Abstract
Inflammatory bowel disease (IBD), encompassing Crohn’s disease (CD), ulcerative colitis (UC) and unclassified IBD (IBDU), is characterized by chronic intestinal inflammation, and has a multi-factorial etiology with complex interactions between genetic and environmental factors. The genetics of IBD are believed to be common and complex with over 163 associated genetic loci. However, the genetic contribution of the majority of these common loci is small and the effect sizes are low. Although childhood onset IBD represents only 10–25% of all IBD cases, in depth research into the genetic networks of pediatric IBD has revealed exciting new developments and unsuspected pathways. Recent pediatric studies have revealed an increasing spectrum of human monogenic diseases with high effect sizes/penetrance that can present with IBD or IBD-like intestinal inflammation. A substantial proportion of patients with these genetic defects present with very early onset of intestinal inflammation, with onset of IBD at less than 10 years of age. There is also considerable overlap with primary immunodeficiencies and very early onset IBD. This review summarizes the current understanding of the genetics of pediatric inflammatory bowel disease with a focus on the very early onset population, and discusses the promising results from the effort of finding missing heritability of IBD from studying pediatric population.
Introduction
Inflammatory bowel disease (IBD) primarily includes Crohn’s disease (CD) and ulcerative colitis (UC), two chronic, debilitating inflammatory disorders of the gastrointestinal tract that can lead to life threatening complications, severe impairment in quality of life, growth failure and high risk for needing surgical resections in children. The incidence of IBD peaks in late adolescents to young adults (second and third decade of life). Approximately 10–25% of incident cases of IBD occur during childhood (1). The definition of childhood or pediatric-onset IBD can be arbitrary. The age at which childhood ends and adulthood begins represents a continuum and accurate classification of pediatric phenotype is essential to determining genotype to phenotype correlation. Recently, the Paris classification was adopted (2) to sufficiently capture the dynamic features of the phenotype (change in disease location and behavior over time, growth failure), improve classification of children, and implement uniform standards for defining IBD phenotypes. Accordingly, age at diagnosis is used to classify IBD as very early onset (VEO - 0 to < 10 years) and early onset (EO −10 to < 17 years). VEO is characterized by a higher tendency for disease manifestation in the colon, more disease extension, and a change in disease location over time, while EO clinical characteristics are very similar to adult onset IBD.
The exact determinants of the age of onset and disease course remain largely unexplained but IBD is believed to result from a dysregulation of the immune response to the gut environmental agents housed in a genetically susceptible host. Recent genome-wide association studies (GWAS) within large IBD cohorts have identified 163 genetic loci (3). These 163 loci in total explain 13.6% of CD and 7.5% of UC total disease variance with the majority of loci contributing only a little towards the explained IBD heritability. While two GWAS focused on pediatric IBD (3, 4), they included only a small sample size with age less than 10 years old at disease onset. Clinical heterogeneity within IBD, particularly based on VEO has long been known (5–7), and recent evidence suggests that this heterogeneity is explained by different mechanism-based disease subsets (8). Growing evidence now supports that these phenotypic differences seen among the childhood onset IBD reflect variations in genetics, composition of the microbiome and perhaps environmental risk factors (10, 11). This review summarizes the current understanding of the genetics of pediatric IBD across the age spectrum with a focus on the very early onset population, and discusses the promising results from the effort of finding missing heritability of IBD from studying pediatric populations.
Decades of progress in genetic studies of IBD
There is enough evidence to support the strong contribution of genetic factors to the pathogenesis of IBD (9–16) and some of the most exciting recent developments in our understanding of the pathogenesis of IBD have been in the field of genetics.
Since the human genome has been sequenced and given the development of technologies for rapid and cost-effective genome analysis, it became possible to identify IBD susceptibility loci using genotyping and the hypothesis free method of genome-wide association studies (GWAS) in CD and UC (17, 18). In all, GWAS have identified over 163 IBD loci for both CD and UC that explain only a small proportion of the inherited disease risk, mostly in Caucasians of European descent, therefore the genetics of IBD with adult-onset disease as the primary phenotype have been extensively describe (19–23). Although most investigators are fatigued and getting tired of GWAS, the GWAS efforts on IBD have revealed many unsuspected pathways and focused the IBD pathogenesis on innate immunity responses involving the integrity of the intestinal epithelial barrier, autophagy, the innate recognition and response towards the gut microbiome (17, 18, 24–26). The common pathways and genes associated with IBD revealed by GWAS are summarized in table 1.
