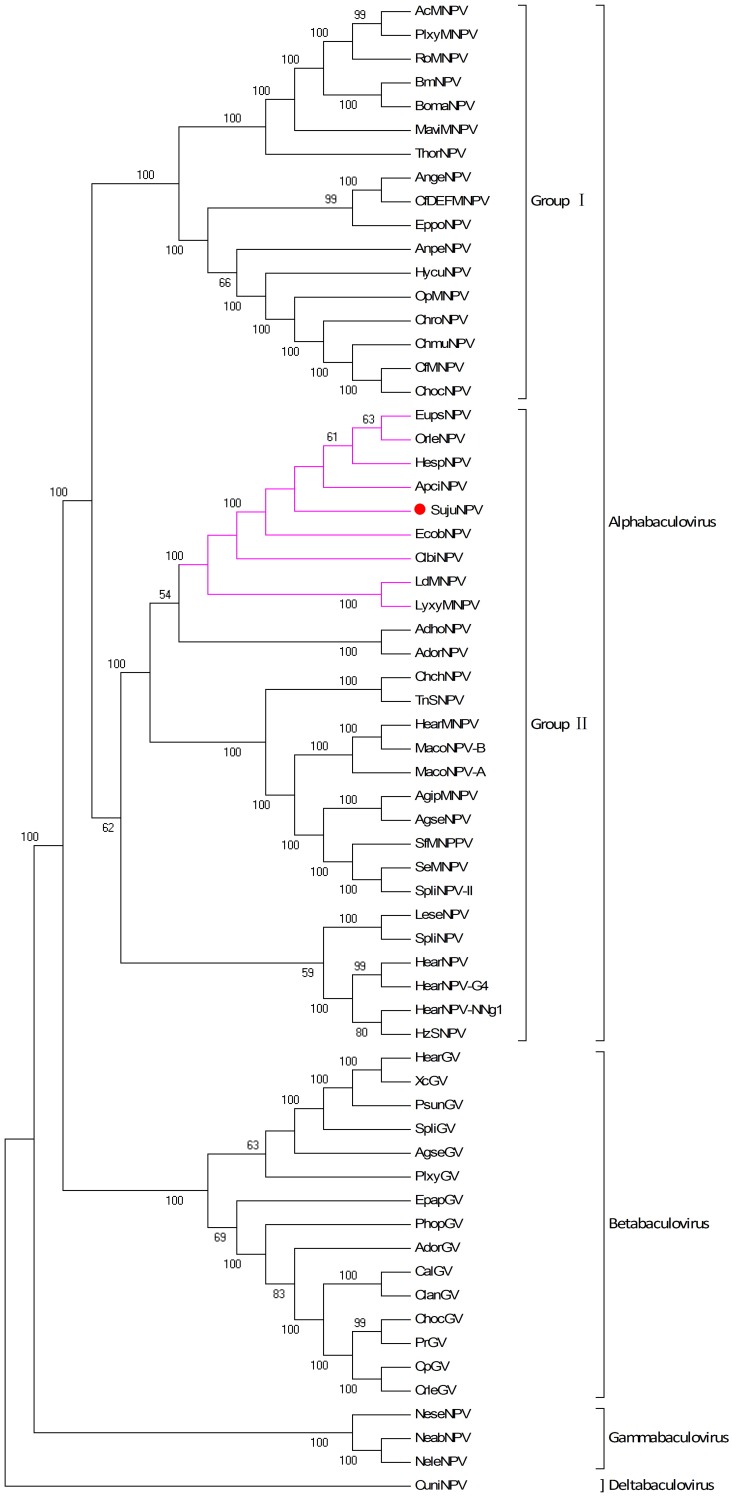
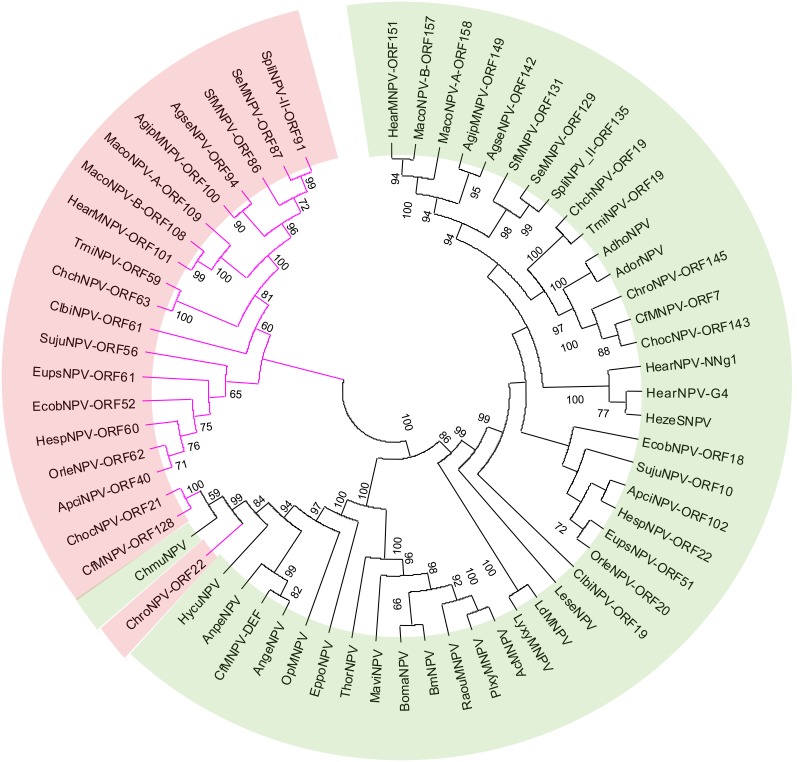
Phyogenetic analysis of the 37 core genes of the 62 reference baculoviruses revealed that SujuNPV is a group II alphabaculovirus (Fig. 2). The virus is a novel member of a subclade containing eight other baculoviruses, including Apocheima cinerarium NPV (ApciNPV), Clanis bilineata NPV (ClbiNPV) [18], Ectropis obliqua NPV (EcobNPV) [19], Euproctis pseudoconspersa NPV (EupsNPV) [20], Hemileuca sp. NPV (HespNPV) [21], Lymantria dispar MNPV (LdMNPV) [22], Lymantria xylina MNPV (LyxyMNPV) [23] and Orgyia leucostigma NPV (OrleNPV) [24].
