Abstract
Endometrioid endometrial carcinoma (EEC) is the most dominant subtype of endometrial cancer. Aberrant transcriptional regulation has been implicated in EEC. Herein, we characterized mRNA and miRNA transcriptomes by RNA sequencing in EEC to investigate potential molecular mechanisms underlying the pathogenesis. Total mRNA and small RNA were simultaneously sequenced by next generation sequencing technology for 3 pairs of stage I EEC and adjacent non-tumorous tissues. On average, 52,716,765 pair-end 100 bp mRNA reads and 1,669,602 single-end 50 bp miRNA reads were generated. Further analysis indicated that 7 miRNAs and 320 corresponding target genes were differentially expressed in the three stage I EEC patients. Six of all the seven differentially expressed miRNAs were targeting on eleven differentially expressed genes in the cell cycle pathway. Real-time quantitative PCR in sequencing samples and other independent 21 pairs of samples validated the miRNA-mRNA differential co-expression, which were involved in cell cycle pathway, in the stage I EEC. Thus, we confirmed the involvement of hsa-let-7c-5p and hsa-miR-99a-3p in EEC and firstly found dysregulation of hsa-miR-196a-5p, hsa-miR-328-3p, hsa-miR-337-3p, and hsa-miR-181c-3p in EEC. Moreover, synergistic regulations among these miRNAs were detected. Transcript sequence variants such as single nucleotide variant (SNV) and short insertions and deletions (Indels) were also characterized. Our results provide insights on dysregulated miRNA-mRNA co-expression and valuable resources on transcript variation in stage I EEC, which implies the new molecular mechanisms that underlying pathogenesis of stage I EEC and supplies opportunity for further in depth investigations.
Introduction
Endometrial cancers are the most frequent cancer occurring in the female genital tract, and its incidence has increased in recent years [1]. Endometrioid endometrial carcinoma (EEC) is the most dominant subtype, accounting for ∼80% of cases [2]. To date, most clinical trials of chemotherapeutics for advanced and recurrent EEC have shown limited benefits [3], [4] and information regarding the molecular mechanisms of EEC etiology is still limited. Searching for innovative diagnosis markers and therapeutic targets for EEC is thus necessary.
Aberrant transcript expression levels are commonly observed in cancer and these aberrations could alter biological pathways and disease phenotypes. Next-generation sequencing (NGS) of RNA (RNA-seq) is a powerful tool to investigate the comprehensive transcriptome. RNA-seq has greatly enhanced our knowledge of the transcriptome in cancer [5] recently. MicroRNAs (miRNAs) are a class of small non-coding RNAs that can regulate mRNA expression and control various biological processes [6] through binding to the 3′ untranslated region of mRNAs. It is reported that about 30% of human genes [7] and virtually all cellular processes are regulated by miRNAs [8]. Studies on miRNA dysregulation in cancers have risen rapidly recently, including those in EEC. MiRNAs such as hsa-miR-503, hsa-miR-205, and hsa-miR-200b are dysregulated in EEC and they could regulate cell proliferation, differentiation, apoptosis, and carcinogenesis [9]–[11]. However, studies on concurrent transcriptome characterization of both mRNA and miRNA in EEC are still lacking.
In this study, we applied an integrative approach by simultaneously sequencing both miRNA and mRNA for International Federation of Gynecology and Obstetrics (FIGO) Stage I EEC and adjacent non-tumorous tissues to investigate the mechanisms responsible for the pathogenesis of EEC. Quantitative real-time reverse transcription PCR (qRT-PCR) in other independent patients was performed on the selected miRNA and mRNAs. Transcript sequence variants such as single nucleotide variant (SNV) and short insertions and deletions (Indels) were also characterized. Our investigation may shed light on the molecular pathogenesis of EEC and offer new possibilities for early diagnosis and systemic treatment.
Materials and Methods
Ethics Statement
Our study design received approval from the institutional review board of the third affiliated hospital of Guangzhou medical college (Guangzhou, China). Written informed consent was obtained from all patients.
Sample collection and RNA extraction
Three female patients aged from 32 to 47 were diagnosed as FIGO stage I EEC (two stage IA patients and one stage IB patient) in the third affiliated hospital of Guangzhou medical college. All primary tumor and adjacent non-tumorous samples were obtained from these three patients who underwent surgical tumor resection. All samples were frozen at −80°C until RNA extraction. Total RNA was isolated by using RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies, Carlsbad, CA, USA) for mRNA and miRNA sequencing, according to the manufacturer’s instructions. Integrity of RNA was checked by Agilent 2100 bioanalyzer.