Table 1.
| Pathways | Genes |
|---|---|
| Cellular responses | |
| Autophagy | ATG16L1, IRGM, NOD2, LRRK2, CUL2, PARK7, DAP |
| Apoptosis/necroptosis | FASLG, THADA, DAP, PUS10, MST1 |
| ER stress | CPEB4, ORMDL3, SERINC3, XBP1 |
| Carbohydrate metabolism | GCKR, SLC2A4RG |
| Oxidative stress | PRDX5, BACH2, ADO, GPX4, GPX1, SLC22A4, LRRK2, NOD2, CARD9, HSPA6, DLD, PARK7, UTS2, PEX13 |
| Intracellular logistics | VAMP3, KIF21B, TTLL8, FGFR1OP, CEP72, TPPP |
| Inflammatory response | PTPN2, PTPN22 |
| Cell migration | ARPC2, LSP1, AAMP |
| Viral response | IFIH1 |
| IBD-related processes | |
| Epithelial barrier | GNA12, HNF4A, CDH1, ERRFI1, MUC19, ITLN1 |
| Restitution | REL, PTGER4, NKX2-3, STAT3, ERRFI1, HNF4A, PLA2G2A/E |
| Solute transport | SLC9A4, SLC22A5, SLC22A4, AQP12A/B, SLC9A3, SLC26A3 |
| Paneth cells | ITLN1, NOD2, ATG16L1, XBP1 |
| Innate mucosal defense | NOD2, ITLN1, CARD9, REL, SLC11A1, FCGR2A/B |
| NF-κB | REL, TNFAIP3, NFKB1 |
| Ubiquitylation | UBE2L3 |
| Immune cell recruitment | CCL11/CCL2/CCL7/CCL8, CCR6, IL8RA/IL8RB, MST1 |
| Antigen presentation | ERAP2, LNPEP, DENND1B |
| IL-23/TH17 | IL23R, IL12B, JAK2, TYK2, STAT3, STAT4, ICOSLG, IL21, TNFSF15, IL27 |
| T-cell regulation | NDFIP1, TNFSF8, TAGAP, IL2, IL2RA, TNFRSF9, PIM3, IL7R, IL12B, IL23R, PRDM1, ICOSLG, TNFSF8, IFNG, IL21 |
| B-cell regulation | IL5, IKZF1, BACH2, IL7R, IRF5, IRF8 |
| Immune tolerance | IL10, IL27, SBNO2, CREM, IL1R1/IL1R2, NOD2 |
| Other | ZMIZ1, YDJC, TAGAP, IL18RAP |
Although GWAS have highlighted the overlap of IBD susceptibility loci with other immune-related diseases, the functional impact of most genes ascertain by GWAS is subtle or not clear, and causative loci bearing variants have not been identified.
GWAS have also outlined important observations, useful in guiding IBD studies. GWAS subscribes to the common disease-common variant model that has been the primary focus of human genomics over the last decade. Using GWAS, only a small fraction of the total heritability of IBD has been explained by identifying common alleles (with minor allele frequency > 5%) with small effect sizes. With the exception of NOD2, common alleles of large effect size have yet to be reported for IBD, and additional larger GWAS studies are highly unlikely to do so. Furthermore, highly penetrant variants of higher effect size are not detectable by GWAS. In addition, most GWAS studies involved mainly adult or adolescent onset IBD cases, and VEO cases were not included. The VEO group has a more severe disease course and shows positive family history for IBD, in support of higher genetic load and common genetic background (27–29). This also suggests a high likelihood that VEO cases will display Mendelian-like forms of IBD characterized by highly penetrant variants of higher effect size. The monogenetic form of VEO IBD was first confirmed by mutations in IL10R gene (30). In order to expand our understanding of the contribution of genetics to IBD, it is imperative to study VEO populations to ascertain low frequency variations (major allele frequency of 0.5 to 1%). Rare genetic variants have not only been predicted to vastly outnumber common variants in the human genome (31), but low frequency variants have been hypothesized to explain a substantial fraction of common complex diseases (32).