Figure 2. Phylogenic analysis of 62 complete baculovirus genomes.
The maximum likelihood (ML) tree was generated based on the concatenated protein sequences of 37 core genes with default parametes and 1000 randoms. The SujuNPV was labeled by a red point and the number on the branch means bootstrap values (only the values over 50 were shown). Pink branches indicate the unique subclade containing a second copy of dbp.
Five representative baculoviruses were chosen for the comparative study of SujuNPV: Autographa californica MNPV (AcMNPV, group I alphabaculovirus) [25], Helicoverpa armigera SNPV (HearNPV, group II alphabaculovirus) [26], Cydia pomonella GV (CpGV, betabaculovirus), Neodiprion lecontei NPV (NeleNPV, gammabaculovirus) and Culex nigripalpus NPV (CuniNPV, deltabaculovirus). SujuNPV shared 102 ORFs with AcMNPV, 108 with HearNPV, 78 with CpGV, 43 with NeleNPV, and 39 with CuniNPV, with an average amino acid (aa) identity of 36.0%, 39.0%, 28.4%, 23.0% and 16.3%, respectively.
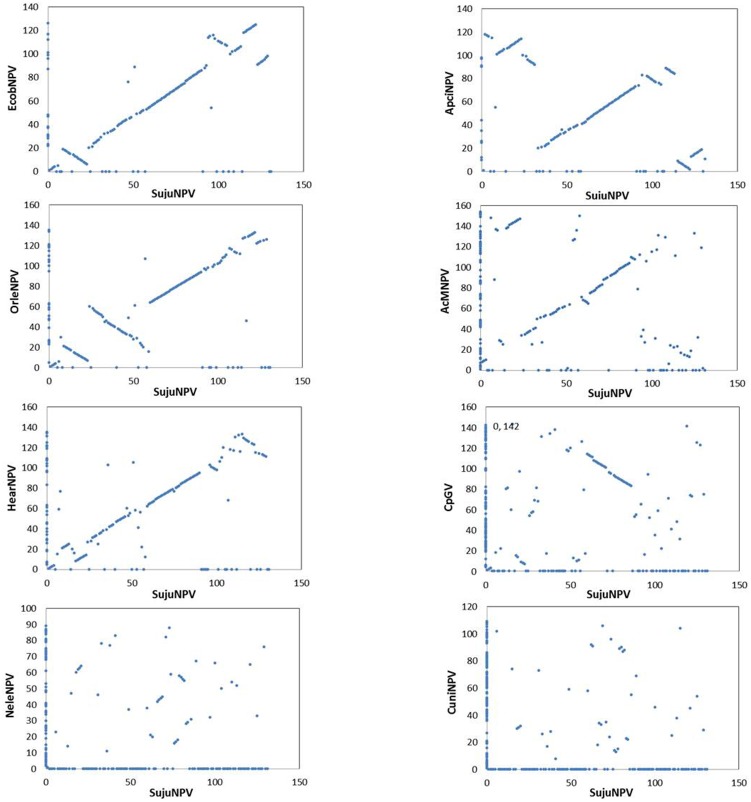
Gene-parity plots of SujuNPV against three viruses in the same subclade and the five representative baculoviruses are shown in Fig. 3. The gene order between SujuNPV and ApciNPV, EcobNPV or OrleNPV revealed a high collinearity along the genomes, with some inversions and drifts. The plots of SujuNPV with representative lepidopteran baculoviruses (AcMNPV, HearNPV and CpGV) showed that SujuNPV is largely collinear with AcMNPV and HearNPV, less collinear with CpGV, but all contains a collinear region from Suju60 to Suju86, containing 20 core genes and five additional lepidopteran baculovirus conserved genes. This region has been suggested to exist in the ancestor of lepidopteran baculoviruses [27]. No obvious collinear region could be found between SujuNPV and NeleNPV or CuniNPV (Fig. 3).
Figure 3. Gene-parity plot analysis.
Gene-parity plots of SujuNPV against three close viruses (EcobNPV, ApciNPV and OrleNPV) and five representative baculoviruses (AcMNPV, HearNPV, CpGV, NeleNPV and CuniNPV).
Homologous regions
Homologous regions (hrs) are common elements in many baculoviruses, with characteristically high A+T contents, tandem repeats and imperfect palindromes. Hrs vary in location within genomes, number of copies and nucleotide sequences between different baculoviruses. These regions are suggested to act as replication origins and transcription enhancers [28], [29].