Sequencing
The sequencing library was prepared according to the standard protocol. Briefly, for mRNA sequencing, total RNA was firstly poly-A-selected followed by fragmentation of RNA into small pieces. The cleaved RNA fragments were reverse transcribed to cDNA end-repaired and ligated with Illumina adapters using Quick ligation TM kit (NEB) and DNA ligase. The libraries were then fractionated on agarose gel; 200-bp fragments were excised and amplified by PCR. After purification, the quality of libraries was checked by using Bioanalyzer 2100 (Agilent). MRNA sequencing was then performed on an Illumina HiSeq 2000 sequencer with 100 bp pair-end reads. For small RNA sequencing, libraries were prepared by ligating different adaptors to the total RNA followed by reverse transcription and PCR amplification. Small RNA libraries were sequenced on the Illumina HiSeq 2000 sequencer with 50 bp single-end reads, according to the standard manufacturer’s protocol. All raw data have been deposited in the NIH Short Read Archive database (SRP045645).
Reads processing
Raw mRNA sequencing reads were filtered for adapters and ribosomal RNA. Reads containing five or more positions with a quality score less than 19 were also removed from further analysis. Remained high-quality sequencing reads were then aligned to human genome (hg19) by using Tophat [12]. The matched reads were aligned to Human transcriptome (Ensembl, GRCh37.73). Cufflinks [13] was used to calculate mRNA expression level which was measured by RPKM (reads per kilobase per million).
For small RNA sequencing reads, all low quality reads, such as reads with adapter contaminants or poly A sequences were filtered. Sequences shorter than 18 nt after trimming adapters were removed. The high-quality clean reads were mapped to o the human genome using the bowtie software [14]. Small RNA tags were aligned to the miRNA precursor/mature miRNA in miRBase [15] (release 20). To identify small RNA tags originating from rRNA, tRNA, snRNA, and snoRNA, NCBI GenBank data and Rfam data were used.
We applied the pair wise t-test to filter differentially expressed miRNAs and mRNAs for the two groups. False discovery rate (FDR)-adjusted P values (P<0.05) and absolute fold change >1 were set as the cutoff.
MiRNA target prediction
The miRNA target prediction was done by three current available methods: Targetscan [16], PITA [17], and miRanda [18]. For each prediction method, high efficacy targets were selected by the following criteria: (1) Targetscan: sum of context score −0.2; (2) PITA: ΔΔG≤−10 kcal/mol; (3) miRanda: score ≥155 and energy ≤−20 kcal/mol. Predicted miRNA–mRNA pairs were further selected according to the integrating mRNA and miRNA transcript expression data.
QRT-PCR of miRNA and mRNA expression
Expression of selected miRNAs and mRNAs were validated by qRT-PCR. Another 21 pairs of samples from stage I EEC patients (12 stage IA and 9 stage IB patients) were collected in addition to the original three pairs of sequencing samples. For mRNA expression, 2 µg total RNA was reverse transcribed and QPCR was done in the real-time cyclers (Strata Gene MX3000P qPCR system). The relative fold changes were determined using the 2−ΔΔCt method with GAPDH as endogenous controls.
For miRNA expression, 5 ng of total RNA per miRNA was reverse transcribed into cDNA with SYBR-Green PCR Master Mix (Takara). QPCR was done in real-time cyclers (Strata Gene MX3000P qPCR system). The relative expression levels were determined using the 2−ΔΔCt method with U6 small nuclear RNA as endogenous controls.
Each reaction was done in triplicate. Pair wise t-test was used to determine the expression difference between the two groups.
SNV/Indels detection
SNV/Indels detection was carried out by using VarScan [19] based on the following filtering criteria: 1) depth of coverage over 3×; 2) variation frequency over 20%; 3) base quality over 20. Detected SNVs and Indels were then annotated by using ANNOVAR [20].
Results
Overview of transcriptome sequencing results
To reveal the transcriptome of EEC, we sequenced the mRNA and small RNA from the same total RNA samples of stage I EEC patients using Illumina HiSeq 2000 sequencer. From the six mRNA libraries, an average of 52,716,765 pair-end 100 bp clean reads was generated (Table 1). Six small RNA libraries were also sequenced and 11,669,602 single-end 50 bp clean reads were generated on average (Table 2).