Extending the Genetic Studies into the Very Early Onset IBD Population
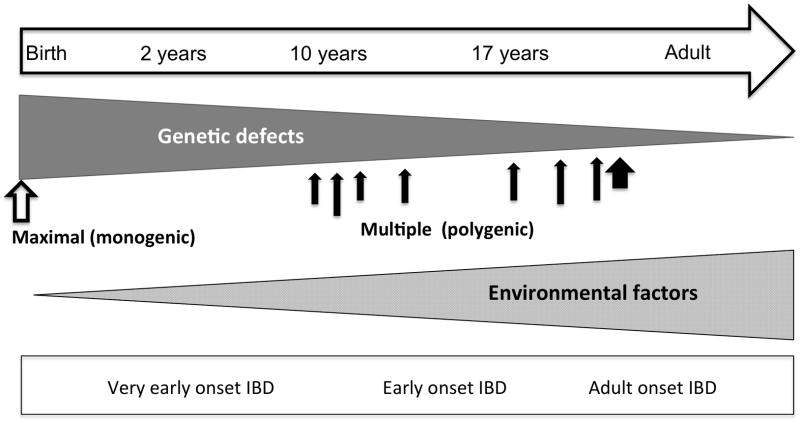
Causative events leading to pediatric IBD can occur any time between birth and the late teens. While the debate on an increased genetic risk at earlier age is ongoing, many studies have outlined an increased rate of family history with very early onset CD and UC (7, 33). Before GWAS, specific variants of genes were linked with early onset IBD; Variations in IBD1 have been associated with early onset IBD (34). Variants in the IBD5 gene have also been shown to be associate with growth indices (35) and a more severe phenotype (36). A specific variant in NOD2 was found more often in early onset than adult CD (37). Because it is difficult to determine when inflammation begins, taking into account the time course of disease when studying pediatric IBD is important. Children constitute a population with distinct physiology and disease risks, and studying them has several advantages compare to adults: (1) the effect of environmental modifiers is minimal or orders of magnitude less (38), (2) the age-dependent gene expression is uniquely relevant, owing to the presence of an active growth phase (39–41), (3) children are more likely to have a family history of IBD than adults (33), and (4) the effect of confounders such as comorbidity and drugs is different (42–44). Consequently, the argument can be made that the disease course and manifestation in VEO cases are subject to higher genetic load (45) and common genetic background (29) as illustrated in figure 1.
Figure 1.
Genetics and environmental contribution to IBD from birth to adulthood
Additionally, children with VEO IBD offer the opportunity to study the initial immune response and the early gut microbiome as well as change over the time, the effects of early therapeutic interventions, the natural history of the disease, and the impact of early environmental modifiers. In short, the pediatric population represents a “virgin” and fertile source of novel and helpful information to understand the triggers and pathogenic mechanisms of IBD in both children and adults alike. To have a better understanding of IBD, its risk and ultimate management, pediatric genomic studies are necessary.
Genetic Studies in Pediatric Onset IBD: Progress
Investigation of the genetic determinants of pediatric IBD has been motivated by the rapid rise in incidence of pediatric-onset IBD in the world, yet data is lacking in VEO and non-European populations. In an attempt to explore their specific implication in children, many established GWAS loci ascertained in adults do not distinguish early from later onset CD (46). Because of the many challenges to performing genomic studies in pediatric populations (47), little effort has been dedicated to identifying IBD genes exclusively in the pediatric-onset IBD population. The need to first identify and characterize the right population as a prerequisite to pediatric IBD studies, led the Crohn’s and Colitis Foundation of America (CCFA) to launch the “Challenges in Pediatric IBD” initiative in 2005 (48). Two pediatric GWAS studies (3, 4) identified novel loci (TNFRSF6B, IL27) genes, but they are not exclusive to pediatric onset, as they have been replicated in GWAS meta-analysis from adult studies.