The SujuNPV genome contains seven homologous regions, covering 3.7% of the genome, as displayed in Fig. 4A. The length of the hrs ranges from 590 bp-971 bp, and each hr consists of four to eight palindromic repeats of 99 bp in length (Fig. 4A and 4B). Fig. 4B shows the arrangement of palindrome repeats in each homologous region. These palindromic repeats share at least 97.6% identity. The predicted secondary structure of the hr1–3 revealed that it contains a core palindrome region, colored by orange in Fig. 4C, and it is highly conserved in all counterparts, with about 99.5% identity on average. While the other two loop were not such conservative, neither on the size nor sequence.
Figure 4. Analysis of SujuNPV hrs.
A. The location and distribution of hrs in the SujuNPV genome. Black bars indicate hrs in the SujuNPV linear map. The number in brackets refers to the number of palindrome repeats in the homologous region. B. The arrangement of palindrome repeats in each homologous region. The rectangle above or below the black line indicates the orientation of repeats and number in the bracket represent the corresponding size of each hr. Different colors means the different fragments within the repeats, and orange indicates the core palindrome region, which is conserved in all the hr repeats. One sequence of each orientation was displayed at the top as a sample. C. The second structure of the hr1–3 palindrome repeats. The background color is in line with the sequence displayed in Fig. 4B.
DNA replication genes
Five core genes, five additional lepidopteran baculovirus conserved genes and eight other common genes involved in DNA replication were found in the SujuNPV genome (Table 2) [30]–[32]. Among these genes: helicase unwinds DNA [32]; dna polymerase is involved in DNA synthesis; late expression factor gene 3 (lef-3) and DNA binding protein gene (dbp) are involved in single-strand binding [33], [34]; dna-ligase in ligation and alkaline exonuclease (alk-exo) in rectification [35], [36], together with some other stimulators are required in the process of replication.
Table 2. Classification of gene function.
| Core genes | Lepidoptera baculovirus conserved genes | Common genes | |
| Replication | alk-exo(Suju125), dna polymerase(Suju63), helicase(Suju79), lef-1(Suju121), lef-2(Suju110) | dbp-1(Suju30), ie-1(Suju23), lef11(Suju28), lef-3(Suju61), me53(Suju16) | dbp-2(Suju13), dnaphotolyase(Suju131), dna-ligase(Suju50), endonuclease(Suju92), helicase-2(Suju57), ie-0(Suju17), nrk1(Suju94), parg(Suju98), |
| Transcription | lef-4(Suju74), lef-5(Suju82), lef8(Suju33), lef-9(Suju49), P47(Suju31), vlf-1(Suju66) | 39k(Suju27), lef-6(Suju12), pk-1(Suju3) | lef12(Suju32) |
| Structure | 38k(Suju81), ac53(Suju38), ac78(Suju67), ac81(Suju69), desmoplakin(Suju62), gp41(Suju68), odv-e18(Suju19), odv-e25(Suju78), odv-ec27(Suju20), odv-ec43(Suju89), p18(Suju77), p33(Suju76), p40(Suju84), p48/p45(Suju86), p49(Suju18), p6.9(Suju83), vp1054(Suju41), vp39(Suju73), vp91(Suju71) | F(Suju115), fp/25k(Suju48), orf1629(Suju2), p12(Suju85), p24(Suju108), polyhedrin(Suju1), tlp-20(Suju70) | calyx/pep(Suju104), cg30-1(Suju72), cg30-2(Suju8), p10(Suju9), pkip(Suju111), vp80(Suju87) |
| Auxiliary | fgf(Suju127) | bro-1(Suju7), bro-2(Suju47), bro-3(Suju51), bro-4(Suju130), chitinase(Suju54), egt(Suju119), gp37(Suju52), iap-1(Suju36), iap-2(Suju59), iap-3(Suju96), sod(Suju102), ubiquitin(Suju26), v-cath(Suju55) | |
| Pifs | p74(Suju15), pif-1(Suju129), pif-2(Suju113), pif-3(Suju100), pif-4(Suju80), pif-5(Suju6), pif-6(Suju60) | ||
| Unknown | 38.7k(Suju122), ac106(Suju97), ac110(Suju88),ac75(Suju64), ac76(Suju65), chtb(Suju21), ep23(Suju22), bv-e31(Suju29) | ac108(Suju90), ac111(Suju114), ac112(Suju93), ac117(Suju103), ac150(Suju58), ac17(Suju117), ac19(Suju123), ac29(Suju11), ac34(Suju24), ac52(Suju37), ac55(Suju42), ac56(Suju43), ac57(Suju44), chaB(Suju45), chaB(Suju46), djbp(Suju35), hoar(Suju4), p26-1 (Suju10), p26-2(Suju56), p43(Suju95), Suju101, Suju105, Suju107, Suju109, Suju112, Suju116, Suju118, Suju120, Suju124, Suju126, Suju128, Suju34, Suju39, Suju40, Suju75, Suju91, Suju99 |
Functional classification of the genes in the SujuNPV genome columns indicate classification by function and rows represent conservatism. Genes in the SujuNPV genome were arranged according to their functions and conservatism in alphabetical order.
Functional classification of the genes in the SujuNPV genome; columns indicate classification by function and rows represent conservatism. Genes in the SujuNPV genome were arranged according to their functions and conservatism in alphabetical order.Some common genes involved in baculovirus replication were not present in SujuNPV. For example, lef-7 which has been shown to be a replication enhancer in baculoviruses [37], was absent from SujuNPV. SujuNPV also lacked certain genes associated with nucleotide biosynthesis, such as the ribonucleotide reductase subunits (rr1, rr2) and dUTPase, which are involved in dTTP biosynthesis [38].