Table 1. Summary of mRNA sequencing.
| Sample id | Clean reads | Mapped reads | Uniquely mapped reads | Paired mapped reads | Unmapped reads | Transcript coverage (×) |
| S1_N | 53,204,178 | 47,891,621 | 45,603,601 | 44,892,334 | 5,312,557 | 40.63 |
| S1_T | 54,527,132 | 49,915,956 | 47,689,962 | 47,086,014 | 4,611,176 | 42.46 |
| S2_N | 52,006,058 | 47,807,313 | 46,016,910 | 45,189,796 | 4,198,745 | 40.82 |
| S2_T | 52,254,225 | 48,152,928 | 45,232,887 | 45,521,896 | 4,101,297 | 40.51 |
| S3_N | 52,891,237 | 48,016,297 | 45,448,760 | 45,323,990 | 4,874,940 | 40.61 |
| S3_T | 51,417,759 | 46,261,459 | 43,257,947 | 43,832,254 | 5,156,300 | 38.94 |
| Average | 52,716,765 | 48,007,596 | 45,541,678 | 45,307,714 | 4,709,169 | 40.66 |
Table 2. Summary of miRNA sequencing.
| ID | Clean reads | miRNA | rRNA | snoRNA | tRNA | Mapped reads | Unmapped reads |
| S1_N | 12,426,115 | 12,385,299 | 36,994 | 1,553 | 2,269 | 10,531,371 | 1,894,744 |
| S1_T | 11,041,505 | 10,994,036 | 42,878 | 1,549 | 3,042 | 9,433,794 | 1,607,711 |
| S2_N | 10,571,133 | 10,557,967 | 11,476 | 136 | 1,554 | 8,905,234 | 1,665,899 |
| S2_T | 10,849,264 | 10,815,517 | 31,412 | 969 | 1,366 | 7,978,261 | 2,871,003 |
| S3_N | 14,584,595 | 14,522,078 | 60,098 | 635 | 1,784 | 9,908,455 | 4,676,140 |
| S3_T | 10,544,999 | 10,424,664 | 111,207 | 2,038 | 7,090 | 7,158,492 | 3,386,507 |
| Average | 11,669,602 | 11,616,594 | 49,011 | 1,147 | 2,851 | 8,985,935 | 2,683,667 |
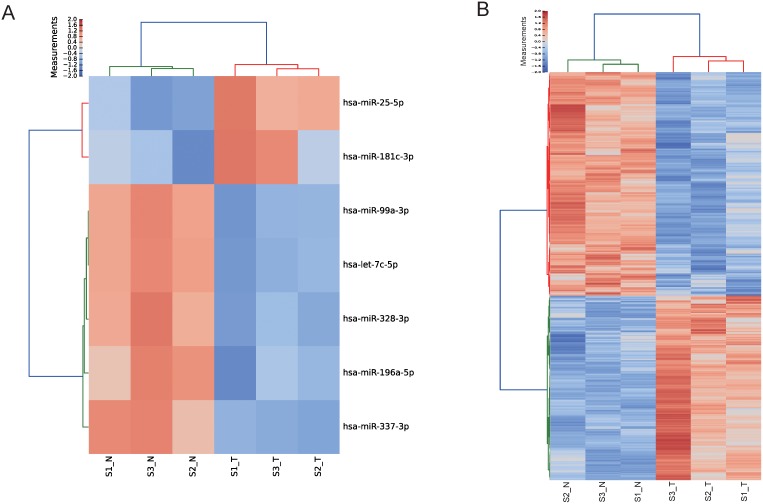
A total of 1079 genes were differentially expressed, including 488 up regulated and 591 down regulated ones (Figure 1). Seven miRNAs were differentially expressed. Among them, hsa-miR-99a-3p, hsa-let-7c-5p, hsa-miR-196a-5p, hsa-miR-328-3p, hsa-miR-337-3p were down regulated while hsa-miR-181c-3p and hsa-miR-25-5p were up regulated (Figure 1).
Figure 1. Hierarchical clustering of differentially expressed miRNAs and mRNAs in endometrioid endometrial carcinoma (S1_T, S2_T, S3_T) and adjacent non-tumorous tissues (S1_N, S2_N, S3_N).
MiRNA target prediction and integration of mRNA and miRNA expression profiles
We ran the three prediction algorithms: Targetscan [16], PITA [17], and miRanda [18] using the selected 7 miRNAs and 1079 genes as input. This resulted in 1098 miRNA-mRNA pairs, which were supported by at least two prediction algorithms. We further integrated the sequencing data into the predicted miRNA-mRNA pairs to validate the target pairs. A total of 438 target pairs were found inversely expressed, including 320 dysregulated genes.