The failure of GWAS loci to distinguish early onset from adult IBD can lead to various conclusions: (1) the similarity in effect of established loci between early and late onset highlight similarity in the overall pathogenicity, or (2) the matching of the IBD phenotype in GWAS interfered with the identification of loci unique to early or late onset IBD. On the other hand, the difficulty associated with the identification of early-onset specific loci by GWAS, suggest that differences in phenotypic characteristics between early and late onset could be driven by variants that are not detectable using GWAS. Therefore, the need to understand the molecular pathogenesis of VEO requires a population with unique or narrow phenotypes and a methodology/technology capable of complementing GWAS beyond the common disease–common variant hypothesis
Sequencing in IBD Genetic Studies: Progress
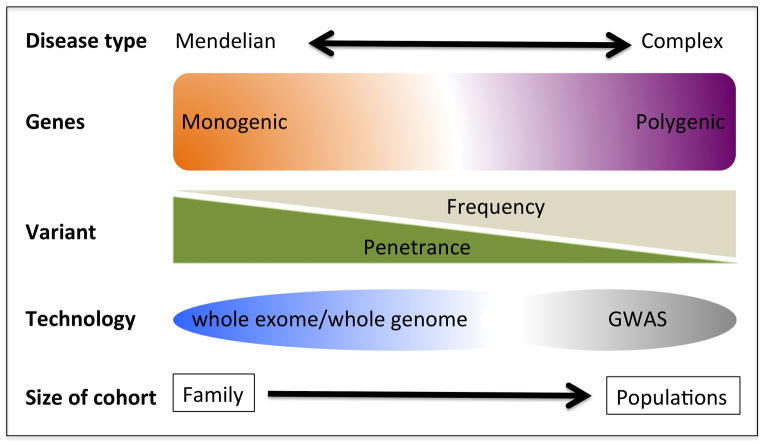
Variants that code for protein are more likely to have greater penetrance and are amenable to functional experimentation. Such variants are usually of low to rare frequency and have not only been predicted to vastly outnumber common variants in the human genome (31), but are also hypothesized to explain a substantial fraction of common complex diseases (32). Sequencing is the method of choice to ascertain low frequency to rare variants that have effect sizes higher than those shown by GWAS (figure 2).
Figure 2.
Identification of disease associated or disease causing genomic variations
An estimated 85% of high effect mutations reside in 1% of the human genome, representing the entire protein coding sequence known as the exome (49). Sequencing of all protein coding regions quickly became a practical method to identify functionally relevant variants. Targeted sequencing represents a hypothesis driven approach to ascertain disease-causing variant in GWAS loci. With rapid advances and decreasing cost of next generation sequencing (NGS) technologies, whole exome sequencing (WES) has become more cost effective than gene panel or multi-gene sequencing to find functional variants in the protein coding regions of the genome. WES represents a hypothesis free and unbiased way of surveying the entire genome for variants (known or novel) that GWAS failed to reveal. Targeted sequencing and WES have been successfully applied to detect causal variants in many genes associated with IBD by GWAS (table 2).
Table 2.
GWAS loci sequenced to identify rare causative variants associated with IBD (genes in bold apply to very early onset studies)
| Gene symbol | Description | Ref. |
|---|---|---|
| ADAM17 | Disintegrin and metalloproteinase domain-containing protein 17 | (50) |
| IL10 | Interleukin 10 | (51) |
| NCF2 | Neutrophil cytosolic factor 2 | (52) |
| C1orf106 | Chromosome 1 open reading frame 106 | |
| CARD9 | Caspase recruitment domain family, member 9 | |
| CUL2 | Cullin 2 | (53) |
| IL18RAP | Interleukin 18 receptor accessory protein | |
| MUC19 | Mucin 19, oligomeric | |
| PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | |
| BACH2 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 | |
| ERAP2 | Endoplasmic reticulum aminopeptidase 2 | (54) |
| IL10 | Interleukin 10 | |
| SEC16A | SEC16 homolog A (S. cerevisiae) | |
| HEATR3 | HEAT repeat containing 3 | (55) |
| IL10RA | Interleukin-10 receptor alpha subunit | (28, 56, 57) |
| IL23R | Interleukin 23 receptor | (58) |
| LRBA | LPS-responsive vesicle trafficking, beach and anchor containing | (59) |
| # NCF4 | Neutrophil cytosolic factor 4 | (60) |
| PRDM1 | PR domain containing 1, with Zinc finger domain | (61) |
| RNF186 | Ring finger protein 186 | (62) |
| SLC22A4 | Solute carrier family 22 (organic cation/ergothioneine transporter), member 4 | (63) |
| TNFRSF6B | Tumor necrosis factor receptor superfamily, member 6b, decoy | (64) |
| TTC7A | tetratricopeptide repeat domain 7 | (65) |
The first application of WES to a VEO case revealed a rare mutation affecting regulatory function of the XIAP gene in a child who presented at 15 months with intractable IBD (66). Since then, WES has successfully identified rare functional variants in novel genes implicated in the pathogenesis of VEO (FOXP3 (67), IL10RB (30, 56) and XIAP (66) genes) or even adult (GSDMB (54) and NDP52 genes (61)) IBD. WES is therefore the most current cost effective technology capable of exposing low to intermediate frequency variants that may have higher effect size than the weaker associations reported by common GWAS variants.