Amongst the DNA replication genes, there are two copies of helicase and dbp in the SujuNPV genome. A full length helicase (Suju79, 1242aa) is a core gene found in all sequenced baculoviruses, whilst a second copy of truncated helicase (helicase-2) (Suju57, 451aa) is present in only six alphabaculoviruses (HearMNPV, LdMNPV, LyxyMNPV, MacoNPV-B, OrleNPV and SpliNPV) and 13 GVs (all sequenced GVs except for ClanGV and CaLGV) [39]. The phylogenetic tree of helicase homologies showed that they can be clearly divided into two groups (Fig. 5A). It is very likely that they were acquired from different sources during evolution. The research of AcMNPV helicase reveals that it belongs to Superfamily 1 helicase, which contain 7 conserved motifs [40], [41]. Motifs I and II are two NPT-binding motifs, together with another four motifs to fulfill the function of helicase [42], [43]. The alignment of the conserved motifs with AcMNPV and E.coli UrvD (representative of Superfamily 1 helicase) reveals that they share the same motifs (Fig. 5B) and that helicase-2 is seemingly more conservative. It appears that the two copies have a common ancestor, but understanding how they evolved and came to balance their specialization and cooperation within one genome requires further research.
Figure 5. Analysis of the duplicated gene helicase and its conservative motifs.
A. The tree was reconstructed based on protein sequences by MEGA5. The second copy was colored by purple branches and pink background and the number on the branch indicates a bootstrap value of 1000 randoms. B. Conservative motifs of E.coli UrvD, AcMNPV, SujuNPV Helicase (SujuNPV-1) and SujuNPV Helicase-2 (SujuNPV-2) were displayed. The blank line indicates the relevant protein with length in the bracket. The colored boxes on the line indicate motifs I-IV and the numbers above and below the box mean the start and end position of each motif in the protein, respectively.
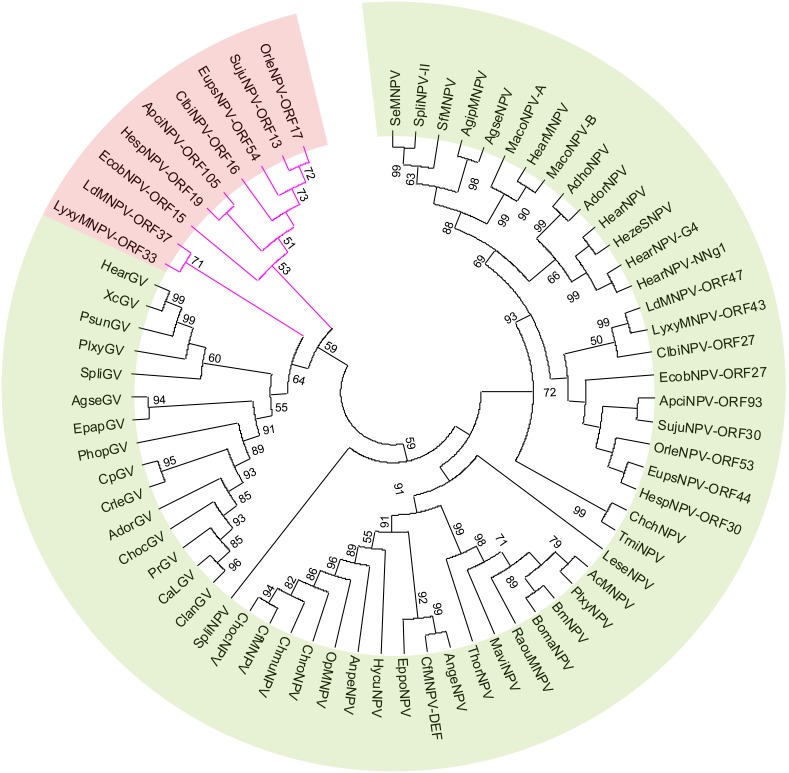
SujuNPV is the ninth baculovirus identified to have double copies of dbp; the other eight are ApciNPV, ClbiNPV, EcobNPV, EupsNPV, HespNPV, LdMNPV, LyxyMNPV and OrleNPV. Interestingly all these viruses belong to the same subclade (Fig. 2). Dbp is a conserved gene in lepidopteran baculoviruses. Phylogenetic analysis indicated that the dbp duplicates of these nine baculoviruses may have evolved separately to the conserved dbp in alphabaculoviruses (Fig. 6). We propose to name the alphabaculovirus-conserved dbp gene as dbp-1, and the second copy as dbp-2. Dbp-2 appears to be more close to the dbp of betabaculovirus. In SujuNPV, dbp-1 (Suju30) and dbp-2 (Suju13) encode 309 aa and 310 aa proteins respectively, with 25% aa identity. Although the significance of SujuNPV and other bacuoviruses carrying two copies of dbp is unclear, it clearly marks out the subclade of these nine group II alhpabaculoviruses.
Figure 6. Analysis of the duplicated gene dbp.
The tree was reconstructed based on protein sequences by MEGA5. The second copy was colored by purple branches and pink background and the number on the branch indicates a bootstrap value of 1000 randoms.
Transcriptional genes
In a baculovirus life cycle, the genes are transcribed in cascades by different polymerase. Early stage genes are transcribed by host RNA polymerase II, while genes expressed during the late period of the life cycle are transcribed by the virus-encoded RNA polymerase, comprising four core gene transcripts: LEF-4, LEF-8, LEF-9, P47 [44]. Two other core genes are involved in late phase transcription: lef-5 and very late factor (vlf-1), acting as an initiation factor [45] and a regulatory factor participating in the hyper-expression of very late genes [46], respectively. These core genes, in addition to genes such as 39k, lef-6, lef-10 and lef-12, are required for late transcription [47]. All of these genes appear in SujuNPV, except for lef-10 (Table 2) and among all the other alphabaculoviruses this gene was only absent from ClbiNPV and OrleNPV.