Functional enrichment
Biological pathway and gene ontology (GO) enrichment were done for the 320 dysregulated genes. Pathway analysis identified 12 pathways overrepresented with dysregulated miRNA target genes. Overall, a genetic cluster summarizing the functions of Cell cycle (hsa04110) was found to have the highest relationship with EEC (Table 3). Considering the pathways involved in cancer development, we identified several significantly related pathways, including p53 signaling pathway, GnRH signaling pathway, Pathways in cancer and Gap junction (Table 3). In addition to these classical pathways, pancreatic cancer and thyroid cancer pathway were also overrepresented with dysregulated genes, implicating a common oncogenic basis. GO enrichment analyses indicated that 9 GO items were significantly enriched with the dysregulated genes (Table 4). Consistent with the pathway analysis, the most significant GO category is mitotic cell cycle (Table 4).
Table 3. Pathway analysis based on miRNA-targeted differentially expressed genes.
| KEGG id | KEGG description | KEGG subclass | p-value |
| hsa04110 | Cell cycle | Cell growth and death | 1.21E-03 |
| hsa04115 | p53 signaling pathway | Cell growth and death | 1.44E-02 |
| hsa04912 | GnRH signaling pathway | Endocrine system | 2.21E-02 |
| hsa04713 | Circadian entrainment | Environmental adaptation | 2.24E-02 |
| hsa05200 | Pathways in cancer | Cancers | 3.41E-02 |
| hsa04973 | Carbohydrate digestion and absorption | Digestive system | 3.94E-02 |
| hsa0480 | Glutathione metabolism | Metabolism of other amino acids | 4.03E-02 |
| hsa05212 | Pancreatic cancer | Cancers | 4.09E-02 |
| hsa04540 | Gap junction | Cell communication | 4.32E-02 |
| hsa04114 | Oocyte meiosis | Cell growth and death | 4.65E-02 |
| hsa05216 | Thyroid cancer | Cancers | 4.72E-02 |
| hsa04918 | Thyroid hormone synthesis | Endocrine system | 4.90E-02 |
Table 4. Gene ontology analysis based on miRNA-targeted differentially expressed genes.
| GO id | GO description | GO class | p-value |
| GO:0000278 | mitotic cell cycle | Process | 1.60E-03 |
| GO:0000777 | condensed chromosome kinetochore | Component | 1.17E-02 |
| GO:0007051 | spindle organization | Component | 1.17E-02 |
| GO:0008156 | negative regulation of DNA replication | Process | 1.17E-02 |
| GO:0032133 | chromosome passenger complex | Component | 2.17E-02 |
| GO:0005876 | spindle microtubule | Process | 2.17E-02 |
| GO:0009636 | response to toxic substance | Process | 2.17E-02 |
| GO:0051301 | cell division | Component | 4.72E-02 |
| GO:0007067 | mitosis | Component | 4.72E-02 |
QRT-PCR validation of miRNA and mRNA co-expression
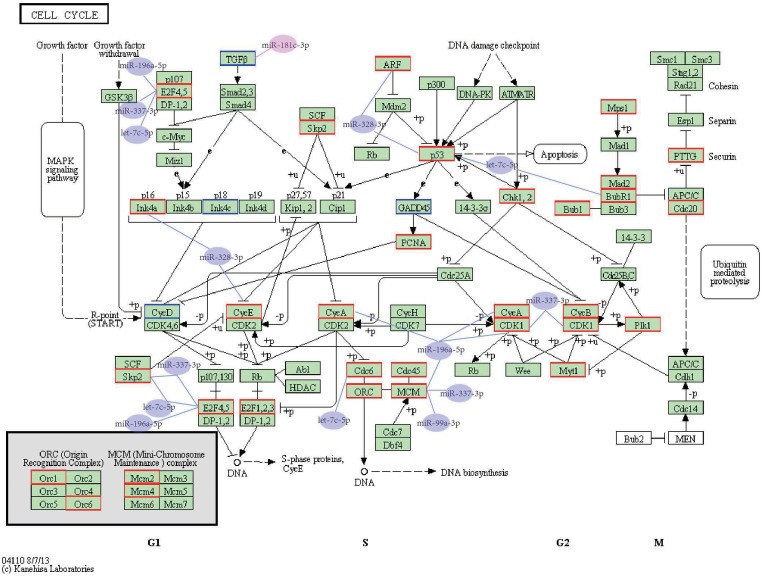
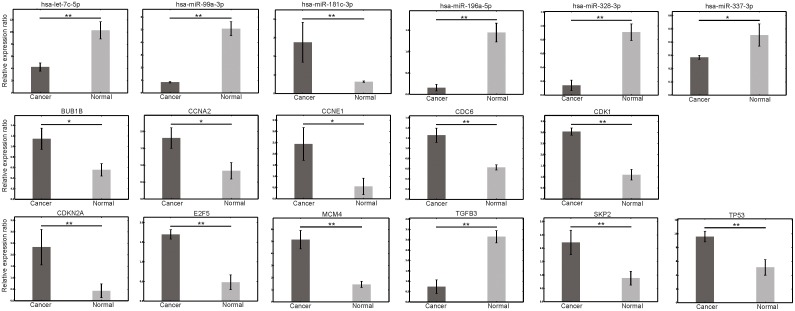