Both targeted sequencing and WES have advance our understanding of IBD by (1) confirming already known IBD risk variants detected by GWAS, (2) showing that previously known loci may harbor rare risk variants with high effect undetected by GWAS, and (3) identifying novel risk and disease causing variants and genes. Studies have shown that VEO cases of IBD experience a distinct and more severe disease course, and show positive family history for IBD. Therefore, the ascertainment of genetic causal variants in VEO IBD cases, clearly lends support to the expectation of higher genetic load or common familial genetic background (27–29). This is reminiscent of Mendelian diseases characterized by highly penetrant variants of higher effect size.
Monogenetic Disorders and Association with Very Early Onset IBD
The distinct and more severe disease phenotypes seen in VEO cases are associated with difficulty in classifying VEO IBD as CD or UC. The rates of unclassified IBD (IBD-U) or indeterminate colitis (IC) are higher in young children (34% in children under 2 years and 21% in children under 7 years) as opposed to adults (6%) (68). The specific VEO phenotypes often include manifestation of known monogenic diseases and the first case of the Mendelian form of VEO IBD was confirmed as a mutation in the IL10R gene (30) that underlined an association of IBD with primary immunodeficiency. Recently, a novel FOXP3 mutation was also identified in a two-generation family with early onset phenotypes similar to IBD (67), thus lending support to the manifestation of Mendelian disease in IBD cases. Many monogenic disease genes have been shown to confer overlapping pathology with IBD (known as IBD-like pathogenesis) and are seen more frequently in VEO cases (69). These diseases represent potential targets for identifying additional VEO heritability using exome sequencing. A list of genes underlying monogenic conditions is detailed in table 3. This confirms previous reports that single gene disorders also predispose to complex disorders (70, 71) and suggest that many Mendelian and complex disorders could share genetic architecture, where Mendelian loci may contain common variants with low effect size (detectable by GWAS) characterized by incomplete penetrance. Such common variants are capable of modifier functions and likely contribute to complex diseases alongside Mendelian, high effect size variants (detectable mainly by sequencing).
Table 3.
Monogenic genes associated with IBD-like presentation. Modified from Uhlig et al. (69)
| Function | Gene | Associated conditions |
|---|---|---|
| Epithelial barrier and epithelial response defects | COL7A1 | Dystrophic epidermolysis bullosa |
| FERMT1 | Kindler syndrome | |
| IKBKG | X linked ectodermal dysplasia and immunodeficiency | |
| ADAM17 | ADAM-17 deficiency | |
| GUCY2C | Familial diarrhoea | |
| Neutropenia and defects in phagocyte bacterial killing | CYBB, CYBA, NCF1, NCF2, NCF4 | Chronic granulomatous disease |
| SLC37A4 | Glycogen storage disease type 1b | |
| G6PC3 | Congenital neutropenia | |
| ITGB2 | Leucocyte adhesion deficiency 1 | |
| Hyper- and autoinflammation | MVK | Mevalonate kinase deficiency |
| PLCG2 | Phospholipase Cγ2 defects | |
| MEFV | Familial Mediterranean fever | |
| STXBP2 | Familial haemophagocytic lymphohistiocytosis type 5 | |
| XIAP | X linked lymphoproliferative syndrome 2 | |
| SH2D1A | X linked lymphoproliferative syndrome 1 | |
| HPS1, HPS4, HPS6 | Hermansky–Pudlak syndrome | |
| ICOS | Common variable immunodeficiency type 1 | |
| LRBA | Common variable immunodeficiency type 8 | |
| BTK, PIK3R1 | Agammaglobulinaemia | |
| CD40LG, AICDA | Hyper-IgM syndrome | |
| WAS | Wiskott–Aldrich syndrome | |
| DCLRE1C | Omenn syndrome | |
| DOCK8 | Hyper IgE syndrome | |
| SKIV2L, TTC37 | Trichohepatoenteric syndrome | |
| PTEN | PTEN hamartoma tumour syndrome | |
| Regulatory T cells and immune regulation | FOXP3, IL2RA | X linked immune dysregulation, polyendocrinopathy, enteropathy |
| IL10RA, IL10RB, IL10 | IL-10 signalling defects | |
| Defects in intestinal innervation | RET | Hirschsprung’s disease |
Future