Structural genes
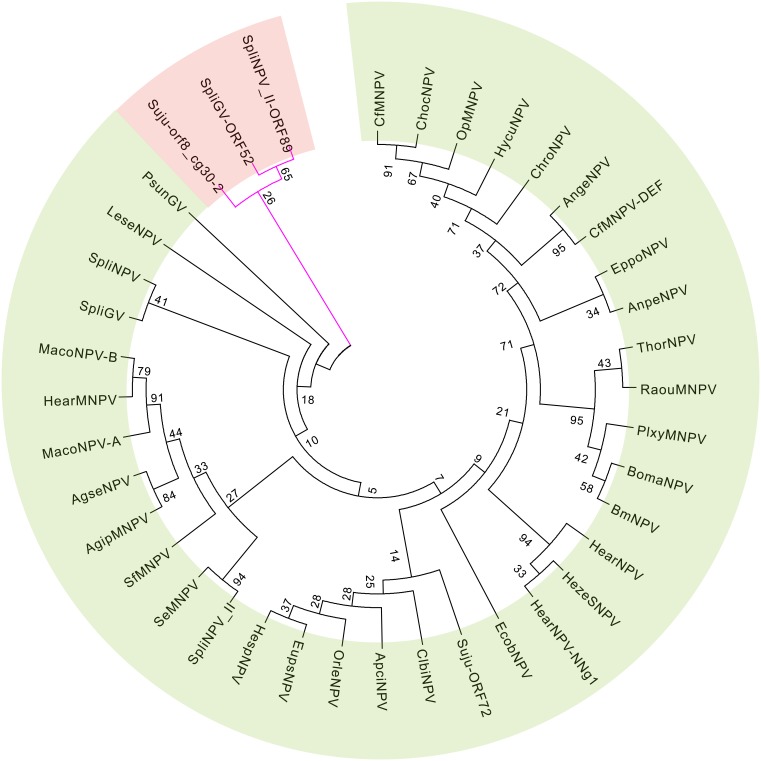
Nineteen core genes and seven additional lepidopteran-conserved genes related to structure were found in the SujuNPV genome (Table 2) [48]–[50]. In addition, six other common genes were also identified in the SujuNPV genome (Table 2). Cg30 is duplicated in SujuNPV: cg30-1 (Suju72, 315 aa) and cg30-2 (Suju8, 272 aa). Among all the baculoviruses sequenced, two copies of cg30 are only present in SpliNPV-(SpliNPV82 and SpliNPV89) and in SpliGV (SpliGV52 and SpliGV124). Cg30-1 of SujuNPV has many homologies with other baculoviruses, while cg30-2 groups with SpliNPV89 and SpliGV52 at the outmost of the phylogenetic tree (Fig. 7), sharing an aa identity of 15% and 14%, respectively.
Figure 7. Analysis of the duplicated gene cg30.
The tree was reconstructed based on protein sequences by MEGA5. The second copy was colored by purple branches and pink background and the number on the branch indicates a bootstrap value of 1000 randoms.
Per os infectivity factors
So far seven genes have been identified as per os infectivity factors (PIFs), including p74, pif1, pif2, pif3, pif4 (odv-e28), pif5 (odv-e56) and pif6, which are essential for the oral infection of insect larvae [51]–[54]. PIF-1, PIF-2 and PIF-3 in association with P74 form a conserved complex on the surface of ODV and were proposed to perform an essential function in the early stages of virus infection [51]. PIF-4 is an envelope-associated protein found in both ODV and BV [52], whereas, PIF-5 and PIF6 have been recently demonstrated to be PIF members [53], [54]. All seven of their genes are conserved within the SujuNPV genome and share 44%–61.2% identity with their homologues in group II representative baculovirus HearNPV.
Auxiliary genes
Auxiliary genes are those not essential for replication, transcription or structures, but provide the virus with the stronger adaptive ability [55], such as affecting the host’s cellular metabolism for successful infection or by promoting the progeny yields of the virus. Examples are fibroblast growth factor (fgf) and gp37, which are proposed to help to spread virions from the primary infection site [56], [57], egt, which promotes viral progeny by delaying larval molting [58], and cathepsin and chitinase, which aid the horizontal spread of viruses [59]. Superoxide dismutase (sod) has been suggested to migrate the effects of free radicals in infected hemocytes [60] and ubiquitin is proposed to stabilize viral proteins against being degraded by hosts [61]. Among these auxiliary genes, no core gene has been found and only fgf is a lepidopteran-conserved gene. SujuNPV was found to contain all the genes above (Table 2).
Anti-apoptosis genes are those encoded by viruses in order to resist the programmed death of infected cells, hence ensuring a successful infection [62]. SujuNPV possesses two types of anti-apoptotic genes: p49 (Suju18) and three copies of inhibitor of apoptosis gene (iaps): iap-1 (Suju36), iap-2 (Suju59) and iap-3 (Suju96). Among the three iaps, iap-2 and iap-3 have a C3HC4 motif at the C terminal, with DNA-binding properties [63].