Our pathway analysis showed that the most significant pathway overrepresented with dysregulated miRNA target genes was the cell cycle pathway (P = 1.21E-03). Since human carcinogenesis is believed to result from uncontrolled cell proliferation and many molecules in the cell cycle pathway have been reported to be associated with endometrial cancer [21], [22], our findings prompted us to validate the expression of miRNAs and their potential target genes in the cell cycle pathway. As shown in Figure 2, among the significantly dysregulated miRNAs, 5 down regulated miRNAs (hsa-let-7c-5p, hsa-miR-196a-5p, hsa-miR-328-3p, hsa-miR-337-3p, and hsa-miR-99a-3p) were predicted to target genes encoding 10 upregulated cell cycle pathway-related mRNAs (E2F5, CDKN2A, CCNA2, TP53, BUB1B, CCNE1, CDK1, MCM4, SKP2, and CDC6). Additionally, one upregulated miRNA, hsa-miR-181c-3p, was predicted to target the downregulated TGFB3 gene in cell cycle pathway (Figure 2). All these 6 miRNAs and 11 genes were present in the three pairs of sequencing samples. Differential expression was confirmed for all the 6 miRNAs and 11 genes in sequencing samples and other independent 21 pairs of samples, as shown in Figure 3. These 6 miRNAs may therefore play a role in the pathogenesis of EEC.
Figure 2. MiRNA-gene network of the cell cycle pathway in endometrioid endometrial carcinoma.
Protein symbols were marked according to gene expression pattern. Those with red frames are up regulated while those with blue frames are down regulated in endometrioid endometrial carcinoma tissues. Similarly, miRNAs with blue background were down regulated while the miRNA with red ground were up regulated in endometrioid endometrial carcinoma tissues.
Figure 3. QPCR expression on QPCR-validated genes and miRNAs in the cell cycle pathway.
*: p<0.05; **: p<0.01.
In addition, for these 6 miRNAs and 11 mRNAs, synergistic regulations among the six miRNAs were detected (Figure 2). For example, E2F5 was predicted as a target of hsa-miR-196a-5p, hsa-miR-337-3p and hsa-let-7c-5p; TP53 was predicted as the target of both hsa-miR-328-3p and hsa-let-7c-5p; MCM4 was targeted by hsa-miR-196a-5p, hsa-miR-337-3p, and hsa-miR-99a-3p; SKP2 was targeted by hsa-miR-337-3p and hsa-let-7c-5p; CDK1 was targeted by hsa-miR-196a-5p and hsa-miR-337-3p.
Sequence variants in EEC transcriptome
Sequence variants analysis detecting SNVs and Indels was carried out using VarScan [19]. The results are summarized in Table 5. A total of 346 variants (17.06%) are from exonic regions of the genome while 1682 (82.93%) are from the non-protein coding regions. Around 75% (260/346) of the exonic variants, including 181 nonsynonymous SNVs, 14 stop gain variants and 65 frameshift Indels, would result in protein coding alterations. Moreover, 17 out of 260 protein-affected variants overlapped with the somatic mutations in the cancer (COSMIC) [23] (Table S1 and Table S2). Of the non-protein coding variants, around half (863/1682) are in the UTR region.