To dissect the complete spectrum of variations that underlie IBD, VEO populations with IBD and Mendelian comorbidity should also be considered because IBD and Mendelian diseases often share phenotypes but involve variants that belong to different part of the spectrum within the same loci. In GWAS, not only are subjects with Mendelian disorders typically excluded by design but also variants that underlie Mendelian forms of VEO IBD are undetected. Substantial IBD heritability remains to be elucidated and it is unlikely that the expansion of GWAS with larger size cohorts will yield any additional missing heritability. It is clear by now that Mendelian loci contain variants that predispose to complex disease such as IBD and this makes next generation resequencing the ideal choice to comprehensively test such a hypothesis.
References
- 1.Loftus EV. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflammatory bowel diseases. 2011;17:1314–1321. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 3.Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gryboski JD. Crohn’s disease in children 10 years old and younger: comparison with ulcerative colitis. J Pediatr Gastroenterol Nutr. 1994;18:174–182. doi: 10.1097/00005176-199402000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Seidman E. Chronic Inflammatory Boeel Disease. In: Roy OCASaDA., editor. Pediatric Clinical Gastroenterology. 4. Mosby; 1995. pp. 427–493. [Google Scholar]
- 7.Polito JM, 2nd, Childs B, Mellits ED, et al. Crohn’s disease: influence of age at diagnosis on site and clinical type of disease. Gastroenterology. 1996;111:580–586. doi: 10.1053/gast.1996.v111.pm8780560. [DOI] [PubMed] [Google Scholar]
- 8.Biank V, Broeckel U, Kugathasan S. Pediatric inflammatory bowel disease: clinical and molecular genetics. Inflamm Bowel Dis. 2007;13:1430–1438. doi: 10.1002/ibd.20213. [DOI] [PubMed] [Google Scholar]
- 9.Tysk C, Lindberg E, Jarnerot G, et al. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson NP, Driscoll R, Pounder RE, et al. Genetics versus environment in inflammatory bowel disease: results of a British twin study. Bmj. 1996;312:95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orholm M, Binder V, Sorensen TI, et al. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35:1075–1081. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 12.Orholm M, Munkholm P, Langholz E, et al. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 13.Peeters M, Nevens H, Baert F, et al. Familial aggregation in Crohn’s disease: increased age-adjusted risk and concordance in clinical characteristics. Gastroenterology. 1996;111:597–603. doi: 10.1053/gast.1996.v111.pm8780562. [DOI] [PubMed] [Google Scholar]
- 14.Halme L, Paavola-Sakki P, Turunen U, et al. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12:3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Rotter JI. Genetic aspects of idiopathic inflammatory bowel disease. In: Kirsner JB, Shorter RG, editors. Inflammatory Bowel Disease. 4. Williams & Wilkins; 1995. pp. 301–331. [Google Scholar]
- 16.Satsangi J, Jewell DP, Bell JI. The genetics of inflammatory bowel disease. Gut. 1997;40:572–574. doi: 10.1136/gut.40.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JC, Parkes M. Genome-wide association studies and Crohn’s disease. Briefings in functional genomics. 2011;10:71–76. doi: 10.1093/bfgp/elr009. [DOI] [PubMed] [Google Scholar]
- 20.Oostenbrug LE, van Dullemen HM, te Meerman GJ, et al. IBD and genetics: new developments. Scand J Gastroenterol Suppl. 2003:63–68. doi: 10.1080/00855920310002717. [DOI] [PubMed] [Google Scholar]
- 21.Parkes M. The genetics universe of Crohn’s disease and ulcerative colitis. Dig Dis. 2012;30 (Suppl 1):78–81. doi: 10.1159/000341130. [DOI] [PubMed] [Google Scholar]
- 22.Doecke JD, Simms LA, Zhao ZZ, et al. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2013;19:240–245. doi: 10.1097/MIB.0b013e3182810041. [DOI] [PubMed] [Google Scholar]
- 23.Khor B, Gardet A, Xavier R. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkes M, Cortes A, van Heel DA, et al. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 27.Cannioto Z, Berti I, Martelossi S, et al. IBD and IBD mimicking enterocolitis in children younger than 2 years of age. European journal of pediatrics. 2009;168:149–155. doi: 10.1007/s00431-008-0721-2. [DOI] [PubMed] [Google Scholar]