Baculovirus repeated ORFs (bros) are repetitive genes, which are widespread in baculoviruses and some other insect virus DNA [64]. Research of BmNPV showed that bros contained DNA-binding activity that could influence host DNA replication and transcription [65]. Four bro genes were identified in SujuNPV, and named bro-1 to bro-4, based upon their order of appearance in the genome (Fig. 1). SujuNPV bro-3 had an aa similarity to its homologues in AcMNPV, OpMNPV, LdMNPV and HearNPV-G4, with 36%, 38.6%, 35.2% and 13.3% sequence identity, respectively. The other three bros only shared a C-terminal region with Ld-bro-m, Ld-bro-p and Ld-bro-n.
Unknown genes
SujuNPV contained an additional eight lepidoptera-conserved genes and 37 common genes with unknown functions (Table 2). P26 is an alphabaculovirus-specific gene. Among the 42 alphabaculoviruses previously sequenced, 19 contained a second copy of p26 and 16 of these belonged to group II. SujuNPV also contains two copies of p26, Suju10 (p26-1, 285 aa) and Suju56 (p26-2, 239 aa), which share 13.8% similarity. We name the one conserved in alphabaculoviruses as p26-1, and the second copy as p26-2. Phylogenetic analysis of p26 showed that the second copies of p26 could be classified into a unique subclade (colored pink in Fig. 8), with the exception of three group I baculoviruses (CfMNPV, ChocNPV and ChroNPV). Interestingly, the group II baculoviruses, except for LeseNPV, all specifically contain a conserved gene cluster that is p10, p26, ac29, lef-6 and dbp (dbp-2 in the 9 dbp-duplicated baculoviruses) in order. Although the significance of this gene cluster is unknown, it can provide us with more information for evolutionary analysis.
Figure 8. Analysis of the duplicated gene p26.
The tree was reconstructed based on protein sequences by MEGA5. The second copy was colored by purple branches and pink background and the number on the branch indicates a bootstrap value of 1000 randoms.
Unique genes
Five genes are unique to the SujuNPV genome, including Suju5 (353 aa), Suju14 (83 aa), Suju25 (192 aa), Suju53 (220 aa) and Suju106 (174 aa) which were not included in Table 2. Suju5 has a similar location and length to the ORF5 of Buzura suppressaria SNPV (BusuNPV) with14.8% aa identity, indicating they may have similar function [27]. Suju25 has an early promoter and a BLAST search showed it to have a slight similarity to the ATP-binding protein of Lysinibacillus sphaericus C3–41 with an E-value of 0.89. No homologues were found in GenBank for the other three ORFs, whether these are functional ORFs of SujuNPV requires further experimentation.
Conclusion
Our analyses revealed that SujuNPV is a novel baculovirus within a unique subclade of group II alphabaculovirues, the members of which all contain a second copy of dbp. The SujuNPV genome contains seven hrs and five unique ORFs, as well as several genes with two or more copies. The presence of duplicated genes in this virus raises the question on the mechanisms of its acquisition (duplication of virus genes or independent horizontal transfer) and maintain, which needs further researches. These findings will facilitate future applications of SujuNPV to pest control and provide new data for the elucidation of the evolutionary pathways of baculoviruses.
Acknowledgments
The authors sincerely thank Bitao Zhu, Leiping Zeng and Yanbo Ye from Wuhan Institute of Virology, CAS for the bioinformatic assistance and instructional advice. We acknowledge the Core Facility and Technical Support of Wuhan Institute of Virology for technical assistants.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No.XDB11030400), and the grants from the National Science Foundation of China (No.31130058 and No.31321001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herniou EA, Arif BM, Becnel JJ, Blissard GW, Bonning B, et al.. (2011) Baculoviridae. In Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Edited by King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. San Diego. Elsevier Academic Press.
- 2. Lucarotti CJ, Morin B, Graham RI, Lapointe R (2007) Production, application, and field performance of Abietiv, the balsam fir sawfly nucleopolyhedrovirus. Virologica Sinica 22: 10. [Google Scholar]
- 3. Yin HY, Zhou XA, Wu HF, Li BA, Zhang YF (2010) Baculovirus vector-mediated transfer of NIS gene into colon tumor cells for radionuclide therapy. World Journal of Gastroenterology 16: 5367–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aucoin MG, Mena JA, Kamen AA (2010) Bioprocessing of Baculovirus Vectors: A Review. Current Gene Therapy 10: 174–186. [DOI] [PubMed] [Google Scholar]
- 5.Fauquet CM, Carstens EB, Estes MK, Lemon SM, Maniloff J, et al.. (2000) Virus Taxonomy-Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press.
- 6. Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, et al. (2006) On the classification and nomenclature of baculoviruses: A proposal for revision. Archives of Virology 151: 1257–1266. [DOI] [PubMed] [Google Scholar]
- 7.Tong DX, Dong H, Li SY, Guo H (2012) Occurence investigation and control technology resarch of Sucra jujuba Chu from mulnerry. China Sericulture: 29–30.
- 8. Fan-ren S (1986) Studies on the nuclear polyhedrosis virus disease of sucra jujuba chu . J Virol 6: 4. [Google Scholar]
- 9. Ji WR, Liu XQ, Shi GL (1999) The bioassay and field efficacy tests of the nuclear polyhedrosis virus of Chihuo zao Yang. Scientia silvae sinicae 35: 81–85. [Google Scholar]
- 10. Yang S, Miller LK (1998) Expression and mutational analysis of the Baculovirus very late factor 1 (vlf-1) gene. Virology 245: 99–109. [DOI] [PubMed] [Google Scholar]
- 11. Vanarsdall AL, Mikhailov VS, Rohrmann GF (2007) Baculovirus DNA replication and processing. Current Drug Targets 8: 1096–1102. [DOI] [PubMed] [Google Scholar]
- 12.Zhang ZY, Xing TX (1986) Study report of Sucra jujuba nucleopolyhedrovirus. Journal of Microbiology: 6–10.