Table 5. Sequence variants identified from transcriptome data.
| Variants | S1 | S2 | S3 | Total | ||||
| SNV | Indel | SNV | Indel | SNV | Indel | SNV | Indel | |
| exonic | 95 | 27 | 81 | 26 | 89 | 28 | 264 | 82 |
| synonymous SNV | 26 | 0 | 26 | 0 | 22 | 0 | 74 | 0 |
| nonsynonymous SNV | 67 | 0 | 50 | 0 | 65 | 0 | 181 | 0 |
| stopgain | 2 | 3 | 5 | 1 | 2 | 1 | 9 | 5 |
| frameshift insertion | 0 | 15 | 0 | 13 | 0 | 10 | 0 | 38 |
| nonframeshift insertion | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 5 |
| frameshift deletion | 0 | 8 | 0 | 7 | 0 | 12 | 0 | 27 |
| nonframeshift deletion | 0 | 0 | 0 | 3 | 0 | 4 | 0 | 7 |
| splicing | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 2 |
| UTR3 | 209 | 59 | 201 | 70 | 180 | 80 | 587 | 209 |
| UTR5 | 23 | 10 | 8 | 9 | 16 | 1 | 47 | 20 |
| downstream | 28 | 4 | 30 | 3 | 16 | 2 | 74 | 9 |
| upstream | 5 | 0 | 2 | 1 | 5 | 0 | 12 | 1 |
| intronic | 185 | 19 | 189 | 27 | 38 | 4 | 404 | 50 |
| intergenic | 114 | 11 | 92 | 8 | 41 | 5 | 244 | 25 |
| Total | 659 | 130 | 603 | 145 | 383 | 121 | 1630 | 398 |
Discussion
Compared with traditional microarrays, NGS enables the identification of novel and more detailed studies of biological pathways in complex diseases. Here with mRNAs and small RNA sequencing for three pairs of stage I EEC and adjacent normal tissues, we provide a global overview of the stage I EEC transcriptome and reveal the miRNA regulation of transcript expression.
MiRNA data analyses identified 7 dysregulated miRNAs (Figure 1) and mRNA sequencing identified a total of 1079 differentially expressed genes (Figure 1). To gain insights into the targets of differentially expressed miRNAs, a stringent prediction of miRNA targets was made by using three different target prediction methods. We further integrated the sequencing data into the predicted miRNA-mRNA pairs to validate the target pairs. A total of 438 target pairs were found inversely expressed, which can serve as references for further studies on miRNA and its targets. Both GO and KEGG pathway enrichment analyses of the putative target genes revealed the dysregulation of the cell cycle process, suggesting that our results were in line with the basic tumor characteristics. We further validated the expression levels of miRNA-target pairs in the cell cycle pathway. As shown in Figure 2, 5 down regulated miRNAs (hsa-let-7c-5p, hsa-miR-196a-5p, hsa-miR-328-3p, hsa-miR-337-3p, and hsa-miR-99a-3p) were predicted to target genes encoding 10 upregulated cell cycle pathway-related mRNAs (E2F5, CDKN2A, CCNA2, TP53, BUB1B, CCNE1, CDK1, MCM4, SKP2, and CDC6). Additionally, one upregulated miRNA, hsa-miR-181c-3p, was predicted to target the downregulated TGFB3 gene. All these 6 miRNAs have previously been suggested to be involved in human cancers. Downregulation of hsa-let-7c has been identified in multiple cancer types, such as non-small cell lung cancer [24], pancreatic cancer [25], prostate cancer [26]. Moreover, let-7c was also found to be down-regulated in endometrial sarcomas when compared to the control samples [27]. Expression of miR-196a was also found to be significantly down-regulated in malignant melanoma cell lines and tissue samples [28]. Decreased expression of miR-328 was also found in multiple cancer types [29], [30]. Loss of has-miR-337-3p expression has been associated with lymph node metastasis of human gastric cancer [31]. Downregulation of miR-99a was discovered in both plasma and tissues of endometrial cancer patients [32]. Overexpression of miR-181c has also been reported in other cancers, such as gastric cancer [33]. In the current study, we confirmed the involvement of hsa-let-7c-5p and hsa-miR-99a-3p in EEC and firstly found dysregulation of hsa-miR-196a-5p, hsa-miR-328-3p, hsa-miR-337-3p, and hsa-miR-181c-3p in EEC. Since all these miRNAs are dysregulated in stage I EEC, they may serve as potential diagnostic markers in clinical practice. In addition, considering the inhibition effects on cancer growth or metastasis of hsa-let-7c [24], [26], hsa-miR-328 [29], hsa-miR-337 [31], these three miRNAs may provide new avenues for the prognosis and therapy for EEC. Among the target genes of these miRNAs in the cell cycle pathway, TP53 [34], CDKN2A [34], SKP2 [35], [36], CCNE1 [36], MCM4 [36], CDK1 [37] and TGFB3 [38] have been extensively studied as anticancer targets in endometrial cancer. Overexpression of CCNA2 was reported to be an indicator of a poor prognosis endometrial carcinoma [39]. Currently, there is no report about the relationship between E2F5, BUB1B, or CDC6 and EEC. E2F5 is a transcription factor predicted as a target of hsa-miR-196a-5p, hsa-miR-337-3p and hsa-let-7c-5p. Human E2F5 gene is oncogenic [40] and overexpression of this gene was associated with worse clinical outcome in other cancers [41]. Both BUB1B and CDC6 are predicted as a target of hsa-let-7c-5p. BUB1B encodes a kinase involved in spindle checkpoint function and involvements of these two genes have been reported in other cancers [42], [43]. Their biological functions in EEC need further study. Moreover, based on our validated co-expression relationships of these miRNA-mRNA pairs, further in vitro experiments may be carried out to further confirm that the differentially expressed miRNAs synergistically affect the expression of the predicted targets.