- 28.Ruemmele FM, El Khoury MG, Talbotec C, et al. Characteristics of inflammatory bowel disease with onset during the first year of life. J Pediatr Gastroenterol Nutr. 2006;43:603–609. doi: 10.1097/01.mpg.0000237938.12674.e3. [DOI] [PubMed] [Google Scholar]
- 29.Bianco AM, Zanin V, Girardelli M, et al. A common genetic background could explain early-onset Crohn’s disease. Medical hypotheses. 2012;78:520–522. doi: 10.1016/j.mehy.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Glocker E-O, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. The New England journal of medicine. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marth GT, Yu F, Indap AR, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012 doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul T, Birnbaum A, Pal DK, et al. Distinct phenotype of early childhood inflammatory bowel disease. J Clin Gastroenterol. 2006;40:583–586. doi: 10.1097/00004836-200608000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Brant SR, Panhuysen CI, Bailey-Wilson JE, et al. Linkage heterogeneity for the IBD1 locus in Crohn’s disease pedigrees by disease onset and severity. Gastroenterology. 2000;119:1483–1490. doi: 10.1053/gast.2000.20245. [DOI] [PubMed] [Google Scholar]
- 35.Russell RK, Drummond HE, Nimmo ER, et al. Analysis of the influence of OCTN1/2 variants within the IBD5 locus on disease susceptibility and growth indices in early onset inflammatory bowel disease. Gut. 2006;55:1114–1123. doi: 10.1136/gut.2005.082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brescianini S, Trinh T, Stoll M, et al. IBD5 is associated with an extensive complicated Crohn’s disease feature: implications from genotype-phenotype analysis. Gut. 2007;56:149–150. doi: 10.1136/gut.2006.102723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Ridder L, Weersma RK, Dijkstra G, et al. Genetic susceptibility has a more important role in pediatric-onset Crohn’s disease than in adult-onset Crohn’s disease. Inflamm Bowel Dis. 2007;13:1083–1092. doi: 10.1002/ibd.20171. [DOI] [PubMed] [Google Scholar]
- 38.Cosnes J, Beaugerie L, Carbonnel F, et al. Smoking cessation and the course of Crohn’s disease: an intervention study. Gastroenterology. 2001;120:1093–1099. doi: 10.1053/gast.2001.23231. [DOI] [PubMed] [Google Scholar]
- 39.Pott J, Stockinger S, Torow N, et al. Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS pathogens. 2012;8:e1002670. doi: 10.1371/journal.ppat.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsey JM, Guest PC, Broek JA, et al. Identification of an age-dependent biomarker signature in children and adolescents with autism spectrum disorders. Molecular autism. 2013;4:27. doi: 10.1186/2040-2392-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanni SB, Smith PB, Benjamin DK, Jr, et al. Higher clearance of micafungin in neonates compared with adults: role of age-dependent micafungin serum binding. Biopharmaceutics & drug disposition. 2011;32:222–232. doi: 10.1002/bdd.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kappelman MD, Galanko JA, Porter CQ, et al. Association of paediatric inflammatory bowel disease with other immune-mediated diseases. Archives of disease in childhood. 2011;96:1042–1046. doi: 10.1136/archdischild-2011-300633. [DOI] [PubMed] [Google Scholar]
- 43.Roman AL, Munoz F. Comorbidity in inflammatory bowel disease. World J Gastroenterol. 2011;17:2723–2733. doi: 10.3748/wjg.v17.i22.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersson B, Almer S, Albertioni F, et al. Differences between children and adults in thiopurine methyltransferase activity and metabolite formation during thiopurine therapy: possible role of concomitant methotrexate. Therapeutic drug monitoring. 2002;24:351–358. doi: 10.1097/00007691-200206000-00005. [DOI] [PubMed] [Google Scholar]