- 13.O’Reilly DR, Miller LK, Luckow VA (1992) Baculovirus Expression Vector: A Laboratory Manual. New York: W H Freeman & Company pp368.
- 14. Solovyev VV, Salamov AA (1999) INFOGENE: a database of known gene structures and predicted genes and proteins in sequences of genome sequencing projects. Nucleic Acids Research 27: 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garavaglia MJ, Miele SA, Iserte JA, Belaich MN, Ghiringhelli PD (2012) The ac53,ac78,ac101 and ac103 genes are newly discovered core genes in the family baculovridae. Journal of Virology 86: 12069–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrelli ML, Salvador R, Biedma ME, Berretta MF, Haase S, et al. (2012) Genome of Epinotia aporema granulovirus (EpapGV), a polyorganotropic fast killing betabaculovirus with a novel thymidylate kinase gene. BMC Genomics 13: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gruber AR, Lorenz R, Bernhart SH, Neuboock R, Hofacker IL (2008) The Vienna RNA Websuite. Nucleic Acids Research 36: 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu SY, Yi JP, Shen WD, Wang LQ, He HG, et al. (2009) Genomic sequence, organization and characteristics of a new nucleopolyhedrovirus isolated from Clanis bilineata larva. BMC Genomics 10: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma XC, Shang JY, Yang ZN, Bao YY, Xiao Q, et al. (2007) Genome sequence and organization of a nucleopolyhedrovirus that infects the tea looper caterpillar, Ectropis obliqua . Virology 360: 235–246. [DOI] [PubMed] [Google Scholar]
- 20. Tang XD, Xiao Q, Ma XC, Zhu ZR, Zhang CX (2009) Morphology and genome of Euproctis pseudoconspersa nucleopolyhedrovirus. Virus Genes 38: 495–506. [DOI] [PubMed] [Google Scholar]
- 21. Rohrmann GF, Erlandson MA, Theilmann DA (2013) The genome of a baculovirus isolated from Hemileuca sp. encodes a serpin ortholog. Virus Genes 47: 357–364. [DOI] [PubMed] [Google Scholar]
- 22. Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, et al. (1999) Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar . Virology 253: 17–34. [DOI] [PubMed] [Google Scholar]
- 23. Nai YS, Wu CY, Wang TC, Chen YR, Lau WH, et al. (2010) Genomic sequencing and analyses of Lymantria xylina multiple nucleopolyhedrovirus. BMC Genomics 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thumbi DK, Eveleigh RJM, Lucarotti CJ, Lapointe R, Graham RI, et al. (2011) Complete Sequence, Analysis and Organization of the Orgyia leucostigma Nucleopolyhedrovirus Genome. Viruses-Basel 3: 2301–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD (1994) The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202: 586–605. [DOI] [PubMed] [Google Scholar]
- 26. Chen X, WF IJ, Tarchini R, Sun X, Sandbrink H, et al. (2001) The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. Journal of General Virology 82: 241–257. [DOI] [PubMed] [Google Scholar]
- 27. Zhu Z, Yin FF, Liu XP, Deng F, Hu ZH, et al. (2014) Genome Sequence and Analysis of Buzura suppressaria Nucleopolyhedrovirus: A Group II Alphabaculovirus. Plos One 9: e86450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pearson MN, Rohrmann GF (1995) Lymantria-Dispar Nuclear Polyhedrosis-Virus Homologous Regions - Characterization of Their Ability to Function as Replication Origins. Journal of Virology 69: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Theilmann DA, Stewart S (1992) Tandemly Repeated Sequence at the 3′ End of the Ie-2 Gene of the Baculovirus Orgyia-Pseudotsugata Multicapsid Nuclear Polyhedrosis-Virus Is an Enhancer Element. Virology 187: 97–106. [DOI] [PubMed] [Google Scholar]
- 30. Evans JT, Leisy DJ, Rohrmann GF (1997) Characterization of the interaction between the Baculovirus replication factors LEF-1 and LEF-2. Journal of Virology 71: 3114–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McDougal VV, Guarino LA (1999) Autographa californica nuclear polyhedrosis virus DNA polymerase: Measurements of processivity and strand displacement. Journal of Virology 73: 4908–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDougal VV, Guarino LA (2000) The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. Journal of Virology 74: 5273–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikhailov VS (2003) Replication of the baculovirus genome. Molecular Biology 37: 250–259. [PubMed] [Google Scholar]
- 34.Rohrmann G (2011) DNA Replication and genome processing. In: Rohrmann G, editor. Baculovirus Molecular Biology. National Library of Medicine (US), Bethesda (MD).
- 35. Mikhailov VS, Okano K, Rohrmann GF (2003) Baculovirus alkaline nuclease possesses a 5′–>3′ exonuclease activity and associates with the DNA-binding protein LEF-3. Journal of Virology 77: 2436–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mikhailov VS, Okano K, Rohrmann GF (2004) Specificity of the endonuclease activity of the baculovirus alkaline nuclease for single-stranded DNA. Journal of Biological Chemistry 279: 14734–14745. [DOI] [PubMed] [Google Scholar]
- 37.Lu A, Miller LK, Krell P, Vlak JM, Rohrmann G (1997) Baculovirus DNA Replication In: LK M, editor. The Baculoviruses. New York and London: PlenumPress.