In addition, for the miRNAs and mRNAs we validated in the cell cycle pathway, synergistic regulations among the six miRNAs were detected (Figure 2). MiRNA–miRNA synergism is important to understand the mechanism of complex post-transcriptional regulations in human [44]. Our results here may provide guide for further validation studies of the regulation mechanisms of these miRNAs.
In addition to expression analysis, we also analyzed the sequence variants. Besides proteins encoding SNV/Indels, a large number of variants are located in UTRs whose function still remains unknown (Table 5). The analysis of SNV, Indels may provide possible targets for further mechanistic studies.
In summary, here we report a comprehensive characterization of mRNA and miRNA transcriptome in stage I EEC from expression level, miRNA/mRNA regulations and sequence variation. We integratively analyzed differentially expressed miRNAs and their target genes. Differential expression levels of target genes in the cell cycle pathway and their regulatory miRNAs were further validated in other independent samples. Our results will provide a valuable reference for future studies in transcript variation and regulation and will potentially be useful for mechanistic and preclinical studies.
Supporting Information
Detail information of the identified single nucleotide variants.
(XLSX)
Detail information of the identified short insertions and deletions.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All sequencing reads are available from the NIH Short Read Archive database (SRP045645). Other relevant data are within the paper and its Supporting Information files.
Funding Statement
This study is supported by the Scientific Research Grant from Guangdong Science and Technology Department (No. 2013B021800193) and the Scientific Research Grant from the Third Affiliated Hospital of Guangzhou Medical University (No. 110201704, 11021403). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Evans T, Sany O, Pearmain P, Ganesan R, Blann A, et al. (2011) Differential trends in the rising incidence of endometrial cancer by type: data from a UK population-based registry from 1994 to 2006. Br J Cancer 104: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, et al. (2005) Endometrial cancer. Lancet 366: 491–505. [DOI] [PubMed] [Google Scholar]
- 3. Obel JC, Friberg G, Fleming GF (2006) Chemotherapy in endometrial cancer. Clin Adv Hematol Oncol 4: 459–468. [PubMed] [Google Scholar]
- 4. Vale CL, Tierney J, Bull SJ, Symonds PR (2012) Chemotherapy for advanced, recurrent or metastatic endometrial carcinoma. Cochrane Database Syst Rev 8: CD003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 7. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 8. Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, et al. (2011) Toward the blood-borne miRNome of human diseases. Nat Methods 8: 841–843. [DOI] [PubMed] [Google Scholar]
- 9. Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y, et al. (2013) MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J 280: 3768–3779. [DOI] [PubMed] [Google Scholar]
- 10. Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW, et al. (2009) Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer 124: 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramon LA, Braza-Boils A, Gilabert J, Chirivella M, Espana F, et al. (2012) microRNAs related to angiogenesis are dysregulated in endometrioid endometrial cancer. Hum Reprod 27: 3036–3045. [DOI] [PubMed] [Google Scholar]
- 12. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- 18. Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cai H, Xiang YB, Qu S, Long J, Cai Q, et al. (2011) Association of genetic polymorphisms in cell-cycle control genes and susceptibility to endometrial cancer among Chinese women. Am J Epidemiol 173: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ray S, Pollard JW (2012) KLF15 negatively regulates estrogen-induced epithelial cell proliferation by inhibition of DNA replication licensing. Proc Natl Acad Sci U S A 109: E1334–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao B, Han H, Chen J, Zhang Z, Li S, et al. (2014) MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett 342: 43–51. [DOI] [PubMed] [Google Scholar]