- 45.Lander ES, Schork NJ. Genetic dissection of complex traits. Science (New York, NY) 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 46.Essers JB, Lee JJ, Kugathasan S, et al. Established genetic risk factors do not distinguish early and later onset Crohn’s disease. Inflamm Bowel Dis. 2009;15:1508–1514. doi: 10.1002/ibd.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohane IS. No small matter: qualitatively distinct challenges of pediatric genomic studies. Genome medicine. 2011;3:62. doi: 10.1186/gm278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bousvaros A, Sylvester F, Kugathasan S, et al. Challenges in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:885–913. doi: 10.1097/01.mib.0000228358.25364.8b. [DOI] [PubMed] [Google Scholar]
- 49.Choi M, Scholl U, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaydon DC, Biancheri P, Di WL, et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N Engl J Med. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
- 51.Kotlarz D, Beier R, Murugan D, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347–355. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 52.Muise A, Xu W, Guo C, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–1035. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature genetics. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christodoulou K, Wiskin AE, Gibson J, et al. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut. 2012 doi: 10.1136/gutjnl-2011-301833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Hui KY, Gusev A, et al. Extended haplotype association study in Crohn’s disease identifies a novel, Ashkenazi Jewish-specific missense mutation in the NF-kappaB pathway gene, HEATR3. Genes Immun. 2013;14:310–316. doi: 10.1038/gene.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moran CJ, Walters TD, Guo CH, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–123. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinwiddie DL, Bracken JM, Bass JA, et al. Molecular diagnosis of infantile onset inflammatory bowel disease by exome sequencing. Genomics. 2013;102:442–447. doi: 10.1016/j.ygeno.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Momozawa Y, Mni M, Nakamura K, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 59.Alangari A, Alsultan A, Adly N, et al. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J Allergy Clin Immunol. 2012;130:481–488. e482. doi: 10.1016/j.jaci.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matute JD, Arias AA, Wright NA, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellinghaus D, Zhang H, Zeissig S, et al. Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339–347. doi: 10.1053/j.gastro.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beaudoin M, Goyette P, Boucher G, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peltekova VD, Wintle RF, Rubin LA, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 64.Cardinale CJ, Wei Z, Panossian S, et al. Targeted resequencing identifies defective variants of decoy receptor 3 in pediatric-onset inflammatory bowel disease. Genes Immun. 2013;14:447–452. doi: 10.1038/gene.2013.43. [DOI] [PubMed] [Google Scholar]
- 65.Avitzur Y, Guo C, Mastropaolo LA, et al. Mutations in Tetratricopeptide Repeat Domain 7A Result in a Severe Form of Very Early Onset Inflammatory Bowel Disease. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Worthey E, Mayer A, Syverson G, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 67.Okou DT, Mondal K, Faubion WA, et al. Exome Sequencing Identifies a Novel FOXP3 Mutation in a Two-Generation Family with Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr. 2014 doi: 10.1097/MPG.0000000000000302. Publish Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prenzel F, Uhlig HH. Frequency of indeterminate colitis in children and adults with IBD -a metaanalysis. J Crohns Colitis. 2009;3:277–281. doi: 10.1016/j.crohns.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Uhlig HH. Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut. 2013;62:1795–1805. doi: 10.1136/gutjnl-2012-303956. [DOI] [PubMed] [Google Scholar]
- 70.Ristow M. Neurodegenerative disorders associated with diabetes mellitus. Journal of molecular medicine. 2004;82:510–529. doi: 10.1007/s00109-004-0552-1. [DOI] [PubMed] [Google Scholar]
- 71.Sinibaldi L, De Luca A, Bellacchio E, et al. Mutations of the Nogo-66 receptor (RTN4R) gene in schizophrenia. Hum Mutat. 2004;24:534–535. doi: 10.1002/humu.9292. [DOI] [PubMed] [Google Scholar]