- 38. Herniou EA, Olszewski JA, Cory JS, O'Reilly DR (2003) The genome sequence and evolution of baculoviruses. Annual Review of Entomology 48: 211–234. [DOI] [PubMed] [Google Scholar]
- 39. Iyer LM, Leipe DD, Koonin EV, Aravind L (2004) Evolutionary history and higher order classification of AAA plus ATPases. Journal of Structural Biology 146: 11–31. [DOI] [PubMed] [Google Scholar]
- 40. Hall MC, Matson SW (1999) Helicase motifs: the engine that powers DNA unwinding. Molecular Microbiology 34: 867–877. [DOI] [PubMed] [Google Scholar]
- 41. Albert Lu, Carstens AB (1991) Nucleotide Sequence of a Gene Essential for Viral DNA Replication in the Baculovirus Autograph califomica Nuclear Polyhedrosis Virus. VIrology 181: 336–347. [DOI] [PubMed] [Google Scholar]
- 42. Hodgman TC (1988) A New Superfamily of Replicative Proteins. Nature 333: 22–23. [DOI] [PubMed] [Google Scholar]
- 43. Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM (1988) A Novel Superfamily of Nucleoside Triphosphate-Binding Motif Containing Proteins Which Are Probably Involved in Duplex Unwinding in DNA and RNA Replication and Recombination. Febs Letters 235: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guarino LA, Xu B, Jin JP, Dong W (1998) A virus-encoded RNA polymerase purified from baculovirus-infected cells. Journal of Virology 72: 7985–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guarino LA, Dong W, Jin JP (2002) In vitro activity of the baculovirus late expression factor LEF-5. Journal of Virology 76: 12663–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mclachlin JR, Miller LK (1994) Identification and Characterization of Vlf-1, a Baculovirus Gene Involved in Very Late Gene-Expression. Journal of Virology 68: 7746–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohrmann G (2011) Baculovirus late transcription. In: Rohrmann G, editor. Baculovirus Molecular Biology. National Library of Medicine (US), NCBI, Bethesda (MD).
- 48.Ferrelli ML, Berretta MF, Belaich MN, Ghiringhelli PD, Romanowski V, et al.. (2012) The Baculoviral Genome; Garcia ML, V R, editors.
- 49. Wang ML, Tuladhar E, Shen S, Wang HL, van Oers MM, et al. (2010) Specificity of Baculovirus P6.9 Basic DNA-Binding Proteins and Critical Role of the C Terminus in Virion Formation. Journal of Virology 84: 8821–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thiem SM, Miller LK (1989) Identification, Sequence, and Transcriptional Mapping of the Major Capsid Protein Gene of the Baculovirus Autographa Californica Nuclear Polyhedrosis-Virus. Journal of Virology 63: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peng K, van Oers MM, Hu ZH, van Lent JWM, Vlak JM (2010) Baculovirus Per Os Infectivity Factors Form a Complex on the Surface of Occlusion-Derived Virus. Journal of Virology 84: 9497–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang MG, Nie YC, Harris S, Erlandson MA, Theilmann DA (2009) Autographa californica Multiple Nucleopolyhedrovirus Core Gene ac96 Encodes a Per Os Infectivity Factor (pif-4). Journal of Virology 83: 12569–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sparks WO, Harrison RL, Bonning BC (2011) Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology 409: 69–76. [DOI] [PubMed] [Google Scholar]
- 54. Nie Y, Fang M, Erlandson MA, Theilmann DA (2012) Analysis of the autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication. Journal of Virology 86: 3985–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange M, Jehle JA (1997) Auxiliary genes of baculoviruses. In: LK M, editor. The Baculoviruses. New York: Plenum Press. pp. 267–300.
- 56. Sutherland D, Samakovlis C, Krasnow MA (1996) Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87: 1091–1101. [DOI] [PubMed] [Google Scholar]
- 57. Mitsuhashi W, Kawakita H, Murakami R, Takemoto Y, Saiki T, et al. (2007) Spindles of an entomopoxvirus facilitate its infection of the host insect by disrupting the peritrophic membrane. Journal of Virology 81: 4235–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oreilly DR, Miller LK (1989) A Baculovirus Blocks Insect Molting by Producing Ecdysteroid Udp-Glucosyl Transferase. Science 245: 1110–1112. [DOI] [PubMed] [Google Scholar]
- 59. Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, et al. (1997) Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238: 243–253. [DOI] [PubMed] [Google Scholar]
- 60.Rohrmann G (2011) The AcMNPV genome: Gene content, conservation, and function. Baculovirus Molecular Biology, 2nd ed National Library of Medicine. US: NCBI, Bethesda (MD).
- 61. Haas AL, Katzung DJ, Reback PM, Guarino LA (1996) Functional characterization of the ubiquitin variant encoded by the baculovirus Autographa californica . Biochemistry 35: 5385–5394. [DOI] [PubMed] [Google Scholar]
- 62. Roulston A, Marcellus RC, Branton PE (1999) Viruses and apoptosis. Annual Review of Microbiology 53: 577–628. [DOI] [PubMed] [Google Scholar]
- 63. Birnbaum MJ, Clem RJ, Miller LK (1994) An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. Journal of Virology 68: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bideshi DK, Renault S, Stasiak K, Federici BA, Bigot Y (2003) Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. Journal of General Virology 84: 2531–2544. [DOI] [PubMed] [Google Scholar]
- 65. Zemskov EA, Kang WY, Maeda S (2000) Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. Journal of Virology 74: 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.