- 25. Ali S, Saleh H, Sethi S, Sarkar FH, Philip PA (2012) MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer 107: 1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nadiminty N, Tummala R, Lou W, Zhu Y, Shi XB, et al. (2012) MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One 7: e32832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowalewska M, Bakula-Zalewska E, Chechlinska M, Goryca K, Nasierowska-Guttmejer A, et al. (2013) microRNAs in uterine sarcomas and mixed epithelial-mesenchymal uterine tumors: a preliminary report. Tumour Biol 34: 2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mueller DW, Bosserhoff AK (2011) MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer 129: 1064–1074. [DOI] [PubMed] [Google Scholar]
- 29. Wu Z, Sun L, Wang H, Yao J, Jiang C, et al. (2012) MiR-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS One 7: e47270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu XT, Xu Q, Tong JL, Zhu MM, Nie F, et al. (2012) MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br J Cancer 106: 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z, Wang J, Yang Y, Hao B, Wang R, et al. (2013) Loss of has-miR-337–3p expression is associated with lymph node metastasis of human gastric cancer. J Exp Clin Cancer Res 32: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, et al. (2012) Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 12: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hashimoto Y, Akiyama Y, Yuasa Y (2013) Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One 8: e62589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buchynska LG, Nesina IP (2006) Expression of the cell cycle regulators p53, p21(WAF1/CIP1) and p16(INK4a) in human endometrial adenocarcinoma. Exp Oncol 28: 152–155. [PubMed] [Google Scholar]
- 35. Kamata Y, Watanabe J, Nishimura Y, Arai T, Kawaguchi M, et al. (2005) High expression of skp2 correlates with poor prognosis in endometrial endometrioid adenocarcinoma. J Cancer Res Clin Oncol 131: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saini S, Hirata H, Majid S, Dahiya R (2009) Functional significance of cytochrome P450 1B1 in endometrial carcinogenesis. Cancer Res 69: 7038–7045. [DOI] [PubMed] [Google Scholar]
- 37. Yan GJ, Yu F, Wang B, Zhou HJ, Ge QY, et al. (2014) MicroRNA miR-302 inhibits the tumorigenicity of endometrial cancer cells by suppression of Cyclin D1 and CDK1. Cancer Lett 345: 39–47. [DOI] [PubMed] [Google Scholar]
- 38. Rodriguez GC, Rimel BJ, Watkin W, Turbov JM, Barry C, et al. (2008) Progestin treatment induces apoptosis and modulates transforming growth factor-beta in the uterine endometrium. Cancer Epidemiol Biomarkers Prev 17: 578–584. [DOI] [PubMed] [Google Scholar]
- 39. Suzuki A, Horiuchi A, Ashida T, Miyamoto T, Kashima H, et al. (2010) Cyclin A2 confers cisplatin resistance to endometrial carcinoma cells via up-regulation of an Akt-binding protein, periplakin. J Cell Mol Med 14: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Polanowska J, Le Cam L, Orsetti B, Valles H, Fabbrizio E, et al. (2000) Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer 28: 126–130. [PubMed] [Google Scholar]
- 41. Umemura S, Shirane M, Takekoshi S, Kusakabe T, Itoh J, et al. (2009) Overexpression of E2F-5 correlates with a pathological basal phenotype and a worse clinical outcome. Br J Cancer 100: 764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen F, Yang G, Xia B (2013) Increased expression of the spindle checkpoint protein BubR1 is associated with high cell proliferation in primary gastrointestinal diffuse large B cell lymphoma. Cell Biochem Biophys 66: 747–752. [DOI] [PubMed] [Google Scholar]
- 43. Murphy N, Ring M, Heffron CC, King B, Killalea AG, et al. (2005) p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol 58: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu J, Li CX, Li YS, Lv JY, Ma Y, et al. (2011) MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res 39: 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detail information of the identified single nucleotide variants.
(XLSX)
Detail information of the identified short insertions and deletions.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All sequencing reads are available from the NIH Short Read Archive database (SRP045645). Other relevant data are within the paper and its Supporting Information files.